Model Deployment : Machine Learning Model Experiment Logging and Tracking Using Open-Source Frameworks¶
1. Table of Contents ¶
This project explores open-source solutions for managing the end-to-end lifecycle of machine learning models, focusing on the use of MLflow for experiment tracking, model selection, and production deployment in Python. The objective was to design a reproducible workflow for evaluating multiple ensemble classifiers, selecting the optimal candidate based on validation and cross-validation metrics, and operationalizing the chosen model for production use. The project workflow involved several stages: first, hyperparameter tuning experiments were conducted across multiple ensemble model types, with all runs logged and organized in the MLflow Tracking UI. Next, optimal candidates were identified for each model family by programmatically querying run metrics, and the best overall model was selected based on a multi-metric ranking strategy. This model was then registered in the MLflow Model Registry, ensuring version-controlled storage alongside associated artifacts and metadata. To enable stable and maintainable production usage, the registered model was assigned a symbolic alias allowing seamless loading of the production model without hardcoding version numbers and simplifying future updates. The selected model was then evaluated on an independent test set to confirm generalization performance before being loaded for production inference via the MLflow PyFunc interface. The entire process—from experiment tracking to production deployment—was automated through the MLflow Python API, demonstrating how open-source MLOps tooling can streamline model governance, reproducibility, and deployment. All results were consolidated in a Summary presented at the end of the document.
Machine Learning Model Lifecycle Management is the systematic process of overseeing a model’s journey from initial development through production deployment, monitoring, and eventual retirement. In the context of MLOps, it encompasses every stage of a model’s evolution — including data preparation, model training, evaluation, version control, deployment, monitoring, retraining, and governance. The goal is to ensure that models remain accurate, reliable, and aligned with evolving business needs over time. Effective lifecycle management addresses challenges such as model drift, reproducibility, compliance, and scalability, while integrating automation to reduce operational overhead. It promotes collaboration between data scientists, engineers, and operations teams by using standardized processes and tooling to ensure smooth transitions between research and production environments. In modern MLOps practices, lifecycle management is supported by platforms and frameworks that provide tracking, registry, deployment orchestration, and monitoring capabilities, enabling continuous delivery and improvement of AI solutions.
MLflow is an open-source platform designed to manage the complete lifecycle of machine learning models, making it a central component in MLOps workflows. It provides four key capabilities: tracking experiments to log parameters, metrics, and artifacts; projects to package and share reproducible ML code; model registry to store, version, and annotate models; and model serving to deploy them in production environments. By offering a unified interface for logging and retrieving experiment data, MLflow helps teams compare results, trace model evolution, and enforce reproducibility. Its Model Registry acts as a centralized hub for managing versions, promoting models to production, and assigning aliases for easy referencing. MLflow supports multiple model formats — including Python functions, scikit-learn models, and deep learning frameworks — and integrates with popular deployment targets. In machine learning lifecycle management, MLflow bridges the gap between experimentation and production, providing transparency, governance, and automation across all stages.
Run Comparison in machine learning model lifecycle management refers to the process of evaluating and contrasting multiple training runs to identify the best-performing model for deployment. This involves analyzing key performance metrics, hyperparameters, and resource usage across different experiments. In MLOps, run comparison ensures that selection decisions are data-driven, reproducible, and aligned with business objectives. Within MLflow, run comparison is facilitated by both the Tracking UI and the Search API. The UI provides sortable tables and visualizations for side-by-side metric analysis, while the Search API allows programmatic filtering and ranking of runs using multiple criteria. This enables teams to systematically evaluate candidate models, pinpoint optimal configurations, and maintain an auditable record of decision-making. By combining interactive visual review with automated selection pipelines, MLflow’s run comparison capabilities streamline the transition from experimentation to registry.
Artifact Storage refers to the practice of preserving all files and outputs generated during the machine learning process, ensuring reproducibility and traceability. Artifacts can include trained model binaries, datasets, configuration files, visualizations, and evaluation reports. In the context of MLOps, centralized artifact storage ensures that every stage of model development is documented and recoverable, enabling consistent retraining, auditing, and compliance verification. In MLflow, artifact storage is integrated into the tracking system: every logged run can store artifacts either locally or on remote storage backends such as Amazon S3, Azure Blob Storage, or Google Cloud Storage. MLflow automatically organizes artifacts by experiment and run ID, allowing quick retrieval for analysis, deployment, or comparison. This eliminates the risk of “lost” models or mismatched dependencies, making artifact storage a foundational pillar of reliable lifecycle management.
Model Registry in machine learning lifecycle management serves as a centralized repository for storing, versioning, and managing machine learning models throughout their lifespan. It enables teams to track which models are in development, testing, or production, ensuring controlled and auditable promotion between stages. In MLOps, a model registry supports governance by maintaining metadata such as version numbers, creation dates, and performance metrics. MLflow’s Model Registry extends this concept by allowing users to register models directly from experiment runs, associate them with descriptions and tags, and manage aliases for flexible referencing. Instead of relying solely on numeric versions or deprecated stage labels, aliases like prod or staging allow CI/CD pipelines to dynamically pull the correct model without manual intervention. This makes the MLflow Model Registry a cornerstone for production-grade MLOps, ensuring both stability and agility in model deployment workflows.
Model Inference is the process of using a trained machine learning model to generate predictions on new, unseen data. In the context of lifecycle management, inference bridges the gap between model development and real-world application, enabling AI systems to deliver value in production environments. Inference workflows must account for input data preprocessing, model loading, execution efficiency, and output formatting. In MLflow, model inference is supported through its PyFunc interface, which provides a standardized API for loading and executing models regardless of their underlying framework. Models can be loaded by URI from local files, remote storage, or directly from the MLflow Model Registry using version numbers or aliases. This abstraction simplifies integration into applications and services, ensuring consistent and scalable predictions across environments. By standardizing inference, MLflow reduces friction in deployment and makes model serving more maintainable.
Model Deployment, in the context of lifecycle management, is the process of making a trained machine learning model available in a production environment where it can serve predictions to end-users or downstream systems. Deployment strategies vary, from batch processing pipelines to real-time APIs, and must consider scalability, latency, monitoring, and security. In MLOps, deployment is not a one-time event — it is part of a continuous delivery process that supports model updates, rollback capabilities, and ongoing performance monitoring. MLflow facilitates deployment through its support for multiple serving options: models can be served locally for testing, deployed as REST APIs using MLflow’s built-in serving tools, or integrated with platforms like Kubernetes, AWS SageMaker, or Azure ML. By coupling deployment with the Model Registry and aliases, MLflow enables automated CI/CD pipelines to update production models without manual configuration changes. This approach ensures rapid, reliable, and traceable promotion of models from experimentation to production use.
1.1. Data Background ¶
An open Thyroid Disease Dataset from Kaggle (with all credits attributed to Jai Naru and Abuchi Onwuegbusi) was used for the analysis as consolidated from the following primary sources:
- Reference Repository entitled Differentiated Thyroid Cancer Recurrence from UC Irvine Machine Learning Repository
- Research Paper entitled Machine Learning for Risk Stratification of Thyroid Cancer Patients: a 15-year Cohort Study from the European Archives of Oto-Rhino-Laryngology
This study hypothesized that the various clinicopathological characteristics influence differentiated thyroid cancer recurrence between patients.
The dichotomous categorical variable for the study is:
- Recurred - Status of the patient (Yes, Recurrence of differentiated thyroid cancer | No, No recurrence of differentiated thyroid cancer)
The predictor variables for the study are:
- Age - Patient's age (Years)
- Gender - Patient's sex (M | F)
- Smoking - Indication of smoking (Yes | No)
- Hx Smoking - Indication of smoking history (Yes | No)
- Hx Radiotherapy - Indication of radiotherapy history for any condition (Yes | No)
- Thyroid Function - Status of thyroid function (Clinical Hyperthyroidism, Hypothyroidism | Subclinical Hyperthyroidism, Hypothyroidism | Euthyroid)
- Physical Examination - Findings from physical examination including palpation of the thyroid gland and surrounding structures (Normal | Diffuse Goiter | Multinodular Goiter | Single Nodular Goiter Left, Right)
- Adenopathy - Indication of enlarged lymph nodes in the neck region (No | Right | Extensive | Left | Bilateral | Posterior)
- Pathology - Specific thyroid cancer type as determined by pathology examination of biopsy samples (Follicular | Hurthel Cell | Micropapillary | Papillary)
- Focality - Indication if the cancer is limited to one location or present in multiple locations (Uni-Focal | Multi-Focal)
- Risk - Risk category of the cancer based on various factors, such as tumor size, extent of spread, and histological type (Low | Intermediate | High)
- T - Tumor classification based on its size and extent of invasion into nearby structures (T1a | T1b | T2 | T3a | T3b | T4a | T4b)
- N - Nodal classification indicating the involvement of lymph nodes (N0 | N1a | N1b)
- M - Metastasis classification indicating the presence or absence of distant metastases (M0 | M1)
- Stage - Overall stage of the cancer, typically determined by combining T, N, and M classifications (I | II | III | IVa | IVb)
- Response - Cancer's response to treatment (Biochemical Incomplete | Indeterminate | Excellent | Structural Incomplete)
1.2. Data Description ¶
- The initial tabular dataset was comprised of 383 observations and 17 variables (including 1 target and 16 predictors).
- 383 rows (observations)
- 17 columns (variables)
- 1/17 target (categorical)
- Recurred
- 1/17 predictor (numeric)
- Age
- 16/17 predictor (categorical)
- Gender
- Smoking
- Hx_Smoking
- Hx_Radiotherapy
- Thyroid_Function
- Physical_Examination
- Adenopathy
- Pathology
- Focality
- Risk
- T
- N
- M
- Stage
- Response
- 1/17 target (categorical)
##################################
# Loading Python Libraries
##################################
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import os
import re
import pickle
%matplotlib inline
import mlflow
from mlflow.tracking import MlflowClient
from mlflow.models.signature import infer_signature
from mlflow.exceptions import RestException
import hashlib
import json
from urllib.parse import urlparse
import logging
from operator import truediv
from sklearn.preprocessing import OrdinalEncoder
from scipy import stats
from scipy.stats import pointbiserialr, chi2_contingency
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
from catboost import CatBoostClassifier
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score, roc_auc_score, confusion_matrix, classification_report
from sklearn.model_selection import train_test_split, ParameterGrid, StratifiedShuffleSplit
from sklearn.base import clone
##################################
# Defining file paths
##################################
DATASETS_ORIGINAL_PATH = r"datasets\original"
DATASETS_FINAL_PATH = r"datasets\final\complete"
DATASETS_FINAL_TRAIN_PATH = r"datasets\final\train"
DATASETS_FINAL_TRAIN_FEATURES_PATH = r"datasets\final\train\features"
DATASETS_FINAL_TRAIN_TARGET_PATH = r"datasets\final\train\target"
DATASETS_FINAL_VALIDATION_PATH = r"datasets\final\validation"
DATASETS_FINAL_VALIDATION_FEATURES_PATH = r"datasets\final\validation\features"
DATASETS_FINAL_VALIDATION_TARGET_PATH = r"datasets\final\validation\target"
DATASETS_FINAL_TEST_PATH = r"datasets\final\test"
DATASETS_FINAL_TEST_FEATURES_PATH = r"datasets\final\test\features"
DATASETS_FINAL_TEST_TARGET_PATH = r"datasets\final\test\target"
DATASETS_PREPROCESSED_PATH = r"datasets\preprocessed"
DATASETS_PREPROCESSED_TRAIN_PATH = r"datasets\preprocessed\train"
DATASETS_PREPROCESSED_TRAIN_FEATURES_PATH = r"datasets\preprocessed\train\features"
DATASETS_PREPROCESSED_TRAIN_TARGET_PATH = r"datasets\preprocessed\train\target"
DATASETS_PREPROCESSED_VALIDATION_PATH = r"datasets\preprocessed\validation"
DATASETS_PREPROCESSED_VALIDATION_FEATURES_PATH = r"datasets\preprocessed\validation\features"
DATASETS_PREPROCESSED_VALIDATION_TARGET_PATH = r"datasets\preprocessed\validation\target"
DATASETS_PREPROCESSED_TEST_PATH = r"datasets\preprocessed\test"
DATASETS_PREPROCESSED_TEST_FEATURES_PATH = r"datasets\preprocessed\test\features"
DATASETS_PREPROCESSED_TEST_TARGET_PATH = r"datasets\preprocessed\test\target"
##################################
# Loading the dataset
# from the DATASETS_ORIGINAL_PATH
##################################
thyroid_cancer = pd.read_csv(os.path.join("..", DATASETS_ORIGINAL_PATH, "Thyroid_Diff.csv"))
##################################
# Performing a general exploration of the dataset
##################################
print('Dataset Dimensions: ')
display(thyroid_cancer.shape)
Dataset Dimensions:
(383, 17)
##################################
# Listing the column names and data types
##################################
print('Column Names and Data Types:')
display(thyroid_cancer.dtypes)
Column Names and Data Types:
Age int64 Gender object Smoking object Hx Smoking object Hx Radiotherapy object Thyroid Function object Physical Examination object Adenopathy object Pathology object Focality object Risk object T object N object M object Stage object Response object Recurred object dtype: object
##################################
# Converting integer columns to float
# for a more convenient downstream processing
##################################
thyroid_cancer = thyroid_cancer.astype({
col: 'float64' for col in thyroid_cancer.select_dtypes(include='int').columns
})
print('Column Names and Data Types:')
display(thyroid_cancer.dtypes)
Column Names and Data Types:
Age float64 Gender object Smoking object Hx Smoking object Hx Radiotherapy object Thyroid Function object Physical Examination object Adenopathy object Pathology object Focality object Risk object T object N object M object Stage object Response object Recurred object dtype: object
##################################
# Renaming and standardizing the column names
# to replace blanks with undercores
##################################
thyroid_cancer.columns = thyroid_cancer.columns.str.replace(" ", "_")
##################################
# Taking a snapshot of the dataset
##################################
thyroid_cancer.head()
| Age | Gender | Smoking | Hx_Smoking | Hx_Radiotherapy | Thyroid_Function | Physical_Examination | Adenopathy | Pathology | Focality | Risk | T | N | M | Stage | Response | Recurred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 27.0 | F | No | No | No | Euthyroid | Single nodular goiter-left | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Indeterminate | No |
| 1 | 34.0 | F | No | Yes | No | Euthyroid | Multinodular goiter | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 2 | 30.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 3 | 62.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 4 | 62.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Micropapillary | Multi-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
##################################
# Selecting categorical columns (both object and categorical types)
# and listing the unique categorical levels
##################################
cat_cols = thyroid_cancer.select_dtypes(include=["object", "category"]).columns
for col in cat_cols:
print(f"Categorical | Object Column: {col}")
print(thyroid_cancer[col].unique())
print("-" * 40)
Categorical | Object Column: Gender ['F' 'M'] ---------------------------------------- Categorical | Object Column: Smoking ['No' 'Yes'] ---------------------------------------- Categorical | Object Column: Hx_Smoking ['No' 'Yes'] ---------------------------------------- Categorical | Object Column: Hx_Radiotherapy ['No' 'Yes'] ---------------------------------------- Categorical | Object Column: Thyroid_Function ['Euthyroid' 'Clinical Hyperthyroidism' 'Clinical Hypothyroidism' 'Subclinical Hyperthyroidism' 'Subclinical Hypothyroidism'] ---------------------------------------- Categorical | Object Column: Physical_Examination ['Single nodular goiter-left' 'Multinodular goiter' 'Single nodular goiter-right' 'Normal' 'Diffuse goiter'] ---------------------------------------- Categorical | Object Column: Adenopathy ['No' 'Right' 'Extensive' 'Left' 'Bilateral' 'Posterior'] ---------------------------------------- Categorical | Object Column: Pathology ['Micropapillary' 'Papillary' 'Follicular' 'Hurthel cell'] ---------------------------------------- Categorical | Object Column: Focality ['Uni-Focal' 'Multi-Focal'] ---------------------------------------- Categorical | Object Column: Risk ['Low' 'Intermediate' 'High'] ---------------------------------------- Categorical | Object Column: T ['T1a' 'T1b' 'T2' 'T3a' 'T3b' 'T4a' 'T4b'] ---------------------------------------- Categorical | Object Column: N ['N0' 'N1b' 'N1a'] ---------------------------------------- Categorical | Object Column: M ['M0' 'M1'] ---------------------------------------- Categorical | Object Column: Stage ['I' 'II' 'IVB' 'III' 'IVA'] ---------------------------------------- Categorical | Object Column: Response ['Indeterminate' 'Excellent' 'Structural Incomplete' 'Biochemical Incomplete'] ---------------------------------------- Categorical | Object Column: Recurred ['No' 'Yes'] ----------------------------------------
##################################
# Correcting a category level
##################################
thyroid_cancer["Pathology"] = thyroid_cancer["Pathology"].replace("Hurthel cell", "Hurthle Cell")
##################################
# Setting the levels of the categorical variables
##################################
thyroid_cancer['Recurred'] = thyroid_cancer['Recurred'].astype('category')
thyroid_cancer['Recurred'] = thyroid_cancer['Recurred'].cat.set_categories(['No', 'Yes'], ordered=True)
thyroid_cancer['Gender'] = thyroid_cancer['Gender'].astype('category')
thyroid_cancer['Gender'] = thyroid_cancer['Gender'].cat.set_categories(['M', 'F'], ordered=True)
thyroid_cancer['Smoking'] = thyroid_cancer['Smoking'].astype('category')
thyroid_cancer['Smoking'] = thyroid_cancer['Smoking'].cat.set_categories(['No', 'Yes'], ordered=True)
thyroid_cancer['Hx_Smoking'] = thyroid_cancer['Hx_Smoking'].astype('category')
thyroid_cancer['Hx_Smoking'] = thyroid_cancer['Hx_Smoking'].cat.set_categories(['No', 'Yes'], ordered=True)
thyroid_cancer['Hx_Radiotherapy'] = thyroid_cancer['Hx_Radiotherapy'].astype('category')
thyroid_cancer['Hx_Radiotherapy'] = thyroid_cancer['Hx_Radiotherapy'].cat.set_categories(['No', 'Yes'], ordered=True)
thyroid_cancer['Thyroid_Function'] = thyroid_cancer['Thyroid_Function'].astype('category')
thyroid_cancer['Thyroid_Function'] = thyroid_cancer['Thyroid_Function'].cat.set_categories(['Euthyroid', 'Subclinical Hypothyroidism', 'Subclinical Hyperthyroidism', 'Clinical Hypothyroidism', 'Clinical Hyperthyroidism'], ordered=True)
thyroid_cancer['Physical_Examination'] = thyroid_cancer['Physical_Examination'].astype('category')
thyroid_cancer['Physical_Examination'] = thyroid_cancer['Physical_Examination'].cat.set_categories(['Normal', 'Single nodular goiter-left', 'Single nodular goiter-right', 'Multinodular goiter', 'Diffuse goiter'], ordered=True)
thyroid_cancer['Adenopathy'] = thyroid_cancer['Adenopathy'].astype('category')
thyroid_cancer['Adenopathy'] = thyroid_cancer['Adenopathy'].cat.set_categories(['No', 'Left', 'Right', 'Bilateral', 'Posterior', 'Extensive'], ordered=True)
thyroid_cancer['Pathology'] = thyroid_cancer['Pathology'].astype('category')
thyroid_cancer['Pathology'] = thyroid_cancer['Pathology'].cat.set_categories(['Hurthle Cell', 'Follicular', 'Micropapillary', 'Papillary'], ordered=True)
thyroid_cancer['Focality'] = thyroid_cancer['Focality'].astype('category')
thyroid_cancer['Focality'] = thyroid_cancer['Focality'].cat.set_categories(['Uni-Focal', 'Multi-Focal'], ordered=True)
thyroid_cancer['Risk'] = thyroid_cancer['Risk'].astype('category')
thyroid_cancer['Risk'] = thyroid_cancer['Risk'].cat.set_categories(['Low', 'Intermediate', 'High'], ordered=True)
thyroid_cancer['T'] = thyroid_cancer['T'].astype('category')
thyroid_cancer['T'] = thyroid_cancer['T'].cat.set_categories(['T1a', 'T1b', 'T2', 'T3a', 'T3b', 'T4a', 'T4b'], ordered=True)
thyroid_cancer['N'] = thyroid_cancer['N'].astype('category')
thyroid_cancer['N'] = thyroid_cancer['N'].cat.set_categories(['N0', 'N1a', 'N1b'], ordered=True)
thyroid_cancer['M'] = thyroid_cancer['M'].astype('category')
thyroid_cancer['M'] = thyroid_cancer['M'].cat.set_categories(['M0', 'M1'], ordered=True)
thyroid_cancer['Stage'] = thyroid_cancer['Stage'].astype('category')
thyroid_cancer['Stage'] = thyroid_cancer['Stage'].cat.set_categories(['I', 'II', 'III', 'IVA', 'IVB'], ordered=True)
thyroid_cancer['Response'] = thyroid_cancer['Response'].astype('category')
thyroid_cancer['Response'] = thyroid_cancer['Response'].cat.set_categories(['Excellent', 'Structural Incomplete', 'Biochemical Incomplete', 'Indeterminate'], ordered=True)
##################################
# Performing a general exploration of the numeric variables
##################################
print('Numeric Variable Summary:')
display(thyroid_cancer.describe(include='number').transpose())
Numeric Variable Summary:
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| Age | 383.0 | 40.866841 | 15.134494 | 15.0 | 29.0 | 37.0 | 51.0 | 82.0 |
##################################
# Performing a general exploration of the categorical variables
##################################
print('Categorical Variable Summary:')
display(thyroid_cancer.describe(include='category').transpose())
Categorical Variable Summary:
| count | unique | top | freq | |
|---|---|---|---|---|
| Gender | 383 | 2 | F | 312 |
| Smoking | 383 | 2 | No | 334 |
| Hx_Smoking | 383 | 2 | No | 355 |
| Hx_Radiotherapy | 383 | 2 | No | 376 |
| Thyroid_Function | 383 | 5 | Euthyroid | 332 |
| Physical_Examination | 383 | 5 | Single nodular goiter-right | 140 |
| Adenopathy | 383 | 6 | No | 277 |
| Pathology | 383 | 4 | Papillary | 287 |
| Focality | 383 | 2 | Uni-Focal | 247 |
| Risk | 383 | 3 | Low | 249 |
| T | 383 | 7 | T2 | 151 |
| N | 383 | 3 | N0 | 268 |
| M | 383 | 2 | M0 | 365 |
| Stage | 383 | 5 | I | 333 |
| Response | 383 | 4 | Excellent | 208 |
| Recurred | 383 | 2 | No | 275 |
##################################
# Performing a general exploration of the categorical variable levels
# based on the ordered categories
##################################
ordered_cat_cols = thyroid_cancer.select_dtypes(include=["category"]).columns
for col in ordered_cat_cols:
print(f"Column: {col}")
print("Absolute Frequencies:")
print(thyroid_cancer[col].value_counts().reindex(thyroid_cancer[col].cat.categories))
print("\nNormalized Frequencies:")
print(thyroid_cancer[col].value_counts(normalize=True).reindex(thyroid_cancer[col].cat.categories))
print("-" * 50)
Column: Gender Absolute Frequencies: M 71 F 312 Name: count, dtype: int64 Normalized Frequencies: M 0.185379 F 0.814621 Name: proportion, dtype: float64 -------------------------------------------------- Column: Smoking Absolute Frequencies: No 334 Yes 49 Name: count, dtype: int64 Normalized Frequencies: No 0.872063 Yes 0.127937 Name: proportion, dtype: float64 -------------------------------------------------- Column: Hx_Smoking Absolute Frequencies: No 355 Yes 28 Name: count, dtype: int64 Normalized Frequencies: No 0.926893 Yes 0.073107 Name: proportion, dtype: float64 -------------------------------------------------- Column: Hx_Radiotherapy Absolute Frequencies: No 376 Yes 7 Name: count, dtype: int64 Normalized Frequencies: No 0.981723 Yes 0.018277 Name: proportion, dtype: float64 -------------------------------------------------- Column: Thyroid_Function Absolute Frequencies: Euthyroid 332 Subclinical Hypothyroidism 14 Subclinical Hyperthyroidism 5 Clinical Hypothyroidism 12 Clinical Hyperthyroidism 20 Name: count, dtype: int64 Normalized Frequencies: Euthyroid 0.866841 Subclinical Hypothyroidism 0.036554 Subclinical Hyperthyroidism 0.013055 Clinical Hypothyroidism 0.031332 Clinical Hyperthyroidism 0.052219 Name: proportion, dtype: float64 -------------------------------------------------- Column: Physical_Examination Absolute Frequencies: Normal 7 Single nodular goiter-left 89 Single nodular goiter-right 140 Multinodular goiter 140 Diffuse goiter 7 Name: count, dtype: int64 Normalized Frequencies: Normal 0.018277 Single nodular goiter-left 0.232376 Single nodular goiter-right 0.365535 Multinodular goiter 0.365535 Diffuse goiter 0.018277 Name: proportion, dtype: float64 -------------------------------------------------- Column: Adenopathy Absolute Frequencies: No 277 Left 17 Right 48 Bilateral 32 Posterior 2 Extensive 7 Name: count, dtype: int64 Normalized Frequencies: No 0.723238 Left 0.044386 Right 0.125326 Bilateral 0.083551 Posterior 0.005222 Extensive 0.018277 Name: proportion, dtype: float64 -------------------------------------------------- Column: Pathology Absolute Frequencies: Hurthle Cell 20 Follicular 28 Micropapillary 48 Papillary 287 Name: count, dtype: int64 Normalized Frequencies: Hurthle Cell 0.052219 Follicular 0.073107 Micropapillary 0.125326 Papillary 0.749347 Name: proportion, dtype: float64 -------------------------------------------------- Column: Focality Absolute Frequencies: Uni-Focal 247 Multi-Focal 136 Name: count, dtype: int64 Normalized Frequencies: Uni-Focal 0.644909 Multi-Focal 0.355091 Name: proportion, dtype: float64 -------------------------------------------------- Column: Risk Absolute Frequencies: Low 249 Intermediate 102 High 32 Name: count, dtype: int64 Normalized Frequencies: Low 0.650131 Intermediate 0.266319 High 0.083551 Name: proportion, dtype: float64 -------------------------------------------------- Column: T Absolute Frequencies: T1a 49 T1b 43 T2 151 T3a 96 T3b 16 T4a 20 T4b 8 Name: count, dtype: int64 Normalized Frequencies: T1a 0.127937 T1b 0.112272 T2 0.394256 T3a 0.250653 T3b 0.041775 T4a 0.052219 T4b 0.020888 Name: proportion, dtype: float64 -------------------------------------------------- Column: N Absolute Frequencies: N0 268 N1a 22 N1b 93 Name: count, dtype: int64 Normalized Frequencies: N0 0.699739 N1a 0.057441 N1b 0.242820 Name: proportion, dtype: float64 -------------------------------------------------- Column: M Absolute Frequencies: M0 365 M1 18 Name: count, dtype: int64 Normalized Frequencies: M0 0.953003 M1 0.046997 Name: proportion, dtype: float64 -------------------------------------------------- Column: Stage Absolute Frequencies: I 333 II 32 III 4 IVA 3 IVB 11 Name: count, dtype: int64 Normalized Frequencies: I 0.869452 II 0.083551 III 0.010444 IVA 0.007833 IVB 0.028721 Name: proportion, dtype: float64 -------------------------------------------------- Column: Response Absolute Frequencies: Excellent 208 Structural Incomplete 91 Biochemical Incomplete 23 Indeterminate 61 Name: count, dtype: int64 Normalized Frequencies: Excellent 0.543081 Structural Incomplete 0.237598 Biochemical Incomplete 0.060052 Indeterminate 0.159269 Name: proportion, dtype: float64 -------------------------------------------------- Column: Recurred Absolute Frequencies: No 275 Yes 108 Name: count, dtype: int64 Normalized Frequencies: No 0.718016 Yes 0.281984 Name: proportion, dtype: float64 --------------------------------------------------
1.3. Data Quality Assessment ¶
Data quality findings based on assessment are as follows:
- A total of 19 duplicated rows were identified.
- In total, 34 observations were affected, consisting of 16 unique occurrences and 19 subsequent duplicates.
- These 19 duplicates spanned 16 distinct variations, meaning some variations had multiple duplicates.
- To clean the dataset, all 19 duplicate rows were removed, retaining only the first occurrence of each of the 16 unique variations.
- No missing data noted for any variable with Null.Count>0 and Fill.Rate<1.0.
- Low variance observed for 8 variables with First.Second.Mode.Ratio>5.
- Hx_Radiotherapy: First.Second.Mode.Ratio = 51.000 (comprised 2 category levels)
- M: First.Second.Mode.Ratio = 19.222 (comprised 2 category levels)
- Thyroid_Function: First.Second.Mode.Ratio = 15.650 (comprised 5 category levels)
- Hx_Smoking: First.Second.Mode.Ratio = 12.000 (comprised 2 category levels)
- Stage: First.Second.Mode.Ratio = 9.812 (comprised 5 category levels)
- Smoking: First.Second.Mode.Ratio = 6.428 (comprised 2 category levels)
- Pathology: First.Second.Mode.Ratio = 6.022 (comprised 4 category levels)
- Adenopathy: First.Second.Mode.Ratio = 5.375 (comprised 5 category levels)
- No low variance observed for any variable with Unique.Count.Ratio>10.
- No high skewness observed for any variable with Skewness>3 or Skewness<(-3).
##################################
# Counting the number of duplicated rows
##################################
thyroid_cancer.duplicated().sum()
np.int64(19)
##################################
# Exploring the duplicated rows
##################################
duplicated_rows = thyroid_cancer[thyroid_cancer.duplicated(keep=False)]
display(duplicated_rows)
| Age | Gender | Smoking | Hx_Smoking | Hx_Radiotherapy | Thyroid_Function | Physical_Examination | Adenopathy | Pathology | Focality | Risk | T | N | M | Stage | Response | Recurred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 51.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 9 | 40.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 22 | 36.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 32 | 36.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 38 | 40.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 40 | 51.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No |
| 61 | 35.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No |
| 66 | 35.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No |
| 67 | 51.0 | F | No | No | No | Euthyroid | Single nodular goiter-left | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No |
| 69 | 51.0 | F | No | No | No | Euthyroid | Single nodular goiter-left | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No |
| 73 | 29.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No |
| 77 | 29.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No |
| 106 | 26.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 110 | 31.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 113 | 32.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 115 | 37.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 119 | 28.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 120 | 37.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 121 | 26.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 123 | 28.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 132 | 32.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 136 | 21.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 137 | 32.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 138 | 26.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 142 | 42.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 161 | 22.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 166 | 31.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 168 | 21.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 170 | 38.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 175 | 34.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 178 | 38.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 183 | 26.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 187 | 34.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 189 | 42.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
| 196 | 22.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No |
##################################
# Checking if duplicated rows have identical values across all columns
##################################
num_unique_dup_rows = duplicated_rows.drop_duplicates().shape[0]
num_total_dup_rows = duplicated_rows.shape[0]
if num_unique_dup_rows == 1:
print("All duplicated rows have the same values across all columns.")
else:
print(f"There are {num_unique_dup_rows} unique versions among the {num_total_dup_rows} duplicated rows.")
There are 16 unique versions among the 35 duplicated rows.
##################################
# Counting the unique variations among duplicated rows
##################################
unique_dup_variations = duplicated_rows.drop_duplicates()
variation_counts = duplicated_rows.value_counts().reset_index(name="Count")
print("Unique duplicated row variations and their counts:")
display(variation_counts)
Unique duplicated row variations and their counts:
| Age | Gender | Smoking | Hx_Smoking | Hx_Radiotherapy | Thyroid_Function | Physical_Examination | Adenopathy | Pathology | Focality | Risk | T | N | M | Stage | Response | Recurred | Count | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 26.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 4 |
| 1 | 32.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 3 |
| 2 | 22.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 2 |
| 3 | 21.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 2 |
| 4 | 28.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 2 |
| 5 | 29.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No | 2 |
| 6 | 31.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 2 |
| 7 | 34.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 2 |
| 8 | 35.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No | 2 |
| 9 | 36.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No | 2 |
| 10 | 37.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 2 |
| 11 | 38.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 2 |
| 12 | 40.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No | 2 |
| 13 | 42.0 | F | No | No | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | M0 | I | Excellent | No | 2 |
| 14 | 51.0 | F | No | No | No | Euthyroid | Single nodular goiter-left | No | Papillary | Uni-Focal | Low | T1b | N0 | M0 | I | Excellent | No | 2 |
| 15 | 51.0 | F | No | No | No | Euthyroid | Single nodular goiter-right | No | Micropapillary | Uni-Focal | Low | T1a | N0 | M0 | I | Excellent | No | 2 |
##################################
# Removing the duplicated rows and
# retaining only the first occurrence
##################################
thyroid_cancer_row_filtered = thyroid_cancer.drop_duplicates(keep="first")
print('Dataset Dimensions: ')
display(thyroid_cancer_row_filtered.shape)
Dataset Dimensions:
(364, 17)
##################################
# Gathering the data types for each column
##################################
data_type_list = list(thyroid_cancer_row_filtered.dtypes)
##################################
# Gathering the variable names for each column
##################################
variable_name_list = list(thyroid_cancer_row_filtered.columns)
##################################
# Gathering the number of observations for each column
##################################
row_count_list = list([len(thyroid_cancer_row_filtered)] * len(thyroid_cancer_row_filtered.columns))
##################################
# Gathering the number of missing data for each column
##################################
null_count_list = list(thyroid_cancer_row_filtered.isna().sum(axis=0))
##################################
# Gathering the number of non-missing data for each column
##################################
non_null_count_list = list(thyroid_cancer_row_filtered.count())
##################################
# Gathering the missing data percentage for each column
##################################
fill_rate_list = map(truediv, non_null_count_list, row_count_list)
##################################
# Formulating the summary
# for all columns
##################################
all_column_quality_summary = pd.DataFrame(zip(variable_name_list,
data_type_list,
row_count_list,
non_null_count_list,
null_count_list,
fill_rate_list),
columns=['Column.Name',
'Column.Type',
'Row.Count',
'Non.Null.Count',
'Null.Count',
'Fill.Rate'])
display(all_column_quality_summary)
| Column.Name | Column.Type | Row.Count | Non.Null.Count | Null.Count | Fill.Rate | |
|---|---|---|---|---|---|---|
| 0 | Age | float64 | 364 | 364 | 0 | 1.0 |
| 1 | Gender | category | 364 | 364 | 0 | 1.0 |
| 2 | Smoking | category | 364 | 364 | 0 | 1.0 |
| 3 | Hx_Smoking | category | 364 | 364 | 0 | 1.0 |
| 4 | Hx_Radiotherapy | category | 364 | 364 | 0 | 1.0 |
| 5 | Thyroid_Function | category | 364 | 364 | 0 | 1.0 |
| 6 | Physical_Examination | category | 364 | 364 | 0 | 1.0 |
| 7 | Adenopathy | category | 364 | 364 | 0 | 1.0 |
| 8 | Pathology | category | 364 | 364 | 0 | 1.0 |
| 9 | Focality | category | 364 | 364 | 0 | 1.0 |
| 10 | Risk | category | 364 | 364 | 0 | 1.0 |
| 11 | T | category | 364 | 364 | 0 | 1.0 |
| 12 | N | category | 364 | 364 | 0 | 1.0 |
| 13 | M | category | 364 | 364 | 0 | 1.0 |
| 14 | Stage | category | 364 | 364 | 0 | 1.0 |
| 15 | Response | category | 364 | 364 | 0 | 1.0 |
| 16 | Recurred | category | 364 | 364 | 0 | 1.0 |
##################################
# Counting the number of columns
# with Fill.Rate < 1.00
##################################
len(all_column_quality_summary[(all_column_quality_summary['Fill.Rate']<1)])
0
##################################
# Identifying the rows
# with Fill.Rate < 0.90
##################################
column_low_fill_rate = all_column_quality_summary[(all_column_quality_summary['Fill.Rate']<0.90)]
##################################
# Gathering the indices for each observation
##################################
row_index_list = thyroid_cancer_row_filtered.index
##################################
# Gathering the number of columns for each observation
##################################
column_count_list = list([len(thyroid_cancer_row_filtered.columns)] * len(thyroid_cancer_row_filtered))
##################################
# Gathering the number of missing data for each row
##################################
null_row_list = list(thyroid_cancer_row_filtered.isna().sum(axis=1))
##################################
# Gathering the missing data percentage for each column
##################################
missing_rate_list = map(truediv, null_row_list, column_count_list)
##################################
# Identifying the rows
# with missing data
##################################
all_row_quality_summary = pd.DataFrame(zip(row_index_list,
column_count_list,
null_row_list,
missing_rate_list),
columns=['Row.Name',
'Column.Count',
'Null.Count',
'Missing.Rate'])
display(all_row_quality_summary)
| Row.Name | Column.Count | Null.Count | Missing.Rate | |
|---|---|---|---|---|
| 0 | 0 | 17 | 0 | 0.0 |
| 1 | 1 | 17 | 0 | 0.0 |
| 2 | 2 | 17 | 0 | 0.0 |
| 3 | 3 | 17 | 0 | 0.0 |
| 4 | 4 | 17 | 0 | 0.0 |
| ... | ... | ... | ... | ... |
| 359 | 378 | 17 | 0 | 0.0 |
| 360 | 379 | 17 | 0 | 0.0 |
| 361 | 380 | 17 | 0 | 0.0 |
| 362 | 381 | 17 | 0 | 0.0 |
| 363 | 382 | 17 | 0 | 0.0 |
364 rows × 4 columns
##################################
# Counting the number of rows
# with Missing.Rate > 0.00
##################################
len(all_row_quality_summary[(all_row_quality_summary['Missing.Rate']>0.00)])
0
##################################
# Formulating the dataset
# with numeric columns only
##################################
thyroid_cancer_numeric = thyroid_cancer_row_filtered.select_dtypes(include='number')
##################################
# Gathering the variable names for each numeric column
##################################
numeric_variable_name_list = thyroid_cancer_numeric.columns
##################################
# Gathering the minimum value for each numeric column
##################################
numeric_minimum_list = thyroid_cancer_numeric.min()
##################################
# Gathering the mean value for each numeric column
##################################
numeric_mean_list = thyroid_cancer_numeric.mean()
##################################
# Gathering the median value for each numeric column
##################################
numeric_median_list = thyroid_cancer_numeric.median()
##################################
# Gathering the maximum value for each numeric column
##################################
numeric_maximum_list = thyroid_cancer_numeric.max()
##################################
# Gathering the first mode values for each numeric column
##################################
numeric_first_mode_list = [thyroid_cancer_row_filtered[x].value_counts(dropna=True).index.tolist()[0] for x in thyroid_cancer_numeric]
##################################
# Gathering the second mode values for each numeric column
##################################
numeric_second_mode_list = [thyroid_cancer_row_filtered[x].value_counts(dropna=True).index.tolist()[1] for x in thyroid_cancer_numeric]
##################################
# Gathering the count of first mode values for each numeric column
##################################
numeric_first_mode_count_list = [thyroid_cancer_numeric[x].isin([thyroid_cancer_row_filtered[x].value_counts(dropna=True).index.tolist()[0]]).sum() for x in thyroid_cancer_numeric]
##################################
# Gathering the count of second mode values for each numeric column
##################################
numeric_second_mode_count_list = [thyroid_cancer_numeric[x].isin([thyroid_cancer_row_filtered[x].value_counts(dropna=True).index.tolist()[1]]).sum() for x in thyroid_cancer_numeric]
##################################
# Gathering the first mode to second mode ratio for each numeric column
##################################
numeric_first_second_mode_ratio_list = map(truediv, numeric_first_mode_count_list, numeric_second_mode_count_list)
##################################
# Gathering the count of unique values for each numeric column
##################################
numeric_unique_count_list = thyroid_cancer_numeric.nunique(dropna=True)
##################################
# Gathering the number of observations for each numeric column
##################################
numeric_row_count_list = list([len(thyroid_cancer_numeric)] * len(thyroid_cancer_numeric.columns))
##################################
# Gathering the unique to count ratio for each numeric column
##################################
numeric_unique_count_ratio_list = map(truediv, numeric_unique_count_list, numeric_row_count_list)
##################################
# Gathering the skewness value for each numeric column
##################################
numeric_skewness_list = thyroid_cancer_numeric.skew()
##################################
# Gathering the kurtosis value for each numeric column
##################################
numeric_kurtosis_list = thyroid_cancer_numeric.kurtosis()
##################################
# Generating a column quality summary for the numeric column
##################################
numeric_column_quality_summary = pd.DataFrame(zip(numeric_variable_name_list,
numeric_minimum_list,
numeric_mean_list,
numeric_median_list,
numeric_maximum_list,
numeric_first_mode_list,
numeric_second_mode_list,
numeric_first_mode_count_list,
numeric_second_mode_count_list,
numeric_first_second_mode_ratio_list,
numeric_unique_count_list,
numeric_row_count_list,
numeric_unique_count_ratio_list,
numeric_skewness_list,
numeric_kurtosis_list),
columns=['Numeric.Column.Name',
'Minimum',
'Mean',
'Median',
'Maximum',
'First.Mode',
'Second.Mode',
'First.Mode.Count',
'Second.Mode.Count',
'First.Second.Mode.Ratio',
'Unique.Count',
'Row.Count',
'Unique.Count.Ratio',
'Skewness',
'Kurtosis'])
display(numeric_column_quality_summary)
| Numeric.Column.Name | Minimum | Mean | Median | Maximum | First.Mode | Second.Mode | First.Mode.Count | Second.Mode.Count | First.Second.Mode.Ratio | Unique.Count | Row.Count | Unique.Count.Ratio | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Age | 15.0 | 41.25 | 38.0 | 82.0 | 31.0 | 27.0 | 21 | 13 | 1.615385 | 65 | 364 | 0.178571 | 0.678269 | -0.359255 |
##################################
# Counting the number of numeric columns
# with First.Second.Mode.Ratio > 5.00
##################################
len(numeric_column_quality_summary[(numeric_column_quality_summary['First.Second.Mode.Ratio']>5)])
0
##################################
# Counting the number of numeric columns
# with Unique.Count.Ratio > 10.00
##################################
len(numeric_column_quality_summary[(numeric_column_quality_summary['Unique.Count.Ratio']>10)])
0
##################################
# Counting the number of numeric columns
# with Skewness > 3.00 or Skewness < -3.00
##################################
len(numeric_column_quality_summary[(numeric_column_quality_summary['Skewness']>3) | (numeric_column_quality_summary['Skewness']<(-3))])
0
##################################
# Formulating the dataset
# with categorical columns only
##################################
thyroid_cancer_categorical = thyroid_cancer_row_filtered.select_dtypes(include='category')
##################################
# Gathering the variable names for the categorical column
##################################
categorical_variable_name_list = thyroid_cancer_categorical.columns
##################################
# Gathering the first mode values for each categorical column
##################################
categorical_first_mode_list = [thyroid_cancer_row_filtered[x].value_counts().index.tolist()[0] for x in thyroid_cancer_categorical]
##################################
# Gathering the second mode values for each categorical column
##################################
categorical_second_mode_list = [thyroid_cancer_row_filtered[x].value_counts().index.tolist()[1] for x in thyroid_cancer_categorical]
##################################
# Gathering the count of first mode values for each categorical column
##################################
categorical_first_mode_count_list = [thyroid_cancer_categorical[x].isin([thyroid_cancer_row_filtered[x].value_counts(dropna=True).index.tolist()[0]]).sum() for x in thyroid_cancer_categorical]
##################################
# Gathering the count of second mode values for each categorical column
##################################
categorical_second_mode_count_list = [thyroid_cancer_categorical[x].isin([thyroid_cancer_row_filtered[x].value_counts(dropna=True).index.tolist()[1]]).sum() for x in thyroid_cancer_categorical]
##################################
# Gathering the first mode to second mode ratio for each categorical column
##################################
categorical_first_second_mode_ratio_list = map(truediv, categorical_first_mode_count_list, categorical_second_mode_count_list)
##################################
# Gathering the count of unique values for each categorical column
##################################
categorical_unique_count_list = thyroid_cancer_categorical.nunique(dropna=True)
##################################
# Gathering the number of observations for each categorical column
##################################
categorical_row_count_list = list([len(thyroid_cancer_categorical)] * len(thyroid_cancer_categorical.columns))
##################################
# Gathering the unique to count ratio for each categorical column
##################################
categorical_unique_count_ratio_list = map(truediv, categorical_unique_count_list, categorical_row_count_list)
##################################
# Generating a column quality summary for the categorical columns
##################################
categorical_column_quality_summary = pd.DataFrame(zip(categorical_variable_name_list,
categorical_first_mode_list,
categorical_second_mode_list,
categorical_first_mode_count_list,
categorical_second_mode_count_list,
categorical_first_second_mode_ratio_list,
categorical_unique_count_list,
categorical_row_count_list,
categorical_unique_count_ratio_list),
columns=['Categorical.Column.Name',
'First.Mode',
'Second.Mode',
'First.Mode.Count',
'Second.Mode.Count',
'First.Second.Mode.Ratio',
'Unique.Count',
'Row.Count',
'Unique.Count.Ratio'])
display(categorical_column_quality_summary)
| Categorical.Column.Name | First.Mode | Second.Mode | First.Mode.Count | Second.Mode.Count | First.Second.Mode.Ratio | Unique.Count | Row.Count | Unique.Count.Ratio | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Gender | F | M | 293 | 71 | 4.126761 | 2 | 364 | 0.005495 |
| 1 | Smoking | No | Yes | 315 | 49 | 6.428571 | 2 | 364 | 0.005495 |
| 2 | Hx_Smoking | No | Yes | 336 | 28 | 12.000000 | 2 | 364 | 0.005495 |
| 3 | Hx_Radiotherapy | No | Yes | 357 | 7 | 51.000000 | 2 | 364 | 0.005495 |
| 4 | Thyroid_Function | Euthyroid | Clinical Hyperthyroidism | 313 | 20 | 15.650000 | 5 | 364 | 0.013736 |
| 5 | Physical_Examination | Multinodular goiter | Single nodular goiter-right | 135 | 127 | 1.062992 | 5 | 364 | 0.013736 |
| 6 | Adenopathy | No | Right | 258 | 48 | 5.375000 | 6 | 364 | 0.016484 |
| 7 | Pathology | Papillary | Micropapillary | 271 | 45 | 6.022222 | 4 | 364 | 0.010989 |
| 8 | Focality | Uni-Focal | Multi-Focal | 228 | 136 | 1.676471 | 2 | 364 | 0.005495 |
| 9 | Risk | Low | Intermediate | 230 | 102 | 2.254902 | 3 | 364 | 0.008242 |
| 10 | T | T2 | T3a | 138 | 96 | 1.437500 | 7 | 364 | 0.019231 |
| 11 | N | N0 | N1b | 249 | 93 | 2.677419 | 3 | 364 | 0.008242 |
| 12 | M | M0 | M1 | 346 | 18 | 19.222222 | 2 | 364 | 0.005495 |
| 13 | Stage | I | II | 314 | 32 | 9.812500 | 5 | 364 | 0.013736 |
| 14 | Response | Excellent | Structural Incomplete | 189 | 91 | 2.076923 | 4 | 364 | 0.010989 |
| 15 | Recurred | No | Yes | 256 | 108 | 2.370370 | 2 | 364 | 0.005495 |
##################################
# Counting the number of categorical columns
# with First.Second.Mode.Ratio > 5.00
##################################
len(categorical_column_quality_summary[(categorical_column_quality_summary['First.Second.Mode.Ratio']>5)])
8
##################################
# Identifying the categorical columns
# with First.Second.Mode.Ratio > 5.00
##################################
display(categorical_column_quality_summary[(categorical_column_quality_summary['First.Second.Mode.Ratio']>5)].sort_values(by=['First.Second.Mode.Ratio'], ascending=False))
| Categorical.Column.Name | First.Mode | Second.Mode | First.Mode.Count | Second.Mode.Count | First.Second.Mode.Ratio | Unique.Count | Row.Count | Unique.Count.Ratio | |
|---|---|---|---|---|---|---|---|---|---|
| 3 | Hx_Radiotherapy | No | Yes | 357 | 7 | 51.000000 | 2 | 364 | 0.005495 |
| 12 | M | M0 | M1 | 346 | 18 | 19.222222 | 2 | 364 | 0.005495 |
| 4 | Thyroid_Function | Euthyroid | Clinical Hyperthyroidism | 313 | 20 | 15.650000 | 5 | 364 | 0.013736 |
| 2 | Hx_Smoking | No | Yes | 336 | 28 | 12.000000 | 2 | 364 | 0.005495 |
| 13 | Stage | I | II | 314 | 32 | 9.812500 | 5 | 364 | 0.013736 |
| 1 | Smoking | No | Yes | 315 | 49 | 6.428571 | 2 | 364 | 0.005495 |
| 7 | Pathology | Papillary | Micropapillary | 271 | 45 | 6.022222 | 4 | 364 | 0.010989 |
| 6 | Adenopathy | No | Right | 258 | 48 | 5.375000 | 6 | 364 | 0.016484 |
##################################
# Counting the number of categorical columns
# with Unique.Count.Ratio > 10.00
##################################
len(categorical_column_quality_summary[(categorical_column_quality_summary['Unique.Count.Ratio']>10)])
0
1.4. Data Preprocessing ¶
1.4.1 Data Splitting¶
- The baseline dataset (with duplicate rows removed from the original dataset) is comprised of:
- 364 rows (observations)
- 256 Recurred=No: 70.33%
- 108 Recurred=Yes: 29.67%
- 17 columns (variables)
- 1/17 target (categorical)
- Recurred
- 1/17 predictor (numeric)
- Age
- 15/17 predictor (categorical)
- Gender
- Smoking
- Hx_Smoking
- Hx_Radiotherapy
- Thyroid_Function
- Physical_Examination
- Adenopathy
- Pathology
- Focality
- Risk
- T
- N
- M
- Stage
- Response
- 1/17 target (categorical)
- 364 rows (observations)
- The baseline dataset was divided into three subsets using a fixed random seed:
- test data: 25% of the original data with class stratification applied
- train data (initial): 75% of the original data with class stratification applied
- train data (final): 75% of the train (initial) data with class stratification applied
- validation data: 25% of the train (initial) data with class stratification applied
- Models were developed from the train data (final). Using the same dataset, a subset of models with optimal hyperparameters were selected, based on cross-validation.
- Among candidate models with optimal hyperparameters, the final model were selected based on performance on the validation data.
- Performance of the selected final model (and other candidate models for post-model selection comparison) were evaluated using the test data.
- The train data (final) subset is comprised of:
- 204 rows (observations)
- 143 Recurred=No: 70.10%
- 61 Recurred=Yes: 29.90%
- 17 columns (variables)
- 204 rows (observations)
- The validation data subset is comprised of:
- 69 rows (observations)
- 49 Recurred=No: 71.01%
- 20 Recurred=Yes: 28.98%
- 17 columns (variables)
- 69 rows (observations)
- The test data subset is comprised of:
- 91 rows (observations)
- 64 Recurred=No: 70.33%
- 27 Recurred=Yes: 29.67%
- 17 columns (variables)
- 91 rows (observations)
##################################
# Creating a dataset copy
# of the row filtered data
##################################
thyroid_cancer_baseline = thyroid_cancer_row_filtered.copy()
##################################
# Performing a general exploration
# of the baseline dataset
##################################
print('Final Dataset Dimensions: ')
display(thyroid_cancer_baseline.shape)
Final Dataset Dimensions:
(364, 17)
##################################
# Obtaining the distribution of
# of the target variable
##################################
print('Target Variable Breakdown: ')
thyroid_cancer_breakdown = thyroid_cancer_baseline.groupby('Recurred', observed=True).size().reset_index(name='Count')
thyroid_cancer_breakdown['Percentage'] = (thyroid_cancer_breakdown['Count'] / len(thyroid_cancer_baseline)) * 100
display(thyroid_cancer_breakdown)
Target Variable Breakdown:
| Recurred | Count | Percentage | |
|---|---|---|---|
| 0 | No | 256 | 70.32967 |
| 1 | Yes | 108 | 29.67033 |
##################################
# Formulating the train and test data
# from the final dataset
# by applying stratification and
# using a 75-25 ratio
##################################
thyroid_cancer_train_initial, thyroid_cancer_test = train_test_split(thyroid_cancer_baseline,
test_size=0.25,
stratify=thyroid_cancer_baseline['Recurred'],
random_state=987654321)
##################################
# Performing a general exploration
# of the initial training dataset
##################################
X_train_initial = thyroid_cancer_train_initial.drop('Recurred', axis = 1)
y_train_initial = thyroid_cancer_train_initial['Recurred']
print('Initial Train Dataset Dimensions: ')
display(X_train_initial.shape)
display(y_train_initial.shape)
print('Initial Train Target Variable Breakdown: ')
display(y_train_initial.value_counts())
print('Initial Train Target Variable Proportion: ')
display(y_train_initial.value_counts(normalize = True))
Initial Train Dataset Dimensions:
(273, 16)
(273,)
Initial Train Target Variable Breakdown:
Recurred No 192 Yes 81 Name: count, dtype: int64
Initial Train Target Variable Proportion:
Recurred No 0.703297 Yes 0.296703 Name: proportion, dtype: float64
##################################
# Performing a general exploration
# of the test dataset
##################################
X_test = thyroid_cancer_test.drop('Recurred', axis = 1)
y_test = thyroid_cancer_test['Recurred']
print('Test Dataset Dimensions: ')
display(X_test.shape)
display(y_test.shape)
print('Test Target Variable Breakdown: ')
display(y_test.value_counts())
print('Test Target Variable Proportion: ')
display(y_test.value_counts(normalize = True))
Test Dataset Dimensions:
(91, 16)
(91,)
Test Target Variable Breakdown:
Recurred No 64 Yes 27 Name: count, dtype: int64
Test Target Variable Proportion:
Recurred No 0.703297 Yes 0.296703 Name: proportion, dtype: float64
##################################
# Formulating the train and validation data
# from the train dataset
# by applying stratification and
# using a 75-25 ratio
##################################
thyroid_cancer_train, thyroid_cancer_validation = train_test_split(thyroid_cancer_train_initial,
test_size=0.25,
stratify=thyroid_cancer_train_initial['Recurred'],
random_state=987654321)
##################################
# Performing a general exploration
# of the final training dataset
##################################
X_train = thyroid_cancer_train.drop('Recurred', axis = 1)
y_train = thyroid_cancer_train['Recurred']
print('Final Train Dataset Dimensions: ')
display(X_train.shape)
display(y_train.shape)
print('Final Train Target Variable Breakdown: ')
display(y_train.value_counts())
print('Final Train Target Variable Proportion: ')
display(y_train.value_counts(normalize = True))
Final Train Dataset Dimensions:
(204, 16)
(204,)
Final Train Target Variable Breakdown:
Recurred No 143 Yes 61 Name: count, dtype: int64
Final Train Target Variable Proportion:
Recurred No 0.70098 Yes 0.29902 Name: proportion, dtype: float64
##################################
# Performing a general exploration
# of the validation dataset
##################################
X_validation = thyroid_cancer_validation.drop('Recurred', axis = 1)
y_validation = thyroid_cancer_validation['Recurred']
print('Validation Dataset Dimensions: ')
display(X_validation.shape)
display(y_validation.shape)
print('Validation Target Variable Breakdown: ')
display(y_validation.value_counts())
print('Validation Target Variable Proportion: ')
display(y_validation.value_counts(normalize = True))
Validation Dataset Dimensions:
(69, 16)
(69,)
Validation Target Variable Breakdown:
Recurred No 49 Yes 20 Name: count, dtype: int64
Validation Target Variable Proportion:
Recurred No 0.710145 Yes 0.289855 Name: proportion, dtype: float64
##################################
# Saving the training data
# to the DATASETS_FINAL_TRAIN_PATH
# and DATASETS_FINAL_TRAIN_FEATURES_PATH
# and DATASETS_FINAL_TRAIN_TARGET_PATH
##################################
thyroid_cancer_train.to_csv(os.path.join("..", DATASETS_FINAL_TRAIN_PATH, "thyroid_cancer_train.csv"), index=False)
X_train.to_csv(os.path.join("..", DATASETS_FINAL_TRAIN_FEATURES_PATH, "X_train.csv"), index=False)
y_train.to_csv(os.path.join("..", DATASETS_FINAL_TRAIN_TARGET_PATH, "y_train.csv"), index=False)
##################################
# Saving the validation data
# to the DATASETS_FINAL_VALIDATION_PATH
# and DATASETS_FINAL_VALIDATION_FEATURE_PATH
# and DATASETS_FINAL_VALIDATION_TARGET_PATH
##################################
thyroid_cancer_validation.to_csv(os.path.join("..", DATASETS_FINAL_VALIDATION_PATH, "thyroid_cancer_validation.csv"), index=False)
X_validation.to_csv(os.path.join("..", DATASETS_FINAL_VALIDATION_FEATURES_PATH, "X_validation.csv"), index=False)
y_validation.to_csv(os.path.join("..", DATASETS_FINAL_VALIDATION_TARGET_PATH, "y_validation.csv"), index=False)
##################################
# Saving the test data
# to the DATASETS_FINAL_TEST_PATH
# and DATASETS_FINAL_TEST_FEATURES_PATH
# and DATASETS_FINAL_TEST_TARGET_PATH
##################################
thyroid_cancer_test.to_csv(os.path.join("..", DATASETS_FINAL_TEST_PATH, "thyroid_cancer_test.csv"), index=False)
X_test.to_csv(os.path.join("..", DATASETS_FINAL_TEST_FEATURES_PATH, "X_test.csv"), index=False)
y_test.to_csv(os.path.join("..", DATASETS_FINAL_TEST_TARGET_PATH, "y_test.csv"), index=False)
1.4.2 Data Profiling¶
- No significant distributional anomalies were observed for the numeric predictor Age.
- 9 categorical predictors were observed with categories consisting of too few cases that risk poor generalization and cross-validation issues:
- Thyroid_Function:
- 171 Thyroid_Function=Euthyroid: 83.82%
- 10 Thyroid_Function=Subclinical Hypothyroidism: 4.90%
- 3 Thyroid_Function=Subclinical Hyperthyroidism: 1.47%
- 7 Thyroid_Function=Clinical Hypothyroidism: 3.43%
- 13 Thyroid_Function=Clinical Hyperthyroidism: 6.37%
- Physical_Examination:
- 4 Physical_Examination=Normal: 1.96%
- 50 Physical_Examination=Single nodular goiter-left: 24.50%
- 68 Physical_Examination=Single nodular goiter-right: 33.33%
- 79 Physical_Examination=Multinodular goiter: 38.72%
- 3 Physical_Examination=Diffuse goiter: 1.47%
- Adenopathy:
- 144 Adenopathy=No: 70.59%
- 14 Adenopathy=Left: 6.86%
- 21 Adenopathy=Right: 10.29%
- 19 Adenopathy=Bilateral: 9.31%
- 2 Adenopathy=Posterior: 9.84%
- 4 Adenopathy=Extensive: 1.96%
- Pathology:
- 15 Pathology=Hurthle Cell: 7.35%
- 14 Pathology=Follicular: 6.86%
- 26 Pathology=Micropapillary: 12.74%
- 149 Pathology=Papillary: 73.03%
- Risk:
- 127 Risk=Low: 62.25%
- 60 Risk=Intermediate: 29.41%
- 17 Risk=High: 8.33%
- T:
- 26 T=T1a: 12.74%
- 21 T=T1b: 10.29%
- 73 T=T2: 35.78%
- 58 T=T3a: 28.43%
- 10 T=T3b: 4.90%
- 12 T=T4a: 5.88%
- 4 T=T4b: 1.96%
- N:
- 139 N=N0: 68.13%
- 11 N=N1a: 5.39%
- 54 N=N1b: 26.47%
- Stage:
- 174 Stage=I: 85.29%
- 21 Stage=II: 10.29%
- 2 Stage=III: 0.98%
- 2 Stage=IVA: 0.98%
- 5 Stage=IVB: 2.45%
- Response:
- 109 Response=Excellent: 53.43%
- 53 Response=Structural Incomplete: 25.98%
- 8 Response=Biochemical Incomplete: 3.92%
- 34 Response=Indeterminate: 16.67%
- Thyroid_Function:
- 3 categorical predictors were excluded from the dataset after having been observed with extremely low variance containing categories with very few or almost no variations across observations that may have limited predictive power or drive increased model complexity without performance gains:
- Hx_Smoking:
- 193 Hx_Smoking=No: 94.61%
- 11 Hx_Smoking=Yes: 5.39%
- Hx_Radiotherapy:
- 202 Hx_Radiotherapy=No: 99.02%
- 2 Hx_Radiotherapy=Yes: 0.98%
- M:
- 194 M=M0: 95.10%
- 10 M=M1: 4.90%
- Hx_Smoking:
##################################
# Segregating the target
# and predictor variables
##################################
thyroid_cancer_train_predictors = thyroid_cancer_train.iloc[:,:-1].columns
thyroid_cancer_train_predictors_numeric = thyroid_cancer_train.iloc[:,:-1].loc[:, thyroid_cancer_train.iloc[:,:-1].columns == 'Age'].columns
thyroid_cancer_train_predictors_categorical = thyroid_cancer_train.iloc[:,:-1].loc[:,thyroid_cancer_train.iloc[:,:-1].columns != 'Age'].columns
##################################
# Gathering the variable names for each numeric column
##################################
numeric_variable_name_list = thyroid_cancer_train_predictors_numeric
##################################
# Segregating the target variable
# and numeric predictors
##################################
histogram_grouping_variable = 'Recurred'
histogram_frequency_variable = numeric_variable_name_list.values[0]
##################################
# Comparing the numeric predictors
# grouped by the target variable
##################################
colors = plt.get_cmap('tab10').colors
plt.figure(figsize=(7, 5))
group_no = thyroid_cancer_train[thyroid_cancer_train[histogram_grouping_variable] == 'No'][histogram_frequency_variable]
group_yes = thyroid_cancer_train[thyroid_cancer_train[histogram_grouping_variable] == 'Yes'][histogram_frequency_variable]
plt.hist(group_no, bins=20, alpha=0.5, color=colors[0], label='No', edgecolor='black')
plt.hist(group_yes, bins=20, alpha=0.5, color=colors[1], label='Yes', edgecolor='black')
plt.title(f'{histogram_grouping_variable} Versus {histogram_frequency_variable}')
plt.xlabel(histogram_frequency_variable)
plt.ylabel('Frequency')
plt.legend()
plt.show()
##################################
# Performing a general exploration of the categorical variable levels
# based on the ordered categories
##################################
ordered_cat_cols = thyroid_cancer_train.select_dtypes(include=["category"]).columns
for col in ordered_cat_cols:
print(f"Column: {col}")
print("Absolute Frequencies:")
print(thyroid_cancer_train[col].value_counts().reindex(thyroid_cancer_train[col].cat.categories))
print("\nNormalized Frequencies:")
print(thyroid_cancer_train[col].value_counts(normalize=True).reindex(thyroid_cancer_train[col].cat.categories))
print("-" * 50)
Column: Gender Absolute Frequencies: M 44 F 160 Name: count, dtype: int64 Normalized Frequencies: M 0.215686 F 0.784314 Name: proportion, dtype: float64 -------------------------------------------------- Column: Smoking Absolute Frequencies: No 177 Yes 27 Name: count, dtype: int64 Normalized Frequencies: No 0.867647 Yes 0.132353 Name: proportion, dtype: float64 -------------------------------------------------- Column: Hx_Smoking Absolute Frequencies: No 193 Yes 11 Name: count, dtype: int64 Normalized Frequencies: No 0.946078 Yes 0.053922 Name: proportion, dtype: float64 -------------------------------------------------- Column: Hx_Radiotherapy Absolute Frequencies: No 202 Yes 2 Name: count, dtype: int64 Normalized Frequencies: No 0.990196 Yes 0.009804 Name: proportion, dtype: float64 -------------------------------------------------- Column: Thyroid_Function Absolute Frequencies: Euthyroid 171 Subclinical Hypothyroidism 10 Subclinical Hyperthyroidism 3 Clinical Hypothyroidism 7 Clinical Hyperthyroidism 13 Name: count, dtype: int64 Normalized Frequencies: Euthyroid 0.838235 Subclinical Hypothyroidism 0.049020 Subclinical Hyperthyroidism 0.014706 Clinical Hypothyroidism 0.034314 Clinical Hyperthyroidism 0.063725 Name: proportion, dtype: float64 -------------------------------------------------- Column: Physical_Examination Absolute Frequencies: Normal 4 Single nodular goiter-left 50 Single nodular goiter-right 68 Multinodular goiter 79 Diffuse goiter 3 Name: count, dtype: int64 Normalized Frequencies: Normal 0.019608 Single nodular goiter-left 0.245098 Single nodular goiter-right 0.333333 Multinodular goiter 0.387255 Diffuse goiter 0.014706 Name: proportion, dtype: float64 -------------------------------------------------- Column: Adenopathy Absolute Frequencies: No 144 Left 14 Right 21 Bilateral 19 Posterior 2 Extensive 4 Name: count, dtype: int64 Normalized Frequencies: No 0.705882 Left 0.068627 Right 0.102941 Bilateral 0.093137 Posterior 0.009804 Extensive 0.019608 Name: proportion, dtype: float64 -------------------------------------------------- Column: Pathology Absolute Frequencies: Hurthle Cell 15 Follicular 14 Micropapillary 26 Papillary 149 Name: count, dtype: int64 Normalized Frequencies: Hurthle Cell 0.073529 Follicular 0.068627 Micropapillary 0.127451 Papillary 0.730392 Name: proportion, dtype: float64 -------------------------------------------------- Column: Focality Absolute Frequencies: Uni-Focal 129 Multi-Focal 75 Name: count, dtype: int64 Normalized Frequencies: Uni-Focal 0.632353 Multi-Focal 0.367647 Name: proportion, dtype: float64 -------------------------------------------------- Column: Risk Absolute Frequencies: Low 127 Intermediate 60 High 17 Name: count, dtype: int64 Normalized Frequencies: Low 0.622549 Intermediate 0.294118 High 0.083333 Name: proportion, dtype: float64 -------------------------------------------------- Column: T Absolute Frequencies: T1a 26 T1b 21 T2 73 T3a 58 T3b 10 T4a 12 T4b 4 Name: count, dtype: int64 Normalized Frequencies: T1a 0.127451 T1b 0.102941 T2 0.357843 T3a 0.284314 T3b 0.049020 T4a 0.058824 T4b 0.019608 Name: proportion, dtype: float64 -------------------------------------------------- Column: N Absolute Frequencies: N0 139 N1a 11 N1b 54 Name: count, dtype: int64 Normalized Frequencies: N0 0.681373 N1a 0.053922 N1b 0.264706 Name: proportion, dtype: float64 -------------------------------------------------- Column: M Absolute Frequencies: M0 194 M1 10 Name: count, dtype: int64 Normalized Frequencies: M0 0.95098 M1 0.04902 Name: proportion, dtype: float64 -------------------------------------------------- Column: Stage Absolute Frequencies: I 174 II 21 III 2 IVA 2 IVB 5 Name: count, dtype: int64 Normalized Frequencies: I 0.852941 II 0.102941 III 0.009804 IVA 0.009804 IVB 0.024510 Name: proportion, dtype: float64 -------------------------------------------------- Column: Response Absolute Frequencies: Excellent 109 Structural Incomplete 53 Biochemical Incomplete 8 Indeterminate 34 Name: count, dtype: int64 Normalized Frequencies: Excellent 0.534314 Structural Incomplete 0.259804 Biochemical Incomplete 0.039216 Indeterminate 0.166667 Name: proportion, dtype: float64 -------------------------------------------------- Column: Recurred Absolute Frequencies: No 143 Yes 61 Name: count, dtype: int64 Normalized Frequencies: No 0.70098 Yes 0.29902 Name: proportion, dtype: float64 --------------------------------------------------
##################################
# Segregating the target variable
# and categorical predictors
##################################
proportion_y_variables = thyroid_cancer_train_predictors_categorical
proportion_x_variable = 'Recurred'
##################################
# Defining the number of
# rows and columns for the subplots
##################################
num_rows = 5
num_cols = 3
##################################
# Formulating the subplot structure
##################################
fig, axes = plt.subplots(num_rows, num_cols, figsize=(15, 25))
##################################
# Flattening the multi-row and
# multi-column axes
##################################
axes = axes.ravel()
##################################
# Formulating the individual stacked column plots
# for all categorical columns
##################################
for i, y_variable in enumerate(proportion_y_variables):
ax = axes[i]
category_counts = thyroid_cancer_train.groupby([proportion_x_variable, y_variable], observed=True).size().unstack(fill_value=0)
category_proportions = category_counts.div(category_counts.sum(axis=1), axis=0)
category_proportions.plot(kind='bar', stacked=True, ax=ax)
ax.set_title(f'{proportion_x_variable} Versus {y_variable}')
ax.set_xlabel(proportion_x_variable)
ax.set_ylabel('Proportions')
ax.legend(loc="lower center")
##################################
# Adjusting the subplot layout
##################################
plt.tight_layout()
##################################
# Presenting the subplots
##################################
plt.show()
##################################
# Removing predictors observed with extreme
# near-zero variance and a limited number of levels
##################################
thyroid_cancer_train_column_filtered = thyroid_cancer_train.drop(columns=['Hx_Radiotherapy','M','Hx_Smoking'])
thyroid_cancer_train_column_filtered.head()
| Age | Gender | Smoking | Thyroid_Function | Physical_Examination | Adenopathy | Pathology | Focality | Risk | T | N | Stage | Response | Recurred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 140 | 28.0 | F | No | Euthyroid | Multinodular goiter | No | Papillary | Uni-Focal | Low | T2 | N0 | I | Excellent | No |
| 205 | 36.0 | F | No | Euthyroid | Single nodular goiter-right | Right | Papillary | Uni-Focal | Low | T2 | N1b | I | Indeterminate | No |
| 277 | 41.0 | M | Yes | Euthyroid | Single nodular goiter-right | No | Hurthle Cell | Multi-Focal | Intermediate | T3a | N0 | I | Excellent | No |
| 294 | 42.0 | M | No | Subclinical Hypothyroidism | Single nodular goiter-right | No | Papillary | Multi-Focal | Intermediate | T3a | N1a | I | Indeterminate | No |
| 268 | 32.0 | F | No | Euthyroid | Single nodular goiter-left | No | Papillary | Uni-Focal | Low | T3a | N0 | I | Excellent | No |
1.4.3 Category Aggregation and Encoding¶
- Category aggregation was applied to the previously identified categorical predictors observed with many levels (high-cardinality) containing only a few observations to improve model stability during cross-validation and enhance generalization:
- Thyroid_Function:
- 171 Thyroid_Function=Euthyroid: 83.82%
- 33 Thyroid_Function=Hypothyroidism or Hyperthyroidism: 16.18%
- Physical_Examination:
- 122 Physical_Examination=Normal or Single Nodular Goiter : 59.80%
- 82 Physical_Examination=Multinodular or Diffuse Goiter: 40.20%
- Adenopathy:
- 144 Adenopathy=No: 70.59%
- 60 Adenopathy=Yes: 29.41%
- Pathology:
- 29 Pathology=Non-Papillary : 14.22%
- 175 Pathology=Papillary: 85.78%
- Risk:
- 127 Risk=Low: 62.25%
- 77 Risk=Intermediate to High: 37.75%
- T:
- 120 T=T1 to T2: 58.82%
- 84 T=T3 to T4b: 41.18%
- N:
- 139 N=N0: 68.14%
- 65 N=N1: 31.86%
- Stage:
- 174 Stage=I: 85.29%
- 30 Stage=II to IVB: 14.71%
- Response:
- 109 Response=Excellent: 53.43%
- 95 Response=Indeterminate or Incomplete: 46.57%
- Thyroid_Function:
##################################
# Merging small categories into broader groups
# for certain categorical predictors
# to ensure sufficient representation in statistical models
# and prevent sparsity issues in cross-validation
##################################
thyroid_cancer_train_column_filtered['Thyroid_Function'] = thyroid_cancer_train_column_filtered['Thyroid_Function'].map(lambda x: 'Euthyroid' if (x in ['Euthyroid']) else 'Hypothyroidism or Hyperthyroidism').astype('category')
thyroid_cancer_train_column_filtered['Physical_Examination'] = thyroid_cancer_train_column_filtered['Physical_Examination'].map(lambda x: 'Normal or Single Nodular Goiter' if (x in ['Normal', 'Single nodular goiter-left', 'Single nodular goiter-right']) else 'Multinodular or Diffuse Goiter').astype('category')
thyroid_cancer_train_column_filtered['Adenopathy'] = thyroid_cancer_train_column_filtered['Adenopathy'].map(lambda x: 'No' if x == 'No' else ('Yes' if pd.notna(x) and x != '' else x)).astype('category')
thyroid_cancer_train_column_filtered['Pathology'] = thyroid_cancer_train_column_filtered['Pathology'].map(lambda x: 'Non-Papillary' if (x in ['Hurthle Cell', 'Follicular']) else 'Papillary').astype('category')
thyroid_cancer_train_column_filtered['Risk'] = thyroid_cancer_train_column_filtered['Risk'].map(lambda x: 'Low' if (x in ['Low']) else 'Intermediate to High').astype('category')
thyroid_cancer_train_column_filtered['T'] = thyroid_cancer_train_column_filtered['T'].map(lambda x: 'T1 to T2' if (x in ['T1a', 'T1b', 'T2']) else 'T3 to T4b').astype('category')
thyroid_cancer_train_column_filtered['N'] = thyroid_cancer_train_column_filtered['N'].map(lambda x: 'N0' if (x in ['N0']) else 'N1').astype('category')
thyroid_cancer_train_column_filtered['Stage'] = thyroid_cancer_train_column_filtered['Stage'].map(lambda x: 'I' if (x in ['I']) else 'II to IVB').astype('category')
thyroid_cancer_train_column_filtered['Response'] = thyroid_cancer_train_column_filtered['Response'].map(lambda x: 'Indeterminate or Incomplete' if (x in ['Indeterminate', 'Structural Incomplete', 'Biochemical Incomplete']) else 'Excellent').astype('category')
thyroid_cancer_train_column_filtered.head()
| Age | Gender | Smoking | Thyroid_Function | Physical_Examination | Adenopathy | Pathology | Focality | Risk | T | N | Stage | Response | Recurred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 140 | 28.0 | F | No | Euthyroid | Multinodular or Diffuse Goiter | No | Papillary | Uni-Focal | Low | T1 to T2 | N0 | I | Excellent | No |
| 205 | 36.0 | F | No | Euthyroid | Normal or Single Nodular Goiter | Yes | Papillary | Uni-Focal | Low | T1 to T2 | N1 | I | Indeterminate or Incomplete | No |
| 277 | 41.0 | M | Yes | Euthyroid | Normal or Single Nodular Goiter | No | Non-Papillary | Multi-Focal | Intermediate to High | T3 to T4b | N0 | I | Excellent | No |
| 294 | 42.0 | M | No | Hypothyroidism or Hyperthyroidism | Normal or Single Nodular Goiter | No | Papillary | Multi-Focal | Intermediate to High | T3 to T4b | N1 | I | Indeterminate or Incomplete | No |
| 268 | 32.0 | F | No | Euthyroid | Normal or Single Nodular Goiter | No | Papillary | Uni-Focal | Low | T3 to T4b | N0 | I | Excellent | No |
##################################
# Performing a general exploration of the categorical variable levels
# based on the ordered categories
##################################
ordered_cat_cols = thyroid_cancer_train_column_filtered.select_dtypes(include=["category"]).columns
for col in ordered_cat_cols:
print(f"Column: {col}")
print("Absolute Frequencies:")
print(thyroid_cancer_train_column_filtered[col].value_counts().reindex(thyroid_cancer_train_column_filtered[col].cat.categories))
print("\nNormalized Frequencies:")
print(thyroid_cancer_train_column_filtered[col].value_counts(normalize=True).reindex(thyroid_cancer_train_column_filtered[col].cat.categories))
print("-" * 50)
Column: Gender Absolute Frequencies: M 44 F 160 Name: count, dtype: int64 Normalized Frequencies: M 0.215686 F 0.784314 Name: proportion, dtype: float64 -------------------------------------------------- Column: Smoking Absolute Frequencies: No 177 Yes 27 Name: count, dtype: int64 Normalized Frequencies: No 0.867647 Yes 0.132353 Name: proportion, dtype: float64 -------------------------------------------------- Column: Thyroid_Function Absolute Frequencies: Euthyroid 171 Hypothyroidism or Hyperthyroidism 33 Name: count, dtype: int64 Normalized Frequencies: Euthyroid 0.838235 Hypothyroidism or Hyperthyroidism 0.161765 Name: proportion, dtype: float64 -------------------------------------------------- Column: Physical_Examination Absolute Frequencies: Multinodular or Diffuse Goiter 82 Normal or Single Nodular Goiter 122 Name: count, dtype: int64 Normalized Frequencies: Multinodular or Diffuse Goiter 0.401961 Normal or Single Nodular Goiter 0.598039 Name: proportion, dtype: float64 -------------------------------------------------- Column: Adenopathy Absolute Frequencies: No 144 Yes 60 Name: count, dtype: int64 Normalized Frequencies: No 0.705882 Yes 0.294118 Name: proportion, dtype: float64 -------------------------------------------------- Column: Pathology Absolute Frequencies: Non-Papillary 29 Papillary 175 Name: count, dtype: int64 Normalized Frequencies: Non-Papillary 0.142157 Papillary 0.857843 Name: proportion, dtype: float64 -------------------------------------------------- Column: Focality Absolute Frequencies: Uni-Focal 129 Multi-Focal 75 Name: count, dtype: int64 Normalized Frequencies: Uni-Focal 0.632353 Multi-Focal 0.367647 Name: proportion, dtype: float64 -------------------------------------------------- Column: Risk Absolute Frequencies: Intermediate to High 77 Low 127 Name: count, dtype: int64 Normalized Frequencies: Intermediate to High 0.377451 Low 0.622549 Name: proportion, dtype: float64 -------------------------------------------------- Column: T Absolute Frequencies: T1 to T2 120 T3 to T4b 84 Name: count, dtype: int64 Normalized Frequencies: T1 to T2 0.588235 T3 to T4b 0.411765 Name: proportion, dtype: float64 -------------------------------------------------- Column: N Absolute Frequencies: N0 139 N1 65 Name: count, dtype: int64 Normalized Frequencies: N0 0.681373 N1 0.318627 Name: proportion, dtype: float64 -------------------------------------------------- Column: Stage Absolute Frequencies: I 174 II to IVB 30 Name: count, dtype: int64 Normalized Frequencies: I 0.852941 II to IVB 0.147059 Name: proportion, dtype: float64 -------------------------------------------------- Column: Response Absolute Frequencies: Excellent 109 Indeterminate or Incomplete 95 Name: count, dtype: int64 Normalized Frequencies: Excellent 0.534314 Indeterminate or Incomplete 0.465686 Name: proportion, dtype: float64 -------------------------------------------------- Column: Recurred Absolute Frequencies: No 143 Yes 61 Name: count, dtype: int64 Normalized Frequencies: No 0.70098 Yes 0.29902 Name: proportion, dtype: float64 --------------------------------------------------
##################################
# Segregating the target
# and predictor variables
##################################
thyroid_cancer_train_predictors = thyroid_cancer_train_column_filtered.iloc[:,:-1].columns
thyroid_cancer_train_predictors_numeric = thyroid_cancer_train_column_filtered.iloc[:,:-1].loc[:, thyroid_cancer_train_column_filtered.iloc[:,:-1].columns == 'Age'].columns
thyroid_cancer_train_predictors_categorical = thyroid_cancer_train_column_filtered.iloc[:,:-1].loc[:,thyroid_cancer_train_column_filtered.iloc[:,:-1].columns != 'Age'].columns
##################################
# Segregating the target variable
# and categorical predictors
##################################
proportion_y_variables = thyroid_cancer_train_predictors_categorical
proportion_x_variable = 'Recurred'
##################################
# Defining the number of
# rows and columns for the subplots
##################################
num_rows = 4
num_cols = 3
##################################
# Formulating the subplot structure
##################################
fig, axes = plt.subplots(num_rows, num_cols, figsize=(15, 20))
##################################
# Flattening the multi-row and
# multi-column axes
##################################
axes = axes.ravel()
##################################
# Formulating the individual stacked column plots
# for all categorical columns
##################################
for i, y_variable in enumerate(proportion_y_variables):
ax = axes[i]
category_counts = thyroid_cancer_train_column_filtered.groupby([proportion_x_variable, y_variable], observed=True).size().unstack(fill_value=0)
category_proportions = category_counts.div(category_counts.sum(axis=1), axis=0)
category_proportions.plot(kind='bar', stacked=True, ax=ax)
ax.set_title(f'{proportion_x_variable} Versus {y_variable}')
ax.set_xlabel(proportion_x_variable)
ax.set_ylabel('Proportions')
ax.legend(loc="lower center")
##################################
# Adjusting the subplot layout
##################################
plt.tight_layout()
##################################
# Presenting the subplots
##################################
plt.show()
1.4.4 Outlier and Distributional Shape Analysis¶
- No outliers (Outlier.Count>0, Outlier.Ratio>0.000), high skewness (Skewness>3 or Skewness<(-3)) or abnormal kurtosis (Skewness>2 or Skewness<(-2)) observed for the numeric predictor.
- Age: Outlier.Count = 0, Outlier.Ratio = 0.000, Skewness = 0.525, Kurtosis = -0.494
##################################
# Formulating the imputed dataset
# with numeric columns only
##################################
thyroid_cancer_train_column_filtered['Age'] = pd.to_numeric(thyroid_cancer_train_column_filtered['Age'])
thyroid_cancer_train_column_filtered_numeric = thyroid_cancer_train_column_filtered.select_dtypes(include='number')
thyroid_cancer_train_column_filtered_numeric = thyroid_cancer_train_column_filtered_numeric.to_frame() if isinstance(thyroid_cancer_train_column_filtered_numeric, pd.Series) else thyroid_cancer_train_column_filtered_numeric
##################################
# Gathering the variable names for each numeric column
##################################
numeric_variable_name_list = list(thyroid_cancer_train_column_filtered_numeric.columns)
##################################
# Gathering the skewness value for each numeric column
##################################
numeric_skewness_list = thyroid_cancer_train_column_filtered_numeric.skew()
##################################
# Computing the interquartile range
# for all columns
##################################
thyroid_cancer_train_column_filtered_numeric_q1 = thyroid_cancer_train_column_filtered_numeric.quantile(0.25)
thyroid_cancer_train_column_filtered_numeric_q3 = thyroid_cancer_train_column_filtered_numeric.quantile(0.75)
thyroid_cancer_train_column_filtered_numeric_iqr = thyroid_cancer_train_column_filtered_numeric_q3 - thyroid_cancer_train_column_filtered_numeric_q1
##################################
# Gathering the outlier count for each numeric column
# based on the interquartile range criterion
##################################
numeric_outlier_count_list = ((thyroid_cancer_train_column_filtered_numeric < (thyroid_cancer_train_column_filtered_numeric_q1 - 1.5 * thyroid_cancer_train_column_filtered_numeric_iqr)) | (thyroid_cancer_train_column_filtered_numeric > (thyroid_cancer_train_column_filtered_numeric_q3 + 1.5 * thyroid_cancer_train_column_filtered_numeric_iqr))).sum()
##################################
# Gathering the number of observations for each column
##################################
numeric_row_count_list = list([len(thyroid_cancer_train_column_filtered_numeric)] * len(thyroid_cancer_train_column_filtered_numeric.columns))
##################################
# Gathering the unique to count ratio for each numeric column
##################################
numeric_outlier_ratio_list = map(truediv, numeric_outlier_count_list, numeric_row_count_list)
##################################
# Gathering the skewness value for each numeric column
##################################
numeric_skewness_list = thyroid_cancer_train_column_filtered_numeric.skew()
##################################
# Gathering the kurtosis value for each numeric column
##################################
numeric_kurtosis_list = thyroid_cancer_train_column_filtered_numeric.kurtosis()
##################################
# Formulating the outlier summary
# for all numeric columns
##################################
numeric_column_outlier_summary = pd.DataFrame(zip(numeric_variable_name_list,
numeric_skewness_list,
numeric_outlier_count_list,
numeric_row_count_list,
numeric_outlier_ratio_list,
numeric_skewness_list,
numeric_kurtosis_list),
columns=['Numeric.Column.Name',
'Skewness',
'Outlier.Count',
'Row.Count',
'Outlier.Ratio',
'Skewness',
'Kurtosis'])
display(numeric_column_outlier_summary)
| Numeric.Column.Name | Skewness | Outlier.Count | Row.Count | Outlier.Ratio | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|---|
| 0 | Age | 0.525218 | 0 | 204 | 0.0 | 0.525218 | -0.494286 |
##################################
# Formulating the individual boxplots
# for all numeric columns
##################################
for column in thyroid_cancer_train_column_filtered_numeric:
plt.figure(figsize=(17,1))
sns.boxplot(data=thyroid_cancer_train_column_filtered_numeric, x=column)
1.4.5 Collinearity¶
- Majority of the predictors reported low (<0.50) to moderate (0.50 to 0.75) correlation.
- Among pairwise combinations of categorical predictors, high Phi.Coefficient values were noted for:
- N and Adenopathy: Phi.Coefficient = +0.805
- N and Risk: Phi.Coefficient = +0.726
- Adenopathy and Risk: Phi.Coefficient = +0.674
##################################
# Creating a dataset copy and
# converting all values to numeric
# for correlation analysis
##################################
pd.set_option('future.no_silent_downcasting', True)
thyroid_cancer_train_correlation = thyroid_cancer_train_column_filtered.copy()
thyroid_cancer_train_correlation_object = thyroid_cancer_train_correlation.iloc[:,1:13].columns
custom_category_orders = {
'Gender': ['M', 'F'],
'Smoking': ['No', 'Yes'],
'Thyroid_Function': ['Euthyroid', 'Hypothyroidism or Hyperthyroidism'],
'Physical_Examination': ['Normal or Single Nodular Goiter', 'Multinodular or Diffuse Goiter'],
'Adenopathy': ['No', 'Yes'],
'Pathology': ['Non-Papillary', 'Papillary'],
'Focality': ['Uni-Focal', 'Multi-Focal'],
'Risk': ['Low', 'Intermediate to High'],
'T': ['T1 to T2', 'T3 to T4b'],
'N': ['N0', 'N1'],
'Stage': ['I', 'II to IVB'],
'Response': ['Excellent', 'Indeterminate or Incomplete']
}
encoder = OrdinalEncoder(categories=[custom_category_orders[col] for col in thyroid_cancer_train_correlation_object])
thyroid_cancer_train_correlation[thyroid_cancer_train_correlation_object] = encoder.fit_transform(
thyroid_cancer_train_correlation[thyroid_cancer_train_correlation_object]
)
thyroid_cancer_train_correlation = thyroid_cancer_train_correlation.drop(['Recurred'], axis=1)
display(thyroid_cancer_train_correlation)
| Age | Gender | Smoking | Thyroid_Function | Physical_Examination | Adenopathy | Pathology | Focality | Risk | T | N | Stage | Response | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 140 | 28.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 205 | 36.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 |
| 277 | 41.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 294 | 42.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 |
| 268 | 32.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 300 | 67.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 |
| 115 | 37.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 67 | 51.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 161 | 22.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 55 | 21.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
204 rows × 13 columns
##################################
# Initializing the correlation matrix
##################################
thyroid_cancer_train_correlation_matrix = pd.DataFrame(np.zeros((len(thyroid_cancer_train_correlation.columns), len(thyroid_cancer_train_correlation.columns))),
columns=thyroid_cancer_train_correlation.columns,
index=thyroid_cancer_train_correlation.columns)
##################################
# Creating an empty correlation matrix
##################################
thyroid_cancer_train_correlation_matrix = pd.DataFrame(
np.zeros((len(thyroid_cancer_train_correlation.columns), len(thyroid_cancer_train_correlation.columns))),
index=thyroid_cancer_train_correlation.columns,
columns=thyroid_cancer_train_correlation.columns
)
##################################
# Calculating different types
# of correlation coefficients
# per variable type
##################################
for i in range(len(thyroid_cancer_train_correlation.columns)):
for j in range(i, len(thyroid_cancer_train_correlation.columns)):
if i == j:
thyroid_cancer_train_correlation_matrix.iloc[i, j] = 1.0
else:
col_i = thyroid_cancer_train_correlation.iloc[:, i]
col_j = thyroid_cancer_train_correlation.iloc[:, j]
# Detecting binary variables (assumes binary variables are coded as 0/1)
is_binary_i = col_i.nunique() == 2
is_binary_j = col_j.nunique() == 2
# Computing the Pearson correlation for two continuous variables
if col_i.dtype in ['int64', 'float64'] and col_j.dtype in ['int64', 'float64']:
corr = col_i.corr(col_j)
# Computing the Point-Biserial correlation for continuous and binary variables
elif (col_i.dtype in ['int64', 'float64'] and is_binary_j) or (col_j.dtype in ['int64', 'float64'] and is_binary_i):
continuous_var = col_i if col_i.dtype in ['int64', 'float64'] else col_j
binary_var = col_j if is_binary_j else col_i
# Convert binary variable to 0/1 (if not already)
binary_var = binary_var.astype('category').cat.codes
corr, _ = pointbiserialr(continuous_var, binary_var)
# Computing the Phi coefficient for two binary variables
elif is_binary_i and is_binary_j:
corr = col_i.corr(col_j)
# Computing the Cramér's V for two categorical variables (if more than 2 categories)
else:
contingency_table = pd.crosstab(col_i, col_j)
chi2, _, _, _ = chi2_contingency(contingency_table)
n = contingency_table.sum().sum()
phi2 = chi2 / n
r, k = contingency_table.shape
corr = np.sqrt(phi2 / min(k - 1, r - 1)) # Cramér's V formula
# Assigning correlation values to the matrix
thyroid_cancer_train_correlation_matrix.iloc[i, j] = corr
thyroid_cancer_train_correlation_matrix.iloc[j, i] = corr
# Displaying the correlation matrix
display(thyroid_cancer_train_correlation_matrix)
| Age | Gender | Smoking | Thyroid_Function | Physical_Examination | Adenopathy | Pathology | Focality | Risk | T | N | Stage | Response | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 1.000000 | -0.185530 | 0.299971 | 0.077845 | 0.012021 | 0.073931 | -0.215274 | 0.195272 | 0.205360 | 0.246838 | 0.013195 | 0.528144 | 0.317978 |
| Gender | -0.185530 | 1.000000 | -0.604101 | -0.093290 | -0.031935 | -0.158480 | 0.127817 | -0.218103 | -0.255507 | -0.215101 | -0.178550 | -0.219727 | -0.179431 |
| Smoking | 0.299971 | -0.604101 | 1.000000 | 0.064124 | 0.004339 | 0.192350 | -0.338086 | 0.182212 | 0.233024 | 0.231679 | 0.105463 | 0.327952 | 0.215362 |
| Thyroid_Function | 0.077845 | -0.093290 | 0.064124 | 1.000000 | 0.019964 | -0.137486 | -0.049893 | 0.051564 | -0.012519 | -0.042960 | -0.043275 | 0.080702 | -0.036498 |
| Physical_Examination | 0.012021 | -0.031935 | 0.004339 | 0.019964 | 1.000000 | 0.063246 | 0.018806 | 0.245779 | 0.166012 | 0.086039 | 0.104553 | 0.054799 | 0.116526 |
| Adenopathy | 0.073931 | -0.158480 | 0.192350 | -0.137486 | 0.063246 | 1.000000 | 0.047117 | 0.288750 | 0.673638 | 0.421762 | 0.805406 | 0.278749 | 0.518887 |
| Pathology | -0.215274 | 0.127817 | -0.338086 | -0.049893 | 0.018806 | 0.047117 | 1.000000 | -0.126299 | -0.117392 | -0.286899 | 0.157869 | -0.187683 | -0.154637 |
| Focality | 0.195272 | -0.218103 | 0.182212 | 0.051564 | 0.245779 | 0.288750 | -0.126299 | 1.000000 | 0.454926 | 0.518864 | 0.307716 | 0.372331 | 0.388741 |
| Risk | 0.205360 | -0.255507 | 0.233024 | -0.012519 | 0.166012 | 0.673638 | -0.117392 | 0.454926 | 1.000000 | 0.622459 | 0.726304 | 0.533264 | 0.631330 |
| T | 0.246838 | -0.215101 | 0.231679 | -0.042960 | 0.086039 | 0.421762 | -0.286899 | 0.518864 | 0.622459 | 1.000000 | 0.368430 | 0.468168 | 0.556742 |
| N | 0.013195 | -0.178550 | 0.105463 | -0.043275 | 0.104553 | 0.805406 | 0.157869 | 0.307716 | 0.726304 | 0.368430 | 1.000000 | 0.310156 | 0.542672 |
| Stage | 0.528144 | -0.219727 | 0.327952 | 0.080702 | 0.054799 | 0.278749 | -0.187683 | 0.372331 | 0.533264 | 0.468168 | 0.310156 | 1.000000 | 0.417025 |
| Response | 0.317978 | -0.179431 | 0.215362 | -0.036498 | 0.116526 | 0.518887 | -0.154637 | 0.388741 | 0.631330 | 0.556742 | 0.542672 | 0.417025 | 1.000000 |
##################################
# Plotting the correlation matrix
# for all pairwise combinations
# of numeric and categorical columns
##################################
plt.figure(figsize=(17, 8))
sns.heatmap(thyroid_cancer_train_correlation_matrix, annot=True, cmap='coolwarm', vmin=-1, vmax=1)
plt.show()
1.5. Data Exploration ¶
1.5.1 Exploratory Data Analysis¶
- Bivariate analysis identified individual predictors with generally positive association to the target variable based on visual inspection.
- Higher values or higher proportions for the following predictors are associated with the Recurred=Yes category:
- Age
- Gender=M
- Smoking=Yes
- Physical_Examination=Multinodular or Diffuse Goiter
- Adenopathy=Yes
- Focality=Multi-Focal
- Risk=Intermediate to High
- T=T3 to T4b
- N=N1
- Stage=II to IVB
- Response=Indeterminate or Incomplete
- Proportions for the following predictors are not associated with the Recurred=Yes or Recurred=No categories:
- Thyroid_Function
- Pathology
##################################
# Segregating the target
# and predictor variables
##################################
thyroid_cancer_train_column_filtered_predictors = thyroid_cancer_train_column_filtered.iloc[:,:-1].columns
thyroid_cancer_train_column_filtered_predictors_numeric = thyroid_cancer_train_column_filtered.iloc[:,:-1].loc[:, thyroid_cancer_train_column_filtered.iloc[:,:-1].columns == 'Age'].columns
thyroid_cancer_train_column_filtered_predictors_categorical = thyroid_cancer_train_column_filtered.iloc[:,:-1].loc[:,thyroid_cancer_train_column_filtered.iloc[:,:-1].columns != 'Age'].columns
##################################
# Gathering the variable names for each numeric column
##################################
numeric_variable_name_list = thyroid_cancer_train_column_filtered_predictors_numeric
##################################
# Segregating the target variable
# and numeric predictors
##################################
boxplot_y_variable = 'Recurred'
boxplot_x_variable = numeric_variable_name_list.values[0]
##################################
# Evaluating the numeric predictors
# against the target variable
##################################
plt.figure(figsize=(7, 5))
plt.boxplot([group[boxplot_x_variable] for name, group in thyroid_cancer_train_column_filtered.groupby(boxplot_y_variable, observed=True)])
plt.title(f'{boxplot_y_variable} Versus {boxplot_x_variable}')
plt.xlabel(boxplot_y_variable)
plt.ylabel(boxplot_x_variable)
plt.xticks(range(1, len(thyroid_cancer_train_column_filtered[boxplot_y_variable].unique()) + 1), ['No', 'Yes'])
plt.show()
##################################
# Segregating the target variable
# and categorical predictors
##################################
proportion_y_variables = thyroid_cancer_train_column_filtered_predictors_categorical
proportion_x_variable = 'Recurred'
##################################
# Defining the number of
# rows and columns for the subplots
##################################
num_rows = 4
num_cols = 3
##################################
# Formulating the subplot structure
##################################
fig, axes = plt.subplots(num_rows, num_cols, figsize=(15, 20))
##################################
# Flattening the multi-row and
# multi-column axes
##################################
axes = axes.ravel()
##################################
# Formulating the individual stacked column plots
# for all categorical columns
##################################
for i, y_variable in enumerate(proportion_y_variables):
ax = axes[i]
category_counts = thyroid_cancer_train_column_filtered.groupby([proportion_x_variable, y_variable], observed=True).size().unstack(fill_value=0)
category_proportions = category_counts.div(category_counts.sum(axis=1), axis=0)
category_proportions.plot(kind='bar', stacked=True, ax=ax)
ax.set_title(f'{proportion_x_variable} Versus {y_variable}')
ax.set_xlabel(proportion_x_variable)
ax.set_ylabel('Proportions')
ax.legend(loc="lower center")
##################################
# Adjusting the subplot layout
##################################
plt.tight_layout()
##################################
# Presenting the subplots
##################################
plt.show()
1.5.2 Hypothesis Testing¶
- The relationship between the numeric predictor to the Recurred target variable was statistically evaluated using the following hypotheses:
- Null: Difference in the means between groups Yes and No is equal to zero
- Alternative: Difference in the means between groups Yes and No is not equal to zero
- There is sufficient evidence to conclude of a statistically significant difference between the means of the numeric measurements obtained from Yes and No groups of the Recurred target variable in 1 of 1 numeric predictor given its high t-test statistic values with reported low p-values less than the significance level of 0.05.
- Age: T.Test.Statistic=-3.791, T.Test.PValue=0.000
- The relationship between the categorical predictors to the Recurred target variable was statistically evaluated using the following hypotheses:
- Null: The categorical predictor is independent of the categorical target variable
- Alternative: The categorical predictor is dependent of the categorical target variable
- There is sufficient evidence to conclude of a statistically significant relationship between the categories of the categorical predictors and the Yes and No groups of the Recurred target variable in 9 of 12 categorical predictors given their high chisquare statistic values with reported low p-values less than the significance level of 0.05.
- Risk: ChiSquare.Test.Statistic=98.599, ChiSquare.Test.PValue=0.000
- Response: ChiSquare.Test.Statistic=90.866, ChiSquare.Test.PValue=0.000
- Adenopathy: ChiSquare.Test.Statistic=73.585, ChiSquare.Test.PValue=0.000
- N: ChiSquare.Test.Statistic=73.176, ChiSquare.Test.PValue=0.000
- T: ChiSquare.Test.Statistic=62.205, ChiSquare.Test.PValue=0.000
- Stage: ChiSquare.Test.Statistic=44.963, ChiSquare.Test.PValue=0.000
- Focality: ChiSquare.Test.Statistic=32.859, ChiSquare.Test.PValue=0.000
- Gender: ChiSquare.Test.Statistic=17.787, ChiSquare.Test.PValue=0.000
- Smoking: ChiSquare.Test.Statistic=14.460, ChiSquare.Test.PValue=0.001
- There is marginal evidence to conclude of a statistically significant relationship between the categories of the categorical predictors and the Yes and No groups of the Recurred target variable in 1 of 12 categorical predictors given its sufficiently high chisquare statistic values with reported low p-values near the significance level of 0.10.
- Physical_Examination: ChiSquare.Test.Statistic=2.413, ChiSquare.Test.PValue=0.120
##################################
# Computing the t-test
# statistic and p-values
# between the target variable
# and numeric predictor columns
##################################
thyroid_cancer_numeric_ttest_target = {}
thyroid_cancer_numeric = thyroid_cancer_train_column_filtered.loc[:,(thyroid_cancer_train_column_filtered.columns == 'Age') | (thyroid_cancer_train_column_filtered.columns == 'Recurred')]
thyroid_cancer_numeric_columns = thyroid_cancer_train_column_filtered_predictors_numeric
for numeric_column in thyroid_cancer_numeric_columns:
group_0 = thyroid_cancer_numeric[thyroid_cancer_numeric.loc[:,'Recurred']=='No']
group_1 = thyroid_cancer_numeric[thyroid_cancer_numeric.loc[:,'Recurred']=='Yes']
thyroid_cancer_numeric_ttest_target['Recurred_' + numeric_column] = stats.ttest_ind(
group_0[numeric_column],
group_1[numeric_column],
equal_var=True)
##################################
# Formulating the pairwise ttest summary
# between the target variable
# and numeric predictor columns
##################################
thyroid_cancer_numeric_summary = thyroid_cancer_numeric.from_dict(thyroid_cancer_numeric_ttest_target, orient='index')
thyroid_cancer_numeric_summary.columns = ['T.Test.Statistic', 'T.Test.PValue']
display(thyroid_cancer_numeric_summary.sort_values(by=['T.Test.PValue'], ascending=True).head(len(thyroid_cancer_train_column_filtered_predictors_numeric)))
| T.Test.Statistic | T.Test.PValue | |
|---|---|---|
| Recurred_Age | -3.747942 | 0.000233 |
##################################
# Computing the chisquare
# statistic and p-values
# between the target variable
# and categorical predictor columns
##################################
thyroid_cancer_categorical_chisquare_target = {}
thyroid_cancer_categorical = thyroid_cancer_train_column_filtered.loc[:,(thyroid_cancer_train_column_filtered.columns != 'Age') | (thyroid_cancer_train_column_filtered.columns == 'Recurred')]
thyroid_cancer_categorical_columns = thyroid_cancer_train_column_filtered_predictors_categorical
for categorical_column in thyroid_cancer_categorical_columns:
contingency_table = pd.crosstab(thyroid_cancer_categorical[categorical_column],
thyroid_cancer_categorical['Recurred'])
thyroid_cancer_categorical_chisquare_target['Recurred_' + categorical_column] = stats.chi2_contingency(
contingency_table)[0:2]
##################################
# Formulating the pairwise chisquare summary
# between the target variable
# and categorical predictor columns
##################################
thyroid_cancer_categorical_summary = thyroid_cancer_categorical.from_dict(thyroid_cancer_categorical_chisquare_target, orient='index')
thyroid_cancer_categorical_summary.columns = ['ChiSquare.Test.Statistic', 'ChiSquare.Test.PValue']
display(thyroid_cancer_categorical_summary.sort_values(by=['ChiSquare.Test.PValue'], ascending=True).head(len(thyroid_cancer_train_column_filtered_predictors_categorical)))
| ChiSquare.Test.Statistic | ChiSquare.Test.PValue | |
|---|---|---|
| Recurred_Risk | 98.599608 | 3.090804e-23 |
| Recurred_Response | 90.866461 | 1.537030e-21 |
| Recurred_Adenopathy | 73.585561 | 9.636704e-18 |
| Recurred_N | 73.176134 | 1.185810e-17 |
| Recurred_T | 62.205367 | 3.094435e-15 |
| Recurred_Stage | 44.963917 | 2.006987e-11 |
| Recurred_Focality | 32.859398 | 9.907099e-09 |
| Recurred_Gender | 17.787641 | 2.469824e-05 |
| Recurred_Smoking | 14.460357 | 1.431406e-04 |
| Recurred_Physical_Examination | 2.413115 | 1.203227e-01 |
| Recurred_Thyroid_Function | 0.966826 | 3.254729e-01 |
| Recurred_Pathology | 0.131614 | 7.167646e-01 |
1.6. Premodelling Data Preparation ¶
1.6.1 Preprocessed Data Description¶
- A total of 6 of the 16 predictors were excluded from the dataset based on the data preprocessing and exploration findings
- There were 3 categorical predictors excluded from the dataset after having been observed with extremely low variance containing categories with very few or almost no variations across observations that may have limited predictive power or drive increased model complexity without performance gains:
- Hx_Smoking:
- 193 Hx_Smoking=No: 94.61%
- 11 Hx_Smoking=Yes: 5.39%
- Hx_Radiotherapy:
- 202 Hx_Radiotherapy=No: 99.02%
- 2 Hx_Radiotherapy=Yes: 0.98%
- M:
- 194 M=M0: 95.10%
- 10 M=M1: 4.90%
- Hx_Smoking:
- There was 1 categorical predictor excluded from the dataset after having been observed with high pairwise collinearity (Phi.Coefficient>0.70) with other 2 predictors that might provide redundant information, leading to potential instability in regression models.
- N and Adenopathy: Phi.Coefficient = +0.805
- N and Risk: Phi.Coefficient = +0.726
- Another 2 categorical predictors were excluded from the dataset for not exhibiting a statistically significant association with the Yes and No groups of the Recurred target variable, indicating weak predictive value.
- Thyroid_Function: ChiSquare.Test.Statistic=0.967, ChiSquare.Test.PValue=0.325
- Pathology: ChiSquare.Test.Statistic=0.132, ChiSquare.Test.PValue=0.717
- The preprocessed train data (final) subset is comprised of:
- 204 rows (observations)
- 143 Recurred=No: 70.10%
- 61 Recurred=Yes: 29.90%
- 11 columns (variables)
- 1/11 target (categorical)
- Recurred
- 1/11 predictor (numeric)
- Age
- 9/11 predictor (categorical)
- Gender
- Smoking
- Physical_Examination
- Adenopathy
- Focality
- Risk
- T
- M
- Stage
- Response
- 1/11 target (categorical)
- 204 rows (observations)
1.6.2 Preprocessing Pipeline Development¶
- A preprocessing pipeline was formulated and applied to the train data (final), validation data and test data with the following actions:
- Excluded specified columns noted with low variance, high collinearity and weak predictive power
- Aggregated categories in multiclass categorical variables into binary levels
- Converted categorical columns to the appropriate type
- Set the order of category levels for ordinal encoding during modeling pipeline creation
##################################
# Formulating a preprocessing pipeline
# that removes the specified columns,
# aggregates categories in multiclass categorical variables,
# converts categorical columns to the appropriate type, and
# sets the order of category levels
##################################
def preprocess_dataset(df):
# Removing the specified columns
columns_to_remove = ['Hx_Smoking', 'Hx_Radiotherapy', 'M', 'N', 'Thyroid_Function', 'Pathology']
df = df.drop(columns=columns_to_remove)
# Applying category aggregation
df['Physical_Examination'] = df['Physical_Examination'].map(
lambda x: 'Normal or Single Nodular Goiter' if x in ['Normal', 'Single nodular goiter-left', 'Single nodular goiter-right']
else 'Multinodular or Diffuse Goiter').astype('category')
df['Adenopathy'] = df['Adenopathy'].map(
lambda x: 'No' if x == 'No' else ('Yes' if pd.notna(x) and x != '' else x)).astype('category')
df['Risk'] = df['Risk'].map(
lambda x: 'Low' if x == 'Low' else 'Intermediate to High').astype('category')
df['T'] = df['T'].map(
lambda x: 'T1 to T2' if x in ['T1a', 'T1b', 'T2'] else 'T3 to T4b').astype('category')
df['Stage'] = df['Stage'].map(
lambda x: 'I' if x == 'I' else 'II to IVB').astype('category')
df['Response'] = df['Response'].map(
lambda x: 'Indeterminate or Incomplete' if x in ['Indeterminate', 'Structural Incomplete', 'Biochemical Incomplete']
else 'Excellent').astype('category')
# Setting category levels
category_mappings = {
'Gender': ['M', 'F'],
'Smoking': ['No', 'Yes'],
'Physical_Examination': ['Normal or Single Nodular Goiter', 'Multinodular or Diffuse Goiter'],
'Adenopathy': ['No', 'Yes'],
'Focality': ['Uni-Focal', 'Multi-Focal'],
'Risk': ['Low', 'Intermediate to High'],
'T': ['T1 to T2', 'T3 to T4b'],
'Stage': ['I', 'II to IVB'],
'Response': ['Excellent', 'Indeterminate or Incomplete']
}
for col, categories in category_mappings.items():
df[col] = df[col].astype('category')
df[col] = df[col].cat.set_categories(categories, ordered=True)
return df
##################################
# Applying the preprocessing pipeline
# to the train data
##################################
thyroid_cancer_preprocessed_train = preprocess_dataset(thyroid_cancer_train)
X_preprocessed_train = thyroid_cancer_preprocessed_train.drop('Recurred', axis = 1)
y_preprocessed_train = thyroid_cancer_preprocessed_train['Recurred']
thyroid_cancer_preprocessed_train.to_csv(os.path.join("..", DATASETS_PREPROCESSED_TRAIN_PATH, "thyroid_cancer_preprocessed_train.csv"), index=False)
X_preprocessed_train.to_csv(os.path.join("..", DATASETS_PREPROCESSED_TRAIN_FEATURES_PATH, "X_preprocessed_train.csv"), index=False)
y_preprocessed_train.to_csv(os.path.join("..", DATASETS_PREPROCESSED_TRAIN_TARGET_PATH, "y_preprocessed_train.csv"), index=False)
train_csv_path = os.path.join("..", DATASETS_PREPROCESSED_TRAIN_PATH, "thyroid_cancer_preprocessed_train.csv")
print('Final Preprocessed Train Dataset Dimensions: ')
display(X_preprocessed_train.shape)
display(y_preprocessed_train.shape)
print('Final Preprocessed Train Target Variable Breakdown: ')
display(y_preprocessed_train.value_counts())
print('Final Preprocessed Train Target Variable Proportion: ')
display(y_preprocessed_train.value_counts(normalize = True))
thyroid_cancer_preprocessed_train.head()
Final Preprocessed Train Dataset Dimensions:
(204, 10)
(204,)
Final Preprocessed Train Target Variable Breakdown:
Recurred No 143 Yes 61 Name: count, dtype: int64
Final Preprocessed Train Target Variable Proportion:
Recurred No 0.70098 Yes 0.29902 Name: proportion, dtype: float64
| Age | Gender | Smoking | Physical_Examination | Adenopathy | Focality | Risk | T | Stage | Response | Recurred | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 140 | 28.0 | F | No | Multinodular or Diffuse Goiter | No | Uni-Focal | Low | T1 to T2 | I | Excellent | No |
| 205 | 36.0 | F | No | Normal or Single Nodular Goiter | Yes | Uni-Focal | Low | T1 to T2 | I | Indeterminate or Incomplete | No |
| 277 | 41.0 | M | Yes | Normal or Single Nodular Goiter | No | Multi-Focal | Intermediate to High | T3 to T4b | I | Excellent | No |
| 294 | 42.0 | M | No | Normal or Single Nodular Goiter | No | Multi-Focal | Intermediate to High | T3 to T4b | I | Indeterminate or Incomplete | No |
| 268 | 32.0 | F | No | Normal or Single Nodular Goiter | No | Uni-Focal | Low | T3 to T4b | I | Excellent | No |
##################################
# Applying the preprocessing pipeline
# to the validation data
##################################
thyroid_cancer_preprocessed_validation = preprocess_dataset(thyroid_cancer_validation)
X_preprocessed_validation = thyroid_cancer_preprocessed_validation.drop('Recurred', axis = 1)
y_preprocessed_validation = thyroid_cancer_preprocessed_validation['Recurred']
thyroid_cancer_preprocessed_validation.to_csv(os.path.join("..", DATASETS_PREPROCESSED_VALIDATION_PATH, "thyroid_cancer_preprocessed_validation.csv"), index=False)
X_preprocessed_validation.to_csv(os.path.join("..", DATASETS_PREPROCESSED_VALIDATION_FEATURES_PATH, "X_preprocessed_validation.csv"), index=False)
y_preprocessed_validation.to_csv(os.path.join("..", DATASETS_PREPROCESSED_VALIDATION_TARGET_PATH, "y_preprocessed_validation.csv"), index=False)
validation_csv_path = os.path.join("..", DATASETS_PREPROCESSED_VALIDATION_PATH, "thyroid_cancer_preprocessed_validation.csv")
print('Final Preprocessed Validation Dataset Dimensions: ')
display(X_preprocessed_validation.shape)
display(y_preprocessed_validation.shape)
print('Final Preprocessed Validation Target Variable Breakdown: ')
display(y_preprocessed_validation.value_counts())
print('Final Preprocessed Validation Target Variable Proportion: ')
display(y_preprocessed_validation.value_counts(normalize = True))
thyroid_cancer_preprocessed_validation.head()
Final Preprocessed Validation Dataset Dimensions:
(69, 10)
(69,)
Final Preprocessed Validation Target Variable Breakdown:
Recurred No 49 Yes 20 Name: count, dtype: int64
Final Preprocessed Validation Target Variable Proportion:
Recurred No 0.710145 Yes 0.289855 Name: proportion, dtype: float64
| Age | Gender | Smoking | Physical_Examination | Adenopathy | Focality | Risk | T | Stage | Response | Recurred | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 173 | 30.0 | F | No | Normal or Single Nodular Goiter | No | Uni-Focal | Low | T1 to T2 | I | Indeterminate or Incomplete | No |
| 164 | 29.0 | F | No | Normal or Single Nodular Goiter | No | Multi-Focal | Low | T1 to T2 | I | Excellent | No |
| 256 | 21.0 | M | Yes | Normal or Single Nodular Goiter | No | Uni-Focal | Low | T3 to T4b | I | Indeterminate or Incomplete | No |
| 348 | 58.0 | F | No | Multinodular or Diffuse Goiter | Yes | Multi-Focal | Intermediate to High | T3 to T4b | II to IVB | Indeterminate or Incomplete | Yes |
| 131 | 31.0 | F | No | Normal or Single Nodular Goiter | No | Uni-Focal | Low | T1 to T2 | I | Excellent | No |
##################################
# Applying the preprocessing pipeline
# to the test data
##################################
thyroid_cancer_preprocessed_test = preprocess_dataset(thyroid_cancer_test)
X_preprocessed_test = thyroid_cancer_preprocessed_test.drop('Recurred', axis = 1)
y_preprocessed_test = thyroid_cancer_preprocessed_test['Recurred']
thyroid_cancer_preprocessed_test.to_csv(os.path.join("..", DATASETS_PREPROCESSED_TEST_PATH, "thyroid_cancer_preprocessed_test.csv"), index=False)
X_preprocessed_test.to_csv(os.path.join("..", DATASETS_PREPROCESSED_TEST_FEATURES_PATH, "X_preprocessed_test.csv"), index=False)
y_preprocessed_test.to_csv(os.path.join("..", DATASETS_PREPROCESSED_TEST_TARGET_PATH, "y_preprocessed_test.csv"), index=False)
print('Final Preprocessed Test Dataset Dimensions: ')
display(X_preprocessed_test.shape)
display(y_preprocessed_test.shape)
print('Final Preprocessed Test Target Variable Breakdown: ')
display(y_preprocessed_test.value_counts())
print('Final Preprocessed Test Target Variable Proportion: ')
display(y_preprocessed_test.value_counts(normalize = True))
thyroid_cancer_preprocessed_test.head()
Final Preprocessed Test Dataset Dimensions:
(91, 10)
(91,)
Final Preprocessed Test Target Variable Breakdown:
Recurred No 64 Yes 27 Name: count, dtype: int64
Final Preprocessed Test Target Variable Proportion:
Recurred No 0.703297 Yes 0.296703 Name: proportion, dtype: float64
| Age | Gender | Smoking | Physical_Examination | Adenopathy | Focality | Risk | T | Stage | Response | Recurred | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 345 | 25.0 | F | No | Multinodular or Diffuse Goiter | Yes | Multi-Focal | Intermediate to High | T3 to T4b | I | Indeterminate or Incomplete | Yes |
| 249 | 46.0 | F | No | Normal or Single Nodular Goiter | No | Multi-Focal | Low | T3 to T4b | I | Excellent | No |
| 83 | 40.0 | F | No | Normal or Single Nodular Goiter | No | Uni-Focal | Intermediate to High | T1 to T2 | I | Excellent | No |
| 184 | 67.0 | F | No | Normal or Single Nodular Goiter | No | Uni-Focal | Low | T1 to T2 | I | Excellent | No |
| 146 | 25.0 | F | No | Multinodular or Diffuse Goiter | No | Uni-Focal | Low | T1 to T2 | I | Indeterminate or Incomplete | No |
##################################
# Defining a function to compute
# model performance
##################################
def model_performance_evaluation(y_true, y_pred):
metric_name = ['Accuracy','Precision','Recall','F1','AUROC']
metric_value = [accuracy_score(y_true, y_pred),
precision_score(y_true, y_pred),
recall_score(y_true, y_pred),
f1_score(y_true, y_pred),
roc_auc_score(y_true, y_pred)]
metric_summary = pd.DataFrame(zip(metric_name, metric_value),
columns=['metric_name','metric_value'])
return(metric_summary)
##################################
# Defining a function to compute
# model performance
##################################
def evaluate_binary_metrics(y_true, y_pred):
acc = accuracy_score(y_true, y_pred)
f1 = f1_score(y_true, y_pred)
recall = recall_score(y_true, y_pred) # Sensitivity
tn, fp, fn, tp = confusion_matrix(y_true, y_pred).ravel()
specificity = tn / (tn + fp) if (tn + fp) > 0 else 0
return {'accuracy': acc, 'sensitivity': recall, 'specificity': specificity, 'f1': f1}
##################################
# Defining a function to
# create a hash representing a specific experiment configuration
##################################
def generate_run_hash(params: dict, mccv_seed: int) -> str:
signature = {
"params": params,
"mccv_seed": mccv_seed # ensures same splits produce same hash
}
run_string = json.dumps(signature, sort_keys=True)
return hashlib.md5(run_string.encode("utf-8")).hexdigest()
##################################
# Defining a function to
# add utility to search MLflow for a matching hash
##################################
def find_existing_run_by_hash(experiment_name: str, run_hash: str):
client = MlflowClient()
experiment = client.get_experiment_by_name(experiment_name)
if not experiment:
return None
runs = client.search_runs(experiment_ids=[experiment.experiment_id])
for run in runs:
if run.data.tags.get("run_hash") == run_hash:
return run.info.run_id
return None
##################################
# Defining a function to
# delete an existing MlFlow run before logging and
# keeping only the latest
##################################
def delete_run(run_id: str):
client = MlflowClient()
client.set_terminated(run_id, status="KILLED")
client.delete_run(run_id)
##################################
# Defining a function to
# delete all MLflow runs in an experiment
# in case needed
##################################
def delete_all_runs_in_experiment(experiment_name: str):
client = MlflowClient()
experiment = client.get_experiment_by_name(experiment_name)
if experiment is None:
print(f"No experiment found with name '{experiment_name}'")
return
experiment_id = experiment.experiment_id
runs = client.search_runs([experiment_id], max_results=10000)
if not runs:
print(f"No runs found in experiment '{experiment_name}'")
return
print(f"Found {len(runs)} runs in experiment '{experiment_name}'. Deleting...")
for run in runs:
run_id = run.info.run_id
client.set_terminated(run_id, status="KILLED") # Optional, but safe
client.delete_run(run_id)
print("All runs deleted.")
##################################
# Defining a function to
# completely remove the MLflow experiment
# including the name and all metadata
##################################
def remove_experiment_name_metadata(experiment_name: str):
client = MlflowClient()
experiment = client.get_experiment_by_name(experiment_name)
if experiment:
client.delete_experiment(experiment.experiment_id)
print(f"Deleted experiment '{experiment.name}'.")
##################################
# Defining a function to
# generate a confusion matrix
# and log as an MLflow artifact
##################################
def log_confusion_matrix(y_true, y_pred, labels, output_path):
cm = confusion_matrix(y_true, y_pred, labels=labels)
plt.figure(figsize=(6, 5))
sns.heatmap(cm, annot=True, fmt='d', cmap='Blues',
xticklabels=labels, yticklabels=labels)
plt.xlabel('Predicted')
plt.ylabel('True')
plt.title('Confusion Matrix')
plt.tight_layout()
plt.savefig(output_path)
plt.close()
##################################
# Defining a function to
# generate a classification report
# and log as an MLflow artifact
##################################
def log_classification_report(y_true, y_pred, labels, output_path):
report = classification_report(y_true, y_pred, target_names=[str(l) for l in labels])
with open(output_path, 'w') as f:
f.write(report)
1.7. Ensemble Model Development, Logging and Tracking using the MLflow Framework ¶
1.7.1 Random Forest¶
Random Forest is an ensemble learning method that builds multiple decision trees and combines their outputs to improve prediction accuracy and robustness in binary classification. Instead of relying on a single decision tree, it aggregates multiple trees, reducing overfitting and increasing generalizability. The algorithm works by training individual decision trees on bootstrapped samples of the dataset, where each tree is trained on a slightly different subset of data. Additionally, at each decision node, a random subset of features is considered for splitting, adding further diversity among the trees. The final classification is determined by majority voting across all trees. The main advantages of Random Forest include its resilience to overfitting, ability to handle high-dimensional data, and robustness against noisy data. However, it has limitations, such as higher computational cost due to multiple trees and reduced interpretability compared to a single decision tree. It can also struggle with highly imbalanced data unless additional techniques like class weighting are applied.
- The random forest model from the sklearn.ensemble Python library API was implemented. Combined with a preprocessing operation into a pipeline, the mlflow.sklearn module was used for loading and logging the model during experiment tracking.
- The model contains 4 hyperparameters for tuning:
- criterion = function to measure the quality of a split made to vary between gini and entropy
- max_depth = maximum depth of the tree made to vary between 3 and 6
- min_samples_leaf = minimum number of samples required to be at a leaf node made to vary between 5 and 10
- n_estimators = number of base estimators in the ensemble made to vary between 100 and 200
- A special hyperparameter (class_weight = balanced) was fixed to address the minimal 2:1 class imbalance observed between the No and Yes Recurred categories.
- Hyperparameter tuning was conducted using a 25-cycle 70-30 stratified train-test split Monte Carlo cross-validation with the results logged using MLflow experiment tracking and organized under bagged_rf_experiment.
- The optimal model performance evaluated using the F1 score, sensitivity and specificity metrics, respectively, was determined for the following hyperparameter combination:
- criterion = entropy
- max_depth = 3
- min_samples_leaf = 5
- n_estimators = 200
- The MCCV model performance of the optimal model as accessed from the MLflow user interface is summarized as follows:
- F1 Score = 0.8310
- Sensitivity = 0.8778
- Specificity = 0.8948
- Accuracy = 0.8896
- The independent validation model performance as accessed from the MLflow user interface of the optimal model is summarized as follows:
- F1 Score = 0.8571
- Sensitivity = 0.9000
- Specificity = 0.9183
- Accuracy = 0.9130
##################################
# Setting the model naming conventions
##################################
model_name = "bagged_rf"
experiment_name = f"{model_name}_experiment"
artifact_dir = f"artifacts/{model_name}"
os.makedirs(artifact_dir, exist_ok=True)
##################################
# Setting the MLflow experiment
# for the Random Forest model
##################################
mlflow.set_experiment(experiment_name)
client = MlflowClient()
##################################
# Defining the categorical preprocessing parameters
##################################
categorical_features = ['Gender','Smoking','Physical_Examination','Adenopathy','Focality','Risk','T','Stage','Response']
categorical_transformer = OrdinalEncoder()
categorical_preprocessor = ColumnTransformer(transformers=[
('cat', categorical_transformer, categorical_features)],
remainder='passthrough',
force_int_remainder_cols=False)
##################################
# Defining the preprocessing and modeling pipeline parameters
##################################
bagged_rf_pipeline = Pipeline([
('categorical_preprocessor', categorical_preprocessor),
('bagged_rf_model', RandomForestClassifier(class_weight='balanced',
random_state=987654321))
])
##################################
# Defining hyperparameter grid
##################################
bagged_rf_hyperparameter_grid = {
'bagged_rf_model__criterion': ['gini', 'entropy'],
'bagged_rf_model__max_depth': [3, 6],
'bagged_rf_model__min_samples_leaf': [5, 10],
'bagged_rf_model__n_estimators': [100, 200]
}
param_combos = list(ParameterGrid(bagged_rf_hyperparameter_grid))
##################################
# Encoding the response variables
# for model evaluation
##################################
y_encoder = OrdinalEncoder()
y_encoder.fit(y_preprocessed_train.values.reshape(-1, 1))
y_preprocessed_train_encoded = y_encoder.transform(y_preprocessed_train.values.reshape(-1, 1)).ravel()
class_labels = list(y_encoder.categories_[0])
y_preprocessed_validation_encoded = y_encoder.transform(y_preprocessed_validation.values.reshape(-1, 1)).ravel()
##################################
# Defining the cross-validation strategy
# using Monte Carlo Cross-Validation Setup (25 cycles of 70-30 stratified splits))
##################################
mccv = StratifiedShuffleSplit(n_splits=25,
test_size=0.3,
random_state=987654321)
##################################
# Creating containers for the best hyperparameter combination
# based on the F1 Score, Sensitivity and Specificity metrics
##################################
best_combo = None
best_metrics = {'f1': -1, 'sensitivity': -1, 'specificity': -1}
all_combos_metrics = []
##################################
# Implementing experiment tracking
# of the hyperparameter tuning exercise
# using MLflow for the Random Forest model
##################################
for combo_index, param_set in enumerate(param_combos):
print(f"Evaluating param combo {combo_index + 1}/{len(param_combos)}: {param_set}")
# Generating hash and checking for duplicates
run_hash = generate_run_hash(param_set, mccv_seed=987654321)
existing_run_id = find_existing_run_by_hash("bagged_rf_experiment", run_hash)
if existing_run_id:
print(f"Duplicate found for this combo (hash={run_hash}). Removing old run: {existing_run_id}")
delete_run(existing_run_id)
else:
print(f"No duplicate found. Proceeding to evaluate and log.")
all_metrics = []
for i, (train_idx, test_idx) in enumerate(mccv.split(X_preprocessed_train, y_preprocessed_train_encoded)):
X_train, X_test = X_preprocessed_train.iloc[train_idx], X_preprocessed_train.iloc[test_idx]
y_train, y_test = y_preprocessed_train_encoded[train_idx], y_preprocessed_train_encoded[test_idx]
# Cloning the pipeline and setting the hyperparameters
model = clone(bagged_rf_pipeline)
model.set_params(**param_set)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
all_metrics.append(evaluate_binary_metrics(y_test, y_pred))
# Aggregating the metrics across MCCV
df_metrics = pd.DataFrame(all_metrics)
avg_metrics = df_metrics.mean().to_dict()
avg_metrics_prefixed = {f"avg_mccv_{k}": v for k, v in avg_metrics.items()}
all_combos_metrics.append({**param_set, **avg_metrics_prefixed})
# Logging the hyperparameter combinations and metrics to MLflow
with mlflow.start_run(run_name=f"RF_HyperparameterCombo_{combo_index+1}"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", run_hash)
mlflow.log_params(param_set)
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in avg_metrics.items()})
mlflow.set_tag("param_index", combo_index)
# Tracking the best hyperparameter combination
better = False
if avg_metrics['f1'] > best_metrics['f1']:
better = True
elif avg_metrics['f1'] == best_metrics['f1']:
if avg_metrics['sensitivity'] > best_metrics['sensitivity']:
better = True
elif avg_metrics['sensitivity'] == best_metrics['sensitivity']:
if avg_metrics['specificity'] > best_metrics['specificity']:
better = True
if better:
best_combo = param_set
best_combo_index = combo_index
best_metrics = avg_metrics
Evaluating param combo 1/16: {'bagged_rf_model__criterion': 'gini', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 100}
Duplicate found for this combo (hash=a5d432cd7e9c3b18c9027bc72eb73a11). Removing old run: ada7c87379da4bcf8a9c14713db6079d
Evaluating param combo 2/16: {'bagged_rf_model__criterion': 'gini', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 200}
Duplicate found for this combo (hash=5598839645a2547b53af6e59276f6ef6). Removing old run: 1908d94563f94f108c108787e293d22a
Evaluating param combo 3/16: {'bagged_rf_model__criterion': 'gini', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 10, 'bagged_rf_model__n_estimators': 100}
Duplicate found for this combo (hash=956ffce5827717d1916515f23ab21a66). Removing old run: 1cde73db5b7d4b79859329badaaf4399
Evaluating param combo 4/16: {'bagged_rf_model__criterion': 'gini', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 10, 'bagged_rf_model__n_estimators': 200}
Duplicate found for this combo (hash=eed7147b6316d00f992ec99413e153ec). Removing old run: 7aaa58643c2342b7b2fe1ed3d7f61144
Evaluating param combo 5/16: {'bagged_rf_model__criterion': 'gini', 'bagged_rf_model__max_depth': 6, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 100}
Duplicate found for this combo (hash=1900e2c31ed46c7a59addf558a3f89b9). Removing old run: c1145c63aa054fb69729713edf60ea38
Evaluating param combo 6/16: {'bagged_rf_model__criterion': 'gini', 'bagged_rf_model__max_depth': 6, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 200}
Duplicate found for this combo (hash=45bfa5bc1d7ae7d9757753f8f5ff108b). Removing old run: 22beacd9a264412688005a86a76592dd
Evaluating param combo 7/16: {'bagged_rf_model__criterion': 'gini', 'bagged_rf_model__max_depth': 6, 'bagged_rf_model__min_samples_leaf': 10, 'bagged_rf_model__n_estimators': 100}
Duplicate found for this combo (hash=c03a488f1ead20eb6657be0995d3c22a). Removing old run: 20552af9cfd34819b1a1fa24b06518d0
Evaluating param combo 8/16: {'bagged_rf_model__criterion': 'gini', 'bagged_rf_model__max_depth': 6, 'bagged_rf_model__min_samples_leaf': 10, 'bagged_rf_model__n_estimators': 200}
Duplicate found for this combo (hash=0b7bcf590451eb776df6dbd593cab3ab). Removing old run: fa42ae73cf044589a854800ddf9785d3
Evaluating param combo 9/16: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 100}
Duplicate found for this combo (hash=925c3c35a791f723107cc08fe6e75da7). Removing old run: 47fae6bd42b6407f947ac62612ec872b
Evaluating param combo 10/16: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 200}
Duplicate found for this combo (hash=31afe18fea6914252e1d87a0f76b9901). Removing old run: c8313358afc84cfab1a64b793da9d917
Evaluating param combo 11/16: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 10, 'bagged_rf_model__n_estimators': 100}
Duplicate found for this combo (hash=8209f8af91543888e31ffef715fbaf28). Removing old run: bd14f9b9cf5845068e2cd13187785101
Evaluating param combo 12/16: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 10, 'bagged_rf_model__n_estimators': 200}
Duplicate found for this combo (hash=eef7e43b885e6c1da3df21a385b913a9). Removing old run: 2db04d02932e46f5a3f0b8b364bccf93
Evaluating param combo 13/16: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 6, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 100}
Duplicate found for this combo (hash=78ade1ff4986105c5c8ea5b4f258114b). Removing old run: c79ca00f4410441099de39f5c32432cd
Evaluating param combo 14/16: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 6, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 200}
Duplicate found for this combo (hash=41c726def174a1b7e7b2b48b032270b7). Removing old run: 995786e4bcf341ccad65a317b87d24ab
Evaluating param combo 15/16: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 6, 'bagged_rf_model__min_samples_leaf': 10, 'bagged_rf_model__n_estimators': 100}
Duplicate found for this combo (hash=e244553ff66fd707f7fb78e4835d5544). Removing old run: 04903691a5154848bfaf6460729bef9c
Evaluating param combo 16/16: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 6, 'bagged_rf_model__min_samples_leaf': 10, 'bagged_rf_model__n_estimators': 200}
Duplicate found for this combo (hash=36578547d5fa720aece0885694e3d15d). Removing old run: 1f880d9b217846f6bba739dd2ad3104e
##################################
# Defining a function to
# generate a unique hash for the selected optimal model
##################################
final_model_hash = generate_run_hash(best_combo, mccv_seed=987654321)
existing_final_run_id = find_existing_run_by_hash("bagged_rf_experiment", final_model_hash)
if existing_final_run_id:
print(f"Duplicate final model found (hash={final_model_hash}). Removing previous run: {existing_final_run_id}")
delete_run(existing_final_run_id)
else:
print(f"No duplicate final model found. Proceeding to log new final model.")
Duplicate final model found (hash=31afe18fea6914252e1d87a0f76b9901). Removing previous run: a07d03c52a1f4516bc023cdbc59b3118
##################################
# Retraining the most optimal Random Forest model
# on the full training data
##################################
print("Optimal Hyperparameter Combination for Random Forest:", best_combo)
final_model = clone(bagged_rf_pipeline)
final_model.set_params(**best_combo)
final_model.fit(X_preprocessed_train, y_preprocessed_train_encoded)
Optimal Hyperparameter Combination for Random Forest: {'bagged_rf_model__criterion': 'entropy', 'bagged_rf_model__max_depth': 3, 'bagged_rf_model__min_samples_leaf': 5, 'bagged_rf_model__n_estimators': 200}
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('bagged_rf_model',
RandomForestClassifier(class_weight='balanced',
criterion='entropy', max_depth=3,
min_samples_leaf=5, n_estimators=200,
random_state=987654321))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('bagged_rf_model',
RandomForestClassifier(class_weight='balanced',
criterion='entropy', max_depth=3,
min_samples_leaf=5, n_estimators=200,
random_state=987654321))])ColumnTransformer(force_int_remainder_cols=False, remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking', 'Physical_Examination',
'Adenopathy', 'Focality', 'Risk', 'T',
'Stage', 'Response'])])['Gender', 'Smoking', 'Physical_Examination', 'Adenopathy', 'Focality', 'Risk', 'T', 'Stage', 'Response']
OrdinalEncoder()
['Age']
passthrough
RandomForestClassifier(class_weight='balanced', criterion='entropy',
max_depth=3, min_samples_leaf=5, n_estimators=200,
random_state=987654321)##################################
# Evaluating the most optimal Random Forest model
# on the validation data
##################################
y_validation_pred = final_model.predict(X_preprocessed_validation)
validation_metrics = evaluate_binary_metrics(y_preprocessed_validation_encoded, y_validation_pred)
##################################
# Consolidating the optimal hyperparameter combination
# and its corresponding MCCV and validation metrics
##################################
consolidated_table_1 = pd.DataFrame({
'hyperparameter': list(best_combo.keys()),
'mccv.optimal.value': list(best_combo.values())
})
consolidated_table_2 = pd.DataFrame({
'metric': list(best_metrics.keys()),
'mccv': [round(v, 4) for v in best_metrics.values()],
'validation': [round(validation_metrics[k], 4) for k in best_metrics.keys()]
})
print(f"{experiment_name} optimal hyperparameter combination:")
display(consolidated_table_1)
print(f"{experiment_name} optimal hyperparameter combination mccv versus validation performance metrics:")
display(consolidated_table_2)
bagged_rf_experiment optimal hyperparameter combination:
| hyperparameter | mccv.optimal.value | |
|---|---|---|
| 0 | bagged_rf_model__criterion | entropy |
| 1 | bagged_rf_model__max_depth | 3 |
| 2 | bagged_rf_model__min_samples_leaf | 5 |
| 3 | bagged_rf_model__n_estimators | 200 |
bagged_rf_experiment optimal hyperparameter combination mccv versus validation performance metrics:
| metric | mccv | validation | |
|---|---|---|---|
| 0 | accuracy | 0.8897 | 0.9130 |
| 1 | sensitivity | 0.8779 | 0.9000 |
| 2 | specificity | 0.8949 | 0.9184 |
| 3 | f1 | 0.8310 | 0.8571 |
##################################
# Decoding the true and predicted labels
# for artifact storage
##################################
y_validation_true_decoded = y_encoder.inverse_transform(y_preprocessed_validation_encoded.reshape(-1, 1)).ravel()
y_validation_pred_decoded = y_encoder.inverse_transform(y_validation_pred.reshape(-1, 1)).ravel()
##################################
# Saving the artifacts from the Random Forest model
# hyperparameter tuning exercise
##################################
mccv_csv_path = os.path.join(artifact_dir, f"{model_name}_MCCV_metrics.csv")
cm_path = os.path.join(artifact_dir, f"{model_name}_validation_confusion_matrix.png")
report_path = os.path.join(artifact_dir, f"{model_name}_validation_classification_report.txt")
df_all_combos = pd.DataFrame(all_combos_metrics)
df_all_combos.to_csv(mccv_csv_path, index=False)
log_confusion_matrix(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=cm_path)
log_classification_report(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=report_path)
pickle_model_path = os.path.join(artifact_dir, f"{model_name}_final_model.pkl")
with open(pickle_model_path, "wb") as f:
pickle.dump(final_model, f)
train_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_train_dataset.csv")
thyroid_cancer_preprocessed_train.to_csv(train_dataset_csv_path, index=False)
validation_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_validation_dataset.csv")
thyroid_cancer_preprocessed_validation.to_csv(validation_dataset_csv_path, index=False)
##################################
# Logging the most optimal Random Forest model
# developed from the full training data
# and evaluated on the validation data
##################################
# Casting integer columns in preprocessed training data to float64
X_logged_input = X_preprocessed_train.astype({
col: 'float64' for col in X_preprocessed_train.select_dtypes(include='int').columns
})
# Preparing the input signature for model logging
input_example = X_logged_input.sample(n=10, random_state=987654321)
signature = infer_signature(input_example, final_model.predict(input_example))
final_run_name = f"RF_HyperparameterCombo_{best_combo_index + 1}_Optimal"
with mlflow.start_run(run_name=final_run_name, description="Retrained on Full Training Set"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", final_model_hash)
mlflow.set_tag("model_type", "final_bagged_rf")
mlflow.set_tag("best_combo_index", best_combo_index)
mlflow.log_params(best_combo)
# Logging the average MCCV metrics computed previously
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in best_metrics.items()})
# Logging validation set metrics
mlflow.log_metrics({f"validation_{k}": v for k, v in validation_metrics.items()})
# Logging the model with input signature
mlflow.sklearn.log_model(
sk_model=final_model,
name="best_bagged_rf_model",
input_example=input_example,
signature=signature
)
# Logging artifacts
mlflow.log_artifact(mccv_csv_path)
mlflow.log_artifact(cm_path)
mlflow.log_artifact(report_path)
mlflow.log_artifact(pickle_model_path)
mlflow.log_artifact(train_dataset_csv_path)
mlflow.log_artifact(validation_dataset_csv_path)
# Logging datasets with metadata (MLflow dataset tracking)
train_dataset = mlflow.data.from_pandas(
pd.read_csv(train_csv_path),
name="mccv_data"
)
mlflow.log_input(train_dataset, context="training")
print("Final RF Model Trained and Logged with Validation Set Evaluation.")
Final RF Model Trained and Logged with Validation Set Evaluation.
1.7.2 AdaBoost¶
AdaBoost (Adaptive Boosting) is a boosting technique that combines multiple weak learners — typically decision stumps (shallow trees) — to form a strong classifier. It works by iteratively training weak models, assigning higher weights to misclassified instances so that subsequent models focus on difficult cases. At each iteration, a new weak model is trained, and its predictions are combined using a weighted voting mechanism. This process continues until a stopping criterion is met, such as a predefined number of iterations or performance threshold. AdaBoost is advantageous because it improves accuracy without overfitting if regularized properly. It performs well with clean data and can transform weak classifiers into strong ones. However, it is sensitive to noisy data and outliers, as misclassified points receive higher importance, leading to potential overfitting. Additionally, training can be slow for large datasets, and performance depends on the choice of base learner, typically decision trees.
- The adaboost model from the sklearn.ensemble Python library API was implemented. Combined with a preprocessing operation into a pipeline, the mlflow.sklearn module was used for loading and logging the model during experiment tracking.
- The model contains 3 hyperparameters for tuning:
- estimator_max_depth = maximum depth of the tree made to vary between 1 and 2
- learning_rate = weight applied to each classifier at each boosting iteration made to vary between 0.01 and 0.10
- n_estimators = maximum number of estimators at which boosting is terminated made to vary between 50 and 100
- No any hyperparameter was defined in the model to address the minimal 2:1 class imbalance observed between the No and Yes Recurred categories.
- Hyperparameter tuning was conducted using a 25-cycle 70-30 stratified train-test split Monte Carlo cross-validation with the results logged using MLflow experiment tracking and organized under boosted_ab_experiment.
- The optimal model performance evaluated using the F1 score, sensitivity and specificity metrics, respectively, was determined for the following hyperparameter combination:
- estimator_max_depth = 1
- learning_rate = 0.10
- n_estimators = 50
- The MCCV model performance of the optimal model as accessed from the MLflow user interface is summarized as follows:
- F1 Score = 0.8403
- Sensitivity = 0.8757
- Specificity = 0.9069
- Accuracy = 0.8974
- The independent validation model performance as accessed from the MLflow user interface of the optimal model is summarized as follows:
- F1 Score = 0.8571
- Sensitivity = 0.9000
- Specificity = 0.9183
- Accuracy = 0.9130
##################################
# Setting the model naming conventions
##################################
model_name = "boosted_ab"
experiment_name = f"{model_name}_experiment"
artifact_dir = f"artifacts/{model_name}"
os.makedirs(artifact_dir, exist_ok=True)
##################################
# Setting the MLflow experiment
# for the AdaBoost model
##################################
mlflow.set_experiment(experiment_name)
client = MlflowClient()
##################################
# Defining the categorical preprocessing parameters
##################################
categorical_features = ['Gender','Smoking','Physical_Examination','Adenopathy','Focality','Risk','T','Stage','Response']
categorical_transformer = OrdinalEncoder()
categorical_preprocessor = ColumnTransformer(transformers=[
('cat', categorical_transformer, categorical_features)],
remainder='passthrough',
force_int_remainder_cols=False)
##################################
# Defining the preprocessing and modeling pipeline parameters
##################################
boosted_ab_pipeline = Pipeline([
('categorical_preprocessor', categorical_preprocessor),
('boosted_ab_model', AdaBoostClassifier(estimator=DecisionTreeClassifier(random_state=987654321),
random_state=987654321))
])
##################################
# Defining hyperparameter grid
##################################
boosted_ab_hyperparameter_grid = {
'boosted_ab_model__learning_rate': [0.01, 0.10],
'boosted_ab_model__estimator__max_depth': [1, 2],
'boosted_ab_model__n_estimators': [50, 100]
}
param_combos = list(ParameterGrid(boosted_ab_hyperparameter_grid))
##################################
# Encoding the response variables
# for model evaluation
##################################
y_encoder = OrdinalEncoder()
y_encoder.fit(y_preprocessed_train.values.reshape(-1, 1))
y_preprocessed_train_encoded = y_encoder.transform(y_preprocessed_train.values.reshape(-1, 1)).ravel()
class_labels = list(y_encoder.categories_[0])
y_preprocessed_validation_encoded = y_encoder.transform(y_preprocessed_validation.values.reshape(-1, 1)).ravel()
##################################
# Defining the cross-validation strategy
# using Monte Carlo Cross-Validation Setup (25 cycles of 70-30 stratified splits))
##################################
mccv = StratifiedShuffleSplit(n_splits=25,
test_size=0.3,
random_state=987654321)
##################################
# Creating containers for the best hyperparameter combination
# based on the F1 Score, Sensitivity and Specificity metrics
##################################
best_combo = None
best_metrics = {'f1': -1, 'sensitivity': -1, 'specificity': -1}
all_combos_metrics = []
##################################
# Implementing experiment tracking
# of the hyperparameter tuning exercise
# using MLflow for the AdaBoost model
##################################
for combo_index, param_set in enumerate(param_combos):
print(f"Evaluating param combo {combo_index + 1}/{len(param_combos)}: {param_set}")
# Generating hash and checking for duplicates
run_hash = generate_run_hash(param_set, mccv_seed=987654321)
existing_run_id = find_existing_run_by_hash("boosted_ab_experiment", run_hash)
if existing_run_id:
print(f"Duplicate found for this combo (hash={run_hash}). Removing old run: {existing_run_id}")
delete_run(existing_run_id)
else:
print(f"No duplicate found. Proceeding to evaluate and log.")
all_metrics = []
for i, (train_idx, test_idx) in enumerate(mccv.split(X_preprocessed_train, y_preprocessed_train_encoded)):
X_train, X_test = X_preprocessed_train.iloc[train_idx], X_preprocessed_train.iloc[test_idx]
y_train, y_test = y_preprocessed_train_encoded[train_idx], y_preprocessed_train_encoded[test_idx]
# Cloning the pipeline and setting the hyperparameters
model = clone(boosted_ab_pipeline)
model.set_params(**param_set)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
all_metrics.append(evaluate_binary_metrics(y_test, y_pred))
# Aggregating the metrics across MCCV
df_metrics = pd.DataFrame(all_metrics)
avg_metrics = df_metrics.mean().to_dict()
avg_metrics_prefixed = {f"avg_mccv_{k}": v for k, v in avg_metrics.items()}
all_combos_metrics.append({**param_set, **avg_metrics_prefixed})
# Logging the hyperparameter combinations and metrics to MLflow
with mlflow.start_run(run_name=f"AB_HyperparameterCombo_{combo_index+1}"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", run_hash)
mlflow.log_params(param_set)
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in avg_metrics.items()})
mlflow.set_tag("param_index", combo_index)
# Tracking the best hyperparameter combination
better = False
if avg_metrics['f1'] > best_metrics['f1']:
better = True
elif avg_metrics['f1'] == best_metrics['f1']:
if avg_metrics['sensitivity'] > best_metrics['sensitivity']:
better = True
elif avg_metrics['sensitivity'] == best_metrics['sensitivity']:
if avg_metrics['specificity'] > best_metrics['specificity']:
better = True
if better:
best_combo = param_set
best_combo_index = combo_index
best_metrics = avg_metrics
Evaluating param combo 1/8: {'boosted_ab_model__estimator__max_depth': 1, 'boosted_ab_model__learning_rate': 0.01, 'boosted_ab_model__n_estimators': 50}
Duplicate found for this combo (hash=adea3f30682c1a791bcc7fc7796ca262). Removing old run: f9a5db8919814eb4bc28b18eb5d20a5b
Evaluating param combo 2/8: {'boosted_ab_model__estimator__max_depth': 1, 'boosted_ab_model__learning_rate': 0.01, 'boosted_ab_model__n_estimators': 100}
Duplicate found for this combo (hash=f4837ebcef67ff0f6e069c7073c2ecce). Removing old run: f5cadff9e7d349688223f3aefc33133b
Evaluating param combo 3/8: {'boosted_ab_model__estimator__max_depth': 1, 'boosted_ab_model__learning_rate': 0.1, 'boosted_ab_model__n_estimators': 50}
Duplicate found for this combo (hash=c02c4c97a7ec2b4be4455df6acde4afb). Removing old run: 37cd7f0c3d24431eade3c9ced4fb5e75
Evaluating param combo 4/8: {'boosted_ab_model__estimator__max_depth': 1, 'boosted_ab_model__learning_rate': 0.1, 'boosted_ab_model__n_estimators': 100}
Duplicate found for this combo (hash=ce0ce0096ec2c8adf9a8eb7478b4cf63). Removing old run: dcb9b413d9e84714815e86262b556446
Evaluating param combo 5/8: {'boosted_ab_model__estimator__max_depth': 2, 'boosted_ab_model__learning_rate': 0.01, 'boosted_ab_model__n_estimators': 50}
Duplicate found for this combo (hash=624b641891c9f4c8f74d87201a813457). Removing old run: 92811c7cb81a4d50af546a343e328d40
Evaluating param combo 6/8: {'boosted_ab_model__estimator__max_depth': 2, 'boosted_ab_model__learning_rate': 0.01, 'boosted_ab_model__n_estimators': 100}
Duplicate found for this combo (hash=a772b2c54f162164349bb3c9f5c48c5c). Removing old run: 9e1cd0faeee84083afcbcf6798132b1e
Evaluating param combo 7/8: {'boosted_ab_model__estimator__max_depth': 2, 'boosted_ab_model__learning_rate': 0.1, 'boosted_ab_model__n_estimators': 50}
Duplicate found for this combo (hash=c66d31c515042c9e49ad8bc9806cf270). Removing old run: d242dc15097d4ad2aff662a8f5f6a527
Evaluating param combo 8/8: {'boosted_ab_model__estimator__max_depth': 2, 'boosted_ab_model__learning_rate': 0.1, 'boosted_ab_model__n_estimators': 100}
Duplicate found for this combo (hash=2a1fc0742630d8e650385636269ed1b8). Removing old run: 21930d9861cc4e4dbc6717275b2cffe0
##################################
# Defining a function to
# generate a unique hash for the selected optimal model
##################################
final_model_hash = generate_run_hash(best_combo, mccv_seed=987654321)
existing_final_run_id = find_existing_run_by_hash("boosted_ab_experiment", final_model_hash)
if existing_final_run_id:
print(f"Duplicate final model found (hash={final_model_hash}). Removing previous run: {existing_final_run_id}")
delete_run(existing_final_run_id)
else:
print(f"No duplicate final model found. Proceeding to log new final model.")
Duplicate final model found (hash=c02c4c97a7ec2b4be4455df6acde4afb). Removing previous run: 54a4e5f658f44a5fad66716b8483aa86
##################################
# Retraining the most optimal AdaBoost model
# on the full training data
##################################
print("Optimal Hyperparameter Combination for AdaBoost:", best_combo)
final_model = clone(boosted_ab_pipeline)
final_model.set_params(**best_combo)
final_model.fit(X_preprocessed_train, y_preprocessed_train_encoded)
Optimal Hyperparameter Combination for AdaBoost: {'boosted_ab_model__estimator__max_depth': 1, 'boosted_ab_model__learning_rate': 0.1, 'boosted_ab_model__n_estimators': 50}
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_ab_model',
AdaBoostClassifier(estimator=DecisionTreeClassifier(max_depth=1,
random_state=987654321),
learning_rate=0.1,
random_state=987654321))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_ab_model',
AdaBoostClassifier(estimator=DecisionTreeClassifier(max_depth=1,
random_state=987654321),
learning_rate=0.1,
random_state=987654321))])ColumnTransformer(force_int_remainder_cols=False, remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking', 'Physical_Examination',
'Adenopathy', 'Focality', 'Risk', 'T',
'Stage', 'Response'])])['Gender', 'Smoking', 'Physical_Examination', 'Adenopathy', 'Focality', 'Risk', 'T', 'Stage', 'Response']
OrdinalEncoder()
['Age']
passthrough
AdaBoostClassifier(estimator=DecisionTreeClassifier(max_depth=1,
random_state=987654321),
learning_rate=0.1, random_state=987654321)DecisionTreeClassifier(max_depth=1, random_state=987654321)
DecisionTreeClassifier(max_depth=1, random_state=987654321)
##################################
# Evaluating the most optimal AdaBoost model
# on the validation data
##################################
y_validation_pred = final_model.predict(X_preprocessed_validation)
validation_metrics = evaluate_binary_metrics(y_preprocessed_validation_encoded, y_validation_pred)
##################################
# Consolidating the optimal hyperparameter combination
# and its corresponding MCCV and validation metrics
##################################
consolidated_table_1 = pd.DataFrame({
'hyperparameter': list(best_combo.keys()),
'mccv.optimal.value': list(best_combo.values())
})
consolidated_table_2 = pd.DataFrame({
'metric': list(best_metrics.keys()),
'mccv': [round(v, 4) for v in best_metrics.values()],
'validation': [round(validation_metrics[k], 4) for k in best_metrics.keys()]
})
print(f"{experiment_name} optimal hyperparameter combination:")
display(consolidated_table_1)
print(f"{experiment_name} optimal hyperparameter combination mccv versus validation performance metrics:")
display(consolidated_table_2)
boosted_ab_experiment optimal hyperparameter combination:
| hyperparameter | mccv.optimal.value | |
|---|---|---|
| 0 | boosted_ab_model__estimator__max_depth | 1.0 |
| 1 | boosted_ab_model__learning_rate | 0.1 |
| 2 | boosted_ab_model__n_estimators | 50.0 |
boosted_ab_experiment optimal hyperparameter combination mccv versus validation performance metrics:
| metric | mccv | validation | |
|---|---|---|---|
| 0 | accuracy | 0.8974 | 0.9130 |
| 1 | sensitivity | 0.8758 | 0.9000 |
| 2 | specificity | 0.9070 | 0.9184 |
| 3 | f1 | 0.8404 | 0.8571 |
##################################
# Decoding the true and predicted labels
# for artifact storage
##################################
y_validation_true_decoded = y_encoder.inverse_transform(y_preprocessed_validation_encoded.reshape(-1, 1)).ravel()
y_validation_pred_decoded = y_encoder.inverse_transform(y_validation_pred.reshape(-1, 1)).ravel()
##################################
# Saving the artifacts from the AdaBoost model
# hyperparameter tuning exercise
##################################
mccv_csv_path = os.path.join(artifact_dir, f"{model_name}_MCCV_metrics.csv")
cm_path = os.path.join(artifact_dir, f"{model_name}_validation_confusion_matrix.png")
report_path = os.path.join(artifact_dir, f"{model_name}_validation_classification_report.txt")
df_all_combos = pd.DataFrame(all_combos_metrics)
df_all_combos.to_csv(mccv_csv_path, index=False)
log_confusion_matrix(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=cm_path)
log_classification_report(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=report_path)
pickle_model_path = os.path.join(artifact_dir, f"{model_name}_final_model.pkl")
with open(pickle_model_path, "wb") as f:
pickle.dump(final_model, f)
train_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_train_dataset.csv")
thyroid_cancer_preprocessed_train.to_csv(train_dataset_csv_path, index=False)
validation_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_validation_dataset.csv")
thyroid_cancer_preprocessed_validation.to_csv(validation_dataset_csv_path, index=False)
##################################
# Logging the most optimal AdaBoost model
# developed from the full training data
# and evaluated on the validation data
##################################
# Casting integer columns in preprocessed training data to float64
X_logged_input = X_preprocessed_train.astype({
col: 'float64' for col in X_preprocessed_train.select_dtypes(include='int').columns
})
# Preparing the input signature for model logging
input_example = X_logged_input.sample(n=10, random_state=987654321)
signature = infer_signature(input_example, final_model.predict(input_example))
final_run_name = f"AB_HyperparameterCombo_{best_combo_index + 1}_Optimal"
with mlflow.start_run(run_name=final_run_name, description="Retrained on Full Training Set"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", final_model_hash)
mlflow.set_tag("model_type", "final_boosted_ab")
mlflow.set_tag("best_combo_index", best_combo_index)
mlflow.log_params(best_combo)
# Logging the average MCCV metrics computed previously
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in best_metrics.items()})
# Logging validation set metrics
mlflow.log_metrics({f"validation_{k}": v for k, v in validation_metrics.items()})
# Logging the model with input signature
mlflow.sklearn.log_model(
sk_model=final_model,
name="best_boosted_ab_model",
input_example=input_example,
signature=signature
)
# Logging artifacts
mlflow.log_artifact(mccv_csv_path)
mlflow.log_artifact(cm_path)
mlflow.log_artifact(report_path)
mlflow.log_artifact(pickle_model_path)
mlflow.log_artifact(train_dataset_csv_path)
mlflow.log_artifact(validation_dataset_csv_path)
# Logging datasets with metadata (MLflow dataset tracking)
train_dataset = mlflow.data.from_pandas(
pd.read_csv(train_csv_path),
name="mccv_data"
)
mlflow.log_input(train_dataset, context="training")
print("Final AB Model Trained and Logged with Validation Set Evaluation.")
Final AB Model Trained and Logged with Validation Set Evaluation.
1.7.3 Gradient Boosting¶
Gradient Boosting builds an ensemble of decision trees sequentially, where each new tree corrects the mistakes of the previous ones by optimizing a loss function. Unlike AdaBoost, which reweights misclassified instances, Gradient Boosting fits each new tree to the residual errors of the previous model, gradually improving predictions. This process continues until a stopping criterion, such as a set number of trees, is met. The key advantages of Gradient Boosting include its flexibility to model complex relationships and strong predictive performance, often outperforming bagging methods. It can handle both numeric and categorical data well. However, it is prone to overfitting if not carefully tuned, especially with deep trees and too many iterations. It is also computationally expensive due to sequential training, and hyperparameter tuning (e.g., learning rate, number of trees, tree depth) can be challenging and time-consuming.
- The gradient boosting model from the sklearn.ensemble Python library API was implemented. Combined with a preprocessing operation into a pipeline, the mlflow.sklearn module was used for loading and logging the model during experiment tracking.
- The model contains 4 hyperparameters for tuning:
- learning_rate = shrinking proportion of the contribution from each tree made to vary between 0.01 and 0.10
- max_depth = maximum depth of the tree made to vary between 3 and 6
- min_samples_leaf = minimum number of samples required to be at a leaf node made to vary between 5 and 10
- n_estimators = number of boosting stages to perform made to vary between 50 and 100
- No any hyperparameter was defined in the model to address the minimal 2:1 class imbalance observed between the No and Yes Recurred categories.
- Hyperparameter tuning was conducted using a 25-cycle 70-30 stratified train-test split Monte Carlo cross-validation with the results logged using MLflow experiment tracking and organized under boosted_gb_experiment.
- The optimal model performance evaluated using the F1 score, sensitivity and specificity metrics, respectively, was determined for the following hyperparameter combination:
- learning_rate = 0.10
- max_depth = 3
- min_samples_leaf = 10
- n_estimators = 50
- The MCCV model performance of the optimal model as accessed from the MLflow user interface is summarized as follows:
- F1 Score = 0.7850
- Sensitivity = 0.7705
- Specificity = 0.9181
- Accuracy = 0.8729
- The independent validation model performance as accessed from the MLflow user interface of the optimal model is summarized as follows:
- F1 Score = 0.8292
- Sensitivity = 0.8500
- Specificity = 0.9183
- Accuracy = 0.8985
##################################
# Setting the model naming conventions
##################################
model_name = "boosted_gb"
experiment_name = f"{model_name}_experiment"
artifact_dir = f"artifacts/{model_name}"
os.makedirs(artifact_dir, exist_ok=True)
##################################
# Setting the MLflow experiment
# for the Gradient Boosting model
##################################
mlflow.set_experiment(experiment_name)
client = MlflowClient()
##################################
# Defining the categorical preprocessing parameters
##################################
categorical_features = ['Gender','Smoking','Physical_Examination','Adenopathy','Focality','Risk','T','Stage','Response']
categorical_transformer = OrdinalEncoder()
categorical_preprocessor = ColumnTransformer(transformers=[
('cat', categorical_transformer, categorical_features)],
remainder='passthrough',
force_int_remainder_cols=False)
##################################
# Defining the preprocessing and modeling pipeline parameters
##################################
boosted_gb_pipeline = Pipeline([
('categorical_preprocessor', categorical_preprocessor),
('boosted_gb_model', GradientBoostingClassifier(random_state=987654321))
])
##################################
# Defining hyperparameter grid
##################################
boosted_gb_hyperparameter_grid = {
'boosted_gb_model__learning_rate': [0.01, 0.10],
'boosted_gb_model__max_depth': [3, 6],
'boosted_gb_model__min_samples_leaf': [5, 10],
'boosted_gb_model__n_estimators': [50, 100]
}
param_combos = list(ParameterGrid(boosted_gb_hyperparameter_grid))
##################################
# Encoding the response variables
# for model evaluation
##################################
y_encoder = OrdinalEncoder()
y_encoder.fit(y_preprocessed_train.values.reshape(-1, 1))
y_preprocessed_train_encoded = y_encoder.transform(y_preprocessed_train.values.reshape(-1, 1)).ravel()
class_labels = list(y_encoder.categories_[0])
y_preprocessed_validation_encoded = y_encoder.transform(y_preprocessed_validation.values.reshape(-1, 1)).ravel()
##################################
# Defining the cross-validation strategy
# using Monte Carlo Cross-Validation Setup (25 cycles of 70-30 stratified splits))
##################################
mccv = StratifiedShuffleSplit(n_splits=25,
test_size=0.3,
random_state=987654321)
##################################
# Creating containers for the best hyperparameter combination
# based on the F1 Score, Sensitivity and Specificity metrics
##################################
best_combo = None
best_metrics = {'f1': -1, 'sensitivity': -1, 'specificity': -1}
all_combos_metrics = []
##################################
# Implementing experiment tracking
# of the hyperparameter tuning exercise
# using MLflow for the Gradient Boosting model
##################################
for combo_index, param_set in enumerate(param_combos):
print(f"Evaluating param combo {combo_index + 1}/{len(param_combos)}: {param_set}")
# Generating hash and checking for duplicates
run_hash = generate_run_hash(param_set, mccv_seed=987654321)
existing_run_id = find_existing_run_by_hash("boosted_gb_experiment", run_hash)
if existing_run_id:
print(f"Duplicate found for this combo (hash={run_hash}). Removing old run: {existing_run_id}")
delete_run(existing_run_id)
else:
print(f"No duplicate found. Proceeding to evaluate and log.")
all_metrics = []
for i, (train_idx, test_idx) in enumerate(mccv.split(X_preprocessed_train, y_preprocessed_train_encoded)):
X_train, X_test = X_preprocessed_train.iloc[train_idx], X_preprocessed_train.iloc[test_idx]
y_train, y_test = y_preprocessed_train_encoded[train_idx], y_preprocessed_train_encoded[test_idx]
# Cloning the pipeline and setting the hyperparameters
model = clone(boosted_gb_pipeline)
model.set_params(**param_set)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
all_metrics.append(evaluate_binary_metrics(y_test, y_pred))
# Aggregating the metrics across MCCV
df_metrics = pd.DataFrame(all_metrics)
avg_metrics = df_metrics.mean().to_dict()
avg_metrics_prefixed = {f"avg_mccv_{k}": v for k, v in avg_metrics.items()}
all_combos_metrics.append({**param_set, **avg_metrics_prefixed})
# Logging the hyperparameter combinations and metrics to MLflow
with mlflow.start_run(run_name=f"GB_HyperparameterCombo_{combo_index+1}"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", run_hash)
mlflow.log_params(param_set)
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in avg_metrics.items()})
mlflow.set_tag("param_index", combo_index)
# Tracking the best hyperparameter combination
better = False
if avg_metrics['f1'] > best_metrics['f1']:
better = True
elif avg_metrics['f1'] == best_metrics['f1']:
if avg_metrics['sensitivity'] > best_metrics['sensitivity']:
better = True
elif avg_metrics['sensitivity'] == best_metrics['sensitivity']:
if avg_metrics['specificity'] > best_metrics['specificity']:
better = True
if better:
best_combo = param_set
best_combo_index = combo_index
best_metrics = avg_metrics
Evaluating param combo 1/16: {'boosted_gb_model__learning_rate': 0.01, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 5, 'boosted_gb_model__n_estimators': 50}
Duplicate found for this combo (hash=c9d3fa21df6427a54ccc941a57acd173). Removing old run: fbe351fa3ba44c949ca861745c6fa881
Evaluating param combo 2/16: {'boosted_gb_model__learning_rate': 0.01, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 5, 'boosted_gb_model__n_estimators': 100}
Duplicate found for this combo (hash=747ceea9a2b1b393e8301b3d47968555). Removing old run: 1930907a61c642f3ba37103be72394c4
Evaluating param combo 3/16: {'boosted_gb_model__learning_rate': 0.01, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 50}
Duplicate found for this combo (hash=1f02e54eb3b4a4b0bfc6483cb5052932). Removing old run: 02154de744ad4bea83c9cfcb423b3479
Evaluating param combo 4/16: {'boosted_gb_model__learning_rate': 0.01, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 100}
Duplicate found for this combo (hash=12b685edf4bb7030dd7a86a0a9ff4ceb). Removing old run: 198d7e19588d4d87ad22e9c1cff55e81
Evaluating param combo 5/16: {'boosted_gb_model__learning_rate': 0.01, 'boosted_gb_model__max_depth': 6, 'boosted_gb_model__min_samples_leaf': 5, 'boosted_gb_model__n_estimators': 50}
Duplicate found for this combo (hash=483f8b7e4542c941ab5ffee6f7fb94b6). Removing old run: 23fc2ad42bf94e4ba57a3d4454c1e798
Evaluating param combo 6/16: {'boosted_gb_model__learning_rate': 0.01, 'boosted_gb_model__max_depth': 6, 'boosted_gb_model__min_samples_leaf': 5, 'boosted_gb_model__n_estimators': 100}
Duplicate found for this combo (hash=b8bffc559956896b38a7575a2f468731). Removing old run: 045d5f4763cb4c3e905e662bc3d4b0fa
Evaluating param combo 7/16: {'boosted_gb_model__learning_rate': 0.01, 'boosted_gb_model__max_depth': 6, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 50}
Duplicate found for this combo (hash=ab334f1aaa60442be90e6bd94f8361ea). Removing old run: b4062ef4c9734ec0ae496fdcf23d581c
Evaluating param combo 8/16: {'boosted_gb_model__learning_rate': 0.01, 'boosted_gb_model__max_depth': 6, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 100}
Duplicate found for this combo (hash=1e3a55a8491a0921132412cdbf74e50f). Removing old run: 6f1fad7d0e124dacb75de77ce96eaa4d
Evaluating param combo 9/16: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 5, 'boosted_gb_model__n_estimators': 50}
Duplicate found for this combo (hash=6721b717a57a2bcbaaaf417ef6c4eb76). Removing old run: 38c978d1a3874cf3a4b31051ca67377c
Evaluating param combo 10/16: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 5, 'boosted_gb_model__n_estimators': 100}
Duplicate found for this combo (hash=3ced802f49f4f725a1669f212d8ee972). Removing old run: 4f111a0f2541469e8102c92c33aad2aa
Evaluating param combo 11/16: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 50}
Duplicate found for this combo (hash=d574d23109eb11783038ded97ebde691). Removing old run: dcc62fc45a6a459b840919d7048aece1
Evaluating param combo 12/16: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 100}
Duplicate found for this combo (hash=3d25b57cd37ca09f0549cac6fc47db53). Removing old run: 37509ba6cd424715902b53bbdcc8915a
Evaluating param combo 13/16: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 6, 'boosted_gb_model__min_samples_leaf': 5, 'boosted_gb_model__n_estimators': 50}
Duplicate found for this combo (hash=0475e97274cbb16b0a8cf69d502d3365). Removing old run: a1bac09414434cd0a6fc110c30d81d83
Evaluating param combo 14/16: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 6, 'boosted_gb_model__min_samples_leaf': 5, 'boosted_gb_model__n_estimators': 100}
Duplicate found for this combo (hash=59697fe5055c3fa1484468b7605d4488). Removing old run: 7d4f5aa9e4a9470697a5ccd26db48819
Evaluating param combo 15/16: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 6, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 50}
Duplicate found for this combo (hash=b68d8eedf5857028c655e52f681d763d). Removing old run: ac5b0aaffe4d49059fdaf23267012359
Evaluating param combo 16/16: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 6, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 100}
Duplicate found for this combo (hash=6b840a2524b1bd5c9cfdc1b548a4c246). Removing old run: d5dceedbf4b046cdae6143c0419247b5
##################################
# Defining a function to
# generate a unique hash for the selected optimal model
##################################
final_model_hash = generate_run_hash(best_combo, mccv_seed=987654321)
existing_final_run_id = find_existing_run_by_hash("boosted_gb_experiment", final_model_hash)
if existing_final_run_id:
print(f"Duplicate final model found (hash={final_model_hash}). Removing previous run: {existing_final_run_id}")
delete_run(existing_final_run_id)
else:
print(f"No duplicate final model found. Proceeding to log new final model.")
Duplicate final model found (hash=d574d23109eb11783038ded97ebde691). Removing previous run: fa4a4c55454542ec860b7fec4be1ac0c
##################################
# Retraining the most optimal Gradient Boosting model
# on the full training data
##################################
print("Optimal Hyperparameter Combination for Gradient Boosting:", best_combo)
final_model = clone(boosted_gb_pipeline)
final_model.set_params(**best_combo)
final_model.fit(X_preprocessed_train, y_preprocessed_train_encoded)
Optimal Hyperparameter Combination for Gradient Boosting: {'boosted_gb_model__learning_rate': 0.1, 'boosted_gb_model__max_depth': 3, 'boosted_gb_model__min_samples_leaf': 10, 'boosted_gb_model__n_estimators': 50}
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_gb_model',
GradientBoostingClassifier(min_samples_leaf=10,
n_estimators=50,
random_state=987654321))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_gb_model',
GradientBoostingClassifier(min_samples_leaf=10,
n_estimators=50,
random_state=987654321))])ColumnTransformer(force_int_remainder_cols=False, remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking', 'Physical_Examination',
'Adenopathy', 'Focality', 'Risk', 'T',
'Stage', 'Response'])])['Gender', 'Smoking', 'Physical_Examination', 'Adenopathy', 'Focality', 'Risk', 'T', 'Stage', 'Response']
OrdinalEncoder()
['Age']
passthrough
GradientBoostingClassifier(min_samples_leaf=10, n_estimators=50,
random_state=987654321)##################################
# Evaluating the most optimal Gradient Boosting model
# on the validation data
##################################
y_validation_pred = final_model.predict(X_preprocessed_validation)
validation_metrics = evaluate_binary_metrics(y_preprocessed_validation_encoded, y_validation_pred)
##################################
# Consolidating the optimal hyperparameter combination
# and its corresponding MCCV and validation metrics
##################################
consolidated_table_1 = pd.DataFrame({
'hyperparameter': list(best_combo.keys()),
'mccv.optimal.value': list(best_combo.values())
})
consolidated_table_2 = pd.DataFrame({
'metric': list(best_metrics.keys()),
'mccv': [round(v, 4) for v in best_metrics.values()],
'validation': [round(validation_metrics[k], 4) for k in best_metrics.keys()]
})
print(f"{experiment_name} optimal hyperparameter combination:")
display(consolidated_table_1)
print(f"{experiment_name} optimal hyperparameter combination mccv versus validation performance metrics:")
display(consolidated_table_2)
boosted_gb_experiment optimal hyperparameter combination:
| hyperparameter | mccv.optimal.value | |
|---|---|---|
| 0 | boosted_gb_model__learning_rate | 0.1 |
| 1 | boosted_gb_model__max_depth | 3.0 |
| 2 | boosted_gb_model__min_samples_leaf | 10.0 |
| 3 | boosted_gb_model__n_estimators | 50.0 |
boosted_gb_experiment optimal hyperparameter combination mccv versus validation performance metrics:
| metric | mccv | validation | |
|---|---|---|---|
| 0 | accuracy | 0.8729 | 0.8986 |
| 1 | sensitivity | 0.7705 | 0.8500 |
| 2 | specificity | 0.9181 | 0.9184 |
| 3 | f1 | 0.7851 | 0.8293 |
##################################
# Decoding the true and predicted labels
# for artifact storage
##################################
y_validation_true_decoded = y_encoder.inverse_transform(y_preprocessed_validation_encoded.reshape(-1, 1)).ravel()
y_validation_pred_decoded = y_encoder.inverse_transform(y_validation_pred.reshape(-1, 1)).ravel()
##################################
# Saving the artifacts from the Gradient Boosting model
# hyperparameter tuning exercise
##################################
mccv_csv_path = os.path.join(artifact_dir, f"{model_name}_MCCV_metrics.csv")
cm_path = os.path.join(artifact_dir, f"{model_name}_validation_confusion_matrix.png")
report_path = os.path.join(artifact_dir, f"{model_name}_validation_classification_report.txt")
df_all_combos = pd.DataFrame(all_combos_metrics)
df_all_combos.to_csv(mccv_csv_path, index=False)
log_confusion_matrix(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=cm_path)
log_classification_report(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=report_path)
pickle_model_path = os.path.join(artifact_dir, f"{model_name}_final_model.pkl")
with open(pickle_model_path, "wb") as f:
pickle.dump(final_model, f)
train_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_train_dataset.csv")
thyroid_cancer_preprocessed_train.to_csv(train_dataset_csv_path, index=False)
validation_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_validation_dataset.csv")
thyroid_cancer_preprocessed_validation.to_csv(validation_dataset_csv_path, index=False)
##################################
# Logging the most optimal Gradient Boosting model
# developed from the full training data
# and evaluated on the validation data
##################################
# Casting integer columns in preprocessed training data to float64
X_logged_input = X_preprocessed_train.astype({
col: 'float64' for col in X_preprocessed_train.select_dtypes(include='int').columns
})
# Preparing the input signature for model logging
input_example = X_logged_input.sample(n=10, random_state=987654321)
signature = infer_signature(input_example, final_model.predict(input_example))
final_run_name = f"GB_HyperparameterCombo_{best_combo_index + 1}_Optimal"
with mlflow.start_run(run_name=final_run_name, description="Retrained on Full Training Set"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", final_model_hash)
mlflow.set_tag("model_type", "final_boosted_gb")
mlflow.set_tag("best_combo_index", best_combo_index)
mlflow.log_params(best_combo)
# Logging the average MCCV metrics computed previously
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in best_metrics.items()})
# Logging validation set metrics
mlflow.log_metrics({f"validation_{k}": v for k, v in validation_metrics.items()})
# Logging the model with input signature
mlflow.sklearn.log_model(
sk_model=final_model,
name="best_boosted_gb_model",
input_example=input_example,
signature=signature
)
# Logging artifacts
mlflow.log_artifact(mccv_csv_path)
mlflow.log_artifact(cm_path)
mlflow.log_artifact(report_path)
mlflow.log_artifact(pickle_model_path)
mlflow.log_artifact(train_dataset_csv_path)
mlflow.log_artifact(validation_dataset_csv_path)
# Logging datasets with metadata (MLflow dataset tracking)
train_dataset = mlflow.data.from_pandas(
pd.read_csv(train_csv_path),
name="mccv_data"
)
mlflow.log_input(train_dataset, context="training")
print("Final GB Model Trained and Logged with Validation Set Evaluation.")
Final GB Model Trained and Logged with Validation Set Evaluation.
1.7.4 XGBoost¶
XGBoost (Extreme Gradient Boosting) is an optimized version of Gradient Boosting that introduces additional regularization and computational efficiencies. It builds decision trees sequentially, with each new tree correcting the residual errors of the previous ones, but it incorporates advanced techniques such as shrinkage (learning rate), column subsampling, and L1/L2 regularization to prevent overfitting. Additionally, XGBoost employs parallelization, reducing training time significantly compared to standard Gradient Boosting. It is widely used in machine learning competitions due to its superior accuracy and efficiency. The key advantages include its ability to handle missing data, built-in regularization for better generalization, and fast training through parallelization. However, XGBoost requires careful hyperparameter tuning to achieve optimal performance, and the model can become overly complex, making interpretation difficult. It is also memory-intensive, especially for large datasets, and can be challenging to deploy efficiently in real-time applications.
- The xgboost model from the xgboost Python library API was implemented.Combined with a preprocessing operation into a pipeline, the mlflow.sklearn module was used for loading and logging the model during experiment tracking.
- The model contains 4 hyperparameters for tuning:
- learning_rate = step size at which weights are updated during training made to vary between 0.01 and 0.10
- max_depth = maximum depth of the tree made to vary between 3 and 6
- gamma = minimum loss reduction required to make a further split in a tree made to vary between 0.10 and 0.20
- n_estimators = number of boosting stages to perform made to vary between 50 and 100
- A special hyperparameter (scale_pos_weight = 2.0) was fixed to address the minimal 2:1 class imbalance observed between the No and Yes Recurred categories.
- Hyperparameter tuning was conducted using a 25-cycle 70-30 stratified train-test split Monte Carlo cross-validation with the results logged using MLflow experiment tracking and organized under bagged_rf_experiment.
- The optimal model performance evaluated using the F1 score, sensitivity and specificity metrics, respectively, was determined for the following hyperparameter combination:
- learning_rate = 0.01
- max_depth = 3
- gamma 0.10
- n_estimators = 50
- The MCCV model performance of the optimal model as accessed from the MLflow user interface is summarized as follows:
- F1 Score = 0.8264
- Sensitivity = 0.8442
- Specificity = 0.9125
- Accuracy = 0.8916
- The independent validation model performance as accessed from the MLflow user interface of the optimal model is summarized as follows:
- F1 Score = 0.8571
- Sensitivity = 0.9000
- Specificity = 0.9183
- Accuracy = 0.9130
##################################
# Setting the model naming conventions
##################################
model_name = "boosted_xgb"
experiment_name = f"{model_name}_experiment"
artifact_dir = f"artifacts/{model_name}"
os.makedirs(artifact_dir, exist_ok=True)
##################################
# Setting the MLflow experiment
# for the XGBoost model
##################################
mlflow.set_experiment(experiment_name)
client = MlflowClient()
##################################
# Defining the categorical preprocessing parameters
##################################
categorical_features = ['Gender','Smoking','Physical_Examination','Adenopathy','Focality','Risk','T','Stage','Response']
categorical_transformer = OrdinalEncoder()
categorical_preprocessor = ColumnTransformer(transformers=[
('cat', categorical_transformer, categorical_features)],
remainder='passthrough',
force_int_remainder_cols=False)
##################################
# Defining the preprocessing and modeling pipeline parameters
##################################
boosted_xgb_pipeline = Pipeline([
('categorical_preprocessor', categorical_preprocessor),
('boosted_xgb_model', XGBClassifier(scale_pos_weight=2.0,
random_state=987654321,
subsample=0.7,
colsample_bytree=0.7,
eval_metric='logloss'))
])
##################################
# Defining hyperparameter grid
##################################
boosted_xgb_hyperparameter_grid = {
'boosted_xgb_model__learning_rate': [0.01, 0.10],
'boosted_xgb_model__max_depth': [3, 6],
'boosted_xgb_model__gamma': [0.1, 0.2],
'boosted_xgb_model__n_estimators': [50, 100]
}
param_combos = list(ParameterGrid(boosted_xgb_hyperparameter_grid))
##################################
# Encoding the response variables
# for model evaluation
##################################
y_encoder = OrdinalEncoder()
y_encoder.fit(y_preprocessed_train.values.reshape(-1, 1))
y_preprocessed_train_encoded = y_encoder.transform(y_preprocessed_train.values.reshape(-1, 1)).ravel()
class_labels = list(y_encoder.categories_[0])
y_preprocessed_validation_encoded = y_encoder.transform(y_preprocessed_validation.values.reshape(-1, 1)).ravel()
##################################
# Defining the cross-validation strategy
# using Monte Carlo Cross-Validation Setup (25 cycles of 70-30 stratified splits))
##################################
mccv = StratifiedShuffleSplit(n_splits=25,
test_size=0.3,
random_state=987654321)
##################################
# Creating containers for the best hyperparameter combination
# based on the F1 Score, Sensitivity and Specificity metrics
##################################
best_combo = None
best_metrics = {'f1': -1, 'sensitivity': -1, 'specificity': -1}
all_combos_metrics = []
##################################
# Implementing experiment tracking
# of the hyperparameter tuning exercise
# using MLflow for the XGBoost model
##################################
for combo_index, param_set in enumerate(param_combos):
print(f"Evaluating param combo {combo_index + 1}/{len(param_combos)}: {param_set}")
# Generating hash and checking for duplicates
run_hash = generate_run_hash(param_set, mccv_seed=987654321)
existing_run_id = find_existing_run_by_hash("boosted_xgb_experiment", run_hash)
if existing_run_id:
print(f"Duplicate found for this combo (hash={run_hash}). Removing old run: {existing_run_id}")
delete_run(existing_run_id)
else:
print(f"No duplicate found. Proceeding to evaluate and log.")
all_metrics = []
for i, (train_idx, test_idx) in enumerate(mccv.split(X_preprocessed_train, y_preprocessed_train_encoded)):
X_train, X_test = X_preprocessed_train.iloc[train_idx], X_preprocessed_train.iloc[test_idx]
y_train, y_test = y_preprocessed_train_encoded[train_idx], y_preprocessed_train_encoded[test_idx]
# Cloning the pipeline and setting the hyperparameters
model = clone(boosted_xgb_pipeline)
model.set_params(**param_set)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
all_metrics.append(evaluate_binary_metrics(y_test, y_pred))
# Aggregating the metrics across MCCV
df_metrics = pd.DataFrame(all_metrics)
avg_metrics = df_metrics.mean().to_dict()
avg_metrics_prefixed = {f"avg_mccv_{k}": v for k, v in avg_metrics.items()}
all_combos_metrics.append({**param_set, **avg_metrics_prefixed})
# Logging the hyperparameter combinations and metrics to MLflow
with mlflow.start_run(run_name=f"XGB_HyperparameterCombo_{combo_index+1}"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", run_hash)
mlflow.log_params(param_set)
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in avg_metrics.items()})
mlflow.set_tag("param_index", combo_index)
# Tracking the best hyperparameter combination
better = False
if avg_metrics['f1'] > best_metrics['f1']:
better = True
elif avg_metrics['f1'] == best_metrics['f1']:
if avg_metrics['sensitivity'] > best_metrics['sensitivity']:
better = True
elif avg_metrics['sensitivity'] == best_metrics['sensitivity']:
if avg_metrics['specificity'] > best_metrics['specificity']:
better = True
if better:
best_combo = param_set
best_combo_index = combo_index
best_metrics = avg_metrics
Evaluating param combo 1/16: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 50}
Duplicate found for this combo (hash=df9bf45aef69ddc89baf1f83e486d043). Removing old run: 7af4d16091984f7c8f98af10c67a9a15
Evaluating param combo 2/16: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 100}
Duplicate found for this combo (hash=58a7168720c1fd4c1b467cd3ec42014b). Removing old run: 1f9946005a0b4827a7f1eb9aa633dcad
Evaluating param combo 3/16: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 6, 'boosted_xgb_model__n_estimators': 50}
Duplicate found for this combo (hash=d9bd415cbfb544dc2ba3328ecc368bc2). Removing old run: aada902547974da2ad7a6e938368ac5f
Evaluating param combo 4/16: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 6, 'boosted_xgb_model__n_estimators': 100}
Duplicate found for this combo (hash=86636543044ed99f3976539cb2e7cb7a). Removing old run: 3136acaca5bb4671990b6e70b1556868
Evaluating param combo 5/16: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.1, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 50}
Duplicate found for this combo (hash=ba3bd6ea2a327793b620b81909bc1fab). Removing old run: 77b2b1cc4063476aaa2442612de73a53
Evaluating param combo 6/16: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.1, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 100}
Duplicate found for this combo (hash=d0180dc353587aead8d2997540b88c2a). Removing old run: c15f4685f7a14f5eaa7730d4a7b152e6
Evaluating param combo 7/16: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.1, 'boosted_xgb_model__max_depth': 6, 'boosted_xgb_model__n_estimators': 50}
Duplicate found for this combo (hash=d792ba10379e693efb9832224ce91848). Removing old run: 2362543d431a4d3985452b91c67231a3
Evaluating param combo 8/16: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.1, 'boosted_xgb_model__max_depth': 6, 'boosted_xgb_model__n_estimators': 100}
Duplicate found for this combo (hash=20a3678a3c262893f56cc2e33467177e). Removing old run: 69cf87b5dbbd4191af62d232311f18f6
Evaluating param combo 9/16: {'boosted_xgb_model__gamma': 0.2, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 50}
Duplicate found for this combo (hash=df12dfc48250c887d56fe46af95147aa). Removing old run: 82231d334f2e4a9fb9cea2e4ec4eaac1
Evaluating param combo 10/16: {'boosted_xgb_model__gamma': 0.2, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 100}
Duplicate found for this combo (hash=4483bdff4370a7d66aa505909511fa25). Removing old run: 87759d7da3e541008d713b2ac7402508
Evaluating param combo 11/16: {'boosted_xgb_model__gamma': 0.2, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 6, 'boosted_xgb_model__n_estimators': 50}
Duplicate found for this combo (hash=8a1fbd418727bc373189c75e024f3b4a). Removing old run: e0aea9c2951e45e5b22dd1d888e231c5
Evaluating param combo 12/16: {'boosted_xgb_model__gamma': 0.2, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 6, 'boosted_xgb_model__n_estimators': 100}
Duplicate found for this combo (hash=b1cafdf9c54c1a0e355dcac35b9ab6e9). Removing old run: f2849915170147ad8161219192caf5b4
Evaluating param combo 13/16: {'boosted_xgb_model__gamma': 0.2, 'boosted_xgb_model__learning_rate': 0.1, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 50}
Duplicate found for this combo (hash=0b4bde63f1a9b1f6be5e869e42926d76). Removing old run: 8d20b0f0a77b4561aa62bb11e48dc6f0
Evaluating param combo 14/16: {'boosted_xgb_model__gamma': 0.2, 'boosted_xgb_model__learning_rate': 0.1, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 100}
Duplicate found for this combo (hash=dff5b1d2039084bdb41a870901fd39e7). Removing old run: 6a1abc27de5d49d8b337469799cbfcca
Evaluating param combo 15/16: {'boosted_xgb_model__gamma': 0.2, 'boosted_xgb_model__learning_rate': 0.1, 'boosted_xgb_model__max_depth': 6, 'boosted_xgb_model__n_estimators': 50}
Duplicate found for this combo (hash=736a4c39a2c030bf9b7e84f2aeaaddd0). Removing old run: b23fe078e41343be96a236221ba207ab
Evaluating param combo 16/16: {'boosted_xgb_model__gamma': 0.2, 'boosted_xgb_model__learning_rate': 0.1, 'boosted_xgb_model__max_depth': 6, 'boosted_xgb_model__n_estimators': 100}
Duplicate found for this combo (hash=ea71e5a334e7dd2bb0e17a28b5cc1ddc). Removing old run: d667638941f5475a9d68255d108fcaed
##################################
# Defining a function to
# generate a unique hash for the selected optimal model
##################################
final_model_hash = generate_run_hash(best_combo, mccv_seed=987654321)
existing_final_run_id = find_existing_run_by_hash("boosted_xgb_experiment", final_model_hash)
if existing_final_run_id:
print(f"Duplicate final model found (hash={final_model_hash}). Removing previous run: {existing_final_run_id}")
delete_run(existing_final_run_id)
else:
print(f"No duplicate final model found. Proceeding to log new final model.")
Duplicate final model found (hash=df9bf45aef69ddc89baf1f83e486d043). Removing previous run: 89df041719d8428984f32f34eee7ae79
##################################
# Retraining the most optimal XGBoost model
# on the full training data
##################################
print("Optimal Hyperparameter Combination for XGBoost:", best_combo)
final_model = clone(boosted_xgb_pipeline)
final_model.set_params(**best_combo)
final_model.fit(X_preprocessed_train, y_preprocessed_train_encoded)
Optimal Hyperparameter Combination for XGBoost: {'boosted_xgb_model__gamma': 0.1, 'boosted_xgb_model__learning_rate': 0.01, 'boosted_xgb_model__max_depth': 3, 'boosted_xgb_model__n_estimators': 50}
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_xgb_model',
XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_byle...
feature_types=None, feature_weights=None,
gamma=0.1, grow_policy=None,
importance_type=None,
interaction_constraints=None, learning_rate=0.01,
max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None,
max_depth=3, max_leaves=None,
min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None,
n_estimators=50, n_jobs=None,
num_parallel_tree=None, ...))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_xgb_model',
XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_byle...
feature_types=None, feature_weights=None,
gamma=0.1, grow_policy=None,
importance_type=None,
interaction_constraints=None, learning_rate=0.01,
max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None,
max_depth=3, max_leaves=None,
min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None,
n_estimators=50, n_jobs=None,
num_parallel_tree=None, ...))])ColumnTransformer(force_int_remainder_cols=False, remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking', 'Physical_Examination',
'Adenopathy', 'Focality', 'Risk', 'T',
'Stage', 'Response'])])['Gender', 'Smoking', 'Physical_Examination', 'Adenopathy', 'Focality', 'Risk', 'T', 'Stage', 'Response']
OrdinalEncoder()
['Age']
passthrough
XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=0.7, device=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric='logloss',
feature_types=None, feature_weights=None, gamma=0.1,
grow_policy=None, importance_type=None,
interaction_constraints=None, learning_rate=0.01, max_bin=None,
max_cat_threshold=None, max_cat_to_onehot=None,
max_delta_step=None, max_depth=3, max_leaves=None,
min_child_weight=None, missing=nan, monotone_constraints=None,
multi_strategy=None, n_estimators=50, n_jobs=None,
num_parallel_tree=None, ...)##################################
# Evaluating the most optimal XGBoost model
# on the validation data
##################################
y_validation_pred = final_model.predict(X_preprocessed_validation)
validation_metrics = evaluate_binary_metrics(y_preprocessed_validation_encoded, y_validation_pred)
##################################
# Consolidating the optimal hyperparameter combination
# and its corresponding MCCV and validation metrics
##################################
consolidated_table_1 = pd.DataFrame({
'hyperparameter': list(best_combo.keys()),
'mccv.optimal.value': list(best_combo.values())
})
consolidated_table_2 = pd.DataFrame({
'metric': list(best_metrics.keys()),
'mccv': [round(v, 4) for v in best_metrics.values()],
'validation': [round(validation_metrics[k], 4) for k in best_metrics.keys()]
})
print(f"{experiment_name} optimal hyperparameter combination:")
display(consolidated_table_1)
print(f"{experiment_name} optimal hyperparameter combination mccv versus validation performance metrics:")
display(consolidated_table_2)
boosted_xgb_experiment optimal hyperparameter combination:
| hyperparameter | mccv.optimal.value | |
|---|---|---|
| 0 | boosted_xgb_model__gamma | 0.10 |
| 1 | boosted_xgb_model__learning_rate | 0.01 |
| 2 | boosted_xgb_model__max_depth | 3.00 |
| 3 | boosted_xgb_model__n_estimators | 50.00 |
boosted_xgb_experiment optimal hyperparameter combination mccv versus validation performance metrics:
| metric | mccv | validation | |
|---|---|---|---|
| 0 | accuracy | 0.8916 | 0.9130 |
| 1 | sensitivity | 0.8442 | 0.9000 |
| 2 | specificity | 0.9126 | 0.9184 |
| 3 | f1 | 0.8264 | 0.8571 |
##################################
# Decoding the true and predicted labels
# for artifact storage
##################################
y_validation_true_decoded = y_encoder.inverse_transform(y_preprocessed_validation_encoded.reshape(-1, 1)).ravel()
y_validation_pred_decoded = y_encoder.inverse_transform(y_validation_pred.reshape(-1, 1)).ravel()
##################################
# Saving the artifacts from the XGBoost model
# hyperparameter tuning exercise
##################################
mccv_csv_path = os.path.join(artifact_dir, f"{model_name}_MCCV_metrics.csv")
cm_path = os.path.join(artifact_dir, f"{model_name}_validation_confusion_matrix.png")
report_path = os.path.join(artifact_dir, f"{model_name}_validation_classification_report.txt")
df_all_combos = pd.DataFrame(all_combos_metrics)
df_all_combos.to_csv(mccv_csv_path, index=False)
log_confusion_matrix(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=cm_path)
log_classification_report(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=report_path)
pickle_model_path = os.path.join(artifact_dir, f"{model_name}_final_model.pkl")
with open(pickle_model_path, "wb") as f:
pickle.dump(final_model, f)
train_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_train_dataset.csv")
thyroid_cancer_preprocessed_train.to_csv(train_dataset_csv_path, index=False)
validation_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_validation_dataset.csv")
thyroid_cancer_preprocessed_validation.to_csv(validation_dataset_csv_path, index=False)
##################################
# Logging the most optimal XGBoost model
# developed from the full training data
# and evaluated on the validation data
##################################
# Casting integer columns in preprocessed training data to float64
X_logged_input = X_preprocessed_train.astype({
col: 'float64' for col in X_preprocessed_train.select_dtypes(include='int').columns
})
# Preparing the input signature for model logging
input_example = X_logged_input.sample(n=10, random_state=987654321)
signature = infer_signature(input_example, final_model.predict(input_example))
final_run_name = f"XGB_HyperparameterCombo_{best_combo_index + 1}_Optimal"
with mlflow.start_run(run_name=final_run_name, description="Retrained on Full Training Set"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", final_model_hash)
mlflow.set_tag("model_type", "final_boosted_xgb")
mlflow.set_tag("best_combo_index", best_combo_index)
mlflow.log_params(best_combo)
# Logging the average MCCV metrics computed previously
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in best_metrics.items()})
# Logging validation set metrics
mlflow.log_metrics({f"validation_{k}": v for k, v in validation_metrics.items()})
# Logging the model with input signature
mlflow.sklearn.log_model(
sk_model=final_model,
name="best_boosted_xgb_model",
input_example=input_example,
signature=signature
)
# Logging artifacts
mlflow.log_artifact(mccv_csv_path)
mlflow.log_artifact(cm_path)
mlflow.log_artifact(report_path)
mlflow.log_artifact(pickle_model_path)
mlflow.log_artifact(train_dataset_csv_path)
mlflow.log_artifact(validation_dataset_csv_path)
# Logging datasets with metadata (MLflow dataset tracking)
train_dataset = mlflow.data.from_pandas(
pd.read_csv(train_csv_path),
name="mccv_data"
)
mlflow.log_input(train_dataset, context="training")
print("Final XGB Model Trained and Logged with Validation Set Evaluation.")
Final XGB Model Trained and Logged with Validation Set Evaluation.
1.7.5 Light GBM¶
Light GBM (Light Gradient Boosting Machine) is a variation of Gradient Boosting designed for efficiency and scalability. Unlike traditional boosting methods that grow trees level by level, LightGBM grows trees leaf-wise, choosing the most informative splits, leading to faster convergence. It also uses histogram-based binning to speed up computations. These optimizations allow LightGBM to train on large datasets efficiently while maintaining high accuracy. Its advantages include faster training speed, reduced memory usage, and strong predictive performance, particularly for large datasets with many features. However, LightGBM can overfit more easily than XGBoost if not properly tuned, and it may not perform as well on small datasets. Additionally, its handling of categorical variables requires careful preprocessing, and the leaf-wise tree growth can sometimes lead to instability if not controlled properly.
- The light gbm model from the light Python library API was implemented. Combined with a preprocessing operation into a pipeline, the mlflow.sklearn module was used for loading and logging the model during experiment tracking.
- The model contains 4 hyperparameters for tuning:
- learning_rate = step size at which weights are updated during training made to vary between 0.01 and 0.10
- min_child_samples = minimum number of data needed in a child 3 and 6
- num_leaves = maximum tree leaves for base learners made to vary between 8 and 16
- n_estimators = number of boosted trees to fit made to vary between 50 and 100
- A special hyperparameter (scale_pos_weight = 2.0) was fixed to address the minimal 2:1 class imbalance observed between the No and Yes Recurred categories.
- Hyperparameter tuning was conducted using a 25-cycle 70-30 stratified train-test split Monte Carlo cross-validation with the results logged using MLflow experiment tracking and organized under bagged_rf_experiment.
- The optimal model performance evaluated using the F1 score, sensitivity and specificity metrics, respectively, was determined for the following hyperparameter combination:
- learning_rate = 0.01
- min_child_samples = 3
- num_leaves 8
- n_estimators = 100
- The MCCV model performance of the optimal model as accessed from the MLflow user interface is summarized as follows:
- F1 Score = 0.8025
- Sensitivity = 0.8000
- Specificity = 0.9162
- Accuracy = 0.8806
- The independent validation model performance as accessed from the MLflow user interface of the optimal model is summarized as follows:
- F1 Score = 0.8292
- Sensitivity = 0.8500
- Specificity = 0.9183
- Accuracy = 0.8985
##################################
# Setting the model naming conventions
##################################
model_name = "boosted_lgbm"
experiment_name = f"{model_name}_experiment"
artifact_dir = f"artifacts/{model_name}"
os.makedirs(artifact_dir, exist_ok=True)
##################################
# Setting the MLflow experiment
# for the Light GBM model
##################################
mlflow.set_experiment(experiment_name)
client = MlflowClient()
##################################
# Defining the categorical preprocessing parameters
##################################
categorical_features = ['Gender','Smoking','Physical_Examination','Adenopathy','Focality','Risk','T','Stage','Response']
categorical_transformer = OrdinalEncoder()
categorical_preprocessor = ColumnTransformer(transformers=[
('cat', categorical_transformer, categorical_features)],
remainder='passthrough',
force_int_remainder_cols=False)
##################################
# Defining the preprocessing and modeling pipeline parameters
##################################
boosted_lgbm_pipeline = Pipeline([
('categorical_preprocessor', categorical_preprocessor),
('boosted_lgbm_model', LGBMClassifier(scale_pos_weight=2.0,
random_state=987654321,
max_depth=-1,
feature_fraction =0.7,
bagging_fraction=0.7,
verbose=-1))
])
##################################
# Defining hyperparameter grid
##################################
boosted_lgbm_hyperparameter_grid = {
'boosted_lgbm_model__learning_rate': [0.01, 0.10],
'boosted_lgbm_model__min_child_samples': [3, 6],
'boosted_lgbm_model__num_leaves': [8, 16],
'boosted_lgbm_model__n_estimators': [50, 100]
}
param_combos = list(ParameterGrid(boosted_lgbm_hyperparameter_grid))
##################################
# Encoding the response variables
# for model evaluation
##################################
y_encoder = OrdinalEncoder()
y_encoder.fit(y_preprocessed_train.values.reshape(-1, 1))
y_preprocessed_train_encoded = y_encoder.transform(y_preprocessed_train.values.reshape(-1, 1)).ravel()
class_labels = list(y_encoder.categories_[0])
y_preprocessed_validation_encoded = y_encoder.transform(y_preprocessed_validation.values.reshape(-1, 1)).ravel()
##################################
# Defining the cross-validation strategy
# using Monte Carlo Cross-Validation Setup (25 cycles of 70-30 stratified splits))
##################################
mccv = StratifiedShuffleSplit(n_splits=25,
test_size=0.3,
random_state=987654321)
##################################
# Creating containers for the best hyperparameter combination
# based on the F1 Score, Sensitivity and Specificity metrics
##################################
best_combo = None
best_metrics = {'f1': -1, 'sensitivity': -1, 'specificity': -1}
all_combos_metrics = []
##################################
# Implementing experiment tracking
# of the hyperparameter tuning exercise
# using MLflow for the Light GBM model
##################################
import warnings
warnings.filterwarnings('ignore', category=UserWarning, module='sklearn.utils.validation')
for combo_index, param_set in enumerate(param_combos):
print(f"Evaluating param combo {combo_index + 1}/{len(param_combos)}: {param_set}")
# Generating hash and checking for duplicates
run_hash = generate_run_hash(param_set, mccv_seed=987654321)
existing_run_id = find_existing_run_by_hash("boosted_lgbm_experiment", run_hash)
if existing_run_id:
print(f"Duplicate found for this combo (hash={run_hash}). Removing old run: {existing_run_id}")
delete_run(existing_run_id)
else:
print(f"No duplicate found. Proceeding to evaluate and log.")
all_metrics = []
for i, (train_idx, test_idx) in enumerate(mccv.split(X_preprocessed_train, y_preprocessed_train_encoded)):
X_train, X_test = X_preprocessed_train.iloc[train_idx], X_preprocessed_train.iloc[test_idx]
y_train, y_test = y_preprocessed_train_encoded[train_idx], y_preprocessed_train_encoded[test_idx]
# Cloning the pipeline and setting the hyperparameters
model = clone(boosted_lgbm_pipeline)
model.set_params(**param_set)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
all_metrics.append(evaluate_binary_metrics(y_test, y_pred))
# Aggregating the metrics across MCCV
df_metrics = pd.DataFrame(all_metrics)
avg_metrics = df_metrics.mean().to_dict()
avg_metrics_prefixed = {f"avg_mccv_{k}": v for k, v in avg_metrics.items()}
all_combos_metrics.append({**param_set, **avg_metrics_prefixed})
# Logging the hyperparameter combinations and metrics to MLflow
with mlflow.start_run(run_name=f"LGBM_HyperparameterCombo_{combo_index+1}"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", run_hash)
mlflow.log_params(param_set)
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in avg_metrics.items()})
mlflow.set_tag("param_index", combo_index)
# Tracking the best hyperparameter combination
better = False
if avg_metrics['f1'] > best_metrics['f1']:
better = True
elif avg_metrics['f1'] == best_metrics['f1']:
if avg_metrics['sensitivity'] > best_metrics['sensitivity']:
better = True
elif avg_metrics['sensitivity'] == best_metrics['sensitivity']:
if avg_metrics['specificity'] > best_metrics['specificity']:
better = True
if better:
best_combo = param_set
best_combo_index = combo_index
best_metrics = avg_metrics
Evaluating param combo 1/16: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 50, 'boosted_lgbm_model__num_leaves': 8}
Duplicate found for this combo (hash=cfbcfc69d01483171c99767848e6ff6c). Removing old run: 870c3d634a1b47e299ce3c6e3ac3cd0a
Evaluating param combo 2/16: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 50, 'boosted_lgbm_model__num_leaves': 16}
Duplicate found for this combo (hash=365bd16ade45a726de60dbfbf3131994). Removing old run: 2ef4ab1627a947149f6731552784b4d6
Evaluating param combo 3/16: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 8}
Duplicate found for this combo (hash=c5ec25839801fb3bc539c05d4def3cef). Removing old run: efd66c45a47c404aa876bdacdbd305f9
Evaluating param combo 4/16: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 16}
Duplicate found for this combo (hash=7db4d8d0b67551a6de17a401d1a4275d). Removing old run: 3a722675877f4b31a1cde1549ae9ed86
Evaluating param combo 5/16: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 6, 'boosted_lgbm_model__n_estimators': 50, 'boosted_lgbm_model__num_leaves': 8}
Duplicate found for this combo (hash=44f22c5b7808559db6fa9f87c58f8f9f). Removing old run: c34c503daf044f29b4269a16c5d35b5c
Evaluating param combo 6/16: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 6, 'boosted_lgbm_model__n_estimators': 50, 'boosted_lgbm_model__num_leaves': 16}
Duplicate found for this combo (hash=97c39002bc2d9c31c49bb580c559d1f0). Removing old run: 790e88c74ffe43c8a345be5d3123dcd2
Evaluating param combo 7/16: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 6, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 8}
Duplicate found for this combo (hash=bf15c0f4bc63a1ce68fac4c0c89d897e). Removing old run: b0dc667f3889437c8a573ded8f93d14a
Evaluating param combo 8/16: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 6, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 16}
Duplicate found for this combo (hash=9dcec3542ac55d6e75728a7fac5a600a). Removing old run: e98c8202e89c441d992c95d8d2112753
Evaluating param combo 9/16: {'boosted_lgbm_model__learning_rate': 0.1, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 50, 'boosted_lgbm_model__num_leaves': 8}
Duplicate found for this combo (hash=d7f2aee7bf3b04785c4bfd53c9fffd23). Removing old run: bcf438f1960f4855807879b46b97cb71
Evaluating param combo 10/16: {'boosted_lgbm_model__learning_rate': 0.1, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 50, 'boosted_lgbm_model__num_leaves': 16}
Duplicate found for this combo (hash=94f72ec03b1151dfbf497a6c7e1c6307). Removing old run: c7fa5f1dac8b495da44649d6d338eb64
Evaluating param combo 11/16: {'boosted_lgbm_model__learning_rate': 0.1, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 8}
Duplicate found for this combo (hash=b2c57e527890facb410bca76851e5cb7). Removing old run: cf706490fc1843919ac3b4dddb1a98e6
Evaluating param combo 12/16: {'boosted_lgbm_model__learning_rate': 0.1, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 16}
Duplicate found for this combo (hash=eefe23e5a8f59bc89c4e737e95b13c9e). Removing old run: 119bdacf93824ae4ac8923060ddaa79c
Evaluating param combo 13/16: {'boosted_lgbm_model__learning_rate': 0.1, 'boosted_lgbm_model__min_child_samples': 6, 'boosted_lgbm_model__n_estimators': 50, 'boosted_lgbm_model__num_leaves': 8}
Duplicate found for this combo (hash=7e206eaf832238b8cc6375d8890f0311). Removing old run: 1d673cdff73749199fd219ec76ecb9db
Evaluating param combo 14/16: {'boosted_lgbm_model__learning_rate': 0.1, 'boosted_lgbm_model__min_child_samples': 6, 'boosted_lgbm_model__n_estimators': 50, 'boosted_lgbm_model__num_leaves': 16}
Duplicate found for this combo (hash=1e567c4bab4eae2b6edb1a262e5895aa). Removing old run: fe3debaf44b541f9ba65daa744c82020
Evaluating param combo 15/16: {'boosted_lgbm_model__learning_rate': 0.1, 'boosted_lgbm_model__min_child_samples': 6, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 8}
Duplicate found for this combo (hash=1de53d811c7746ae5aae65c7ef6f0fe6). Removing old run: 7a9ad98800244f80a3d27acb0576ab5c
Evaluating param combo 16/16: {'boosted_lgbm_model__learning_rate': 0.1, 'boosted_lgbm_model__min_child_samples': 6, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 16}
Duplicate found for this combo (hash=70874e474e04a087b85a5babba5b7101). Removing old run: ca61d949e34a4ba5ae1ff2359953ff05
##################################
# Defining a function to
# generate a unique hash for the selected optimal model
##################################
final_model_hash = generate_run_hash(best_combo, mccv_seed=987654321)
existing_final_run_id = find_existing_run_by_hash("boosted_lgbm_experiment", final_model_hash)
if existing_final_run_id:
print(f"Duplicate final model found (hash={final_model_hash}). Removing previous run: {existing_final_run_id}")
delete_run(existing_final_run_id)
else:
print(f"No duplicate final model found. Proceeding to log new final model.")
Duplicate final model found (hash=c5ec25839801fb3bc539c05d4def3cef). Removing previous run: 19eee4dd8d0f4c5b90166d67957d9740
##################################
# Retraining the most optimal Light GBM model
# on the full training data
##################################
print("Optimal Hyperparameter Combination for Light GBM:", best_combo)
final_model = clone(boosted_lgbm_pipeline)
final_model.set_params(**best_combo)
final_model.fit(X_preprocessed_train, y_preprocessed_train_encoded)
Optimal Hyperparameter Combination for Light GBM: {'boosted_lgbm_model__learning_rate': 0.01, 'boosted_lgbm_model__min_child_samples': 3, 'boosted_lgbm_model__n_estimators': 100, 'boosted_lgbm_model__num_leaves': 8}
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_lgbm_model',
LGBMClassifier(bagging_fraction=0.7, feature_fraction=0.7,
learning_rate=0.01, min_child_samples=3,
num_leaves=8, random_state=987654321,
scale_pos_weight=2.0, verbose=-1))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_lgbm_model',
LGBMClassifier(bagging_fraction=0.7, feature_fraction=0.7,
learning_rate=0.01, min_child_samples=3,
num_leaves=8, random_state=987654321,
scale_pos_weight=2.0, verbose=-1))])ColumnTransformer(force_int_remainder_cols=False, remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking', 'Physical_Examination',
'Adenopathy', 'Focality', 'Risk', 'T',
'Stage', 'Response'])])['Gender', 'Smoking', 'Physical_Examination', 'Adenopathy', 'Focality', 'Risk', 'T', 'Stage', 'Response']
OrdinalEncoder()
['Age']
passthrough
LGBMClassifier(bagging_fraction=0.7, feature_fraction=0.7, learning_rate=0.01,
min_child_samples=3, num_leaves=8, random_state=987654321,
scale_pos_weight=2.0, verbose=-1)##################################
# Evaluating the most optimal Light GBM model
# on the validation data
##################################
y_validation_pred = final_model.predict(X_preprocessed_validation)
validation_metrics = evaluate_binary_metrics(y_preprocessed_validation_encoded, y_validation_pred)
##################################
# Consolidating the optimal hyperparameter combination
# and its corresponding MCCV and validation metrics
##################################
consolidated_table_1 = pd.DataFrame({
'hyperparameter': list(best_combo.keys()),
'mccv.optimal.value': list(best_combo.values())
})
consolidated_table_2 = pd.DataFrame({
'metric': list(best_metrics.keys()),
'mccv': [round(v, 4) for v in best_metrics.values()],
'validation': [round(validation_metrics[k], 4) for k in best_metrics.keys()]
})
print(f"{experiment_name} optimal hyperparameter combination:")
display(consolidated_table_1)
print(f"{experiment_name} optimal hyperparameter combination mccv versus validation performance metrics:")
display(consolidated_table_2)
boosted_lgbm_experiment optimal hyperparameter combination:
| hyperparameter | mccv.optimal.value | |
|---|---|---|
| 0 | boosted_lgbm_model__learning_rate | 0.01 |
| 1 | boosted_lgbm_model__min_child_samples | 3.00 |
| 2 | boosted_lgbm_model__n_estimators | 100.00 |
| 3 | boosted_lgbm_model__num_leaves | 8.00 |
boosted_lgbm_experiment optimal hyperparameter combination mccv versus validation performance metrics:
| metric | mccv | validation | |
|---|---|---|---|
| 0 | accuracy | 0.8806 | 0.8986 |
| 1 | sensitivity | 0.8000 | 0.8500 |
| 2 | specificity | 0.9163 | 0.9184 |
| 3 | f1 | 0.8025 | 0.8293 |
##################################
# Decoding the true and predicted labels
# for artifact storage
##################################
y_validation_true_decoded = y_encoder.inverse_transform(y_preprocessed_validation_encoded.reshape(-1, 1)).ravel()
y_validation_pred_decoded = y_encoder.inverse_transform(y_validation_pred.reshape(-1, 1)).ravel()
##################################
# Saving the artifacts from the Light GBM model
# hyperparameter tuning exercise
##################################
mccv_csv_path = os.path.join(artifact_dir, f"{model_name}_MCCV_metrics.csv")
cm_path = os.path.join(artifact_dir, f"{model_name}_validation_confusion_matrix.png")
report_path = os.path.join(artifact_dir, f"{model_name}_validation_classification_report.txt")
df_all_combos = pd.DataFrame(all_combos_metrics)
df_all_combos.to_csv(mccv_csv_path, index=False)
log_confusion_matrix(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=cm_path)
log_classification_report(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=report_path)
pickle_model_path = os.path.join(artifact_dir, f"{model_name}_final_model.pkl")
with open(pickle_model_path, "wb") as f:
pickle.dump(final_model, f)
train_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_train_dataset.csv")
thyroid_cancer_preprocessed_train.to_csv(train_dataset_csv_path, index=False)
validation_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_validation_dataset.csv")
thyroid_cancer_preprocessed_validation.to_csv(validation_dataset_csv_path, index=False)
##################################
# Logging the most optimal Light GBM model
# developed from the full training data
# and evaluated on the validation data
##################################
# Casting integer columns in preprocessed training data to float64
X_logged_input = X_preprocessed_train.astype({
col: 'float64' for col in X_preprocessed_train.select_dtypes(include='int').columns
})
# Preparing the input signature for model logging
input_example = X_logged_input.sample(n=10, random_state=987654321)
signature = infer_signature(input_example, final_model.predict(input_example))
final_run_name = f"LGBM_HyperparameterCombo_{best_combo_index + 1}_Optimal"
with mlflow.start_run(run_name=final_run_name, description="Retrained on Full Training Set"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", final_model_hash)
mlflow.set_tag("model_type", "final_boosted_lgbm")
mlflow.set_tag("best_combo_index", best_combo_index)
mlflow.log_params(best_combo)
# Logging the average MCCV metrics computed previously
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in best_metrics.items()})
# Logging validation set metrics
mlflow.log_metrics({f"validation_{k}": v for k, v in validation_metrics.items()})
# Logging the model with input signature
mlflow.sklearn.log_model(
sk_model=final_model,
name="best_boosted_lgbm_model",
input_example=input_example,
signature=signature
)
# Logging artifacts
mlflow.log_artifact(mccv_csv_path)
mlflow.log_artifact(cm_path)
mlflow.log_artifact(report_path)
mlflow.log_artifact(pickle_model_path)
mlflow.log_artifact(train_dataset_csv_path)
mlflow.log_artifact(validation_dataset_csv_path)
# Logging datasets with metadata (MLflow dataset tracking)
train_dataset = mlflow.data.from_pandas(
pd.read_csv(train_csv_path),
name="mccv_data"
)
mlflow.log_input(train_dataset, context="training")
print("Final LGBM Model Trained and Logged with Validation Set Evaluation.")
Final LGBM Model Trained and Logged with Validation Set Evaluation.
1.7.6 CatBoost¶
CatBoost (Categorical Boosting) is a boosting algorithm optimized for categorical data. Unlike other gradient boosting methods that require categorical variables to be manually encoded, CatBoost handles them natively, reducing preprocessing effort and improving performance. It builds decision trees iteratively, like other boosting methods, but uses ordered boosting to prevent target leakage and enhance generalization. The main advantages of CatBoost are its ability to handle categorical data without extensive preprocessing, high accuracy with minimal tuning, and robustness against overfitting due to built-in regularization. Additionally, it is relatively fast and memory-efficient. However, CatBoost can still be slower than LightGBM on very large datasets, and while it requires less tuning, improper parameter selection can lead to suboptimal performance. Its internal mechanics, such as ordered boosting, make interpretation more complex compared to simpler models.
- The catboost model from the catboost Python library API was implemented. Combined with a preprocessing operation into a pipeline, the mlflow.sklearn module was used for loading and logging the model during experiment tracking.
- The model contains 4 hyperparameters for tuning:
- learning_rate = step size at which weights are updated during training made to vary between 0.01 and 0.10
- max_depth = maximum depth of each decision tree in the boosting process made to vary between 3 and 6
- num_leaves = maximum tree leaves for base learners made to vary between 8 and 16
- iterations = number of boosted trees to fit made to vary between 50 and 100
- A special hyperparameter (scale_pos_weight = 2.0) was fixed to address the minimal 2:1 class imbalance observed between the No and Yes Recurred categories.
- Hyperparameter tuning was conducted using a 25-cycle 70-30 stratified train-test split Monte Carlo cross-validation with the results logged using MLflow experiment tracking and organized under bagged_rf_experiment.
- The optimal model performance evaluated using the F1 score, sensitivity and specificity metrics, respectively, was determined for the following hyperparameter combination:
- learning_rate = 0.01
- max_depth = 3
- num_leaves = 8
- iterations = 50
- The MCCV model performance of the optimal model as accessed from the MLflow user interface is summarized as follows:
- F1 Score = 0.8348
- Sensitivity = 0.8799
- Specificity = 0.8976
- Accuracy = 0.8922
- The independent validation model performance as accessed from the MLflow user interface of the optimal model is summarized as follows:
- F1 Score = 0.8571
- Sensitivity = 0.9000
- Specificity = 0.9183
- Accuracy = 0.9130
##################################
# Setting the model naming conventions
##################################
model_name = "boosted_cb"
experiment_name = f"{model_name}_experiment"
artifact_dir = f"artifacts/{model_name}"
os.makedirs(artifact_dir, exist_ok=True)
##################################
# Setting the MLflow experiment
# for the CatBoost model
##################################
mlflow.set_experiment(experiment_name)
client = MlflowClient()
##################################
# Defining the categorical preprocessing parameters
##################################
categorical_features = ['Gender','Smoking','Physical_Examination','Adenopathy','Focality','Risk','T','Stage','Response']
categorical_transformer = OrdinalEncoder()
categorical_preprocessor = ColumnTransformer(transformers=[
('cat', categorical_transformer, categorical_features)],
remainder='passthrough',
force_int_remainder_cols=False)
##################################
# Defining the preprocessing and modeling pipeline parameters
##################################
boosted_cb_pipeline = Pipeline([
('categorical_preprocessor', categorical_preprocessor),
('boosted_cb_model', CatBoostClassifier(scale_pos_weight=2.0,
random_state=987654321,
subsample =0.7,
colsample_bylevel=0.7,
grow_policy='Lossguide'))
])
##################################
# Defining hyperparameter grid
##################################
boosted_cb_hyperparameter_grid = {
'boosted_cb_model__learning_rate': [0.01, 0.10],
'boosted_cb_model__max_depth': [3, 6],
'boosted_cb_model__num_leaves': [8, 16],
'boosted_cb_model__iterations': [50, 100]
}
param_combos = list(ParameterGrid(boosted_cb_hyperparameter_grid))
##################################
# Encoding the response variables
# for model evaluation
##################################
y_encoder = OrdinalEncoder()
y_encoder.fit(y_preprocessed_train.values.reshape(-1, 1))
y_preprocessed_train_encoded = y_encoder.transform(y_preprocessed_train.values.reshape(-1, 1)).ravel()
class_labels = list(y_encoder.categories_[0])
y_preprocessed_validation_encoded = y_encoder.transform(y_preprocessed_validation.values.reshape(-1, 1)).ravel()
##################################
# Defining the cross-validation strategy
# using Monte Carlo Cross-Validation Setup (25 cycles of 70-30 stratified splits))
##################################
mccv = StratifiedShuffleSplit(n_splits=25,
test_size=0.3,
random_state=987654321)
##################################
# Creating containers for the best hyperparameter combination
# based on the F1 Score, Sensitivity and Specificity metrics
##################################
best_combo = None
best_metrics = {'f1': -1, 'sensitivity': -1, 'specificity': -1}
all_combos_metrics = []
##################################
# Implementing experiment tracking
# of the hyperparameter tuning exercise
# using MLflow for the CatBoost model
##################################
for combo_index, param_set in enumerate(param_combos):
print(f"Evaluating param combo {combo_index + 1}/{len(param_combos)}: {param_set}")
# Generating hash and checking for duplicates
run_hash = generate_run_hash(param_set, mccv_seed=987654321)
existing_run_id = find_existing_run_by_hash("boosted_cb_experiment", run_hash)
if existing_run_id:
print(f"Duplicate found for this combo (hash={run_hash}). Removing old run: {existing_run_id}")
delete_run(existing_run_id)
else:
print(f"No duplicate found. Proceeding to evaluate and log.")
all_metrics = []
for i, (train_idx, test_idx) in enumerate(mccv.split(X_preprocessed_train, y_preprocessed_train_encoded)):
X_train, X_test = X_preprocessed_train.iloc[train_idx], X_preprocessed_train.iloc[test_idx]
y_train, y_test = y_preprocessed_train_encoded[train_idx], y_preprocessed_train_encoded[test_idx]
# Cloning the pipeline and setting the hyperparameters
model = clone(boosted_cb_pipeline)
model.set_params(**param_set)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
all_metrics.append(evaluate_binary_metrics(y_test, y_pred))
# Aggregating the metrics across MCCV
df_metrics = pd.DataFrame(all_metrics)
avg_metrics = df_metrics.mean().to_dict()
avg_metrics_prefixed = {f"avg_mccv_{k}": v for k, v in avg_metrics.items()}
all_combos_metrics.append({**param_set, **avg_metrics_prefixed})
# Logging the hyperparameter combinations and metrics to MLflow
with mlflow.start_run(run_name=f"CB_HyperparameterCombo_{combo_index+1}"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", run_hash)
mlflow.log_params(param_set)
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in avg_metrics.items()})
mlflow.set_tag("param_index", combo_index)
# Tracking the best hyperparameter combination
better = False
if avg_metrics['f1'] > best_metrics['f1']:
better = True
elif avg_metrics['f1'] == best_metrics['f1']:
if avg_metrics['sensitivity'] > best_metrics['sensitivity']:
better = True
elif avg_metrics['sensitivity'] == best_metrics['sensitivity']:
if avg_metrics['specificity'] > best_metrics['specificity']:
better = True
if better:
best_combo = param_set
best_combo_index = combo_index
best_metrics = avg_metrics
Evaluating param combo 1/16: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 8}
Duplicate found for this combo (hash=09ee965f4973379bda050e0af8b07bb7). Removing old run: 9a1a5e71144545648fa6c234814c7ad4
0: learn: 0.6882794 total: 93ms remaining: 4.56s
1: learn: 0.6829408 total: 93.9ms remaining: 2.25s
2: learn: 0.6779769 total: 94.6ms remaining: 1.48s
3: learn: 0.6731590 total: 95ms remaining: 1.09s
4: learn: 0.6685559 total: 95.3ms remaining: 858ms
5: learn: 0.6636779 total: 95.8ms remaining: 702ms
6: learn: 0.6591763 total: 96ms remaining: 590ms
7: learn: 0.6551357 total: 96.3ms remaining: 506ms
8: learn: 0.6504101 total: 96.8ms remaining: 441ms
9: learn: 0.6464580 total: 97.4ms remaining: 390ms
10: learn: 0.6425359 total: 98ms remaining: 347ms
11: learn: 0.6387708 total: 99.1ms remaining: 314ms
12: learn: 0.6346059 total: 101ms remaining: 286ms
13: learn: 0.6304314 total: 101ms remaining: 260ms
14: learn: 0.6268009 total: 102ms remaining: 237ms
15: learn: 0.6229881 total: 102ms remaining: 218ms
16: learn: 0.6190015 total: 104ms remaining: 202ms
17: learn: 0.6151072 total: 105ms remaining: 186ms
18: learn: 0.6113699 total: 105ms remaining: 171ms
19: learn: 0.6075444 total: 106ms remaining: 158ms
20: learn: 0.6037564 total: 107ms remaining: 147ms
21: learn: 0.5999778 total: 108ms remaining: 137ms
22: learn: 0.5958785 total: 108ms remaining: 127ms
23: learn: 0.5922311 total: 109ms remaining: 118ms
24: learn: 0.5892157 total: 110ms remaining: 110ms
25: learn: 0.5861289 total: 111ms remaining: 102ms
26: learn: 0.5829753 total: 112ms remaining: 95.2ms
27: learn: 0.5798042 total: 113ms remaining: 88.8ms
28: learn: 0.5767399 total: 114ms remaining: 82.5ms
29: learn: 0.5739181 total: 115ms remaining: 76.4ms
30: learn: 0.5712722 total: 116ms remaining: 70.8ms
31: learn: 0.5679889 total: 116ms remaining: 65.3ms
32: learn: 0.5647182 total: 117ms remaining: 60.3ms
33: learn: 0.5613468 total: 118ms remaining: 55.4ms
34: learn: 0.5577149 total: 118ms remaining: 50.6ms
35: learn: 0.5547437 total: 119ms remaining: 46.2ms
36: learn: 0.5517524 total: 119ms remaining: 41.9ms
37: learn: 0.5484946 total: 120ms remaining: 37.9ms
38: learn: 0.5451312 total: 120ms remaining: 33.9ms
39: learn: 0.5421510 total: 121ms remaining: 30.2ms
40: learn: 0.5387318 total: 122ms remaining: 26.7ms
41: learn: 0.5355684 total: 122ms remaining: 23.2ms
42: learn: 0.5324215 total: 122ms remaining: 19.9ms
43: learn: 0.5298921 total: 123ms remaining: 16.8ms
44: learn: 0.5271650 total: 124ms remaining: 13.8ms
45: learn: 0.5248260 total: 125ms remaining: 10.8ms
46: learn: 0.5222534 total: 125ms remaining: 7.99ms
47: learn: 0.5193675 total: 126ms remaining: 5.24ms
48: learn: 0.5168600 total: 127ms remaining: 2.58ms
49: learn: 0.5145437 total: 127ms remaining: 0us
0: learn: 0.6880102 total: 633us remaining: 31ms
1: learn: 0.6822828 total: 1.09ms remaining: 26.1ms
2: learn: 0.6772106 total: 1.48ms remaining: 23.2ms
3: learn: 0.6726730 total: 2.05ms remaining: 23.6ms
4: learn: 0.6682095 total: 2.46ms remaining: 22.1ms
5: learn: 0.6635268 total: 2.74ms remaining: 20.1ms
6: learn: 0.6588811 total: 2.96ms remaining: 18.2ms
7: learn: 0.6545703 total: 3.34ms remaining: 17.5ms
8: learn: 0.6497856 total: 3.63ms remaining: 16.5ms
9: learn: 0.6448858 total: 4.06ms remaining: 16.2ms
10: learn: 0.6405230 total: 4.35ms remaining: 15.4ms
11: learn: 0.6361860 total: 4.84ms remaining: 15.3ms
12: learn: 0.6314977 total: 5.34ms remaining: 15.2ms
13: learn: 0.6272630 total: 5.77ms remaining: 14.8ms
14: learn: 0.6237927 total: 6.26ms remaining: 14.6ms
15: learn: 0.6202946 total: 6.92ms remaining: 14.7ms
16: learn: 0.6160996 total: 7.42ms remaining: 14.4ms
17: learn: 0.6121987 total: 7.69ms remaining: 13.7ms
18: learn: 0.6085887 total: 8.08ms remaining: 13.2ms
19: learn: 0.6053875 total: 8.57ms remaining: 12.9ms
20: learn: 0.6012749 total: 8.91ms remaining: 12.3ms
21: learn: 0.5970959 total: 9.48ms remaining: 12.1ms
22: learn: 0.5929697 total: 9.97ms remaining: 11.7ms
23: learn: 0.5894176 total: 10.4ms remaining: 11.3ms
24: learn: 0.5848522 total: 10.7ms remaining: 10.7ms
25: learn: 0.5821273 total: 11.1ms remaining: 10.3ms
26: learn: 0.5789455 total: 11.5ms remaining: 9.76ms
27: learn: 0.5754969 total: 11.9ms remaining: 9.37ms
28: learn: 0.5721862 total: 12.3ms remaining: 8.93ms
29: learn: 0.5691136 total: 12.7ms remaining: 8.45ms
30: learn: 0.5660278 total: 13.1ms remaining: 8.04ms
31: learn: 0.5628754 total: 13.3ms remaining: 7.47ms
32: learn: 0.5589497 total: 13.6ms remaining: 7.03ms
33: learn: 0.5553905 total: 13.9ms remaining: 6.55ms
34: learn: 0.5515082 total: 14.2ms remaining: 6.1ms
35: learn: 0.5482866 total: 14.6ms remaining: 5.69ms
36: learn: 0.5449903 total: 15ms remaining: 5.27ms
37: learn: 0.5423647 total: 15.4ms remaining: 4.86ms
38: learn: 0.5386898 total: 15.7ms remaining: 4.41ms
39: learn: 0.5350966 total: 16ms remaining: 4ms
40: learn: 0.5319492 total: 16.3ms remaining: 3.58ms
41: learn: 0.5288603 total: 16.6ms remaining: 3.17ms
42: learn: 0.5260202 total: 17ms remaining: 2.77ms
43: learn: 0.5230047 total: 17.4ms remaining: 2.37ms
44: learn: 0.5198876 total: 17.8ms remaining: 1.98ms
45: learn: 0.5168284 total: 18.2ms remaining: 1.58ms
46: learn: 0.5136389 total: 18.5ms remaining: 1.18ms
47: learn: 0.5112892 total: 18.9ms remaining: 787us
48: learn: 0.5085121 total: 19.2ms remaining: 392us
49: learn: 0.5054328 total: 19.6ms remaining: 0us
0: learn: 0.6881915 total: 271us remaining: 13.3ms
1: learn: 0.6830903 total: 484us remaining: 11.6ms
2: learn: 0.6779156 total: 659us remaining: 10.3ms
3: learn: 0.6727870 total: 862us remaining: 9.92ms
4: learn: 0.6680191 total: 1.06ms remaining: 9.58ms
5: learn: 0.6632566 total: 1.19ms remaining: 8.71ms
6: learn: 0.6589348 total: 1.6ms remaining: 9.84ms
7: learn: 0.6548497 total: 1.8ms remaining: 9.45ms
8: learn: 0.6499422 total: 2.17ms remaining: 9.87ms
9: learn: 0.6456086 total: 2.6ms remaining: 10.4ms
10: learn: 0.6416798 total: 2.79ms remaining: 9.89ms
11: learn: 0.6376258 total: 3.09ms remaining: 9.79ms
12: learn: 0.6328452 total: 3.34ms remaining: 9.52ms
13: learn: 0.6291112 total: 3.93ms remaining: 10.1ms
14: learn: 0.6252126 total: 4.21ms remaining: 9.82ms
15: learn: 0.6213698 total: 4.68ms remaining: 9.94ms
16: learn: 0.6172627 total: 5.13ms remaining: 9.97ms
17: learn: 0.6136354 total: 5.32ms remaining: 9.46ms
18: learn: 0.6099157 total: 5.48ms remaining: 8.94ms
19: learn: 0.6065275 total: 5.69ms remaining: 8.54ms
20: learn: 0.6026046 total: 6.07ms remaining: 8.39ms
21: learn: 0.5986205 total: 6.52ms remaining: 8.29ms
22: learn: 0.5943580 total: 6.72ms remaining: 7.89ms
23: learn: 0.5906964 total: 6.85ms remaining: 7.42ms
24: learn: 0.5869638 total: 7.04ms remaining: 7.04ms
25: learn: 0.5838294 total: 7.45ms remaining: 6.87ms
26: learn: 0.5809812 total: 7.8ms remaining: 6.64ms
27: learn: 0.5776571 total: 8.14ms remaining: 6.39ms
28: learn: 0.5743650 total: 8.35ms remaining: 6.04ms
29: learn: 0.5714944 total: 8.64ms remaining: 5.76ms
30: learn: 0.5687434 total: 8.87ms remaining: 5.44ms
31: learn: 0.5653821 total: 8.97ms remaining: 5.04ms
32: learn: 0.5620526 total: 9.26ms remaining: 4.77ms
33: learn: 0.5584545 total: 9.52ms remaining: 4.48ms
34: learn: 0.5550520 total: 9.75ms remaining: 4.18ms
35: learn: 0.5514777 total: 9.99ms remaining: 3.89ms
36: learn: 0.5483220 total: 10.3ms remaining: 3.6ms
37: learn: 0.5453927 total: 10.5ms remaining: 3.3ms
38: learn: 0.5418823 total: 10.7ms remaining: 3.02ms
39: learn: 0.5394460 total: 11ms remaining: 2.75ms
40: learn: 0.5362960 total: 11.3ms remaining: 2.48ms
41: learn: 0.5328204 total: 11.4ms remaining: 2.18ms
42: learn: 0.5296236 total: 11.8ms remaining: 1.93ms
43: learn: 0.5272600 total: 12.1ms remaining: 1.64ms
44: learn: 0.5244234 total: 12.3ms remaining: 1.36ms
45: learn: 0.5224995 total: 12.5ms remaining: 1.09ms
46: learn: 0.5199309 total: 12.8ms remaining: 816us
47: learn: 0.5169806 total: 13ms remaining: 541us
48: learn: 0.5142186 total: 13.2ms remaining: 269us
49: learn: 0.5113786 total: 13.5ms remaining: 0us
0: learn: 0.6883581 total: 322us remaining: 15.8ms
1: learn: 0.6829490 total: 652us remaining: 15.7ms
2: learn: 0.6777105 total: 884us remaining: 13.9ms
3: learn: 0.6724965 total: 1.14ms remaining: 13.1ms
4: learn: 0.6678869 total: 1.6ms remaining: 14.4ms
5: learn: 0.6630227 total: 2.28ms remaining: 16.8ms
6: learn: 0.6580845 total: 2.8ms remaining: 17.2ms
7: learn: 0.6539384 total: 3.77ms remaining: 19.8ms
8: learn: 0.6488837 total: 4.66ms remaining: 21.2ms
9: learn: 0.6442866 total: 4.92ms remaining: 19.7ms
10: learn: 0.6404207 total: 5.09ms remaining: 18.1ms
11: learn: 0.6359348 total: 5.9ms remaining: 18.7ms
12: learn: 0.6315682 total: 7.01ms remaining: 20ms
13: learn: 0.6273393 total: 7.73ms remaining: 19.9ms
14: learn: 0.6234264 total: 8.47ms remaining: 19.8ms
15: learn: 0.6197946 total: 9.34ms remaining: 19.9ms
16: learn: 0.6155398 total: 10.1ms remaining: 19.6ms
17: learn: 0.6113889 total: 10.9ms remaining: 19.3ms
18: learn: 0.6079857 total: 11.9ms remaining: 19.4ms
19: learn: 0.6040984 total: 12.3ms remaining: 18.5ms
20: learn: 0.6001675 total: 13.1ms remaining: 18.1ms
21: learn: 0.5959995 total: 13.6ms remaining: 17.4ms
22: learn: 0.5919796 total: 14.2ms remaining: 16.7ms
23: learn: 0.5884148 total: 14.5ms remaining: 15.7ms
24: learn: 0.5841205 total: 14.6ms remaining: 14.6ms
25: learn: 0.5809535 total: 15.2ms remaining: 14ms
26: learn: 0.5776310 total: 15.8ms remaining: 13.5ms
27: learn: 0.5743591 total: 16.5ms remaining: 13ms
28: learn: 0.5712718 total: 17.3ms remaining: 12.5ms
29: learn: 0.5684340 total: 17.8ms remaining: 11.9ms
30: learn: 0.5657478 total: 18.6ms remaining: 11.4ms
31: learn: 0.5623463 total: 18.9ms remaining: 10.6ms
32: learn: 0.5591812 total: 19.4ms remaining: 10ms
33: learn: 0.5555487 total: 19.9ms remaining: 9.34ms
34: learn: 0.5525218 total: 20.4ms remaining: 8.76ms
35: learn: 0.5495863 total: 21ms remaining: 8.16ms
36: learn: 0.5463044 total: 21.5ms remaining: 7.54ms
37: learn: 0.5429413 total: 22.2ms remaining: 7ms
38: learn: 0.5394540 total: 22.6ms remaining: 6.38ms
39: learn: 0.5363956 total: 23.2ms remaining: 5.8ms
40: learn: 0.5330491 total: 23.5ms remaining: 5.16ms
41: learn: 0.5306820 total: 24.1ms remaining: 4.59ms
42: learn: 0.5275766 total: 24.8ms remaining: 4.03ms
43: learn: 0.5246236 total: 25.5ms remaining: 3.47ms
44: learn: 0.5218134 total: 26.1ms remaining: 2.9ms
45: learn: 0.5189606 total: 26.7ms remaining: 2.33ms
46: learn: 0.5159627 total: 27.4ms remaining: 1.75ms
47: learn: 0.5128221 total: 28.1ms remaining: 1.17ms
48: learn: 0.5096912 total: 28.6ms remaining: 582us
49: learn: 0.5072024 total: 29.1ms remaining: 0us
0: learn: 0.6883175 total: 319us remaining: 15.7ms
1: learn: 0.6833177 total: 608us remaining: 14.6ms
2: learn: 0.6787594 total: 793us remaining: 12.4ms
3: learn: 0.6742785 total: 1.05ms remaining: 12.1ms
4: learn: 0.6698376 total: 1.39ms remaining: 12.5ms
5: learn: 0.6655790 total: 2.14ms remaining: 15.7ms
6: learn: 0.6606753 total: 2.53ms remaining: 15.5ms
7: learn: 0.6567153 total: 2.9ms remaining: 15.3ms
8: learn: 0.6521844 total: 3.25ms remaining: 14.8ms
9: learn: 0.6476670 total: 3.7ms remaining: 14.8ms
10: learn: 0.6436401 total: 4.01ms remaining: 14.2ms
11: learn: 0.6392506 total: 4.34ms remaining: 13.8ms
12: learn: 0.6352223 total: 4.73ms remaining: 13.5ms
13: learn: 0.6314012 total: 5.2ms remaining: 13.4ms
14: learn: 0.6271130 total: 5.65ms remaining: 13.2ms
15: learn: 0.6228527 total: 5.91ms remaining: 12.6ms
16: learn: 0.6193636 total: 6.23ms remaining: 12.1ms
17: learn: 0.6149698 total: 6.63ms remaining: 11.8ms
18: learn: 0.6115084 total: 6.78ms remaining: 11.1ms
19: learn: 0.6080555 total: 7.2ms remaining: 10.8ms
20: learn: 0.6044442 total: 7.47ms remaining: 10.3ms
21: learn: 0.6011192 total: 7.71ms remaining: 9.81ms
22: learn: 0.5974142 total: 8.12ms remaining: 9.53ms
23: learn: 0.5941231 total: 8.53ms remaining: 9.24ms
24: learn: 0.5900927 total: 8.81ms remaining: 8.81ms
25: learn: 0.5870085 total: 9.27ms remaining: 8.56ms
26: learn: 0.5840114 total: 9.71ms remaining: 8.27ms
27: learn: 0.5808997 total: 10.1ms remaining: 7.96ms
28: learn: 0.5778788 total: 10.5ms remaining: 7.62ms
29: learn: 0.5750842 total: 10.9ms remaining: 7.24ms
30: learn: 0.5722879 total: 11.2ms remaining: 6.87ms
31: learn: 0.5693116 total: 11.4ms remaining: 6.39ms
32: learn: 0.5661397 total: 11.8ms remaining: 6.09ms
33: learn: 0.5627994 total: 12.1ms remaining: 5.71ms
34: learn: 0.5597978 total: 12.4ms remaining: 5.31ms
35: learn: 0.5564847 total: 12.7ms remaining: 4.92ms
36: learn: 0.5535813 total: 12.9ms remaining: 4.54ms
37: learn: 0.5511907 total: 13.2ms remaining: 4.17ms
38: learn: 0.5477524 total: 13.4ms remaining: 3.79ms
39: learn: 0.5446551 total: 13.7ms remaining: 3.43ms
40: learn: 0.5418243 total: 14ms remaining: 3.07ms
41: learn: 0.5384306 total: 14.2ms remaining: 2.7ms
42: learn: 0.5355158 total: 14.4ms remaining: 2.35ms
43: learn: 0.5330304 total: 14.8ms remaining: 2.01ms
44: learn: 0.5303464 total: 15ms remaining: 1.67ms
45: learn: 0.5282582 total: 15.3ms remaining: 1.33ms
46: learn: 0.5258402 total: 15.5ms remaining: 992us
47: learn: 0.5235785 total: 15.8ms remaining: 656us
48: learn: 0.5207689 total: 16ms remaining: 325us
49: learn: 0.5183663 total: 16.2ms remaining: 0us
0: learn: 0.6886824 total: 364us remaining: 17.9ms
1: learn: 0.6836439 total: 741us remaining: 17.8ms
2: learn: 0.6783803 total: 1.05ms remaining: 16.5ms
3: learn: 0.6732528 total: 1.48ms remaining: 17ms
4: learn: 0.6684937 total: 1.76ms remaining: 15.8ms
5: learn: 0.6634511 total: 2.07ms remaining: 15.2ms
6: learn: 0.6589192 total: 2.36ms remaining: 14.5ms
7: learn: 0.6551883 total: 2.76ms remaining: 14.5ms
8: learn: 0.6502473 total: 3.01ms remaining: 13.7ms
9: learn: 0.6460178 total: 3.28ms remaining: 13.1ms
10: learn: 0.6419694 total: 3.55ms remaining: 12.6ms
11: learn: 0.6383164 total: 3.92ms remaining: 12.4ms
12: learn: 0.6337982 total: 4.27ms remaining: 12.2ms
13: learn: 0.6295016 total: 4.62ms remaining: 11.9ms
14: learn: 0.6253754 total: 4.92ms remaining: 11.5ms
15: learn: 0.6213047 total: 5.38ms remaining: 11.4ms
16: learn: 0.6172331 total: 5.78ms remaining: 11.2ms
17: learn: 0.6140067 total: 6.12ms remaining: 10.9ms
18: learn: 0.6101040 total: 6.38ms remaining: 10.4ms
19: learn: 0.6066469 total: 6.7ms remaining: 10.1ms
20: learn: 0.6029030 total: 6.98ms remaining: 9.64ms
21: learn: 0.5987959 total: 7.38ms remaining: 9.39ms
22: learn: 0.5948275 total: 7.71ms remaining: 9.05ms
23: learn: 0.5915322 total: 8ms remaining: 8.66ms
24: learn: 0.5872507 total: 8.32ms remaining: 8.32ms
25: learn: 0.5839759 total: 8.7ms remaining: 8.03ms
26: learn: 0.5805313 total: 8.99ms remaining: 7.66ms
27: learn: 0.5778302 total: 9.3ms remaining: 7.3ms
28: learn: 0.5752130 total: 9.6ms remaining: 6.95ms
29: learn: 0.5727888 total: 9.87ms remaining: 6.58ms
30: learn: 0.5702654 total: 10.2ms remaining: 6.25ms
31: learn: 0.5666849 total: 10.3ms remaining: 5.82ms
32: learn: 0.5634739 total: 10.7ms remaining: 5.5ms
33: learn: 0.5599432 total: 10.9ms remaining: 5.15ms
34: learn: 0.5569637 total: 11.2ms remaining: 4.82ms
35: learn: 0.5542632 total: 11.6ms remaining: 4.5ms
36: learn: 0.5511816 total: 11.9ms remaining: 4.19ms
37: learn: 0.5479280 total: 12.3ms remaining: 3.87ms
38: learn: 0.5446114 total: 12.5ms remaining: 3.54ms
39: learn: 0.5416442 total: 12.8ms remaining: 3.2ms
40: learn: 0.5388472 total: 13.1ms remaining: 2.88ms
41: learn: 0.5353803 total: 13.4ms remaining: 2.54ms
42: learn: 0.5323092 total: 13.7ms remaining: 2.23ms
43: learn: 0.5297938 total: 14ms remaining: 1.91ms
44: learn: 0.5270777 total: 14.3ms remaining: 1.58ms
45: learn: 0.5242021 total: 14.6ms remaining: 1.26ms
46: learn: 0.5210269 total: 15ms remaining: 955us
47: learn: 0.5183354 total: 15.3ms remaining: 635us
48: learn: 0.5155134 total: 15.6ms remaining: 317us
49: learn: 0.5125593 total: 15.9ms remaining: 0us
0: learn: 0.6884710 total: 279us remaining: 13.7ms
1: learn: 0.6831483 total: 512us remaining: 12.3ms
2: learn: 0.6781742 total: 686us remaining: 10.8ms
3: learn: 0.6735979 total: 1.47ms remaining: 16.9ms
4: learn: 0.6690825 total: 2.01ms remaining: 18.1ms
5: learn: 0.6643177 total: 2.44ms remaining: 17.9ms
6: learn: 0.6602321 total: 2.7ms remaining: 16.6ms
7: learn: 0.6561625 total: 3.08ms remaining: 16.2ms
8: learn: 0.6513684 total: 3.35ms remaining: 15.3ms
9: learn: 0.6464446 total: 3.79ms remaining: 15.2ms
10: learn: 0.6425160 total: 4.09ms remaining: 14.5ms
11: learn: 0.6379029 total: 4.38ms remaining: 13.9ms
12: learn: 0.6338682 total: 4.7ms remaining: 13.4ms
13: learn: 0.6299456 total: 5.08ms remaining: 13.1ms
14: learn: 0.6261431 total: 5.42ms remaining: 12.7ms
15: learn: 0.6214318 total: 5.75ms remaining: 12.2ms
16: learn: 0.6176888 total: 6.07ms remaining: 11.8ms
17: learn: 0.6136496 total: 6.39ms remaining: 11.4ms
18: learn: 0.6098567 total: 6.53ms remaining: 10.7ms
19: learn: 0.6061197 total: 6.91ms remaining: 10.4ms
20: learn: 0.6021095 total: 7.32ms remaining: 10.1ms
21: learn: 0.5977883 total: 7.67ms remaining: 9.76ms
22: learn: 0.5936190 total: 7.95ms remaining: 9.34ms
23: learn: 0.5899167 total: 8.22ms remaining: 8.9ms
24: learn: 0.5858649 total: 8.51ms remaining: 8.51ms
25: learn: 0.5829470 total: 8.83ms remaining: 8.15ms
26: learn: 0.5798447 total: 9.06ms remaining: 7.72ms
27: learn: 0.5765984 total: 9.34ms remaining: 7.33ms
28: learn: 0.5737242 total: 9.67ms remaining: 7ms
29: learn: 0.5710620 total: 10ms remaining: 6.67ms
30: learn: 0.5684374 total: 10.4ms remaining: 6.36ms
31: learn: 0.5652138 total: 10.6ms remaining: 5.95ms
32: learn: 0.5622089 total: 11ms remaining: 5.66ms
33: learn: 0.5588240 total: 11.3ms remaining: 5.3ms
34: learn: 0.5553290 total: 11.6ms remaining: 4.96ms
35: learn: 0.5523463 total: 11.9ms remaining: 4.61ms
36: learn: 0.5491377 total: 12.2ms remaining: 4.3ms
37: learn: 0.5464623 total: 12.5ms remaining: 3.94ms
38: learn: 0.5430188 total: 12.7ms remaining: 3.59ms
39: learn: 0.5400183 total: 13ms remaining: 3.26ms
40: learn: 0.5375189 total: 13.3ms remaining: 2.92ms
41: learn: 0.5339443 total: 13.5ms remaining: 2.57ms
42: learn: 0.5309167 total: 13.8ms remaining: 2.24ms
43: learn: 0.5285461 total: 14.1ms remaining: 1.92ms
44: learn: 0.5257915 total: 14.4ms remaining: 1.6ms
45: learn: 0.5235061 total: 14.5ms remaining: 1.26ms
46: learn: 0.5208134 total: 14.9ms remaining: 948us
47: learn: 0.5181357 total: 15.1ms remaining: 628us
48: learn: 0.5152125 total: 15.3ms remaining: 312us
49: learn: 0.5127882 total: 15.6ms remaining: 0us
0: learn: 0.6888251 total: 608us remaining: 29.8ms
1: learn: 0.6838743 total: 1.06ms remaining: 25.5ms
2: learn: 0.6790688 total: 1.27ms remaining: 19.9ms
3: learn: 0.6744890 total: 1.62ms remaining: 18.6ms
4: learn: 0.6697935 total: 1.82ms remaining: 16.4ms
5: learn: 0.6652243 total: 2.09ms remaining: 15.4ms
6: learn: 0.6616198 total: 2.64ms remaining: 16.2ms
7: learn: 0.6576694 total: 2.96ms remaining: 15.6ms
8: learn: 0.6532890 total: 3.31ms remaining: 15.1ms
9: learn: 0.6492571 total: 3.64ms remaining: 14.5ms
10: learn: 0.6463276 total: 3.95ms remaining: 14ms
11: learn: 0.6419683 total: 4.31ms remaining: 13.7ms
12: learn: 0.6383123 total: 4.7ms remaining: 13.4ms
13: learn: 0.6346641 total: 5.11ms remaining: 13.1ms
14: learn: 0.6310619 total: 5.56ms remaining: 13ms
15: learn: 0.6268264 total: 5.85ms remaining: 12.4ms
16: learn: 0.6232008 total: 6.13ms remaining: 11.9ms
17: learn: 0.6188903 total: 6.47ms remaining: 11.5ms
18: learn: 0.6154295 total: 6.76ms remaining: 11ms
19: learn: 0.6110983 total: 7.09ms remaining: 10.6ms
20: learn: 0.6074348 total: 7.42ms remaining: 10.3ms
21: learn: 0.6035654 total: 7.77ms remaining: 9.89ms
22: learn: 0.6001629 total: 8.1ms remaining: 9.51ms
23: learn: 0.5972153 total: 8.48ms remaining: 9.19ms
24: learn: 0.5938984 total: 8.78ms remaining: 8.78ms
25: learn: 0.5914189 total: 9.17ms remaining: 8.46ms
26: learn: 0.5879412 total: 9.44ms remaining: 8.04ms
27: learn: 0.5844532 total: 9.71ms remaining: 7.63ms
28: learn: 0.5812474 total: 9.91ms remaining: 7.18ms
29: learn: 0.5780829 total: 10.3ms remaining: 6.85ms
30: learn: 0.5747267 total: 10.6ms remaining: 6.53ms
31: learn: 0.5714414 total: 10.9ms remaining: 6.16ms
32: learn: 0.5686548 total: 11.2ms remaining: 5.77ms
33: learn: 0.5656884 total: 11.4ms remaining: 5.36ms
34: learn: 0.5628498 total: 11.7ms remaining: 5.01ms
35: learn: 0.5601182 total: 12ms remaining: 4.68ms
36: learn: 0.5571363 total: 12.4ms remaining: 4.34ms
37: learn: 0.5543120 total: 12.7ms remaining: 4ms
38: learn: 0.5508372 total: 12.9ms remaining: 3.65ms
39: learn: 0.5482084 total: 13.4ms remaining: 3.34ms
40: learn: 0.5451572 total: 13.7ms remaining: 3ms
41: learn: 0.5418393 total: 13.9ms remaining: 2.65ms
42: learn: 0.5393336 total: 14.2ms remaining: 2.31ms
43: learn: 0.5368743 total: 14.5ms remaining: 1.98ms
44: learn: 0.5341000 total: 14.9ms remaining: 1.66ms
45: learn: 0.5315797 total: 15.2ms remaining: 1.32ms
46: learn: 0.5288963 total: 15.5ms remaining: 986us
47: learn: 0.5262389 total: 15.7ms remaining: 654us
48: learn: 0.5234348 total: 16ms remaining: 325us
49: learn: 0.5209916 total: 16.2ms remaining: 0us
0: learn: 0.6881993 total: 339us remaining: 16.6ms
1: learn: 0.6827008 total: 579us remaining: 13.9ms
2: learn: 0.6774722 total: 787us remaining: 12.3ms
3: learn: 0.6721781 total: 1.83ms remaining: 21ms
4: learn: 0.6675501 total: 2.45ms remaining: 22ms
5: learn: 0.6623863 total: 2.86ms remaining: 21ms
6: learn: 0.6575695 total: 3.87ms remaining: 23.8ms
7: learn: 0.6535074 total: 4.62ms remaining: 24.3ms
8: learn: 0.6485427 total: 4.88ms remaining: 22.3ms
9: learn: 0.6436104 total: 5.29ms remaining: 21.2ms
10: learn: 0.6395245 total: 6.24ms remaining: 22.1ms
11: learn: 0.6347631 total: 7.08ms remaining: 22.4ms
12: learn: 0.6305070 total: 8.17ms remaining: 23.3ms
13: learn: 0.6265698 total: 9.55ms remaining: 24.6ms
14: learn: 0.6229825 total: 11ms remaining: 25.6ms
15: learn: 0.6182567 total: 11.9ms remaining: 25.3ms
16: learn: 0.6144702 total: 12.6ms remaining: 24.5ms
17: learn: 0.6102756 total: 13.4ms remaining: 23.9ms
18: learn: 0.6063688 total: 13.9ms remaining: 22.7ms
19: learn: 0.6025233 total: 14.4ms remaining: 21.7ms
20: learn: 0.5986269 total: 14.7ms remaining: 20.4ms
21: learn: 0.5942607 total: 15.8ms remaining: 20.1ms
22: learn: 0.5899548 total: 16.7ms remaining: 19.6ms
23: learn: 0.5862924 total: 17.2ms remaining: 18.6ms
24: learn: 0.5830002 total: 17.9ms remaining: 17.9ms
25: learn: 0.5798113 total: 18.6ms remaining: 17.2ms
26: learn: 0.5764879 total: 18.8ms remaining: 16.1ms
27: learn: 0.5732837 total: 19.1ms remaining: 15ms
28: learn: 0.5701657 total: 20.3ms remaining: 14.7ms
29: learn: 0.5673144 total: 21.1ms remaining: 14.1ms
30: learn: 0.5645907 total: 22.1ms remaining: 13.6ms
31: learn: 0.5612450 total: 22.6ms remaining: 12.7ms
32: learn: 0.5579183 total: 23.4ms remaining: 12ms
33: learn: 0.5543718 total: 24.2ms remaining: 11.4ms
34: learn: 0.5513997 total: 25.3ms remaining: 10.8ms
35: learn: 0.5484174 total: 26ms remaining: 10.1ms
36: learn: 0.5451814 total: 26.6ms remaining: 9.35ms
37: learn: 0.5421966 total: 26.9ms remaining: 8.5ms
38: learn: 0.5386374 total: 27.4ms remaining: 7.72ms
39: learn: 0.5359418 total: 28.1ms remaining: 7.02ms
40: learn: 0.5328272 total: 28.4ms remaining: 6.22ms
41: learn: 0.5292787 total: 28.8ms remaining: 5.48ms
42: learn: 0.5261061 total: 29.6ms remaining: 4.83ms
43: learn: 0.5237163 total: 30.7ms remaining: 4.19ms
44: learn: 0.5208345 total: 31.8ms remaining: 3.53ms
45: learn: 0.5184355 total: 32.4ms remaining: 2.81ms
46: learn: 0.5158247 total: 33.5ms remaining: 2.14ms
47: learn: 0.5135288 total: 34ms remaining: 1.42ms
48: learn: 0.5104705 total: 34.8ms remaining: 710us
49: learn: 0.5081160 total: 35.7ms remaining: 0us
0: learn: 0.6886959 total: 303us remaining: 14.9ms
1: learn: 0.6838265 total: 538us remaining: 12.9ms
2: learn: 0.6793184 total: 820us remaining: 12.8ms
3: learn: 0.6746587 total: 1.12ms remaining: 12.9ms
4: learn: 0.6698654 total: 1.29ms remaining: 11.6ms
5: learn: 0.6655969 total: 1.77ms remaining: 13ms
6: learn: 0.6613624 total: 2.16ms remaining: 13.3ms
7: learn: 0.6574805 total: 2.47ms remaining: 13ms
8: learn: 0.6531921 total: 2.79ms remaining: 12.7ms
9: learn: 0.6488752 total: 3.13ms remaining: 12.5ms
10: learn: 0.6451668 total: 3.47ms remaining: 12.3ms
11: learn: 0.6411035 total: 3.8ms remaining: 12ms
12: learn: 0.6370053 total: 4.21ms remaining: 12ms
13: learn: 0.6329368 total: 4.61ms remaining: 11.8ms
14: learn: 0.6292349 total: 4.92ms remaining: 11.5ms
15: learn: 0.6251452 total: 5.27ms remaining: 11.2ms
16: learn: 0.6212621 total: 5.6ms remaining: 10.9ms
17: learn: 0.6170321 total: 6.01ms remaining: 10.7ms
18: learn: 0.6132722 total: 6.3ms remaining: 10.3ms
19: learn: 0.6099714 total: 6.67ms remaining: 10ms
20: learn: 0.6066178 total: 7.09ms remaining: 9.79ms
21: learn: 0.6027816 total: 7.5ms remaining: 9.54ms
22: learn: 0.5990405 total: 7.83ms remaining: 9.2ms
23: learn: 0.5957426 total: 8.06ms remaining: 8.73ms
24: learn: 0.5922791 total: 8.37ms remaining: 8.37ms
25: learn: 0.5893376 total: 8.76ms remaining: 8.08ms
26: learn: 0.5861754 total: 9.09ms remaining: 7.74ms
27: learn: 0.5829950 total: 9.44ms remaining: 7.42ms
28: learn: 0.5794016 total: 9.66ms remaining: 6.99ms
29: learn: 0.5765496 total: 10ms remaining: 6.67ms
30: learn: 0.5736307 total: 10.4ms remaining: 6.35ms
31: learn: 0.5704244 total: 10.6ms remaining: 5.97ms
32: learn: 0.5676356 total: 10.9ms remaining: 5.64ms
33: learn: 0.5646941 total: 11.2ms remaining: 5.25ms
34: learn: 0.5620194 total: 11.5ms remaining: 4.91ms
35: learn: 0.5600655 total: 11.6ms remaining: 4.53ms
36: learn: 0.5572839 total: 12ms remaining: 4.21ms
37: learn: 0.5547866 total: 12.2ms remaining: 3.86ms
38: learn: 0.5521352 total: 12.6ms remaining: 3.55ms
39: learn: 0.5493307 total: 12.9ms remaining: 3.23ms
40: learn: 0.5467472 total: 13.3ms remaining: 2.91ms
41: learn: 0.5446479 total: 13.6ms remaining: 2.59ms
42: learn: 0.5426825 total: 14ms remaining: 2.27ms
43: learn: 0.5399677 total: 14.3ms remaining: 1.95ms
44: learn: 0.5379476 total: 14.6ms remaining: 1.62ms
45: learn: 0.5354392 total: 14.9ms remaining: 1.3ms
46: learn: 0.5325750 total: 15.3ms remaining: 975us
47: learn: 0.5300571 total: 15.5ms remaining: 647us
48: learn: 0.5276968 total: 15.8ms remaining: 322us
49: learn: 0.5250418 total: 16.1ms remaining: 0us
0: learn: 0.6888430 total: 538us remaining: 26.4ms
1: learn: 0.6844696 total: 966us remaining: 23.2ms
2: learn: 0.6801295 total: 1.64ms remaining: 25.7ms
3: learn: 0.6752420 total: 2.56ms remaining: 29.5ms
4: learn: 0.6701626 total: 3.44ms remaining: 31ms
5: learn: 0.6655182 total: 4.27ms remaining: 31.3ms
6: learn: 0.6620344 total: 5.19ms remaining: 31.9ms
7: learn: 0.6584874 total: 5.9ms remaining: 31ms
8: learn: 0.6539240 total: 7.02ms remaining: 32ms
9: learn: 0.6499828 total: 7.84ms remaining: 31.4ms
10: learn: 0.6455269 total: 8.77ms remaining: 31.1ms
11: learn: 0.6414986 total: 9.54ms remaining: 30.2ms
12: learn: 0.6374259 total: 10.3ms remaining: 29.3ms
13: learn: 0.6333841 total: 11.1ms remaining: 28.5ms
14: learn: 0.6287552 total: 11.6ms remaining: 27ms
15: learn: 0.6255913 total: 12.4ms remaining: 26.4ms
16: learn: 0.6228745 total: 13.2ms remaining: 25.6ms
17: learn: 0.6192158 total: 14ms remaining: 24.8ms
18: learn: 0.6151053 total: 14.5ms remaining: 23.7ms
19: learn: 0.6117255 total: 15.1ms remaining: 22.6ms
20: learn: 0.6077871 total: 16.1ms remaining: 22.2ms
21: learn: 0.6036369 total: 17.1ms remaining: 21.8ms
22: learn: 0.5997223 total: 18.3ms remaining: 21.5ms
23: learn: 0.5957430 total: 19ms remaining: 20.5ms
24: learn: 0.5924717 total: 19.8ms remaining: 19.8ms
25: learn: 0.5891835 total: 20.8ms remaining: 19.2ms
26: learn: 0.5859911 total: 21.9ms remaining: 18.7ms
27: learn: 0.5830599 total: 22.6ms remaining: 17.8ms
28: learn: 0.5804551 total: 23ms remaining: 16.6ms
29: learn: 0.5777595 total: 24.1ms remaining: 16.1ms
30: learn: 0.5746008 total: 24.6ms remaining: 15.1ms
31: learn: 0.5713796 total: 25.5ms remaining: 14.3ms
32: learn: 0.5679259 total: 26ms remaining: 13.4ms
33: learn: 0.5650754 total: 26.4ms remaining: 12.4ms
34: learn: 0.5622872 total: 27.6ms remaining: 11.8ms
35: learn: 0.5591263 total: 29ms remaining: 11.3ms
36: learn: 0.5563465 total: 29.7ms remaining: 10.4ms
37: learn: 0.5529493 total: 30.3ms remaining: 9.57ms
38: learn: 0.5501690 total: 31.7ms remaining: 8.94ms
39: learn: 0.5469190 total: 32.2ms remaining: 8.05ms
40: learn: 0.5432410 total: 32.5ms remaining: 7.13ms
41: learn: 0.5401389 total: 32.9ms remaining: 6.27ms
42: learn: 0.5378797 total: 33.8ms remaining: 5.5ms
43: learn: 0.5350410 total: 35.1ms remaining: 4.79ms
44: learn: 0.5326489 total: 35.9ms remaining: 3.98ms
45: learn: 0.5300776 total: 36.8ms remaining: 3.2ms
46: learn: 0.5270868 total: 38.4ms remaining: 2.45ms
47: learn: 0.5249808 total: 39.2ms remaining: 1.64ms
48: learn: 0.5227876 total: 40.2ms remaining: 820us
49: learn: 0.5200878 total: 41.5ms remaining: 0us
0: learn: 0.6886167 total: 348us remaining: 17.1ms
1: learn: 0.6839296 total: 640us remaining: 15.4ms
2: learn: 0.6790130 total: 883us remaining: 13.8ms
3: learn: 0.6745981 total: 1.14ms remaining: 13.1ms
4: learn: 0.6700984 total: 1.33ms remaining: 12ms
5: learn: 0.6652168 total: 1.6ms remaining: 11.7ms
6: learn: 0.6606527 total: 1.81ms remaining: 11.2ms
7: learn: 0.6566447 total: 2.03ms remaining: 10.7ms
8: learn: 0.6519140 total: 2.19ms remaining: 9.99ms
9: learn: 0.6475038 total: 2.4ms remaining: 9.58ms
10: learn: 0.6444133 total: 2.53ms remaining: 8.98ms
11: learn: 0.6400321 total: 2.67ms remaining: 8.47ms
12: learn: 0.6361821 total: 2.84ms remaining: 8.09ms
13: learn: 0.6324678 total: 3.05ms remaining: 7.85ms
14: learn: 0.6284970 total: 3.26ms remaining: 7.61ms
15: learn: 0.6241548 total: 3.42ms remaining: 7.28ms
16: learn: 0.6212588 total: 3.59ms remaining: 6.97ms
17: learn: 0.6181208 total: 3.81ms remaining: 6.77ms
18: learn: 0.6149727 total: 4.02ms remaining: 6.55ms
19: learn: 0.6112675 total: 4.2ms remaining: 6.3ms
20: learn: 0.6076084 total: 4.4ms remaining: 6.07ms
21: learn: 0.6038614 total: 4.62ms remaining: 5.88ms
22: learn: 0.5996882 total: 4.78ms remaining: 5.61ms
23: learn: 0.5962705 total: 4.96ms remaining: 5.37ms
24: learn: 0.5923546 total: 5.08ms remaining: 5.08ms
25: learn: 0.5894756 total: 5.28ms remaining: 4.88ms
26: learn: 0.5864478 total: 5.47ms remaining: 4.66ms
27: learn: 0.5833874 total: 5.65ms remaining: 4.44ms
28: learn: 0.5804593 total: 5.96ms remaining: 4.32ms
29: learn: 0.5777455 total: 6.31ms remaining: 4.21ms
30: learn: 0.5751136 total: 6.61ms remaining: 4.05ms
31: learn: 0.5718058 total: 6.76ms remaining: 3.81ms
32: learn: 0.5689155 total: 7.13ms remaining: 3.67ms
33: learn: 0.5658437 total: 7.5ms remaining: 3.53ms
34: learn: 0.5625370 total: 7.7ms remaining: 3.3ms
35: learn: 0.5600906 total: 7.99ms remaining: 3.11ms
36: learn: 0.5574840 total: 8.25ms remaining: 2.9ms
37: learn: 0.5548446 total: 8.59ms remaining: 2.71ms
38: learn: 0.5521241 total: 8.84ms remaining: 2.49ms
39: learn: 0.5493534 total: 9.16ms remaining: 2.29ms
40: learn: 0.5464643 total: 9.36ms remaining: 2.05ms
41: learn: 0.5439402 total: 9.65ms remaining: 1.84ms
42: learn: 0.5407503 total: 9.85ms remaining: 1.6ms
43: learn: 0.5383021 total: 10.2ms remaining: 1.39ms
44: learn: 0.5357105 total: 10.4ms remaining: 1.16ms
45: learn: 0.5332736 total: 10.6ms remaining: 923us
46: learn: 0.5308820 total: 11ms remaining: 699us
47: learn: 0.5280463 total: 11.2ms remaining: 465us
48: learn: 0.5251939 total: 11.5ms remaining: 234us
49: learn: 0.5226601 total: 11.8ms remaining: 0us
0: learn: 0.6885339 total: 441us remaining: 21.6ms
1: learn: 0.6834227 total: 1.46ms remaining: 35ms
2: learn: 0.6786232 total: 2.06ms remaining: 32.3ms
3: learn: 0.6739003 total: 2.83ms remaining: 32.5ms
4: learn: 0.6693001 total: 3.47ms remaining: 31.2ms
5: learn: 0.6644451 total: 4.08ms remaining: 29.9ms
6: learn: 0.6598747 total: 4.7ms remaining: 28.9ms
7: learn: 0.6558602 total: 5.5ms remaining: 28.9ms
8: learn: 0.6512912 total: 6.02ms remaining: 27.4ms
9: learn: 0.6467205 total: 6.77ms remaining: 27.1ms
10: learn: 0.6428079 total: 7.47ms remaining: 26.5ms
11: learn: 0.6384207 total: 8.07ms remaining: 25.6ms
12: learn: 0.6344698 total: 8.73ms remaining: 24.8ms
13: learn: 0.6304828 total: 9.52ms remaining: 24.5ms
14: learn: 0.6266173 total: 10.1ms remaining: 23.6ms
15: learn: 0.6223299 total: 10.9ms remaining: 23.1ms
16: learn: 0.6186546 total: 11.6ms remaining: 22.5ms
17: learn: 0.6147792 total: 12.4ms remaining: 22ms
18: learn: 0.6110091 total: 12.6ms remaining: 20.6ms
19: learn: 0.6073387 total: 13.4ms remaining: 20.1ms
20: learn: 0.6038272 total: 14.1ms remaining: 19.4ms
21: learn: 0.6001991 total: 14.7ms remaining: 18.7ms
22: learn: 0.5964357 total: 15.2ms remaining: 17.9ms
23: learn: 0.5929511 total: 15.4ms remaining: 16.7ms
24: learn: 0.5893719 total: 16ms remaining: 16ms
25: learn: 0.5864672 total: 16.6ms remaining: 15.3ms
26: learn: 0.5837256 total: 17.2ms remaining: 14.7ms
27: learn: 0.5805511 total: 17.9ms remaining: 14.1ms
28: learn: 0.5772513 total: 18.5ms remaining: 13.4ms
29: learn: 0.5745393 total: 18.8ms remaining: 12.5ms
30: learn: 0.5717751 total: 19.3ms remaining: 11.8ms
31: learn: 0.5685652 total: 19.7ms remaining: 11.1ms
32: learn: 0.5654609 total: 20.3ms remaining: 10.4ms
33: learn: 0.5622746 total: 20.9ms remaining: 9.82ms
34: learn: 0.5588724 total: 21.4ms remaining: 9.19ms
35: learn: 0.5555821 total: 21.9ms remaining: 8.5ms
36: learn: 0.5527355 total: 22.5ms remaining: 7.91ms
37: learn: 0.5500608 total: 23.2ms remaining: 7.32ms
38: learn: 0.5467145 total: 23.7ms remaining: 6.68ms
39: learn: 0.5441104 total: 24.2ms remaining: 6.06ms
40: learn: 0.5417590 total: 24.5ms remaining: 5.39ms
41: learn: 0.5389472 total: 24.7ms remaining: 4.71ms
42: learn: 0.5365060 total: 25.2ms remaining: 4.11ms
43: learn: 0.5333535 total: 25.7ms remaining: 3.5ms
44: learn: 0.5309882 total: 26.3ms remaining: 2.93ms
45: learn: 0.5286618 total: 26.8ms remaining: 2.33ms
46: learn: 0.5259226 total: 27.3ms remaining: 1.74ms
47: learn: 0.5235263 total: 27.9ms remaining: 1.16ms
48: learn: 0.5210188 total: 28.3ms remaining: 578us
49: learn: 0.5185356 total: 28.8ms remaining: 0us
0: learn: 0.6883550 total: 315us remaining: 15.5ms
1: learn: 0.6836109 total: 625us remaining: 15ms
2: learn: 0.6787323 total: 813us remaining: 12.7ms
3: learn: 0.6738335 total: 1.29ms remaining: 14.9ms
4: learn: 0.6693032 total: 1.65ms remaining: 14.9ms
5: learn: 0.6644562 total: 2.03ms remaining: 14.9ms
6: learn: 0.6602992 total: 2.34ms remaining: 14.4ms
7: learn: 0.6565796 total: 2.73ms remaining: 14.3ms
8: learn: 0.6517652 total: 2.96ms remaining: 13.5ms
9: learn: 0.6474219 total: 3.39ms remaining: 13.6ms
10: learn: 0.6444322 total: 3.63ms remaining: 12.9ms
11: learn: 0.6397700 total: 3.97ms remaining: 12.6ms
12: learn: 0.6359370 total: 4.37ms remaining: 12.4ms
13: learn: 0.6324723 total: 4.79ms remaining: 12.3ms
14: learn: 0.6285007 total: 5.2ms remaining: 12.1ms
15: learn: 0.6241371 total: 5.51ms remaining: 11.7ms
16: learn: 0.6205886 total: 5.98ms remaining: 11.6ms
17: learn: 0.6165610 total: 6.37ms remaining: 11.3ms
18: learn: 0.6127567 total: 6.64ms remaining: 10.8ms
19: learn: 0.6088093 total: 7.12ms remaining: 10.7ms
20: learn: 0.6051968 total: 7.53ms remaining: 10.4ms
21: learn: 0.6014306 total: 7.93ms remaining: 10.1ms
22: learn: 0.5974358 total: 8.31ms remaining: 9.76ms
23: learn: 0.5939927 total: 8.61ms remaining: 9.33ms
24: learn: 0.5909375 total: 9.04ms remaining: 9.04ms
25: learn: 0.5873963 total: 9.44ms remaining: 8.71ms
26: learn: 0.5844064 total: 9.84ms remaining: 8.38ms
27: learn: 0.5813962 total: 10.2ms remaining: 8.04ms
28: learn: 0.5785490 total: 10.6ms remaining: 7.66ms
29: learn: 0.5758908 total: 10.8ms remaining: 7.23ms
30: learn: 0.5732910 total: 11.2ms remaining: 6.88ms
31: learn: 0.5700190 total: 11.4ms remaining: 6.42ms
32: learn: 0.5669492 total: 11.8ms remaining: 6.06ms
33: learn: 0.5635924 total: 12.1ms remaining: 5.69ms
34: learn: 0.5600908 total: 12.5ms remaining: 5.36ms
35: learn: 0.5572768 total: 12.9ms remaining: 5ms
36: learn: 0.5542501 total: 13.3ms remaining: 4.67ms
37: learn: 0.5515929 total: 13.6ms remaining: 4.3ms
38: learn: 0.5480918 total: 13.9ms remaining: 3.92ms
39: learn: 0.5449589 total: 14.3ms remaining: 3.58ms
40: learn: 0.5420133 total: 14.6ms remaining: 3.21ms
41: learn: 0.5389790 total: 14.9ms remaining: 2.83ms
42: learn: 0.5360152 total: 15.2ms remaining: 2.47ms
43: learn: 0.5336202 total: 15.5ms remaining: 2.12ms
44: learn: 0.5308643 total: 15.8ms remaining: 1.76ms
45: learn: 0.5285430 total: 16ms remaining: 1.4ms
46: learn: 0.5260419 total: 16.4ms remaining: 1.05ms
47: learn: 0.5236528 total: 16.7ms remaining: 694us
48: learn: 0.5206597 total: 16.9ms remaining: 344us
49: learn: 0.5182717 total: 17.2ms remaining: 0us
0: learn: 0.6889933 total: 301us remaining: 14.8ms
1: learn: 0.6836164 total: 586us remaining: 14.1ms
2: learn: 0.6784902 total: 831us remaining: 13ms
3: learn: 0.6738344 total: 1.1ms remaining: 12.7ms
4: learn: 0.6690759 total: 1.34ms remaining: 12ms
5: learn: 0.6641179 total: 1.59ms remaining: 11.7ms
6: learn: 0.6609183 total: 1.89ms remaining: 11.6ms
7: learn: 0.6564302 total: 2.11ms remaining: 11.1ms
8: learn: 0.6518274 total: 2.39ms remaining: 10.9ms
9: learn: 0.6472279 total: 2.57ms remaining: 10.3ms
10: learn: 0.6439145 total: 2.79ms remaining: 9.91ms
11: learn: 0.6400426 total: 3.09ms remaining: 9.79ms
12: learn: 0.6356838 total: 3.35ms remaining: 9.53ms
13: learn: 0.6314445 total: 3.63ms remaining: 9.35ms
14: learn: 0.6280712 total: 3.88ms remaining: 9.04ms
15: learn: 0.6240877 total: 4.18ms remaining: 8.88ms
16: learn: 0.6200849 total: 4.47ms remaining: 8.69ms
17: learn: 0.6168229 total: 4.67ms remaining: 8.31ms
18: learn: 0.6129056 total: 4.88ms remaining: 7.96ms
19: learn: 0.6101052 total: 5.14ms remaining: 7.71ms
20: learn: 0.6064010 total: 5.41ms remaining: 7.47ms
21: learn: 0.6022683 total: 5.68ms remaining: 7.23ms
22: learn: 0.5980306 total: 5.9ms remaining: 6.93ms
23: learn: 0.5949120 total: 6.08ms remaining: 6.59ms
24: learn: 0.5912501 total: 6.37ms remaining: 6.37ms
25: learn: 0.5879653 total: 6.63ms remaining: 6.12ms
26: learn: 0.5845873 total: 6.92ms remaining: 5.9ms
27: learn: 0.5814684 total: 7.17ms remaining: 5.63ms
28: learn: 0.5788440 total: 7.44ms remaining: 5.38ms
29: learn: 0.5759054 total: 7.7ms remaining: 5.13ms
30: learn: 0.5723048 total: 7.95ms remaining: 4.87ms
31: learn: 0.5690872 total: 8.22ms remaining: 4.63ms
32: learn: 0.5660702 total: 8.49ms remaining: 4.37ms
33: learn: 0.5628655 total: 8.74ms remaining: 4.11ms
34: learn: 0.5599171 total: 9ms remaining: 3.86ms
35: learn: 0.5573603 total: 9.26ms remaining: 3.6ms
36: learn: 0.5551515 total: 9.48ms remaining: 3.33ms
37: learn: 0.5521042 total: 9.74ms remaining: 3.07ms
38: learn: 0.5486314 total: 9.99ms remaining: 2.82ms
39: learn: 0.5453812 total: 10.2ms remaining: 2.56ms
40: learn: 0.5422646 total: 10.5ms remaining: 2.31ms
41: learn: 0.5396314 total: 10.8ms remaining: 2.06ms
42: learn: 0.5369313 total: 11ms remaining: 1.8ms
43: learn: 0.5341681 total: 11.3ms remaining: 1.54ms
44: learn: 0.5311134 total: 11.5ms remaining: 1.28ms
45: learn: 0.5279754 total: 11.8ms remaining: 1.02ms
46: learn: 0.5252587 total: 12ms remaining: 767us
47: learn: 0.5224699 total: 12.2ms remaining: 510us
48: learn: 0.5196144 total: 12.5ms remaining: 254us
49: learn: 0.5168594 total: 12.7ms remaining: 0us
0: learn: 0.6890031 total: 329us remaining: 16.1ms
1: learn: 0.6839941 total: 647us remaining: 15.5ms
2: learn: 0.6790950 total: 845us remaining: 13.2ms
3: learn: 0.6741892 total: 1.09ms remaining: 12.6ms
4: learn: 0.6699145 total: 1.28ms remaining: 11.5ms
5: learn: 0.6651736 total: 1.49ms remaining: 10.9ms
6: learn: 0.6605127 total: 1.67ms remaining: 10.2ms
7: learn: 0.6563352 total: 1.9ms remaining: 9.99ms
8: learn: 0.6525859 total: 2.02ms remaining: 9.22ms
9: learn: 0.6483100 total: 2.36ms remaining: 9.43ms
10: learn: 0.6443013 total: 2.53ms remaining: 8.96ms
11: learn: 0.6404553 total: 2.78ms remaining: 8.8ms
12: learn: 0.6363750 total: 3ms remaining: 8.55ms
13: learn: 0.6320174 total: 3.23ms remaining: 8.3ms
14: learn: 0.6288432 total: 3.41ms remaining: 7.95ms
15: learn: 0.6252655 total: 3.67ms remaining: 7.79ms
16: learn: 0.6214597 total: 3.93ms remaining: 7.63ms
17: learn: 0.6174034 total: 4.13ms remaining: 7.33ms
18: learn: 0.6143807 total: 4.35ms remaining: 7.09ms
19: learn: 0.6108119 total: 4.54ms remaining: 6.81ms
20: learn: 0.6071982 total: 4.72ms remaining: 6.52ms
21: learn: 0.6035162 total: 4.92ms remaining: 6.26ms
22: learn: 0.5995561 total: 5.09ms remaining: 5.97ms
23: learn: 0.5967885 total: 5.27ms remaining: 5.71ms
24: learn: 0.5928977 total: 5.4ms remaining: 5.4ms
25: learn: 0.5896768 total: 5.59ms remaining: 5.16ms
26: learn: 0.5864794 total: 5.77ms remaining: 4.92ms
27: learn: 0.5831156 total: 5.94ms remaining: 4.67ms
28: learn: 0.5804258 total: 6.1ms remaining: 4.42ms
29: learn: 0.5780639 total: 6.24ms remaining: 4.16ms
30: learn: 0.5757587 total: 7.04ms remaining: 4.31ms
31: learn: 0.5725405 total: 7.28ms remaining: 4.09ms
32: learn: 0.5693987 total: 7.5ms remaining: 3.86ms
33: learn: 0.5660367 total: 7.84ms remaining: 3.69ms
34: learn: 0.5626815 total: 8.06ms remaining: 3.46ms
35: learn: 0.5601347 total: 8.25ms remaining: 3.21ms
36: learn: 0.5570503 total: 8.4ms remaining: 2.95ms
37: learn: 0.5542878 total: 8.59ms remaining: 2.71ms
38: learn: 0.5511386 total: 8.76ms remaining: 2.47ms
39: learn: 0.5481846 total: 8.97ms remaining: 2.24ms
40: learn: 0.5450162 total: 9.12ms remaining: 2ms
41: learn: 0.5426760 total: 9.29ms remaining: 1.77ms
42: learn: 0.5395370 total: 9.43ms remaining: 1.53ms
43: learn: 0.5373680 total: 9.68ms remaining: 1.32ms
44: learn: 0.5345015 total: 9.85ms remaining: 1.09ms
45: learn: 0.5318743 total: 10ms remaining: 872us
46: learn: 0.5292160 total: 10.2ms remaining: 649us
47: learn: 0.5267250 total: 10.8ms remaining: 448us
48: learn: 0.5243110 total: 11ms remaining: 224us
49: learn: 0.5214591 total: 11.6ms remaining: 0us
0: learn: 0.6884444 total: 611us remaining: 30ms
1: learn: 0.6833777 total: 1.04ms remaining: 25.1ms
2: learn: 0.6789418 total: 1.32ms remaining: 20.8ms
3: learn: 0.6740883 total: 1.98ms remaining: 22.7ms
4: learn: 0.6695991 total: 2.71ms remaining: 24.4ms
5: learn: 0.6649560 total: 3.33ms remaining: 24.4ms
6: learn: 0.6607099 total: 3.98ms remaining: 24.5ms
7: learn: 0.6568861 total: 4.75ms remaining: 24.9ms
8: learn: 0.6526495 total: 4.94ms remaining: 22.5ms
9: learn: 0.6486747 total: 5.95ms remaining: 23.8ms
10: learn: 0.6446790 total: 6.94ms remaining: 24.6ms
11: learn: 0.6408419 total: 7.78ms remaining: 24.6ms
12: learn: 0.6366769 total: 8.8ms remaining: 25.1ms
13: learn: 0.6327758 total: 10.1ms remaining: 26.1ms
14: learn: 0.6289897 total: 11.4ms remaining: 26.5ms
15: learn: 0.6254446 total: 12.9ms remaining: 27.3ms
16: learn: 0.6214642 total: 13.6ms remaining: 26.5ms
17: learn: 0.6179222 total: 15ms remaining: 26.7ms
18: learn: 0.6145156 total: 16ms remaining: 26.2ms
19: learn: 0.6111632 total: 16.8ms remaining: 25.3ms
20: learn: 0.6075448 total: 17.9ms remaining: 24.8ms
21: learn: 0.6041593 total: 19ms remaining: 24.2ms
22: learn: 0.6005087 total: 20ms remaining: 23.5ms
23: learn: 0.5967117 total: 21.2ms remaining: 23ms
24: learn: 0.5937134 total: 22.5ms remaining: 22.5ms
25: learn: 0.5902819 total: 23ms remaining: 21.3ms
26: learn: 0.5876279 total: 23.8ms remaining: 20.3ms
27: learn: 0.5847773 total: 24.5ms remaining: 19.2ms
28: learn: 0.5817003 total: 24.8ms remaining: 18ms
29: learn: 0.5787036 total: 26ms remaining: 17.3ms
30: learn: 0.5760609 total: 26.8ms remaining: 16.4ms
31: learn: 0.5734735 total: 27.2ms remaining: 15.3ms
32: learn: 0.5707002 total: 27.8ms remaining: 14.3ms
33: learn: 0.5675329 total: 28.8ms remaining: 13.5ms
34: learn: 0.5643493 total: 29.6ms remaining: 12.7ms
35: learn: 0.5610712 total: 30.7ms remaining: 11.9ms
36: learn: 0.5582207 total: 32ms remaining: 11.2ms
37: learn: 0.5559611 total: 33.2ms remaining: 10.5ms
38: learn: 0.5537050 total: 33.6ms remaining: 9.48ms
39: learn: 0.5503936 total: 33.9ms remaining: 8.48ms
40: learn: 0.5478983 total: 34.2ms remaining: 7.51ms
41: learn: 0.5451844 total: 35ms remaining: 6.66ms
42: learn: 0.5418663 total: 35.8ms remaining: 5.83ms
43: learn: 0.5390675 total: 36.9ms remaining: 5.03ms
44: learn: 0.5366144 total: 38.3ms remaining: 4.26ms
45: learn: 0.5341339 total: 39.3ms remaining: 3.42ms
46: learn: 0.5320371 total: 40.1ms remaining: 2.56ms
47: learn: 0.5297100 total: 40.9ms remaining: 1.7ms
48: learn: 0.5275120 total: 41.8ms remaining: 852us
49: learn: 0.5249431 total: 42.6ms remaining: 0us
0: learn: 0.6885566 total: 442us remaining: 21.7ms
1: learn: 0.6836071 total: 717us remaining: 17.2ms
2: learn: 0.6782460 total: 905us remaining: 14.2ms
3: learn: 0.6743132 total: 1.7ms remaining: 19.6ms
4: learn: 0.6702237 total: 2.31ms remaining: 20.8ms
5: learn: 0.6660752 total: 2.8ms remaining: 20.6ms
6: learn: 0.6616702 total: 3.7ms remaining: 22.7ms
7: learn: 0.6566263 total: 4.48ms remaining: 23.5ms
8: learn: 0.6518664 total: 5.22ms remaining: 23.8ms
9: learn: 0.6490527 total: 5.46ms remaining: 21.8ms
10: learn: 0.6444509 total: 5.79ms remaining: 20.5ms
11: learn: 0.6403671 total: 6.69ms remaining: 21.2ms
12: learn: 0.6363304 total: 7.54ms remaining: 21.5ms
13: learn: 0.6327678 total: 8.48ms remaining: 21.8ms
14: learn: 0.6282780 total: 9.31ms remaining: 21.7ms
15: learn: 0.6244872 total: 10.2ms remaining: 21.6ms
16: learn: 0.6203105 total: 10.6ms remaining: 20.5ms
17: learn: 0.6165432 total: 10.7ms remaining: 19ms
18: learn: 0.6128502 total: 11.3ms remaining: 18.5ms
19: learn: 0.6090656 total: 12.2ms remaining: 18.3ms
20: learn: 0.6050572 total: 12.9ms remaining: 17.8ms
21: learn: 0.6007740 total: 13.6ms remaining: 17.4ms
22: learn: 0.5970249 total: 14.6ms remaining: 17.1ms
23: learn: 0.5935481 total: 15.3ms remaining: 16.5ms
24: learn: 0.5906288 total: 15.6ms remaining: 15.6ms
25: learn: 0.5877041 total: 16ms remaining: 14.8ms
26: learn: 0.5839478 total: 17ms remaining: 14.5ms
27: learn: 0.5811081 total: 17.8ms remaining: 14ms
28: learn: 0.5773239 total: 18.6ms remaining: 13.5ms
29: learn: 0.5742893 total: 19.5ms remaining: 13ms
30: learn: 0.5706335 total: 20.2ms remaining: 12.4ms
31: learn: 0.5674696 total: 21ms remaining: 11.8ms
32: learn: 0.5645132 total: 21.7ms remaining: 11.2ms
33: learn: 0.5608553 total: 22.7ms remaining: 10.7ms
34: learn: 0.5579249 total: 23.7ms remaining: 10.1ms
35: learn: 0.5548740 total: 24.7ms remaining: 9.6ms
36: learn: 0.5521033 total: 25.8ms remaining: 9.05ms
37: learn: 0.5487798 total: 26.7ms remaining: 8.42ms
38: learn: 0.5458292 total: 27.7ms remaining: 7.8ms
39: learn: 0.5434128 total: 28.5ms remaining: 7.13ms
40: learn: 0.5398872 total: 29.1ms remaining: 6.4ms
41: learn: 0.5372327 total: 30ms remaining: 5.72ms
42: learn: 0.5348076 total: 31.1ms remaining: 5.06ms
43: learn: 0.5319729 total: 32.2ms remaining: 4.38ms
44: learn: 0.5296587 total: 32.7ms remaining: 3.64ms
45: learn: 0.5271298 total: 33.8ms remaining: 2.94ms
46: learn: 0.5243558 total: 34.5ms remaining: 2.21ms
47: learn: 0.5216964 total: 35.5ms remaining: 1.48ms
48: learn: 0.5191863 total: 36.7ms remaining: 748us
49: learn: 0.5165567 total: 37.7ms remaining: 0us
0: learn: 0.6896203 total: 402us remaining: 19.7ms
1: learn: 0.6850714 total: 757us remaining: 18.2ms
2: learn: 0.6803862 total: 977us remaining: 15.3ms
3: learn: 0.6758516 total: 1.25ms remaining: 14.4ms
4: learn: 0.6716479 total: 1.48ms remaining: 13.4ms
5: learn: 0.6663871 total: 1.77ms remaining: 13ms
6: learn: 0.6619637 total: 2.04ms remaining: 12.5ms
7: learn: 0.6576289 total: 2.19ms remaining: 11.5ms
8: learn: 0.6535165 total: 2.36ms remaining: 10.7ms
9: learn: 0.6494451 total: 2.61ms remaining: 10.4ms
10: learn: 0.6454696 total: 2.81ms remaining: 9.96ms
11: learn: 0.6411167 total: 3ms remaining: 9.51ms
12: learn: 0.6375680 total: 3.26ms remaining: 9.29ms
13: learn: 0.6336478 total: 3.53ms remaining: 9.07ms
14: learn: 0.6296995 total: 3.81ms remaining: 8.9ms
15: learn: 0.6252287 total: 4.05ms remaining: 8.61ms
16: learn: 0.6216951 total: 4.31ms remaining: 8.36ms
17: learn: 0.6177813 total: 4.57ms remaining: 8.13ms
18: learn: 0.6140884 total: 4.7ms remaining: 7.67ms
19: learn: 0.6108468 total: 4.98ms remaining: 7.47ms
20: learn: 0.6072031 total: 5.26ms remaining: 7.26ms
21: learn: 0.6030750 total: 5.52ms remaining: 7.02ms
22: learn: 0.5992488 total: 5.74ms remaining: 6.74ms
23: learn: 0.5955617 total: 5.95ms remaining: 6.45ms
24: learn: 0.5922429 total: 6.13ms remaining: 6.13ms
25: learn: 0.5884927 total: 6.41ms remaining: 5.92ms
26: learn: 0.5856491 total: 6.63ms remaining: 5.64ms
27: learn: 0.5820865 total: 6.89ms remaining: 5.41ms
28: learn: 0.5795715 total: 7.12ms remaining: 5.15ms
29: learn: 0.5761366 total: 7.36ms remaining: 4.91ms
30: learn: 0.5728400 total: 7.58ms remaining: 4.64ms
31: learn: 0.5697248 total: 7.84ms remaining: 4.41ms
32: learn: 0.5668872 total: 8.15ms remaining: 4.2ms
33: learn: 0.5633521 total: 8.44ms remaining: 3.97ms
34: learn: 0.5601400 total: 8.72ms remaining: 3.73ms
35: learn: 0.5574012 total: 9.02ms remaining: 3.51ms
36: learn: 0.5543751 total: 9.29ms remaining: 3.26ms
37: learn: 0.5520800 total: 9.59ms remaining: 3.03ms
38: learn: 0.5495015 total: 9.82ms remaining: 2.77ms
39: learn: 0.5462074 total: 10.1ms remaining: 2.53ms
40: learn: 0.5434947 total: 10.3ms remaining: 2.27ms
41: learn: 0.5412224 total: 10.6ms remaining: 2.02ms
42: learn: 0.5385021 total: 11ms remaining: 1.78ms
43: learn: 0.5355544 total: 11.2ms remaining: 1.53ms
44: learn: 0.5328038 total: 11.5ms remaining: 1.28ms
45: learn: 0.5305187 total: 11.8ms remaining: 1.02ms
46: learn: 0.5281554 total: 12ms remaining: 765us
47: learn: 0.5251024 total: 12.3ms remaining: 510us
48: learn: 0.5225982 total: 12.4ms remaining: 253us
49: learn: 0.5203373 total: 12.7ms remaining: 0us
0: learn: 0.6894871 total: 357us remaining: 17.5ms
1: learn: 0.6842252 total: 600us remaining: 14.4ms
2: learn: 0.6798605 total: 861us remaining: 13.5ms
3: learn: 0.6746510 total: 1.06ms remaining: 12.2ms
4: learn: 0.6695916 total: 1.3ms remaining: 11.7ms
5: learn: 0.6646934 total: 1.48ms remaining: 10.8ms
6: learn: 0.6608368 total: 1.76ms remaining: 10.8ms
7: learn: 0.6560198 total: 1.95ms remaining: 10.2ms
8: learn: 0.6514894 total: 2.25ms remaining: 10.3ms
9: learn: 0.6484876 total: 2.48ms remaining: 9.93ms
10: learn: 0.6438394 total: 2.69ms remaining: 9.54ms
11: learn: 0.6400430 total: 2.95ms remaining: 9.33ms
12: learn: 0.6364403 total: 3.26ms remaining: 9.27ms
13: learn: 0.6328070 total: 3.59ms remaining: 9.22ms
14: learn: 0.6285253 total: 3.82ms remaining: 8.92ms
15: learn: 0.6244703 total: 4.09ms remaining: 8.69ms
16: learn: 0.6203785 total: 4.39ms remaining: 8.53ms
17: learn: 0.6170779 total: 4.53ms remaining: 8.05ms
18: learn: 0.6134879 total: 4.84ms remaining: 7.91ms
19: learn: 0.6098244 total: 5.14ms remaining: 7.71ms
20: learn: 0.6058554 total: 5.42ms remaining: 7.49ms
21: learn: 0.6020761 total: 5.67ms remaining: 7.22ms
22: learn: 0.5989954 total: 5.83ms remaining: 6.85ms
23: learn: 0.5950739 total: 6.1ms remaining: 6.61ms
24: learn: 0.5921883 total: 6.39ms remaining: 6.39ms
25: learn: 0.5889009 total: 6.67ms remaining: 6.16ms
26: learn: 0.5860139 total: 6.94ms remaining: 5.91ms
27: learn: 0.5833418 total: 7.2ms remaining: 5.65ms
28: learn: 0.5810032 total: 7.4ms remaining: 5.36ms
29: learn: 0.5784543 total: 7.68ms remaining: 5.12ms
30: learn: 0.5750472 total: 7.84ms remaining: 4.8ms
31: learn: 0.5717814 total: 8.11ms remaining: 4.56ms
32: learn: 0.5686437 total: 8.33ms remaining: 4.29ms
33: learn: 0.5653167 total: 8.56ms remaining: 4.03ms
34: learn: 0.5630180 total: 8.84ms remaining: 3.79ms
35: learn: 0.5603828 total: 9.14ms remaining: 3.55ms
36: learn: 0.5575529 total: 9.42ms remaining: 3.31ms
37: learn: 0.5553556 total: 9.66ms remaining: 3.05ms
38: learn: 0.5525472 total: 9.94ms remaining: 2.8ms
39: learn: 0.5495731 total: 10.2ms remaining: 2.54ms
40: learn: 0.5473859 total: 10.4ms remaining: 2.29ms
41: learn: 0.5444052 total: 10.7ms remaining: 2.03ms
42: learn: 0.5422100 total: 10.9ms remaining: 1.77ms
43: learn: 0.5393793 total: 11.2ms remaining: 1.52ms
44: learn: 0.5367392 total: 11.4ms remaining: 1.27ms
45: learn: 0.5339108 total: 11.7ms remaining: 1.01ms
46: learn: 0.5312177 total: 11.9ms remaining: 762us
47: learn: 0.5287355 total: 12.3ms remaining: 513us
48: learn: 0.5265024 total: 12.6ms remaining: 257us
49: learn: 0.5235980 total: 12.9ms remaining: 0us
0: learn: 0.6895315 total: 342us remaining: 16.8ms
1: learn: 0.6844180 total: 629us remaining: 15.1ms
2: learn: 0.6797461 total: 978us remaining: 15.3ms
3: learn: 0.6750861 total: 1.29ms remaining: 14.8ms
4: learn: 0.6707944 total: 1.49ms remaining: 13.4ms
5: learn: 0.6661817 total: 1.95ms remaining: 14.3ms
6: learn: 0.6616283 total: 2.19ms remaining: 13.4ms
7: learn: 0.6576498 total: 2.44ms remaining: 12.8ms
8: learn: 0.6531931 total: 2.64ms remaining: 12ms
9: learn: 0.6487036 total: 3.06ms remaining: 12.3ms
10: learn: 0.6449099 total: 3.33ms remaining: 11.8ms
11: learn: 0.6406385 total: 3.65ms remaining: 11.6ms
12: learn: 0.6364890 total: 3.9ms remaining: 11.1ms
13: learn: 0.6327426 total: 4.2ms remaining: 10.8ms
14: learn: 0.6293871 total: 4.59ms remaining: 10.7ms
15: learn: 0.6251434 total: 4.82ms remaining: 10.2ms
16: learn: 0.6214766 total: 5.1ms remaining: 9.91ms
17: learn: 0.6178709 total: 5.47ms remaining: 9.72ms
18: learn: 0.6143560 total: 5.62ms remaining: 9.16ms
19: learn: 0.6108690 total: 6.01ms remaining: 9.01ms
20: learn: 0.6073974 total: 6.31ms remaining: 8.71ms
21: learn: 0.6036627 total: 6.63ms remaining: 8.43ms
22: learn: 0.5997537 total: 6.9ms remaining: 8.11ms
23: learn: 0.5964076 total: 7.13ms remaining: 7.72ms
24: learn: 0.5934630 total: 7.4ms remaining: 7.4ms
25: learn: 0.5903772 total: 7.71ms remaining: 7.12ms
26: learn: 0.5873292 total: 8.08ms remaining: 6.88ms
27: learn: 0.5843486 total: 8.37ms remaining: 6.58ms
28: learn: 0.5815989 total: 8.7ms remaining: 6.3ms
29: learn: 0.5790148 total: 8.94ms remaining: 5.96ms
30: learn: 0.5764376 total: 9.31ms remaining: 5.71ms
31: learn: 0.5734280 total: 9.47ms remaining: 5.33ms
32: learn: 0.5703529 total: 9.78ms remaining: 5.04ms
33: learn: 0.5671999 total: 10ms remaining: 4.72ms
34: learn: 0.5637969 total: 10.3ms remaining: 4.43ms
35: learn: 0.5609537 total: 10.6ms remaining: 4.14ms
36: learn: 0.5583443 total: 10.9ms remaining: 3.84ms
37: learn: 0.5554906 total: 11.3ms remaining: 3.56ms
38: learn: 0.5523799 total: 11.5ms remaining: 3.26ms
39: learn: 0.5496638 total: 11.9ms remaining: 2.97ms
40: learn: 0.5467338 total: 12.1ms remaining: 2.66ms
41: learn: 0.5437414 total: 12.4ms remaining: 2.35ms
42: learn: 0.5410515 total: 12.6ms remaining: 2.06ms
43: learn: 0.5385706 total: 12.9ms remaining: 1.76ms
44: learn: 0.5359205 total: 13.2ms remaining: 1.47ms
45: learn: 0.5335821 total: 13.4ms remaining: 1.17ms
46: learn: 0.5311304 total: 13.8ms remaining: 879us
47: learn: 0.5287105 total: 14ms remaining: 583us
48: learn: 0.5262265 total: 14.3ms remaining: 290us
49: learn: 0.5235670 total: 14.5ms remaining: 0us
0: learn: 0.6884251 total: 417us remaining: 20.5ms
1: learn: 0.6837882 total: 690us remaining: 16.6ms
2: learn: 0.6788893 total: 1.09ms remaining: 17.1ms
3: learn: 0.6742531 total: 1.36ms remaining: 15.7ms
4: learn: 0.6694941 total: 1.58ms remaining: 14.2ms
5: learn: 0.6644974 total: 1.82ms remaining: 13.4ms
6: learn: 0.6602396 total: 2.15ms remaining: 13.2ms
7: learn: 0.6546575 total: 2.45ms remaining: 12.9ms
8: learn: 0.6495477 total: 2.81ms remaining: 12.8ms
9: learn: 0.6460980 total: 3.05ms remaining: 12.2ms
10: learn: 0.6407818 total: 3.29ms remaining: 11.7ms
11: learn: 0.6358886 total: 3.65ms remaining: 11.6ms
12: learn: 0.6319770 total: 4.03ms remaining: 11.5ms
13: learn: 0.6280995 total: 4.45ms remaining: 11.5ms
14: learn: 0.6233202 total: 4.75ms remaining: 11.1ms
15: learn: 0.6192099 total: 5ms remaining: 10.6ms
16: learn: 0.6147762 total: 5.29ms remaining: 10.3ms
17: learn: 0.6103238 total: 5.49ms remaining: 9.77ms
18: learn: 0.6060995 total: 5.88ms remaining: 9.6ms
19: learn: 0.6020556 total: 6.22ms remaining: 9.33ms
20: learn: 0.5977037 total: 6.61ms remaining: 9.13ms
21: learn: 0.5931581 total: 6.96ms remaining: 8.85ms
22: learn: 0.5890359 total: 7.22ms remaining: 8.48ms
23: learn: 0.5846377 total: 7.5ms remaining: 8.12ms
24: learn: 0.5814286 total: 7.82ms remaining: 7.82ms
25: learn: 0.5780560 total: 8.21ms remaining: 7.58ms
26: learn: 0.5739783 total: 8.54ms remaining: 7.28ms
27: learn: 0.5709059 total: 8.83ms remaining: 6.94ms
28: learn: 0.5681418 total: 9.06ms remaining: 6.56ms
29: learn: 0.5650585 total: 9.32ms remaining: 6.21ms
30: learn: 0.5612944 total: 9.51ms remaining: 5.83ms
31: learn: 0.5579837 total: 9.82ms remaining: 5.52ms
32: learn: 0.5545437 total: 10.1ms remaining: 5.19ms
33: learn: 0.5508624 total: 10.3ms remaining: 4.86ms
34: learn: 0.5476947 total: 10.6ms remaining: 4.56ms
35: learn: 0.5443277 total: 11ms remaining: 4.26ms
36: learn: 0.5410425 total: 11.3ms remaining: 3.98ms
37: learn: 0.5381051 total: 11.6ms remaining: 3.67ms
38: learn: 0.5349005 total: 11.9ms remaining: 3.36ms
39: learn: 0.5320597 total: 12.2ms remaining: 3.06ms
40: learn: 0.5288238 total: 12.5ms remaining: 2.74ms
41: learn: 0.5252659 total: 12.7ms remaining: 2.42ms
42: learn: 0.5227562 total: 12.9ms remaining: 2.1ms
43: learn: 0.5199700 total: 13.2ms remaining: 1.8ms
44: learn: 0.5171837 total: 13.6ms remaining: 1.51ms
45: learn: 0.5143990 total: 13.8ms remaining: 1.2ms
46: learn: 0.5116150 total: 14ms remaining: 895us
47: learn: 0.5085206 total: 14.4ms remaining: 598us
48: learn: 0.5052346 total: 14.6ms remaining: 298us
49: learn: 0.5021518 total: 14.9ms remaining: 0us
0: learn: 0.6878984 total: 271us remaining: 13.3ms
1: learn: 0.6825637 total: 482us remaining: 11.6ms
2: learn: 0.6778136 total: 662us remaining: 10.4ms
3: learn: 0.6730922 total: 857us remaining: 9.86ms
4: learn: 0.6683979 total: 1.01ms remaining: 9.13ms
5: learn: 0.6639298 total: 1.19ms remaining: 8.72ms
6: learn: 0.6588845 total: 1.36ms remaining: 8.34ms
7: learn: 0.6547090 total: 1.53ms remaining: 8.02ms
8: learn: 0.6501215 total: 1.68ms remaining: 7.64ms
9: learn: 0.6455656 total: 1.89ms remaining: 7.58ms
10: learn: 0.6411502 total: 2.22ms remaining: 7.88ms
11: learn: 0.6367748 total: 2.38ms remaining: 7.53ms
12: learn: 0.6324957 total: 2.57ms remaining: 7.31ms
13: learn: 0.6282916 total: 2.94ms remaining: 7.57ms
14: learn: 0.6244643 total: 3.61ms remaining: 8.43ms
15: learn: 0.6197827 total: 3.86ms remaining: 8.2ms
16: learn: 0.6162918 total: 4.11ms remaining: 7.97ms
17: learn: 0.6117805 total: 4.31ms remaining: 7.67ms
18: learn: 0.6076094 total: 4.7ms remaining: 7.66ms
19: learn: 0.6039850 total: 5.01ms remaining: 7.52ms
20: learn: 0.6001150 total: 5.39ms remaining: 7.44ms
21: learn: 0.5961289 total: 5.94ms remaining: 7.56ms
22: learn: 0.5919174 total: 6.29ms remaining: 7.39ms
23: learn: 0.5880261 total: 6.45ms remaining: 6.98ms
24: learn: 0.5845686 total: 6.64ms remaining: 6.64ms
25: learn: 0.5814502 total: 6.83ms remaining: 6.31ms
26: learn: 0.5783653 total: 7.08ms remaining: 6.03ms
27: learn: 0.5749444 total: 7.44ms remaining: 5.84ms
28: learn: 0.5717356 total: 8.12ms remaining: 5.88ms
29: learn: 0.5686947 total: 8.29ms remaining: 5.53ms
30: learn: 0.5658574 total: 8.74ms remaining: 5.36ms
31: learn: 0.5627563 total: 8.91ms remaining: 5.01ms
32: learn: 0.5594251 total: 9.12ms remaining: 4.7ms
33: learn: 0.5561764 total: 9.32ms remaining: 4.39ms
34: learn: 0.5529952 total: 10.1ms remaining: 4.33ms
35: learn: 0.5508336 total: 10.3ms remaining: 4.01ms
36: learn: 0.5478067 total: 10.5ms remaining: 3.7ms
37: learn: 0.5449646 total: 11.1ms remaining: 3.5ms
38: learn: 0.5420459 total: 11.4ms remaining: 3.21ms
39: learn: 0.5388165 total: 11.6ms remaining: 2.9ms
40: learn: 0.5358141 total: 11.8ms remaining: 2.6ms
41: learn: 0.5331245 total: 12.1ms remaining: 2.3ms
42: learn: 0.5300696 total: 12.4ms remaining: 2.02ms
43: learn: 0.5273135 total: 12.7ms remaining: 1.73ms
44: learn: 0.5247408 total: 13ms remaining: 1.44ms
45: learn: 0.5220816 total: 13.2ms remaining: 1.15ms
46: learn: 0.5198094 total: 13.7ms remaining: 875us
47: learn: 0.5168347 total: 13.9ms remaining: 579us
48: learn: 0.5144139 total: 14.1ms remaining: 287us
49: learn: 0.5116065 total: 14.2ms remaining: 0us
0: learn: 0.6882230 total: 292us remaining: 14.4ms
1: learn: 0.6830130 total: 609us remaining: 14.6ms
2: learn: 0.6777502 total: 775us remaining: 12.1ms
3: learn: 0.6725869 total: 1.02ms remaining: 11.8ms
4: learn: 0.6678779 total: 1.18ms remaining: 10.6ms
5: learn: 0.6627342 total: 1.37ms remaining: 10ms
6: learn: 0.6578700 total: 1.53ms remaining: 9.39ms
7: learn: 0.6539456 total: 1.75ms remaining: 9.22ms
8: learn: 0.6489295 total: 1.93ms remaining: 8.8ms
9: learn: 0.6447199 total: 2.32ms remaining: 9.27ms
10: learn: 0.6410565 total: 2.5ms remaining: 8.87ms
11: learn: 0.6370334 total: 2.82ms remaining: 8.95ms
12: learn: 0.6323019 total: 3.1ms remaining: 8.82ms
13: learn: 0.6278074 total: 3.35ms remaining: 8.61ms
14: learn: 0.6244961 total: 3.53ms remaining: 8.25ms
15: learn: 0.6205365 total: 3.77ms remaining: 8.02ms
16: learn: 0.6164157 total: 4.03ms remaining: 7.81ms
17: learn: 0.6125643 total: 4.21ms remaining: 7.49ms
18: learn: 0.6087596 total: 4.47ms remaining: 7.3ms
19: learn: 0.6051407 total: 4.71ms remaining: 7.06ms
20: learn: 0.6012506 total: 5.03ms remaining: 6.95ms
21: learn: 0.5969953 total: 5.39ms remaining: 6.87ms
22: learn: 0.5926725 total: 5.67ms remaining: 6.65ms
23: learn: 0.5892457 total: 5.84ms remaining: 6.33ms
24: learn: 0.5861760 total: 6.11ms remaining: 6.11ms
25: learn: 0.5827887 total: 6.33ms remaining: 5.84ms
26: learn: 0.5792932 total: 6.66ms remaining: 5.67ms
27: learn: 0.5751741 total: 6.93ms remaining: 5.45ms
28: learn: 0.5722502 total: 7.13ms remaining: 5.16ms
29: learn: 0.5696262 total: 7.35ms remaining: 4.9ms
30: learn: 0.5670091 total: 7.65ms remaining: 4.69ms
31: learn: 0.5635108 total: 7.77ms remaining: 4.37ms
32: learn: 0.5600613 total: 8.06ms remaining: 4.15ms
33: learn: 0.5568582 total: 8.23ms remaining: 3.87ms
34: learn: 0.5533880 total: 8.46ms remaining: 3.63ms
35: learn: 0.5503392 total: 8.78ms remaining: 3.41ms
36: learn: 0.5472394 total: 9.14ms remaining: 3.21ms
37: learn: 0.5443595 total: 9.4ms remaining: 2.97ms
38: learn: 0.5412247 total: 9.78ms remaining: 2.76ms
39: learn: 0.5382340 total: 10ms remaining: 2.51ms
40: learn: 0.5349641 total: 10.3ms remaining: 2.25ms
41: learn: 0.5321799 total: 10.4ms remaining: 1.98ms
42: learn: 0.5289777 total: 10.7ms remaining: 1.73ms
43: learn: 0.5259768 total: 11ms remaining: 1.5ms
44: learn: 0.5230374 total: 11.2ms remaining: 1.25ms
45: learn: 0.5207025 total: 11.4ms remaining: 992us
46: learn: 0.5181586 total: 11.7ms remaining: 744us
47: learn: 0.5151432 total: 11.9ms remaining: 494us
48: learn: 0.5125814 total: 12.1ms remaining: 247us
49: learn: 0.5098965 total: 12.5ms remaining: 0us
0: learn: 0.6886159 total: 355us remaining: 17.4ms
1: learn: 0.6833054 total: 998us remaining: 24ms
2: learn: 0.6784831 total: 1.18ms remaining: 18.5ms
3: learn: 0.6744239 total: 1.41ms remaining: 16.2ms
4: learn: 0.6703678 total: 1.76ms remaining: 15.8ms
5: learn: 0.6657698 total: 2.65ms remaining: 19.4ms
6: learn: 0.6613446 total: 3.39ms remaining: 20.8ms
7: learn: 0.6575104 total: 4.24ms remaining: 22.3ms
8: learn: 0.6529390 total: 4.95ms remaining: 22.5ms
9: learn: 0.6485341 total: 5.38ms remaining: 21.5ms
10: learn: 0.6445752 total: 5.96ms remaining: 21.1ms
11: learn: 0.6403021 total: 6.5ms remaining: 20.6ms
12: learn: 0.6365289 total: 7.33ms remaining: 20.9ms
13: learn: 0.6324526 total: 8.16ms remaining: 21ms
14: learn: 0.6290089 total: 8.96ms remaining: 20.9ms
15: learn: 0.6245414 total: 9.58ms remaining: 20.4ms
16: learn: 0.6210188 total: 10.4ms remaining: 20.2ms
17: learn: 0.6174320 total: 11ms remaining: 19.6ms
18: learn: 0.6140296 total: 11.2ms remaining: 18.2ms
19: learn: 0.6105599 total: 11.5ms remaining: 17.3ms
20: learn: 0.6069129 total: 12.3ms remaining: 16.9ms
21: learn: 0.6030373 total: 13ms remaining: 16.5ms
22: learn: 0.5995989 total: 13.6ms remaining: 16ms
23: learn: 0.5963684 total: 14.4ms remaining: 15.6ms
24: learn: 0.5922892 total: 14.9ms remaining: 14.9ms
25: learn: 0.5894788 total: 15.6ms remaining: 14.4ms
26: learn: 0.5863141 total: 16.4ms remaining: 13.9ms
27: learn: 0.5833325 total: 17.1ms remaining: 13.5ms
28: learn: 0.5804530 total: 17.9ms remaining: 13ms
29: learn: 0.5778179 total: 18.5ms remaining: 12.4ms
30: learn: 0.5753587 total: 18.9ms remaining: 11.6ms
31: learn: 0.5724407 total: 19.2ms remaining: 10.8ms
32: learn: 0.5690258 total: 19.8ms remaining: 10.2ms
33: learn: 0.5657888 total: 20.4ms remaining: 9.58ms
34: learn: 0.5621902 total: 20.9ms remaining: 8.96ms
35: learn: 0.5593933 total: 21.6ms remaining: 8.39ms
36: learn: 0.5564423 total: 22.2ms remaining: 7.8ms
37: learn: 0.5537762 total: 22.9ms remaining: 7.22ms
38: learn: 0.5504946 total: 23.5ms remaining: 6.61ms
39: learn: 0.5481516 total: 24.1ms remaining: 6.02ms
40: learn: 0.5452933 total: 24.4ms remaining: 5.35ms
41: learn: 0.5425361 total: 24.5ms remaining: 4.66ms
42: learn: 0.5398155 total: 25.2ms remaining: 4.1ms
43: learn: 0.5365296 total: 25.6ms remaining: 3.49ms
44: learn: 0.5338784 total: 26.3ms remaining: 2.92ms
45: learn: 0.5312372 total: 26.9ms remaining: 2.34ms
46: learn: 0.5285292 total: 27.6ms remaining: 1.76ms
47: learn: 0.5257266 total: 28.1ms remaining: 1.17ms
48: learn: 0.5232061 total: 28.4ms remaining: 578us
49: learn: 0.5203838 total: 28.7ms remaining: 0us
Evaluating param combo 2/16: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 16}
Duplicate found for this combo (hash=866bf6c611c544b1a16becf0cb7674ab). Removing old run: 43d07b5aea0346a9aba72848f4e0e4e1
0: learn: 0.6882794 total: 469us remaining: 23ms
1: learn: 0.6829408 total: 872us remaining: 20.9ms
2: learn: 0.6779769 total: 1.03ms remaining: 16.1ms
3: learn: 0.6731590 total: 1.3ms remaining: 15ms
4: learn: 0.6685559 total: 1.6ms remaining: 14.4ms
5: learn: 0.6636779 total: 2.22ms remaining: 16.3ms
6: learn: 0.6591763 total: 3.04ms remaining: 18.7ms
7: learn: 0.6551357 total: 4.14ms remaining: 21.7ms
8: learn: 0.6504101 total: 4.86ms remaining: 22.1ms
9: learn: 0.6464580 total: 5.69ms remaining: 22.8ms
10: learn: 0.6425359 total: 6.32ms remaining: 22.4ms
11: learn: 0.6387708 total: 7.74ms remaining: 24.5ms
12: learn: 0.6346059 total: 8.63ms remaining: 24.6ms
13: learn: 0.6304314 total: 8.99ms remaining: 23.1ms
14: learn: 0.6268009 total: 9.82ms remaining: 22.9ms
15: learn: 0.6229881 total: 10.9ms remaining: 23.2ms
16: learn: 0.6190015 total: 12.3ms remaining: 23.8ms
17: learn: 0.6151072 total: 13ms remaining: 23ms
18: learn: 0.6113699 total: 13.3ms remaining: 21.8ms
19: learn: 0.6075444 total: 14.6ms remaining: 22ms
20: learn: 0.6037564 total: 15.7ms remaining: 21.7ms
21: learn: 0.5999778 total: 16.8ms remaining: 21.4ms
22: learn: 0.5958785 total: 17.7ms remaining: 20.7ms
23: learn: 0.5922311 total: 18.3ms remaining: 19.8ms
24: learn: 0.5892157 total: 19.5ms remaining: 19.5ms
25: learn: 0.5861289 total: 21ms remaining: 19.4ms
26: learn: 0.5829753 total: 22.3ms remaining: 19ms
27: learn: 0.5798042 total: 23.5ms remaining: 18.5ms
28: learn: 0.5767399 total: 24.4ms remaining: 17.7ms
29: learn: 0.5739181 total: 25ms remaining: 16.7ms
30: learn: 0.5712722 total: 26.1ms remaining: 16ms
31: learn: 0.5679889 total: 26.8ms remaining: 15.1ms
32: learn: 0.5647182 total: 27.7ms remaining: 14.3ms
33: learn: 0.5613468 total: 28.9ms remaining: 13.6ms
34: learn: 0.5577149 total: 29.7ms remaining: 12.7ms
35: learn: 0.5547437 total: 30.9ms remaining: 12ms
36: learn: 0.5517524 total: 31.9ms remaining: 11.2ms
37: learn: 0.5484946 total: 32.9ms remaining: 10.4ms
38: learn: 0.5451312 total: 33.7ms remaining: 9.5ms
39: learn: 0.5421510 total: 34.3ms remaining: 8.58ms
40: learn: 0.5387318 total: 34.6ms remaining: 7.58ms
41: learn: 0.5355684 total: 35.3ms remaining: 6.72ms
42: learn: 0.5324215 total: 35.9ms remaining: 5.85ms
43: learn: 0.5298921 total: 36.6ms remaining: 4.99ms
44: learn: 0.5271650 total: 37.3ms remaining: 4.14ms
45: learn: 0.5248260 total: 38ms remaining: 3.31ms
46: learn: 0.5222534 total: 38.7ms remaining: 2.47ms
47: learn: 0.5193675 total: 38.9ms remaining: 1.62ms
48: learn: 0.5168600 total: 39.2ms remaining: 799us
49: learn: 0.5145437 total: 39.9ms remaining: 0us
0: learn: 0.6880102 total: 511us remaining: 25.1ms
1: learn: 0.6822828 total: 1.19ms remaining: 28.7ms
2: learn: 0.6772106 total: 1.68ms remaining: 26.3ms
3: learn: 0.6726730 total: 2.26ms remaining: 26ms
4: learn: 0.6682095 total: 2.75ms remaining: 24.7ms
5: learn: 0.6635268 total: 3.09ms remaining: 22.7ms
6: learn: 0.6588811 total: 3.58ms remaining: 22ms
7: learn: 0.6545703 total: 3.79ms remaining: 19.9ms
8: learn: 0.6497856 total: 3.93ms remaining: 17.9ms
9: learn: 0.6448858 total: 4.22ms remaining: 16.9ms
10: learn: 0.6405230 total: 4.66ms remaining: 16.5ms
11: learn: 0.6361860 total: 5.34ms remaining: 16.9ms
12: learn: 0.6314977 total: 5.87ms remaining: 16.7ms
13: learn: 0.6272630 total: 6.49ms remaining: 16.7ms
14: learn: 0.6237927 total: 6.95ms remaining: 16.2ms
15: learn: 0.6202946 total: 7.47ms remaining: 15.9ms
16: learn: 0.6160996 total: 8.1ms remaining: 15.7ms
17: learn: 0.6121987 total: 8.55ms remaining: 15.2ms
18: learn: 0.6085887 total: 8.99ms remaining: 14.7ms
19: learn: 0.6053875 total: 9.6ms remaining: 14.4ms
20: learn: 0.6012749 total: 10.4ms remaining: 14.3ms
21: learn: 0.5970959 total: 10.9ms remaining: 13.9ms
22: learn: 0.5929697 total: 11.6ms remaining: 13.6ms
23: learn: 0.5894176 total: 12.3ms remaining: 13.4ms
24: learn: 0.5848522 total: 12.8ms remaining: 12.8ms
25: learn: 0.5821273 total: 13.3ms remaining: 12.3ms
26: learn: 0.5789455 total: 14ms remaining: 11.9ms
27: learn: 0.5754969 total: 14.5ms remaining: 11.4ms
28: learn: 0.5721862 total: 15.1ms remaining: 10.9ms
29: learn: 0.5691136 total: 15.5ms remaining: 10.3ms
30: learn: 0.5660278 total: 16.1ms remaining: 9.86ms
31: learn: 0.5628754 total: 16.5ms remaining: 9.27ms
32: learn: 0.5589497 total: 17.1ms remaining: 8.8ms
33: learn: 0.5553905 total: 17.6ms remaining: 8.28ms
34: learn: 0.5515082 total: 18.2ms remaining: 7.81ms
35: learn: 0.5482866 total: 18.7ms remaining: 7.26ms
36: learn: 0.5449903 total: 19.2ms remaining: 6.73ms
37: learn: 0.5423647 total: 19.6ms remaining: 6.2ms
38: learn: 0.5386898 total: 20.1ms remaining: 5.66ms
39: learn: 0.5350966 total: 20.5ms remaining: 5.12ms
40: learn: 0.5319492 total: 20.8ms remaining: 4.57ms
41: learn: 0.5288603 total: 21.2ms remaining: 4.04ms
42: learn: 0.5260202 total: 21.8ms remaining: 3.55ms
43: learn: 0.5230047 total: 22.4ms remaining: 3.05ms
44: learn: 0.5198876 total: 23ms remaining: 2.55ms
45: learn: 0.5168284 total: 23.4ms remaining: 2.03ms
46: learn: 0.5136389 total: 23.9ms remaining: 1.53ms
47: learn: 0.5112892 total: 24.4ms remaining: 1.02ms
48: learn: 0.5085121 total: 24.7ms remaining: 504us
49: learn: 0.5054328 total: 25.2ms remaining: 0us
0: learn: 0.6881915 total: 275us remaining: 13.5ms
1: learn: 0.6830903 total: 524us remaining: 12.6ms
2: learn: 0.6779156 total: 681us remaining: 10.7ms
3: learn: 0.6727870 total: 973us remaining: 11.2ms
4: learn: 0.6680191 total: 1.21ms remaining: 10.9ms
5: learn: 0.6632566 total: 1.41ms remaining: 10.4ms
6: learn: 0.6589348 total: 1.62ms remaining: 9.93ms
7: learn: 0.6548497 total: 1.81ms remaining: 9.49ms
8: learn: 0.6499422 total: 1.95ms remaining: 8.87ms
9: learn: 0.6456086 total: 2.12ms remaining: 8.47ms
10: learn: 0.6416798 total: 2.26ms remaining: 8.01ms
11: learn: 0.6376258 total: 2.45ms remaining: 7.76ms
12: learn: 0.6328452 total: 2.63ms remaining: 7.48ms
13: learn: 0.6291112 total: 2.8ms remaining: 7.19ms
14: learn: 0.6252126 total: 2.97ms remaining: 6.92ms
15: learn: 0.6213698 total: 3.14ms remaining: 6.68ms
16: learn: 0.6172627 total: 3.33ms remaining: 6.47ms
17: learn: 0.6136354 total: 3.47ms remaining: 6.17ms
18: learn: 0.6099157 total: 3.61ms remaining: 5.89ms
19: learn: 0.6065275 total: 3.8ms remaining: 5.7ms
20: learn: 0.6026046 total: 4ms remaining: 5.53ms
21: learn: 0.5986205 total: 4.25ms remaining: 5.41ms
22: learn: 0.5943580 total: 4.41ms remaining: 5.18ms
23: learn: 0.5906964 total: 4.56ms remaining: 4.94ms
24: learn: 0.5869638 total: 4.76ms remaining: 4.76ms
25: learn: 0.5838294 total: 4.99ms remaining: 4.61ms
26: learn: 0.5809812 total: 5.18ms remaining: 4.42ms
27: learn: 0.5776571 total: 5.38ms remaining: 4.23ms
28: learn: 0.5743650 total: 5.58ms remaining: 4.04ms
29: learn: 0.5714944 total: 5.76ms remaining: 3.84ms
30: learn: 0.5687434 total: 6.14ms remaining: 3.76ms
31: learn: 0.5653821 total: 6.29ms remaining: 3.54ms
32: learn: 0.5620526 total: 6.6ms remaining: 3.4ms
33: learn: 0.5584545 total: 6.87ms remaining: 3.23ms
34: learn: 0.5550520 total: 7.1ms remaining: 3.04ms
35: learn: 0.5514777 total: 7.51ms remaining: 2.92ms
36: learn: 0.5483220 total: 7.91ms remaining: 2.78ms
37: learn: 0.5453927 total: 8.46ms remaining: 2.67ms
38: learn: 0.5418823 total: 8.7ms remaining: 2.45ms
39: learn: 0.5394460 total: 9.21ms remaining: 2.3ms
40: learn: 0.5362960 total: 9.42ms remaining: 2.07ms
41: learn: 0.5328204 total: 9.56ms remaining: 1.82ms
42: learn: 0.5296236 total: 10ms remaining: 1.63ms
43: learn: 0.5272600 total: 10.6ms remaining: 1.45ms
44: learn: 0.5244234 total: 10.8ms remaining: 1.2ms
45: learn: 0.5224995 total: 11.3ms remaining: 983us
46: learn: 0.5199309 total: 11.9ms remaining: 760us
47: learn: 0.5169806 total: 12.6ms remaining: 524us
48: learn: 0.5142186 total: 12.9ms remaining: 262us
49: learn: 0.5113786 total: 13.1ms remaining: 0us
0: learn: 0.6883581 total: 545us remaining: 26.7ms
1: learn: 0.6829490 total: 863us remaining: 20.7ms
2: learn: 0.6777105 total: 1.12ms remaining: 17.6ms
3: learn: 0.6724965 total: 1.49ms remaining: 17.1ms
4: learn: 0.6678869 total: 1.92ms remaining: 17.3ms
5: learn: 0.6630227 total: 2.32ms remaining: 17ms
6: learn: 0.6580845 total: 2.82ms remaining: 17.3ms
7: learn: 0.6539384 total: 3.31ms remaining: 17.4ms
8: learn: 0.6488837 total: 3.51ms remaining: 16ms
9: learn: 0.6442866 total: 3.75ms remaining: 15ms
10: learn: 0.6404207 total: 4.08ms remaining: 14.5ms
11: learn: 0.6359348 total: 4.62ms remaining: 14.6ms
12: learn: 0.6315682 total: 5.07ms remaining: 14.4ms
13: learn: 0.6273393 total: 5.49ms remaining: 14.1ms
14: learn: 0.6234264 total: 5.87ms remaining: 13.7ms
15: learn: 0.6197946 total: 6.28ms remaining: 13.3ms
16: learn: 0.6155398 total: 6.66ms remaining: 12.9ms
17: learn: 0.6113889 total: 6.93ms remaining: 12.3ms
18: learn: 0.6079857 total: 7.29ms remaining: 11.9ms
19: learn: 0.6040984 total: 7.64ms remaining: 11.5ms
20: learn: 0.6001675 total: 8.02ms remaining: 11.1ms
21: learn: 0.5959995 total: 8.37ms remaining: 10.7ms
22: learn: 0.5919796 total: 8.86ms remaining: 10.4ms
23: learn: 0.5884148 total: 9.3ms remaining: 10.1ms
24: learn: 0.5841205 total: 9.59ms remaining: 9.59ms
25: learn: 0.5809535 total: 10ms remaining: 9.23ms
26: learn: 0.5776310 total: 10.4ms remaining: 8.84ms
27: learn: 0.5743591 total: 10.7ms remaining: 8.38ms
28: learn: 0.5712718 total: 11.1ms remaining: 8.03ms
29: learn: 0.5684340 total: 11.4ms remaining: 7.62ms
30: learn: 0.5657478 total: 11.9ms remaining: 7.29ms
31: learn: 0.5623463 total: 12.1ms remaining: 6.79ms
32: learn: 0.5591812 total: 12.5ms remaining: 6.42ms
33: learn: 0.5555487 total: 12.8ms remaining: 6ms
34: learn: 0.5525218 total: 13ms remaining: 5.58ms
35: learn: 0.5495863 total: 13.3ms remaining: 5.18ms
36: learn: 0.5463044 total: 13.7ms remaining: 4.81ms
37: learn: 0.5429413 total: 14.1ms remaining: 4.46ms
38: learn: 0.5394540 total: 14.5ms remaining: 4.08ms
39: learn: 0.5363956 total: 14.8ms remaining: 3.69ms
40: learn: 0.5330491 total: 15ms remaining: 3.29ms
41: learn: 0.5306820 total: 15.3ms remaining: 2.91ms
42: learn: 0.5275766 total: 15.6ms remaining: 2.54ms
43: learn: 0.5246236 total: 15.9ms remaining: 2.17ms
44: learn: 0.5218134 total: 16.2ms remaining: 1.8ms
45: learn: 0.5189606 total: 16.5ms remaining: 1.44ms
46: learn: 0.5159627 total: 16.9ms remaining: 1.08ms
47: learn: 0.5128221 total: 17.2ms remaining: 717us
48: learn: 0.5096912 total: 17.4ms remaining: 355us
49: learn: 0.5072024 total: 17.8ms remaining: 0us
0: learn: 0.6883175 total: 386us remaining: 18.9ms
1: learn: 0.6833177 total: 657us remaining: 15.8ms
2: learn: 0.6787594 total: 863us remaining: 13.5ms
3: learn: 0.6742785 total: 1.15ms remaining: 13.2ms
4: learn: 0.6698376 total: 1.46ms remaining: 13.1ms
5: learn: 0.6655790 total: 1.79ms remaining: 13.1ms
6: learn: 0.6606753 total: 2.07ms remaining: 12.7ms
7: learn: 0.6567153 total: 2.37ms remaining: 12.5ms
8: learn: 0.6521844 total: 2.65ms remaining: 12.1ms
9: learn: 0.6476670 total: 3ms remaining: 12ms
10: learn: 0.6436401 total: 3.26ms remaining: 11.6ms
11: learn: 0.6392506 total: 3.55ms remaining: 11.2ms
12: learn: 0.6352223 total: 3.89ms remaining: 11.1ms
13: learn: 0.6314012 total: 4.35ms remaining: 11.2ms
14: learn: 0.6271130 total: 4.8ms remaining: 11.2ms
15: learn: 0.6228527 total: 5.32ms remaining: 11.3ms
16: learn: 0.6193636 total: 5.59ms remaining: 10.8ms
17: learn: 0.6149698 total: 5.82ms remaining: 10.3ms
18: learn: 0.6115084 total: 6.04ms remaining: 9.86ms
19: learn: 0.6080555 total: 6.48ms remaining: 9.72ms
20: learn: 0.6044442 total: 6.86ms remaining: 9.47ms
21: learn: 0.6011192 total: 7.2ms remaining: 9.17ms
22: learn: 0.5974142 total: 7.64ms remaining: 8.96ms
23: learn: 0.5941231 total: 8.01ms remaining: 8.68ms
24: learn: 0.5900927 total: 8.32ms remaining: 8.32ms
25: learn: 0.5870085 total: 8.72ms remaining: 8.05ms
26: learn: 0.5840114 total: 9.13ms remaining: 7.78ms
27: learn: 0.5808997 total: 9.46ms remaining: 7.43ms
28: learn: 0.5778788 total: 9.87ms remaining: 7.15ms
29: learn: 0.5750842 total: 10.2ms remaining: 6.79ms
30: learn: 0.5722879 total: 10.7ms remaining: 6.53ms
31: learn: 0.5693116 total: 10.9ms remaining: 6.14ms
32: learn: 0.5661397 total: 11.4ms remaining: 5.87ms
33: learn: 0.5627994 total: 11.8ms remaining: 5.56ms
34: learn: 0.5597978 total: 12.2ms remaining: 5.23ms
35: learn: 0.5564847 total: 12.6ms remaining: 4.88ms
36: learn: 0.5535813 total: 12.9ms remaining: 4.53ms
37: learn: 0.5511907 total: 13.2ms remaining: 4.18ms
38: learn: 0.5477524 total: 13.6ms remaining: 3.83ms
39: learn: 0.5446551 total: 13.9ms remaining: 3.48ms
40: learn: 0.5418243 total: 14.2ms remaining: 3.12ms
41: learn: 0.5384306 total: 14.5ms remaining: 2.76ms
42: learn: 0.5355158 total: 15ms remaining: 2.44ms
43: learn: 0.5330304 total: 15.4ms remaining: 2.1ms
44: learn: 0.5303464 total: 15.8ms remaining: 1.76ms
45: learn: 0.5282582 total: 16.1ms remaining: 1.4ms
46: learn: 0.5258402 total: 16.5ms remaining: 1.05ms
47: learn: 0.5235785 total: 16.7ms remaining: 696us
48: learn: 0.5207689 total: 17ms remaining: 346us
49: learn: 0.5183663 total: 17.3ms remaining: 0us
0: learn: 0.6886824 total: 309us remaining: 15.2ms
1: learn: 0.6836439 total: 590us remaining: 14.2ms
2: learn: 0.6783803 total: 767us remaining: 12ms
3: learn: 0.6732528 total: 1.03ms remaining: 11.9ms
4: learn: 0.6684937 total: 1.27ms remaining: 11.4ms
5: learn: 0.6634511 total: 1.54ms remaining: 11.3ms
6: learn: 0.6589192 total: 1.81ms remaining: 11.1ms
7: learn: 0.6551883 total: 2.05ms remaining: 10.8ms
8: learn: 0.6502473 total: 2.31ms remaining: 10.5ms
9: learn: 0.6460178 total: 2.62ms remaining: 10.5ms
10: learn: 0.6419694 total: 2.92ms remaining: 10.3ms
11: learn: 0.6383164 total: 3.34ms remaining: 10.6ms
12: learn: 0.6337982 total: 3.72ms remaining: 10.6ms
13: learn: 0.6295016 total: 4.14ms remaining: 10.6ms
14: learn: 0.6253754 total: 4.4ms remaining: 10.3ms
15: learn: 0.6213047 total: 4.72ms remaining: 10ms
16: learn: 0.6172331 total: 5.04ms remaining: 9.79ms
17: learn: 0.6140067 total: 5.34ms remaining: 9.5ms
18: learn: 0.6101040 total: 5.62ms remaining: 9.16ms
19: learn: 0.6066469 total: 5.96ms remaining: 8.94ms
20: learn: 0.6029030 total: 6.33ms remaining: 8.75ms
21: learn: 0.5987959 total: 6.74ms remaining: 8.57ms
22: learn: 0.5948275 total: 7.04ms remaining: 8.27ms
23: learn: 0.5915322 total: 7.27ms remaining: 7.88ms
24: learn: 0.5872507 total: 7.57ms remaining: 7.57ms
25: learn: 0.5839759 total: 7.86ms remaining: 7.26ms
26: learn: 0.5805313 total: 8.22ms remaining: 7ms
27: learn: 0.5778302 total: 8.51ms remaining: 6.69ms
28: learn: 0.5752130 total: 8.78ms remaining: 6.36ms
29: learn: 0.5727888 total: 9.09ms remaining: 6.06ms
30: learn: 0.5702654 total: 9.45ms remaining: 5.79ms
31: learn: 0.5666849 total: 9.68ms remaining: 5.45ms
32: learn: 0.5634739 total: 9.99ms remaining: 5.14ms
33: learn: 0.5599432 total: 10.3ms remaining: 4.87ms
34: learn: 0.5569637 total: 10.6ms remaining: 4.56ms
35: learn: 0.5542632 total: 11ms remaining: 4.27ms
36: learn: 0.5511816 total: 11.4ms remaining: 3.99ms
37: learn: 0.5479280 total: 11.7ms remaining: 3.7ms
38: learn: 0.5446114 total: 12ms remaining: 3.38ms
39: learn: 0.5416442 total: 12.3ms remaining: 3.08ms
40: learn: 0.5388472 total: 12.7ms remaining: 2.78ms
41: learn: 0.5353803 total: 12.9ms remaining: 2.45ms
42: learn: 0.5323092 total: 13.2ms remaining: 2.15ms
43: learn: 0.5297938 total: 13.6ms remaining: 1.85ms
44: learn: 0.5270777 total: 13.8ms remaining: 1.53ms
45: learn: 0.5242021 total: 14.2ms remaining: 1.23ms
46: learn: 0.5210269 total: 14.5ms remaining: 926us
47: learn: 0.5183354 total: 14.8ms remaining: 615us
48: learn: 0.5155134 total: 15.1ms remaining: 307us
49: learn: 0.5125593 total: 15.3ms remaining: 0us
0: learn: 0.6884710 total: 313us remaining: 15.3ms
1: learn: 0.6831483 total: 808us remaining: 19.4ms
2: learn: 0.6781742 total: 1.06ms remaining: 16.6ms
3: learn: 0.6735979 total: 1.39ms remaining: 16ms
4: learn: 0.6690825 total: 1.64ms remaining: 14.8ms
5: learn: 0.6643177 total: 1.96ms remaining: 14.3ms
6: learn: 0.6602321 total: 2.15ms remaining: 13.2ms
7: learn: 0.6561625 total: 2.39ms remaining: 12.5ms
8: learn: 0.6513684 total: 2.66ms remaining: 12.1ms
9: learn: 0.6464446 total: 3.17ms remaining: 12.7ms
10: learn: 0.6425160 total: 3.38ms remaining: 12ms
11: learn: 0.6379029 total: 3.64ms remaining: 11.5ms
12: learn: 0.6338682 total: 4.02ms remaining: 11.4ms
13: learn: 0.6299456 total: 4.4ms remaining: 11.3ms
14: learn: 0.6261431 total: 4.7ms remaining: 11ms
15: learn: 0.6214318 total: 4.93ms remaining: 10.5ms
16: learn: 0.6176888 total: 5.26ms remaining: 10.2ms
17: learn: 0.6136496 total: 5.49ms remaining: 9.76ms
18: learn: 0.6098567 total: 5.71ms remaining: 9.31ms
19: learn: 0.6061197 total: 5.96ms remaining: 8.95ms
20: learn: 0.6021095 total: 6.2ms remaining: 8.56ms
21: learn: 0.5977883 total: 6.44ms remaining: 8.19ms
22: learn: 0.5936190 total: 6.72ms remaining: 7.89ms
23: learn: 0.5899167 total: 6.97ms remaining: 7.56ms
24: learn: 0.5858649 total: 7.24ms remaining: 7.24ms
25: learn: 0.5829470 total: 7.53ms remaining: 6.95ms
26: learn: 0.5798447 total: 7.9ms remaining: 6.73ms
27: learn: 0.5765984 total: 8.13ms remaining: 6.39ms
28: learn: 0.5737242 total: 8.5ms remaining: 6.15ms
29: learn: 0.5710620 total: 8.7ms remaining: 5.8ms
30: learn: 0.5684374 total: 9.09ms remaining: 5.57ms
31: learn: 0.5652138 total: 9.25ms remaining: 5.21ms
32: learn: 0.5622089 total: 9.46ms remaining: 4.87ms
33: learn: 0.5588240 total: 9.73ms remaining: 4.58ms
34: learn: 0.5553290 total: 9.97ms remaining: 4.27ms
35: learn: 0.5523463 total: 10.2ms remaining: 3.98ms
36: learn: 0.5491377 total: 10.5ms remaining: 3.68ms
37: learn: 0.5464623 total: 10.8ms remaining: 3.4ms
38: learn: 0.5430188 total: 11ms remaining: 3.11ms
39: learn: 0.5400183 total: 11.4ms remaining: 2.84ms
40: learn: 0.5375189 total: 11.7ms remaining: 2.57ms
41: learn: 0.5339443 total: 11.9ms remaining: 2.27ms
42: learn: 0.5309167 total: 12.2ms remaining: 1.98ms
43: learn: 0.5285461 total: 12.5ms remaining: 1.71ms
44: learn: 0.5257915 total: 12.9ms remaining: 1.43ms
45: learn: 0.5235061 total: 13.2ms remaining: 1.14ms
46: learn: 0.5208134 total: 13.6ms remaining: 868us
47: learn: 0.5181357 total: 13.9ms remaining: 579us
48: learn: 0.5152125 total: 14.1ms remaining: 287us
49: learn: 0.5127882 total: 14.3ms remaining: 0us
0: learn: 0.6888251 total: 350us remaining: 17.2ms
1: learn: 0.6838743 total: 608us remaining: 14.6ms
2: learn: 0.6790688 total: 850us remaining: 13.3ms
3: learn: 0.6744890 total: 1.18ms remaining: 13.6ms
4: learn: 0.6697935 total: 1.44ms remaining: 12.9ms
5: learn: 0.6652243 total: 1.69ms remaining: 12.4ms
6: learn: 0.6616198 total: 2.25ms remaining: 13.9ms
7: learn: 0.6576694 total: 2.52ms remaining: 13.2ms
8: learn: 0.6532890 total: 2.85ms remaining: 13ms
9: learn: 0.6492571 total: 3.14ms remaining: 12.6ms
10: learn: 0.6463276 total: 3.47ms remaining: 12.3ms
11: learn: 0.6419683 total: 3.79ms remaining: 12ms
12: learn: 0.6383123 total: 4.12ms remaining: 11.7ms
13: learn: 0.6346641 total: 4.43ms remaining: 11.4ms
14: learn: 0.6310619 total: 4.72ms remaining: 11ms
15: learn: 0.6268264 total: 5.03ms remaining: 10.7ms
16: learn: 0.6232008 total: 5.24ms remaining: 10.2ms
17: learn: 0.6188903 total: 5.66ms remaining: 10.1ms
18: learn: 0.6154295 total: 6ms remaining: 9.8ms
19: learn: 0.6110983 total: 6.34ms remaining: 9.51ms
20: learn: 0.6074348 total: 6.57ms remaining: 9.07ms
21: learn: 0.6035654 total: 6.82ms remaining: 8.68ms
22: learn: 0.6001629 total: 7.1ms remaining: 8.34ms
23: learn: 0.5972153 total: 7.43ms remaining: 8.05ms
24: learn: 0.5938984 total: 7.7ms remaining: 7.7ms
25: learn: 0.5914189 total: 8.05ms remaining: 7.43ms
26: learn: 0.5879412 total: 8.3ms remaining: 7.07ms
27: learn: 0.5844532 total: 8.5ms remaining: 6.68ms
28: learn: 0.5812474 total: 8.74ms remaining: 6.33ms
29: learn: 0.5780829 total: 9.09ms remaining: 6.06ms
30: learn: 0.5747267 total: 9.31ms remaining: 5.71ms
31: learn: 0.5714414 total: 9.65ms remaining: 5.43ms
32: learn: 0.5686548 total: 10.2ms remaining: 5.23ms
33: learn: 0.5656884 total: 10.3ms remaining: 4.86ms
34: learn: 0.5628498 total: 10.6ms remaining: 4.55ms
35: learn: 0.5601182 total: 10.8ms remaining: 4.21ms
36: learn: 0.5571363 total: 11.1ms remaining: 3.91ms
37: learn: 0.5543120 total: 11.3ms remaining: 3.58ms
38: learn: 0.5508372 total: 11.6ms remaining: 3.27ms
39: learn: 0.5482084 total: 11.9ms remaining: 2.98ms
40: learn: 0.5451572 total: 12.1ms remaining: 2.66ms
41: learn: 0.5418393 total: 12.3ms remaining: 2.34ms
42: learn: 0.5393336 total: 12.6ms remaining: 2.05ms
43: learn: 0.5368743 total: 12.8ms remaining: 1.75ms
44: learn: 0.5341000 total: 13.1ms remaining: 1.46ms
45: learn: 0.5315797 total: 13.3ms remaining: 1.16ms
46: learn: 0.5288963 total: 13.5ms remaining: 864us
47: learn: 0.5262389 total: 13.7ms remaining: 570us
48: learn: 0.5234348 total: 13.9ms remaining: 282us
49: learn: 0.5209916 total: 14.1ms remaining: 0us
0: learn: 0.6881993 total: 356us remaining: 17.5ms
1: learn: 0.6827008 total: 605us remaining: 14.5ms
2: learn: 0.6774722 total: 809us remaining: 12.7ms
3: learn: 0.6721781 total: 1.77ms remaining: 20.4ms
4: learn: 0.6675501 total: 2.65ms remaining: 23.9ms
5: learn: 0.6623863 total: 3.64ms remaining: 26.7ms
6: learn: 0.6575695 total: 4.61ms remaining: 28.3ms
7: learn: 0.6535074 total: 5.33ms remaining: 28ms
8: learn: 0.6485427 total: 5.86ms remaining: 26.7ms
9: learn: 0.6436104 total: 6.85ms remaining: 27.4ms
10: learn: 0.6395245 total: 7.54ms remaining: 26.7ms
11: learn: 0.6347631 total: 8.58ms remaining: 27.2ms
12: learn: 0.6305070 total: 9.31ms remaining: 26.5ms
13: learn: 0.6265698 total: 9.61ms remaining: 24.7ms
14: learn: 0.6229825 total: 10.8ms remaining: 25.2ms
15: learn: 0.6182567 total: 11.5ms remaining: 24.5ms
16: learn: 0.6144702 total: 12.5ms remaining: 24.2ms
17: learn: 0.6102756 total: 13.6ms remaining: 24.2ms
18: learn: 0.6063688 total: 14.1ms remaining: 23ms
19: learn: 0.6025233 total: 15.3ms remaining: 23ms
20: learn: 0.5986269 total: 15.9ms remaining: 22ms
21: learn: 0.5942607 total: 16.2ms remaining: 20.6ms
22: learn: 0.5899548 total: 16.5ms remaining: 19.3ms
23: learn: 0.5862924 total: 17ms remaining: 18.4ms
24: learn: 0.5830002 total: 18ms remaining: 18ms
25: learn: 0.5798113 total: 18.9ms remaining: 17.5ms
26: learn: 0.5764879 total: 19.6ms remaining: 16.7ms
27: learn: 0.5732837 total: 20.3ms remaining: 15.9ms
28: learn: 0.5701657 total: 21.3ms remaining: 15.4ms
29: learn: 0.5673144 total: 22.1ms remaining: 14.7ms
30: learn: 0.5645907 total: 23.3ms remaining: 14.3ms
31: learn: 0.5612450 total: 23.8ms remaining: 13.4ms
32: learn: 0.5579183 total: 24.8ms remaining: 12.8ms
33: learn: 0.5543718 total: 25.5ms remaining: 12ms
34: learn: 0.5513997 total: 26.7ms remaining: 11.4ms
35: learn: 0.5484174 total: 27.8ms remaining: 10.8ms
36: learn: 0.5451814 total: 28.8ms remaining: 10.1ms
37: learn: 0.5421966 total: 29.8ms remaining: 9.4ms
38: learn: 0.5386374 total: 30.4ms remaining: 8.56ms
39: learn: 0.5359418 total: 31ms remaining: 7.75ms
40: learn: 0.5328272 total: 31.9ms remaining: 7.01ms
41: learn: 0.5292787 total: 32.2ms remaining: 6.13ms
42: learn: 0.5261061 total: 32.9ms remaining: 5.36ms
43: learn: 0.5237163 total: 33.8ms remaining: 4.61ms
44: learn: 0.5208345 total: 34.8ms remaining: 3.87ms
45: learn: 0.5184355 total: 35.9ms remaining: 3.12ms
46: learn: 0.5158247 total: 37.2ms remaining: 2.37ms
47: learn: 0.5135288 total: 38.3ms remaining: 1.59ms
48: learn: 0.5104705 total: 39.2ms remaining: 800us
49: learn: 0.5081160 total: 40.2ms remaining: 0us
0: learn: 0.6886959 total: 332us remaining: 16.3ms
1: learn: 0.6838265 total: 720us remaining: 17.3ms
2: learn: 0.6793184 total: 923us remaining: 14.5ms
3: learn: 0.6746587 total: 1.23ms remaining: 14.1ms
4: learn: 0.6698654 total: 1.48ms remaining: 13.3ms
5: learn: 0.6655969 total: 1.72ms remaining: 12.6ms
6: learn: 0.6613624 total: 2.23ms remaining: 13.7ms
7: learn: 0.6574805 total: 3.11ms remaining: 16.3ms
8: learn: 0.6531921 total: 4.36ms remaining: 19.9ms
9: learn: 0.6488752 total: 5.5ms remaining: 22ms
10: learn: 0.6451668 total: 6.36ms remaining: 22.5ms
11: learn: 0.6411035 total: 7.17ms remaining: 22.7ms
12: learn: 0.6370053 total: 7.92ms remaining: 22.6ms
13: learn: 0.6329368 total: 8.63ms remaining: 22.2ms
14: learn: 0.6292349 total: 9.68ms remaining: 22.6ms
15: learn: 0.6251452 total: 10.3ms remaining: 21.8ms
16: learn: 0.6212621 total: 10.7ms remaining: 20.7ms
17: learn: 0.6170321 total: 11.3ms remaining: 20ms
18: learn: 0.6132722 total: 11.6ms remaining: 19ms
19: learn: 0.6099714 total: 12.5ms remaining: 18.8ms
20: learn: 0.6066178 total: 13.8ms remaining: 19.1ms
21: learn: 0.6027816 total: 14.7ms remaining: 18.6ms
22: learn: 0.5990405 total: 14.9ms remaining: 17.5ms
23: learn: 0.5957426 total: 15.7ms remaining: 17ms
24: learn: 0.5922791 total: 16.4ms remaining: 16.4ms
25: learn: 0.5893376 total: 17.4ms remaining: 16.1ms
26: learn: 0.5861754 total: 18.6ms remaining: 15.8ms
27: learn: 0.5829950 total: 19.9ms remaining: 15.6ms
28: learn: 0.5794016 total: 20.9ms remaining: 15.1ms
29: learn: 0.5765496 total: 22.2ms remaining: 14.8ms
30: learn: 0.5736307 total: 23.2ms remaining: 14.2ms
31: learn: 0.5704244 total: 23.9ms remaining: 13.5ms
32: learn: 0.5676356 total: 24.9ms remaining: 12.8ms
33: learn: 0.5646941 total: 25.6ms remaining: 12ms
34: learn: 0.5620194 total: 26.5ms remaining: 11.4ms
35: learn: 0.5600655 total: 27.7ms remaining: 10.8ms
36: learn: 0.5572839 total: 28.9ms remaining: 10.2ms
37: learn: 0.5547866 total: 29.4ms remaining: 9.3ms
38: learn: 0.5521352 total: 29.8ms remaining: 8.42ms
39: learn: 0.5493307 total: 30.7ms remaining: 7.68ms
40: learn: 0.5467472 total: 31.9ms remaining: 7ms
41: learn: 0.5446479 total: 32.7ms remaining: 6.23ms
42: learn: 0.5426825 total: 33.9ms remaining: 5.51ms
43: learn: 0.5399677 total: 34.9ms remaining: 4.76ms
44: learn: 0.5379476 total: 35.7ms remaining: 3.96ms
45: learn: 0.5354392 total: 36.9ms remaining: 3.21ms
46: learn: 0.5325750 total: 37.7ms remaining: 2.4ms
47: learn: 0.5300571 total: 38.6ms remaining: 1.61ms
48: learn: 0.5276968 total: 39.4ms remaining: 804us
49: learn: 0.5250418 total: 40.5ms remaining: 0us
0: learn: 0.6888430 total: 379us remaining: 18.6ms
1: learn: 0.6844696 total: 607us remaining: 14.6ms
2: learn: 0.6801295 total: 1.28ms remaining: 20ms
3: learn: 0.6752420 total: 2.12ms remaining: 24.4ms
4: learn: 0.6701626 total: 2.92ms remaining: 26.2ms
5: learn: 0.6655182 total: 3.62ms remaining: 26.6ms
6: learn: 0.6620344 total: 4.38ms remaining: 26.9ms
7: learn: 0.6584874 total: 4.9ms remaining: 25.7ms
8: learn: 0.6539240 total: 5.67ms remaining: 25.9ms
9: learn: 0.6499828 total: 6.16ms remaining: 24.6ms
10: learn: 0.6455269 total: 6.96ms remaining: 24.7ms
11: learn: 0.6414986 total: 7.59ms remaining: 24ms
12: learn: 0.6374259 total: 7.89ms remaining: 22.5ms
13: learn: 0.6333841 total: 8.64ms remaining: 22.2ms
14: learn: 0.6287552 total: 9.18ms remaining: 21.4ms
15: learn: 0.6255913 total: 9.74ms remaining: 20.7ms
16: learn: 0.6228745 total: 10.2ms remaining: 19.8ms
17: learn: 0.6192158 total: 10.9ms remaining: 19.3ms
18: learn: 0.6151053 total: 11.5ms remaining: 18.7ms
19: learn: 0.6117255 total: 12.2ms remaining: 18.3ms
20: learn: 0.6077871 total: 13ms remaining: 17.9ms
21: learn: 0.6036369 total: 13.5ms remaining: 17.1ms
22: learn: 0.5997223 total: 14ms remaining: 16.5ms
23: learn: 0.5957430 total: 14.2ms remaining: 15.4ms
24: learn: 0.5924717 total: 14.5ms remaining: 14.5ms
25: learn: 0.5891835 total: 15ms remaining: 13.8ms
26: learn: 0.5859911 total: 15.6ms remaining: 13.3ms
27: learn: 0.5830599 total: 16.3ms remaining: 12.8ms
28: learn: 0.5804551 total: 16.9ms remaining: 12.2ms
29: learn: 0.5777595 total: 17.5ms remaining: 11.7ms
30: learn: 0.5746008 total: 17.9ms remaining: 10.9ms
31: learn: 0.5713796 total: 18.4ms remaining: 10.4ms
32: learn: 0.5679259 total: 18.9ms remaining: 9.75ms
33: learn: 0.5650754 total: 19.5ms remaining: 9.16ms
34: learn: 0.5622872 total: 19.9ms remaining: 8.54ms
35: learn: 0.5591263 total: 20.5ms remaining: 7.96ms
36: learn: 0.5563465 total: 21ms remaining: 7.38ms
37: learn: 0.5529493 total: 21.5ms remaining: 6.78ms
38: learn: 0.5501690 total: 22ms remaining: 6.2ms
39: learn: 0.5469190 total: 22.5ms remaining: 5.61ms
40: learn: 0.5432410 total: 22.8ms remaining: 5.01ms
41: learn: 0.5401389 total: 23.4ms remaining: 4.45ms
42: learn: 0.5378797 total: 24ms remaining: 3.9ms
43: learn: 0.5350410 total: 24.5ms remaining: 3.35ms
44: learn: 0.5326489 total: 24.9ms remaining: 2.76ms
45: learn: 0.5300776 total: 25.4ms remaining: 2.21ms
46: learn: 0.5270868 total: 25.8ms remaining: 1.65ms
47: learn: 0.5249808 total: 26.1ms remaining: 1.09ms
48: learn: 0.5227876 total: 26.7ms remaining: 545us
49: learn: 0.5200878 total: 27.3ms remaining: 0us
0: learn: 0.6886167 total: 302us remaining: 14.8ms
1: learn: 0.6839296 total: 546us remaining: 13.1ms
2: learn: 0.6790130 total: 747us remaining: 11.7ms
3: learn: 0.6745981 total: 960us remaining: 11ms
4: learn: 0.6700984 total: 1.31ms remaining: 11.8ms
5: learn: 0.6652168 total: 1.86ms remaining: 13.6ms
6: learn: 0.6606527 total: 2.37ms remaining: 14.6ms
7: learn: 0.6566447 total: 3.46ms remaining: 18.2ms
8: learn: 0.6519140 total: 4.07ms remaining: 18.5ms
9: learn: 0.6475038 total: 4.4ms remaining: 17.6ms
10: learn: 0.6444133 total: 4.83ms remaining: 17.1ms
11: learn: 0.6400321 total: 5.6ms remaining: 17.7ms
12: learn: 0.6361821 total: 6.36ms remaining: 18.1ms
13: learn: 0.6324678 total: 7.19ms remaining: 18.5ms
14: learn: 0.6284970 total: 8.13ms remaining: 19ms
15: learn: 0.6241548 total: 8.82ms remaining: 18.7ms
16: learn: 0.6212588 total: 9.53ms remaining: 18.5ms
17: learn: 0.6181208 total: 11.3ms remaining: 20.1ms
18: learn: 0.6149727 total: 12.2ms remaining: 20ms
19: learn: 0.6112675 total: 13.2ms remaining: 19.7ms
20: learn: 0.6076084 total: 13.9ms remaining: 19.2ms
21: learn: 0.6038614 total: 14.9ms remaining: 19ms
22: learn: 0.5996882 total: 15.6ms remaining: 18.3ms
23: learn: 0.5962705 total: 16.4ms remaining: 17.7ms
24: learn: 0.5923546 total: 16.5ms remaining: 16.5ms
25: learn: 0.5894756 total: 17ms remaining: 15.7ms
26: learn: 0.5864478 total: 18.1ms remaining: 15.4ms
27: learn: 0.5833874 total: 19.1ms remaining: 15ms
28: learn: 0.5804593 total: 19.9ms remaining: 14.4ms
29: learn: 0.5777455 total: 20.6ms remaining: 13.7ms
30: learn: 0.5751136 total: 20.9ms remaining: 12.8ms
31: learn: 0.5718058 total: 21.1ms remaining: 11.8ms
32: learn: 0.5689155 total: 21.7ms remaining: 11.2ms
33: learn: 0.5658437 total: 22ms remaining: 10.4ms
34: learn: 0.5625370 total: 22.5ms remaining: 9.64ms
35: learn: 0.5600906 total: 23.2ms remaining: 9.01ms
36: learn: 0.5574840 total: 23.4ms remaining: 8.23ms
37: learn: 0.5548446 total: 24ms remaining: 7.58ms
38: learn: 0.5521241 total: 24.8ms remaining: 7ms
39: learn: 0.5493534 total: 25.5ms remaining: 6.38ms
40: learn: 0.5464643 total: 26.3ms remaining: 5.77ms
41: learn: 0.5439402 total: 27.3ms remaining: 5.2ms
42: learn: 0.5407503 total: 27.9ms remaining: 4.54ms
43: learn: 0.5383021 total: 28.7ms remaining: 3.92ms
44: learn: 0.5357105 total: 29.7ms remaining: 3.3ms
45: learn: 0.5332736 total: 30.4ms remaining: 2.65ms
46: learn: 0.5308820 total: 31.1ms remaining: 1.99ms
47: learn: 0.5280463 total: 32.1ms remaining: 1.34ms
48: learn: 0.5251939 total: 33.1ms remaining: 674us
49: learn: 0.5226601 total: 34ms remaining: 0us
0: learn: 0.6885339 total: 378us remaining: 18.5ms
1: learn: 0.6834227 total: 651us remaining: 15.6ms
2: learn: 0.6786232 total: 834us remaining: 13.1ms
3: learn: 0.6739003 total: 1.12ms remaining: 12.9ms
4: learn: 0.6693001 total: 1.37ms remaining: 12.4ms
5: learn: 0.6644451 total: 1.66ms remaining: 12.2ms
6: learn: 0.6598747 total: 1.91ms remaining: 11.7ms
7: learn: 0.6558602 total: 2.16ms remaining: 11.3ms
8: learn: 0.6512912 total: 2.36ms remaining: 10.7ms
9: learn: 0.6467205 total: 2.64ms remaining: 10.6ms
10: learn: 0.6428079 total: 2.88ms remaining: 10.2ms
11: learn: 0.6384207 total: 3.11ms remaining: 9.86ms
12: learn: 0.6344698 total: 3.37ms remaining: 9.6ms
13: learn: 0.6304828 total: 3.68ms remaining: 9.46ms
14: learn: 0.6266173 total: 4ms remaining: 9.35ms
15: learn: 0.6223299 total: 4.21ms remaining: 8.94ms
16: learn: 0.6186546 total: 4.48ms remaining: 8.69ms
17: learn: 0.6147792 total: 4.71ms remaining: 8.37ms
18: learn: 0.6110091 total: 4.84ms remaining: 7.89ms
19: learn: 0.6073387 total: 5.24ms remaining: 7.87ms
20: learn: 0.6038272 total: 5.62ms remaining: 7.76ms
21: learn: 0.6001991 total: 5.93ms remaining: 7.55ms
22: learn: 0.5964357 total: 6.2ms remaining: 7.28ms
23: learn: 0.5929511 total: 6.4ms remaining: 6.93ms
24: learn: 0.5893719 total: 6.72ms remaining: 6.72ms
25: learn: 0.5864672 total: 7.05ms remaining: 6.51ms
26: learn: 0.5837256 total: 7.4ms remaining: 6.3ms
27: learn: 0.5805511 total: 7.72ms remaining: 6.06ms
28: learn: 0.5772513 total: 7.96ms remaining: 5.76ms
29: learn: 0.5745393 total: 8.19ms remaining: 5.46ms
30: learn: 0.5717751 total: 8.54ms remaining: 5.24ms
31: learn: 0.5685652 total: 8.74ms remaining: 4.91ms
32: learn: 0.5654609 total: 9.09ms remaining: 4.68ms
33: learn: 0.5622746 total: 9.39ms remaining: 4.42ms
34: learn: 0.5588724 total: 9.74ms remaining: 4.17ms
35: learn: 0.5555821 total: 10ms remaining: 3.9ms
36: learn: 0.5527355 total: 10.4ms remaining: 3.64ms
37: learn: 0.5500608 total: 10.7ms remaining: 3.37ms
38: learn: 0.5467145 total: 11ms remaining: 3.09ms
39: learn: 0.5441104 total: 11.3ms remaining: 2.82ms
40: learn: 0.5417590 total: 11.7ms remaining: 2.56ms
41: learn: 0.5389472 total: 11.9ms remaining: 2.26ms
42: learn: 0.5365060 total: 12.2ms remaining: 1.98ms
43: learn: 0.5333535 total: 12.5ms remaining: 1.71ms
44: learn: 0.5309882 total: 12.9ms remaining: 1.43ms
45: learn: 0.5286618 total: 13.1ms remaining: 1.14ms
46: learn: 0.5259226 total: 13.4ms remaining: 854us
47: learn: 0.5235263 total: 13.7ms remaining: 570us
48: learn: 0.5210188 total: 14ms remaining: 285us
49: learn: 0.5185356 total: 14.3ms remaining: 0us
0: learn: 0.6883550 total: 442us remaining: 21.7ms
1: learn: 0.6836109 total: 754us remaining: 18.1ms
2: learn: 0.6787323 total: 1.02ms remaining: 16ms
3: learn: 0.6738335 total: 1.53ms remaining: 17.6ms
4: learn: 0.6693032 total: 1.8ms remaining: 16.2ms
5: learn: 0.6644562 total: 2.25ms remaining: 16.5ms
6: learn: 0.6602992 total: 2.55ms remaining: 15.6ms
7: learn: 0.6565796 total: 2.94ms remaining: 15.4ms
8: learn: 0.6517652 total: 3.27ms remaining: 14.9ms
9: learn: 0.6474219 total: 3.71ms remaining: 14.8ms
10: learn: 0.6444322 total: 4.08ms remaining: 14.5ms
11: learn: 0.6397700 total: 4.42ms remaining: 14ms
12: learn: 0.6359370 total: 4.8ms remaining: 13.7ms
13: learn: 0.6324723 total: 5.28ms remaining: 13.6ms
14: learn: 0.6285007 total: 5.66ms remaining: 13.2ms
15: learn: 0.6241371 total: 5.98ms remaining: 12.7ms
16: learn: 0.6205886 total: 6.29ms remaining: 12.2ms
17: learn: 0.6165610 total: 6.7ms remaining: 11.9ms
18: learn: 0.6127567 total: 6.87ms remaining: 11.2ms
19: learn: 0.6088093 total: 7.29ms remaining: 10.9ms
20: learn: 0.6051968 total: 7.68ms remaining: 10.6ms
21: learn: 0.6014306 total: 8.01ms remaining: 10.2ms
22: learn: 0.5974358 total: 8.32ms remaining: 9.77ms
23: learn: 0.5939927 total: 8.55ms remaining: 9.27ms
24: learn: 0.5909375 total: 8.99ms remaining: 8.99ms
25: learn: 0.5873963 total: 9.39ms remaining: 8.67ms
26: learn: 0.5844064 total: 9.73ms remaining: 8.29ms
27: learn: 0.5813962 total: 10.1ms remaining: 7.96ms
28: learn: 0.5785490 total: 10.5ms remaining: 7.59ms
29: learn: 0.5758908 total: 10.8ms remaining: 7.2ms
30: learn: 0.5732910 total: 11.2ms remaining: 6.85ms
31: learn: 0.5700190 total: 11.4ms remaining: 6.42ms
32: learn: 0.5669492 total: 11.8ms remaining: 6.07ms
33: learn: 0.5635924 total: 12.1ms remaining: 5.71ms
34: learn: 0.5600908 total: 12.4ms remaining: 5.33ms
35: learn: 0.5572768 total: 12.8ms remaining: 4.98ms
36: learn: 0.5542501 total: 13.2ms remaining: 4.65ms
37: learn: 0.5515929 total: 13.6ms remaining: 4.29ms
38: learn: 0.5480918 total: 13.9ms remaining: 3.93ms
39: learn: 0.5449589 total: 14.3ms remaining: 3.58ms
40: learn: 0.5420133 total: 14.7ms remaining: 3.22ms
41: learn: 0.5389790 total: 15ms remaining: 2.85ms
42: learn: 0.5360152 total: 15.3ms remaining: 2.49ms
43: learn: 0.5336202 total: 15.6ms remaining: 2.13ms
44: learn: 0.5308643 total: 16ms remaining: 1.77ms
45: learn: 0.5285430 total: 16.2ms remaining: 1.41ms
46: learn: 0.5260419 total: 16.5ms remaining: 1.05ms
47: learn: 0.5236528 total: 16.8ms remaining: 699us
48: learn: 0.5206597 total: 17.1ms remaining: 348us
49: learn: 0.5182717 total: 17.4ms remaining: 0us
0: learn: 0.6889933 total: 295us remaining: 14.5ms
1: learn: 0.6836164 total: 515us remaining: 12.4ms
2: learn: 0.6784902 total: 676us remaining: 10.6ms
3: learn: 0.6738344 total: 874us remaining: 10.1ms
4: learn: 0.6690759 total: 1.04ms remaining: 9.41ms
5: learn: 0.6641179 total: 1.24ms remaining: 9.08ms
6: learn: 0.6609183 total: 1.47ms remaining: 9.02ms
7: learn: 0.6564302 total: 1.64ms remaining: 8.61ms
8: learn: 0.6518274 total: 1.86ms remaining: 8.46ms
9: learn: 0.6472279 total: 2.04ms remaining: 8.17ms
10: learn: 0.6439145 total: 2.19ms remaining: 7.77ms
11: learn: 0.6400426 total: 2.63ms remaining: 8.34ms
12: learn: 0.6356838 total: 2.94ms remaining: 8.37ms
13: learn: 0.6314445 total: 3.2ms remaining: 8.22ms
14: learn: 0.6280712 total: 3.4ms remaining: 7.94ms
15: learn: 0.6240877 total: 3.66ms remaining: 7.78ms
16: learn: 0.6200849 total: 4.05ms remaining: 7.87ms
17: learn: 0.6168229 total: 4.41ms remaining: 7.84ms
18: learn: 0.6129056 total: 4.85ms remaining: 7.92ms
19: learn: 0.6101052 total: 5.38ms remaining: 8.08ms
20: learn: 0.6064010 total: 5.6ms remaining: 7.74ms
21: learn: 0.6022683 total: 5.82ms remaining: 7.4ms
22: learn: 0.5980306 total: 6.21ms remaining: 7.29ms
23: learn: 0.5949120 total: 6.35ms remaining: 6.88ms
24: learn: 0.5912501 total: 6.58ms remaining: 6.58ms
25: learn: 0.5879653 total: 7.51ms remaining: 6.94ms
26: learn: 0.5845873 total: 8.3ms remaining: 7.07ms
27: learn: 0.5814684 total: 8.56ms remaining: 6.72ms
28: learn: 0.5788440 total: 8.76ms remaining: 6.34ms
29: learn: 0.5759054 total: 8.96ms remaining: 5.97ms
30: learn: 0.5723048 total: 9.16ms remaining: 5.61ms
31: learn: 0.5690872 total: 9.36ms remaining: 5.26ms
32: learn: 0.5660702 total: 9.55ms remaining: 4.92ms
33: learn: 0.5628655 total: 9.73ms remaining: 4.58ms
34: learn: 0.5599171 total: 9.93ms remaining: 4.25ms
35: learn: 0.5573603 total: 10.1ms remaining: 3.93ms
36: learn: 0.5551515 total: 10.3ms remaining: 3.6ms
37: learn: 0.5521042 total: 10.4ms remaining: 3.3ms
38: learn: 0.5486314 total: 10.7ms remaining: 3ms
39: learn: 0.5453812 total: 10.9ms remaining: 2.72ms
40: learn: 0.5422646 total: 11.1ms remaining: 2.44ms
41: learn: 0.5396314 total: 11.4ms remaining: 2.17ms
42: learn: 0.5369313 total: 11.9ms remaining: 1.93ms
43: learn: 0.5341681 total: 12.5ms remaining: 1.7ms
44: learn: 0.5311134 total: 12.7ms remaining: 1.42ms
45: learn: 0.5279754 total: 13ms remaining: 1.13ms
46: learn: 0.5252587 total: 13.3ms remaining: 850us
47: learn: 0.5224699 total: 13.6ms remaining: 567us
48: learn: 0.5196144 total: 13.8ms remaining: 281us
49: learn: 0.5168594 total: 14ms remaining: 0us
0: learn: 0.6890031 total: 330us remaining: 16.2ms
1: learn: 0.6839941 total: 556us remaining: 13.3ms
2: learn: 0.6790950 total: 745us remaining: 11.7ms
3: learn: 0.6741892 total: 1.12ms remaining: 12.9ms
4: learn: 0.6699145 total: 1.38ms remaining: 12.4ms
5: learn: 0.6651736 total: 1.67ms remaining: 12.3ms
6: learn: 0.6605127 total: 1.94ms remaining: 11.9ms
7: learn: 0.6563352 total: 2.23ms remaining: 11.7ms
8: learn: 0.6525859 total: 2.38ms remaining: 10.8ms
9: learn: 0.6483100 total: 2.67ms remaining: 10.7ms
10: learn: 0.6443013 total: 2.92ms remaining: 10.4ms
11: learn: 0.6404553 total: 3.24ms remaining: 10.3ms
12: learn: 0.6363750 total: 3.55ms remaining: 10.1ms
13: learn: 0.6320174 total: 3.86ms remaining: 9.93ms
14: learn: 0.6288432 total: 4.09ms remaining: 9.54ms
15: learn: 0.6252655 total: 4.4ms remaining: 9.34ms
16: learn: 0.6214597 total: 4.76ms remaining: 9.23ms
17: learn: 0.6174034 total: 5.01ms remaining: 8.91ms
18: learn: 0.6143807 total: 5.3ms remaining: 8.65ms
19: learn: 0.6108119 total: 5.63ms remaining: 8.44ms
20: learn: 0.6071982 total: 5.92ms remaining: 8.17ms
21: learn: 0.6035162 total: 6.14ms remaining: 7.82ms
22: learn: 0.5995561 total: 6.32ms remaining: 7.42ms
23: learn: 0.5967885 total: 6.67ms remaining: 7.23ms
24: learn: 0.5928977 total: 6.86ms remaining: 6.86ms
25: learn: 0.5896768 total: 7.2ms remaining: 6.64ms
26: learn: 0.5864794 total: 7.45ms remaining: 6.34ms
27: learn: 0.5831156 total: 7.75ms remaining: 6.09ms
28: learn: 0.5804258 total: 8.14ms remaining: 5.89ms
29: learn: 0.5780639 total: 8.3ms remaining: 5.54ms
30: learn: 0.5757587 total: 8.62ms remaining: 5.28ms
31: learn: 0.5725405 total: 8.73ms remaining: 4.91ms
32: learn: 0.5693987 total: 9.07ms remaining: 4.67ms
33: learn: 0.5660367 total: 9.3ms remaining: 4.38ms
34: learn: 0.5626815 total: 9.6ms remaining: 4.12ms
35: learn: 0.5601347 total: 9.81ms remaining: 3.82ms
36: learn: 0.5570503 total: 10.1ms remaining: 3.53ms
37: learn: 0.5542878 total: 10.3ms remaining: 3.24ms
38: learn: 0.5511386 total: 10.5ms remaining: 2.96ms
39: learn: 0.5481846 total: 10.7ms remaining: 2.69ms
40: learn: 0.5450162 total: 10.9ms remaining: 2.4ms
41: learn: 0.5426760 total: 11.1ms remaining: 2.12ms
42: learn: 0.5395370 total: 11.3ms remaining: 1.84ms
43: learn: 0.5373680 total: 11.5ms remaining: 1.57ms
44: learn: 0.5345015 total: 11.8ms remaining: 1.31ms
45: learn: 0.5318743 total: 12.1ms remaining: 1.05ms
46: learn: 0.5292160 total: 12.2ms remaining: 781us
47: learn: 0.5267250 total: 12.5ms remaining: 519us
48: learn: 0.5243110 total: 12.8ms remaining: 262us
49: learn: 0.5214591 total: 13.2ms remaining: 0us
0: learn: 0.6884444 total: 449us remaining: 22ms
1: learn: 0.6833777 total: 726us remaining: 17.4ms
2: learn: 0.6789418 total: 1.01ms remaining: 15.8ms
3: learn: 0.6740883 total: 1.36ms remaining: 15.7ms
4: learn: 0.6695991 total: 1.55ms remaining: 13.9ms
5: learn: 0.6649560 total: 1.94ms remaining: 14.3ms
6: learn: 0.6607099 total: 2.4ms remaining: 14.7ms
7: learn: 0.6568861 total: 2.82ms remaining: 14.8ms
8: learn: 0.6526495 total: 3.03ms remaining: 13.8ms
9: learn: 0.6486747 total: 3.25ms remaining: 13ms
10: learn: 0.6446790 total: 3.61ms remaining: 12.8ms
11: learn: 0.6408419 total: 3.88ms remaining: 12.3ms
12: learn: 0.6366769 total: 4.19ms remaining: 11.9ms
13: learn: 0.6327758 total: 4.74ms remaining: 12.2ms
14: learn: 0.6289897 total: 5.24ms remaining: 12.2ms
15: learn: 0.6254446 total: 5.58ms remaining: 11.9ms
16: learn: 0.6214642 total: 6.02ms remaining: 11.7ms
17: learn: 0.6179222 total: 6.47ms remaining: 11.5ms
18: learn: 0.6145156 total: 6.98ms remaining: 11.4ms
19: learn: 0.6111632 total: 7.15ms remaining: 10.7ms
20: learn: 0.6075448 total: 7.47ms remaining: 10.3ms
21: learn: 0.6041593 total: 7.89ms remaining: 10ms
22: learn: 0.6005087 total: 8.37ms remaining: 9.82ms
23: learn: 0.5967117 total: 8.76ms remaining: 9.49ms
24: learn: 0.5937134 total: 9.18ms remaining: 9.18ms
25: learn: 0.5902819 total: 9.37ms remaining: 8.65ms
26: learn: 0.5876279 total: 9.75ms remaining: 8.31ms
27: learn: 0.5847773 total: 10.2ms remaining: 8ms
28: learn: 0.5817003 total: 10.8ms remaining: 7.8ms
29: learn: 0.5787036 total: 11.2ms remaining: 7.49ms
30: learn: 0.5760609 total: 11.6ms remaining: 7.08ms
31: learn: 0.5734735 total: 12.1ms remaining: 6.79ms
32: learn: 0.5707002 total: 12.3ms remaining: 6.33ms
33: learn: 0.5675329 total: 12.6ms remaining: 5.94ms
34: learn: 0.5643493 total: 12.9ms remaining: 5.51ms
35: learn: 0.5610712 total: 13.4ms remaining: 5.21ms
36: learn: 0.5582207 total: 13.8ms remaining: 4.84ms
37: learn: 0.5559611 total: 14.1ms remaining: 4.46ms
38: learn: 0.5537050 total: 14.8ms remaining: 4.17ms
39: learn: 0.5503936 total: 15.2ms remaining: 3.79ms
40: learn: 0.5478983 total: 15.4ms remaining: 3.39ms
41: learn: 0.5451844 total: 15.8ms remaining: 3.01ms
42: learn: 0.5418663 total: 16.2ms remaining: 2.63ms
43: learn: 0.5390675 total: 16.8ms remaining: 2.29ms
44: learn: 0.5366144 total: 17.1ms remaining: 1.9ms
45: learn: 0.5341339 total: 17.4ms remaining: 1.51ms
46: learn: 0.5320371 total: 17.6ms remaining: 1.12ms
47: learn: 0.5297100 total: 17.9ms remaining: 746us
48: learn: 0.5275120 total: 18.1ms remaining: 370us
49: learn: 0.5249431 total: 18.3ms remaining: 0us
0: learn: 0.6885566 total: 297us remaining: 14.6ms
1: learn: 0.6836071 total: 529us remaining: 12.7ms
2: learn: 0.6782460 total: 1.2ms remaining: 18.8ms
3: learn: 0.6743132 total: 2.21ms remaining: 25.4ms
4: learn: 0.6702237 total: 3.48ms remaining: 31.3ms
5: learn: 0.6660752 total: 4.49ms remaining: 32.9ms
6: learn: 0.6616702 total: 5.68ms remaining: 34.9ms
7: learn: 0.6566263 total: 6.72ms remaining: 35.3ms
8: learn: 0.6518664 total: 7.35ms remaining: 33.5ms
9: learn: 0.6490527 total: 7.52ms remaining: 30.1ms
10: learn: 0.6444509 total: 7.72ms remaining: 27.4ms
11: learn: 0.6403671 total: 8.76ms remaining: 27.7ms
12: learn: 0.6363304 total: 9.54ms remaining: 27.2ms
13: learn: 0.6327678 total: 10.5ms remaining: 27.1ms
14: learn: 0.6282780 total: 11.1ms remaining: 25.8ms
15: learn: 0.6244872 total: 11.7ms remaining: 24.9ms
16: learn: 0.6203105 total: 12.6ms remaining: 24.4ms
17: learn: 0.6165432 total: 13.1ms remaining: 23.3ms
18: learn: 0.6128502 total: 14.2ms remaining: 23.1ms
19: learn: 0.6090656 total: 15ms remaining: 22.5ms
20: learn: 0.6050572 total: 15.7ms remaining: 21.7ms
21: learn: 0.6007740 total: 16.7ms remaining: 21.2ms
22: learn: 0.5970249 total: 17.4ms remaining: 20.4ms
23: learn: 0.5935481 total: 18ms remaining: 19.5ms
24: learn: 0.5906288 total: 18.4ms remaining: 18.4ms
25: learn: 0.5877041 total: 18.7ms remaining: 17.2ms
26: learn: 0.5839478 total: 19.6ms remaining: 16.7ms
27: learn: 0.5811081 total: 20.5ms remaining: 16.1ms
28: learn: 0.5773239 total: 21.5ms remaining: 15.6ms
29: learn: 0.5742893 total: 21.8ms remaining: 14.5ms
30: learn: 0.5706335 total: 22ms remaining: 13.5ms
31: learn: 0.5674696 total: 22.3ms remaining: 12.5ms
32: learn: 0.5645132 total: 22.6ms remaining: 11.6ms
33: learn: 0.5608553 total: 23.4ms remaining: 11ms
34: learn: 0.5579249 total: 24.9ms remaining: 10.7ms
35: learn: 0.5548740 total: 25.9ms remaining: 10.1ms
36: learn: 0.5521033 total: 26.2ms remaining: 9.2ms
37: learn: 0.5487798 total: 27ms remaining: 8.54ms
38: learn: 0.5458292 total: 27.7ms remaining: 7.81ms
39: learn: 0.5434128 total: 27.9ms remaining: 6.99ms
40: learn: 0.5398872 total: 28.6ms remaining: 6.27ms
41: learn: 0.5372327 total: 29.2ms remaining: 5.57ms
42: learn: 0.5348076 total: 29.5ms remaining: 4.8ms
43: learn: 0.5319729 total: 30.4ms remaining: 4.14ms
44: learn: 0.5296587 total: 31ms remaining: 3.44ms
45: learn: 0.5271298 total: 31.9ms remaining: 2.78ms
46: learn: 0.5243558 total: 32.9ms remaining: 2.1ms
47: learn: 0.5216964 total: 33.7ms remaining: 1.4ms
48: learn: 0.5191863 total: 34.6ms remaining: 706us
49: learn: 0.5165567 total: 35.6ms remaining: 0us
0: learn: 0.6896203 total: 397us remaining: 19.5ms
1: learn: 0.6850714 total: 661us remaining: 15.9ms
2: learn: 0.6803862 total: 872us remaining: 13.7ms
3: learn: 0.6758516 total: 1.56ms remaining: 18ms
4: learn: 0.6716479 total: 2.17ms remaining: 19.6ms
5: learn: 0.6663871 total: 3.2ms remaining: 23.4ms
6: learn: 0.6619637 total: 3.92ms remaining: 24.1ms
7: learn: 0.6576289 total: 4.09ms remaining: 21.5ms
8: learn: 0.6535165 total: 4.29ms remaining: 19.5ms
9: learn: 0.6494451 total: 4.75ms remaining: 19ms
10: learn: 0.6454696 total: 4.93ms remaining: 17.5ms
11: learn: 0.6411167 total: 5.13ms remaining: 16.2ms
12: learn: 0.6375680 total: 5.92ms remaining: 16.9ms
13: learn: 0.6336478 total: 6.97ms remaining: 17.9ms
14: learn: 0.6296995 total: 7.94ms remaining: 18.5ms
15: learn: 0.6252287 total: 8.58ms remaining: 18.2ms
16: learn: 0.6216951 total: 9.28ms remaining: 18ms
17: learn: 0.6177813 total: 10.2ms remaining: 18.1ms
18: learn: 0.6140884 total: 10.6ms remaining: 17.4ms
19: learn: 0.6108468 total: 11.5ms remaining: 17.3ms
20: learn: 0.6072031 total: 12.4ms remaining: 17.2ms
21: learn: 0.6030750 total: 13.3ms remaining: 16.9ms
22: learn: 0.5992488 total: 13.8ms remaining: 16.2ms
23: learn: 0.5955617 total: 14.3ms remaining: 15.5ms
24: learn: 0.5922429 total: 15ms remaining: 15ms
25: learn: 0.5884927 total: 15.9ms remaining: 14.6ms
26: learn: 0.5856491 total: 16.5ms remaining: 14.1ms
27: learn: 0.5820865 total: 17.3ms remaining: 13.6ms
28: learn: 0.5795715 total: 18.1ms remaining: 13.1ms
29: learn: 0.5761366 total: 18.8ms remaining: 12.5ms
30: learn: 0.5728400 total: 19.4ms remaining: 11.9ms
31: learn: 0.5697248 total: 20ms remaining: 11.3ms
32: learn: 0.5668872 total: 20.9ms remaining: 10.8ms
33: learn: 0.5633521 total: 21.8ms remaining: 10.2ms
34: learn: 0.5601400 total: 22.2ms remaining: 9.53ms
35: learn: 0.5574012 total: 22.6ms remaining: 8.8ms
36: learn: 0.5543751 total: 23.3ms remaining: 8.2ms
37: learn: 0.5520800 total: 24.3ms remaining: 7.66ms
38: learn: 0.5495015 total: 24.9ms remaining: 7.02ms
39: learn: 0.5462074 total: 25.6ms remaining: 6.41ms
40: learn: 0.5434947 total: 26.2ms remaining: 5.74ms
41: learn: 0.5412224 total: 26.9ms remaining: 5.12ms
42: learn: 0.5385021 total: 27.6ms remaining: 4.5ms
43: learn: 0.5355544 total: 28.3ms remaining: 3.86ms
44: learn: 0.5328038 total: 28.9ms remaining: 3.21ms
45: learn: 0.5305187 total: 29.7ms remaining: 2.59ms
46: learn: 0.5281554 total: 30.3ms remaining: 1.93ms
47: learn: 0.5251024 total: 31ms remaining: 1.29ms
48: learn: 0.5225982 total: 31.5ms remaining: 642us
49: learn: 0.5203373 total: 32ms remaining: 0us
0: learn: 0.6894871 total: 424us remaining: 20.8ms
1: learn: 0.6842252 total: 654us remaining: 15.7ms
2: learn: 0.6798605 total: 936us remaining: 14.7ms
3: learn: 0.6746510 total: 1.17ms remaining: 13.4ms
4: learn: 0.6695916 total: 1.4ms remaining: 12.6ms
5: learn: 0.6646934 total: 1.93ms remaining: 14.2ms
6: learn: 0.6608368 total: 2.64ms remaining: 16.2ms
7: learn: 0.6560198 total: 3.14ms remaining: 16.5ms
8: learn: 0.6514894 total: 3.76ms remaining: 17.1ms
9: learn: 0.6484876 total: 4.29ms remaining: 17.2ms
10: learn: 0.6438394 total: 4.78ms remaining: 17ms
11: learn: 0.6400430 total: 5.45ms remaining: 17.3ms
12: learn: 0.6364403 total: 6.04ms remaining: 17.2ms
13: learn: 0.6328070 total: 6.59ms remaining: 16.9ms
14: learn: 0.6285253 total: 7ms remaining: 16.3ms
15: learn: 0.6244703 total: 7.48ms remaining: 15.9ms
16: learn: 0.6203785 total: 8.09ms remaining: 15.7ms
17: learn: 0.6170779 total: 8.35ms remaining: 14.8ms
18: learn: 0.6134879 total: 8.89ms remaining: 14.5ms
19: learn: 0.6098244 total: 9.48ms remaining: 14.2ms
20: learn: 0.6058554 total: 10ms remaining: 13.8ms
21: learn: 0.6020761 total: 10.5ms remaining: 13.4ms
22: learn: 0.5989954 total: 10.9ms remaining: 12.8ms
23: learn: 0.5950739 total: 11.4ms remaining: 12.3ms
24: learn: 0.5921883 total: 11.9ms remaining: 11.9ms
25: learn: 0.5889009 total: 12.5ms remaining: 11.5ms
26: learn: 0.5860139 total: 13ms remaining: 11ms
27: learn: 0.5833418 total: 13.5ms remaining: 10.6ms
28: learn: 0.5810032 total: 13.9ms remaining: 10.1ms
29: learn: 0.5784543 total: 14.5ms remaining: 9.68ms
30: learn: 0.5750472 total: 14.8ms remaining: 9.08ms
31: learn: 0.5717814 total: 15.3ms remaining: 8.62ms
32: learn: 0.5686437 total: 15.7ms remaining: 8.09ms
33: learn: 0.5653167 total: 16.1ms remaining: 7.6ms
34: learn: 0.5630180 total: 16.7ms remaining: 7.16ms
35: learn: 0.5603828 total: 17.2ms remaining: 6.7ms
36: learn: 0.5575529 total: 17.7ms remaining: 6.24ms
37: learn: 0.5553556 total: 18.2ms remaining: 5.74ms
38: learn: 0.5525472 total: 18.7ms remaining: 5.28ms
39: learn: 0.5495731 total: 19.2ms remaining: 4.81ms
40: learn: 0.5473859 total: 19.8ms remaining: 4.34ms
41: learn: 0.5444052 total: 20.5ms remaining: 3.9ms
42: learn: 0.5422100 total: 20.9ms remaining: 3.4ms
43: learn: 0.5393793 total: 21.6ms remaining: 2.94ms
44: learn: 0.5367392 total: 22.2ms remaining: 2.47ms
45: learn: 0.5339108 total: 22.8ms remaining: 1.98ms
46: learn: 0.5312177 total: 23.3ms remaining: 1.49ms
47: learn: 0.5287355 total: 24ms remaining: 1ms
48: learn: 0.5265024 total: 24.7ms remaining: 503us
49: learn: 0.5235980 total: 25.4ms remaining: 0us
0: learn: 0.6895315 total: 322us remaining: 15.8ms
1: learn: 0.6844180 total: 543us remaining: 13.1ms
2: learn: 0.6797461 total: 754us remaining: 11.8ms
3: learn: 0.6750861 total: 1.18ms remaining: 13.6ms
4: learn: 0.6707944 total: 1.84ms remaining: 16.5ms
5: learn: 0.6661817 total: 2.6ms remaining: 19ms
6: learn: 0.6616283 total: 3.19ms remaining: 19.6ms
7: learn: 0.6576498 total: 4.08ms remaining: 21.4ms
8: learn: 0.6531931 total: 4.74ms remaining: 21.6ms
9: learn: 0.6487036 total: 5.64ms remaining: 22.6ms
10: learn: 0.6449099 total: 6.18ms remaining: 21.9ms
11: learn: 0.6406385 total: 6.78ms remaining: 21.5ms
12: learn: 0.6364890 total: 7.73ms remaining: 22ms
13: learn: 0.6327426 total: 8.67ms remaining: 22.3ms
14: learn: 0.6293871 total: 9.55ms remaining: 22.3ms
15: learn: 0.6251434 total: 10.1ms remaining: 21.4ms
16: learn: 0.6214766 total: 10.7ms remaining: 20.8ms
17: learn: 0.6178709 total: 11.5ms remaining: 20.4ms
18: learn: 0.6143560 total: 11.9ms remaining: 19.5ms
19: learn: 0.6108690 total: 12.8ms remaining: 19.2ms
20: learn: 0.6073974 total: 13.5ms remaining: 18.7ms
21: learn: 0.6036627 total: 14.2ms remaining: 18.1ms
22: learn: 0.5997537 total: 14.9ms remaining: 17.4ms
23: learn: 0.5964076 total: 15.4ms remaining: 16.7ms
24: learn: 0.5934630 total: 16.1ms remaining: 16.1ms
25: learn: 0.5903772 total: 16.9ms remaining: 15.6ms
26: learn: 0.5873292 total: 17.6ms remaining: 15ms
27: learn: 0.5843486 total: 18.3ms remaining: 14.4ms
28: learn: 0.5815989 total: 19ms remaining: 13.7ms
29: learn: 0.5790148 total: 19.5ms remaining: 13ms
30: learn: 0.5764376 total: 20.1ms remaining: 12.3ms
31: learn: 0.5734280 total: 20.5ms remaining: 11.5ms
32: learn: 0.5703529 total: 21.2ms remaining: 10.9ms
33: learn: 0.5671999 total: 21.8ms remaining: 10.3ms
34: learn: 0.5637969 total: 22.4ms remaining: 9.6ms
35: learn: 0.5609537 total: 23ms remaining: 8.95ms
36: learn: 0.5583443 total: 23.7ms remaining: 8.32ms
37: learn: 0.5554906 total: 24.2ms remaining: 7.65ms
38: learn: 0.5523799 total: 24.8ms remaining: 6.99ms
39: learn: 0.5496638 total: 25.4ms remaining: 6.34ms
40: learn: 0.5467338 total: 25.9ms remaining: 5.68ms
41: learn: 0.5437414 total: 26.4ms remaining: 5.03ms
42: learn: 0.5410515 total: 27ms remaining: 4.4ms
43: learn: 0.5385706 total: 27.7ms remaining: 3.77ms
44: learn: 0.5359205 total: 28.2ms remaining: 3.14ms
45: learn: 0.5335821 total: 28.8ms remaining: 2.5ms
46: learn: 0.5311304 total: 29.4ms remaining: 1.88ms
47: learn: 0.5287105 total: 29.8ms remaining: 1.24ms
48: learn: 0.5262265 total: 30.2ms remaining: 616us
49: learn: 0.5235670 total: 30.8ms remaining: 0us
0: learn: 0.6884251 total: 550us remaining: 27ms
1: learn: 0.6837882 total: 850us remaining: 20.4ms
2: learn: 0.6788893 total: 1.23ms remaining: 19.2ms
3: learn: 0.6742531 total: 1.5ms remaining: 17.2ms
4: learn: 0.6694941 total: 1.96ms remaining: 17.7ms
5: learn: 0.6644974 total: 2.37ms remaining: 17.4ms
6: learn: 0.6602396 total: 2.74ms remaining: 16.8ms
7: learn: 0.6546575 total: 3.15ms remaining: 16.5ms
8: learn: 0.6495477 total: 3.5ms remaining: 16ms
9: learn: 0.6460980 total: 3.72ms remaining: 14.9ms
10: learn: 0.6407818 total: 4.02ms remaining: 14.3ms
11: learn: 0.6358886 total: 4.49ms remaining: 14.2ms
12: learn: 0.6319770 total: 4.88ms remaining: 13.9ms
13: learn: 0.6280995 total: 5.31ms remaining: 13.6ms
14: learn: 0.6233202 total: 5.66ms remaining: 13.2ms
15: learn: 0.6192099 total: 6ms remaining: 12.8ms
16: learn: 0.6147762 total: 6.28ms remaining: 12.2ms
17: learn: 0.6103238 total: 6.5ms remaining: 11.6ms
18: learn: 0.6060995 total: 6.82ms remaining: 11.1ms
19: learn: 0.6020556 total: 7.16ms remaining: 10.7ms
20: learn: 0.5977037 total: 7.56ms remaining: 10.4ms
21: learn: 0.5931581 total: 7.96ms remaining: 10.1ms
22: learn: 0.5890359 total: 8.3ms remaining: 9.74ms
23: learn: 0.5846377 total: 8.66ms remaining: 9.38ms
24: learn: 0.5814286 total: 9.04ms remaining: 9.04ms
25: learn: 0.5780560 total: 9.43ms remaining: 8.71ms
26: learn: 0.5739783 total: 9.76ms remaining: 8.31ms
27: learn: 0.5709059 total: 10.1ms remaining: 7.9ms
28: learn: 0.5681418 total: 10.3ms remaining: 7.44ms
29: learn: 0.5650585 total: 10.7ms remaining: 7.1ms
30: learn: 0.5612944 total: 10.9ms remaining: 6.65ms
31: learn: 0.5579837 total: 11.3ms remaining: 6.34ms
32: learn: 0.5545437 total: 11.5ms remaining: 5.95ms
33: learn: 0.5508624 total: 11.9ms remaining: 5.58ms
34: learn: 0.5476947 total: 12.2ms remaining: 5.24ms
35: learn: 0.5443277 total: 12.6ms remaining: 4.9ms
36: learn: 0.5410425 total: 12.9ms remaining: 4.55ms
37: learn: 0.5381051 total: 13.2ms remaining: 4.17ms
38: learn: 0.5349005 total: 13.6ms remaining: 3.84ms
39: learn: 0.5320597 total: 13.9ms remaining: 3.48ms
40: learn: 0.5288238 total: 14.3ms remaining: 3.13ms
41: learn: 0.5252659 total: 14.6ms remaining: 2.77ms
42: learn: 0.5227562 total: 14.9ms remaining: 2.42ms
43: learn: 0.5199700 total: 15.2ms remaining: 2.08ms
44: learn: 0.5171837 total: 15.6ms remaining: 1.73ms
45: learn: 0.5143990 total: 15.9ms remaining: 1.38ms
46: learn: 0.5116150 total: 16.3ms remaining: 1.04ms
47: learn: 0.5085206 total: 16.7ms remaining: 697us
48: learn: 0.5052346 total: 17.2ms remaining: 350us
49: learn: 0.5021518 total: 17.6ms remaining: 0us
0: learn: 0.6878984 total: 433us remaining: 21.2ms
1: learn: 0.6825637 total: 796us remaining: 19.1ms
2: learn: 0.6778136 total: 1.08ms remaining: 17ms
3: learn: 0.6730922 total: 1.46ms remaining: 16.8ms
4: learn: 0.6683979 total: 1.75ms remaining: 15.8ms
5: learn: 0.6639298 total: 2.24ms remaining: 16.5ms
6: learn: 0.6588845 total: 2.6ms remaining: 16ms
7: learn: 0.6547090 total: 2.98ms remaining: 15.6ms
8: learn: 0.6501215 total: 3.31ms remaining: 15.1ms
9: learn: 0.6455656 total: 3.71ms remaining: 14.8ms
10: learn: 0.6411502 total: 4.01ms remaining: 14.2ms
11: learn: 0.6367748 total: 4.33ms remaining: 13.7ms
12: learn: 0.6324957 total: 4.73ms remaining: 13.5ms
13: learn: 0.6282916 total: 5.05ms remaining: 13ms
14: learn: 0.6244643 total: 5.43ms remaining: 12.7ms
15: learn: 0.6197827 total: 5.78ms remaining: 12.3ms
16: learn: 0.6162918 total: 6.15ms remaining: 11.9ms
17: learn: 0.6117805 total: 6.66ms remaining: 11.8ms
18: learn: 0.6076094 total: 6.85ms remaining: 11.2ms
19: learn: 0.6039850 total: 7.29ms remaining: 10.9ms
20: learn: 0.6001150 total: 7.64ms remaining: 10.6ms
21: learn: 0.5961289 total: 8.07ms remaining: 10.3ms
22: learn: 0.5919174 total: 8.46ms remaining: 9.93ms
23: learn: 0.5880261 total: 8.8ms remaining: 9.54ms
24: learn: 0.5845686 total: 9.24ms remaining: 9.24ms
25: learn: 0.5814502 total: 9.64ms remaining: 8.9ms
26: learn: 0.5783653 total: 10.1ms remaining: 8.57ms
27: learn: 0.5749444 total: 10.4ms remaining: 8.2ms
28: learn: 0.5717356 total: 10.8ms remaining: 7.8ms
29: learn: 0.5686947 total: 11.1ms remaining: 7.42ms
30: learn: 0.5658574 total: 11.5ms remaining: 7.04ms
31: learn: 0.5627563 total: 11.7ms remaining: 6.6ms
32: learn: 0.5594251 total: 12.2ms remaining: 6.29ms
33: learn: 0.5561764 total: 12.6ms remaining: 5.92ms
34: learn: 0.5529952 total: 12.9ms remaining: 5.51ms
35: learn: 0.5508336 total: 13.2ms remaining: 5.13ms
36: learn: 0.5478067 total: 13.6ms remaining: 4.78ms
37: learn: 0.5449646 total: 14ms remaining: 4.41ms
38: learn: 0.5420459 total: 14.3ms remaining: 4.03ms
39: learn: 0.5388165 total: 14.6ms remaining: 3.66ms
40: learn: 0.5358141 total: 15ms remaining: 3.29ms
41: learn: 0.5331245 total: 15.3ms remaining: 2.92ms
42: learn: 0.5300696 total: 15.7ms remaining: 2.55ms
43: learn: 0.5273135 total: 16ms remaining: 2.19ms
44: learn: 0.5247408 total: 16.5ms remaining: 1.83ms
45: learn: 0.5220816 total: 17ms remaining: 1.47ms
46: learn: 0.5198094 total: 17.3ms remaining: 1.1ms
47: learn: 0.5168347 total: 17.8ms remaining: 742us
48: learn: 0.5144139 total: 18.2ms remaining: 371us
49: learn: 0.5116065 total: 18.5ms remaining: 0us
0: learn: 0.6882230 total: 473us remaining: 23.2ms
1: learn: 0.6830130 total: 936us remaining: 22.5ms
2: learn: 0.6777502 total: 1.34ms remaining: 21ms
3: learn: 0.6725869 total: 1.84ms remaining: 21.2ms
4: learn: 0.6678779 total: 2.19ms remaining: 19.7ms
5: learn: 0.6627342 total: 2.63ms remaining: 19.3ms
6: learn: 0.6578700 total: 3ms remaining: 18.4ms
7: learn: 0.6539456 total: 3.39ms remaining: 17.8ms
8: learn: 0.6489295 total: 3.73ms remaining: 17ms
9: learn: 0.6447199 total: 4.11ms remaining: 16.4ms
10: learn: 0.6410565 total: 4.46ms remaining: 15.8ms
11: learn: 0.6370334 total: 4.91ms remaining: 15.5ms
12: learn: 0.6323019 total: 5.42ms remaining: 15.4ms
13: learn: 0.6278074 total: 5.95ms remaining: 15.3ms
14: learn: 0.6244961 total: 6.38ms remaining: 14.9ms
15: learn: 0.6205365 total: 6.81ms remaining: 14.5ms
16: learn: 0.6164157 total: 7.31ms remaining: 14.2ms
17: learn: 0.6125643 total: 7.84ms remaining: 13.9ms
18: learn: 0.6087596 total: 8.36ms remaining: 13.6ms
19: learn: 0.6051407 total: 8.73ms remaining: 13.1ms
20: learn: 0.6012506 total: 9.07ms remaining: 12.5ms
21: learn: 0.5969953 total: 9.5ms remaining: 12.1ms
22: learn: 0.5926725 total: 9.74ms remaining: 11.4ms
23: learn: 0.5892457 total: 9.87ms remaining: 10.7ms
24: learn: 0.5861760 total: 10.1ms remaining: 10.1ms
25: learn: 0.5827887 total: 10.6ms remaining: 9.77ms
26: learn: 0.5792932 total: 11.1ms remaining: 9.42ms
27: learn: 0.5751741 total: 11.5ms remaining: 9.02ms
28: learn: 0.5722502 total: 11.8ms remaining: 8.54ms
29: learn: 0.5696262 total: 12.1ms remaining: 8.06ms
30: learn: 0.5670091 total: 12.4ms remaining: 7.62ms
31: learn: 0.5635108 total: 12.6ms remaining: 7.08ms
32: learn: 0.5600613 total: 13.1ms remaining: 6.76ms
33: learn: 0.5568582 total: 13.5ms remaining: 6.36ms
34: learn: 0.5533880 total: 13.9ms remaining: 5.94ms
35: learn: 0.5503392 total: 14.2ms remaining: 5.52ms
36: learn: 0.5472394 total: 14.6ms remaining: 5.13ms
37: learn: 0.5443595 total: 15ms remaining: 4.74ms
38: learn: 0.5412247 total: 15.3ms remaining: 4.32ms
39: learn: 0.5382340 total: 15.6ms remaining: 3.91ms
40: learn: 0.5349641 total: 16.1ms remaining: 3.53ms
41: learn: 0.5321799 total: 16.5ms remaining: 3.13ms
42: learn: 0.5289777 total: 16.8ms remaining: 2.74ms
43: learn: 0.5259768 total: 17.3ms remaining: 2.35ms
44: learn: 0.5230374 total: 17.6ms remaining: 1.95ms
45: learn: 0.5207025 total: 17.9ms remaining: 1.55ms
46: learn: 0.5181586 total: 18.2ms remaining: 1.16ms
47: learn: 0.5151432 total: 18.5ms remaining: 772us
48: learn: 0.5125814 total: 18.7ms remaining: 382us
49: learn: 0.5098965 total: 19ms remaining: 0us
0: learn: 0.6886159 total: 400us remaining: 19.6ms
1: learn: 0.6833054 total: 720us remaining: 17.3ms
2: learn: 0.6784831 total: 1.01ms remaining: 15.9ms
3: learn: 0.6744239 total: 1.38ms remaining: 15.8ms
4: learn: 0.6703678 total: 1.63ms remaining: 14.7ms
5: learn: 0.6657698 total: 2.03ms remaining: 14.9ms
6: learn: 0.6613446 total: 2.41ms remaining: 14.8ms
7: learn: 0.6575104 total: 2.74ms remaining: 14.4ms
8: learn: 0.6529390 total: 3.02ms remaining: 13.8ms
9: learn: 0.6485341 total: 3.34ms remaining: 13.4ms
10: learn: 0.6445752 total: 3.72ms remaining: 13.2ms
11: learn: 0.6403021 total: 4.08ms remaining: 12.9ms
12: learn: 0.6365289 total: 4.46ms remaining: 12.7ms
13: learn: 0.6324526 total: 4.89ms remaining: 12.6ms
14: learn: 0.6290089 total: 5.25ms remaining: 12.2ms
15: learn: 0.6245414 total: 5.52ms remaining: 11.7ms
16: learn: 0.6210188 total: 5.83ms remaining: 11.3ms
17: learn: 0.6174320 total: 6.13ms remaining: 10.9ms
18: learn: 0.6140296 total: 6.3ms remaining: 10.3ms
19: learn: 0.6105599 total: 6.63ms remaining: 9.95ms
20: learn: 0.6069129 total: 6.97ms remaining: 9.63ms
21: learn: 0.6030373 total: 7.28ms remaining: 9.27ms
22: learn: 0.5995989 total: 7.59ms remaining: 8.91ms
23: learn: 0.5963684 total: 7.9ms remaining: 8.56ms
24: learn: 0.5922892 total: 8.14ms remaining: 8.14ms
25: learn: 0.5894788 total: 8.49ms remaining: 7.83ms
26: learn: 0.5863141 total: 8.78ms remaining: 7.48ms
27: learn: 0.5833325 total: 9.13ms remaining: 7.17ms
28: learn: 0.5804530 total: 9.45ms remaining: 6.84ms
29: learn: 0.5778179 total: 9.79ms remaining: 6.52ms
30: learn: 0.5753587 total: 10.2ms remaining: 6.25ms
31: learn: 0.5724407 total: 10.4ms remaining: 5.84ms
32: learn: 0.5690258 total: 10.8ms remaining: 5.54ms
33: learn: 0.5657888 total: 11.1ms remaining: 5.23ms
34: learn: 0.5621902 total: 11.6ms remaining: 4.96ms
35: learn: 0.5593933 total: 11.9ms remaining: 4.63ms
36: learn: 0.5564423 total: 12.3ms remaining: 4.32ms
37: learn: 0.5537762 total: 12.6ms remaining: 3.99ms
38: learn: 0.5504946 total: 13ms remaining: 3.66ms
39: learn: 0.5481516 total: 13.4ms remaining: 3.36ms
40: learn: 0.5452933 total: 13.8ms remaining: 3.02ms
41: learn: 0.5425361 total: 14ms remaining: 2.66ms
42: learn: 0.5398155 total: 14.4ms remaining: 2.34ms
43: learn: 0.5365296 total: 14.7ms remaining: 2.01ms
44: learn: 0.5338784 total: 15.1ms remaining: 1.68ms
45: learn: 0.5312372 total: 15.5ms remaining: 1.34ms
46: learn: 0.5285292 total: 15.8ms remaining: 1.01ms
47: learn: 0.5257266 total: 16.1ms remaining: 672us
48: learn: 0.5232061 total: 16.3ms remaining: 333us
49: learn: 0.5203838 total: 16.7ms remaining: 0us
Evaluating param combo 3/16: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 6, 'boosted_cb_model__num_leaves': 8}
Duplicate found for this combo (hash=500d0797d66eb11cdee398dcac2ee0e9). Removing old run: c571e94807d54652b5681faebef3a577
0: learn: 0.6882547 total: 512us remaining: 25.1ms
1: learn: 0.6826413 total: 1.01ms remaining: 24.2ms
2: learn: 0.6787574 total: 1.78ms remaining: 27.9ms
3: learn: 0.6737900 total: 2.56ms remaining: 29.4ms
4: learn: 0.6686791 total: 3.29ms remaining: 29.6ms
5: learn: 0.6643458 total: 4.08ms remaining: 29.9ms
6: learn: 0.6596252 total: 4.99ms remaining: 30.7ms
7: learn: 0.6554987 total: 5.44ms remaining: 28.6ms
8: learn: 0.6509583 total: 6.24ms remaining: 28.4ms
9: learn: 0.6469695 total: 6.85ms remaining: 27.4ms
10: learn: 0.6423582 total: 7.78ms remaining: 27.6ms
11: learn: 0.6374666 total: 8.58ms remaining: 27.2ms
12: learn: 0.6326421 total: 9.26ms remaining: 26.3ms
13: learn: 0.6295545 total: 10.6ms remaining: 27.1ms
14: learn: 0.6256949 total: 11.5ms remaining: 26.9ms
15: learn: 0.6213727 total: 12.3ms remaining: 26.1ms
16: learn: 0.6171218 total: 12.8ms remaining: 24.9ms
17: learn: 0.6139615 total: 13.6ms remaining: 24.2ms
18: learn: 0.6105782 total: 14.4ms remaining: 23.6ms
19: learn: 0.6064788 total: 15.2ms remaining: 22.9ms
20: learn: 0.6038663 total: 16ms remaining: 22.1ms
21: learn: 0.6003718 total: 16.7ms remaining: 21.3ms
22: learn: 0.5963038 total: 17.3ms remaining: 20.3ms
23: learn: 0.5942131 total: 18ms remaining: 19.5ms
24: learn: 0.5901593 total: 18.6ms remaining: 18.6ms
25: learn: 0.5866114 total: 19.2ms remaining: 17.7ms
26: learn: 0.5830859 total: 19.9ms remaining: 17ms
27: learn: 0.5797169 total: 20.7ms remaining: 16.2ms
28: learn: 0.5765771 total: 21.3ms remaining: 15.4ms
29: learn: 0.5731998 total: 21.7ms remaining: 14.5ms
30: learn: 0.5698716 total: 22.3ms remaining: 13.7ms
31: learn: 0.5662749 total: 22.9ms remaining: 12.9ms
32: learn: 0.5633926 total: 23.4ms remaining: 12ms
33: learn: 0.5596447 total: 23.8ms remaining: 11.2ms
34: learn: 0.5563727 total: 24.2ms remaining: 10.4ms
35: learn: 0.5536910 total: 24.7ms remaining: 9.59ms
36: learn: 0.5511013 total: 25.2ms remaining: 8.87ms
37: learn: 0.5473601 total: 25.7ms remaining: 8.13ms
38: learn: 0.5440328 total: 26.3ms remaining: 7.42ms
39: learn: 0.5413512 total: 26.9ms remaining: 6.73ms
40: learn: 0.5382235 total: 27.5ms remaining: 6.03ms
41: learn: 0.5355798 total: 28ms remaining: 5.34ms
42: learn: 0.5320209 total: 28.5ms remaining: 4.63ms
43: learn: 0.5292923 total: 29ms remaining: 3.95ms
44: learn: 0.5264982 total: 29.2ms remaining: 3.25ms
45: learn: 0.5236175 total: 29.8ms remaining: 2.59ms
46: learn: 0.5205947 total: 30.3ms remaining: 1.94ms
47: learn: 0.5178331 total: 30.8ms remaining: 1.28ms
48: learn: 0.5149838 total: 31.3ms remaining: 639us
49: learn: 0.5120349 total: 31.7ms remaining: 0us
0: learn: 0.6879855 total: 530us remaining: 26ms
1: learn: 0.6817990 total: 966us remaining: 23.2ms
2: learn: 0.6768301 total: 1.33ms remaining: 20.8ms
3: learn: 0.6725467 total: 1.78ms remaining: 20.5ms
4: learn: 0.6677060 total: 2.08ms remaining: 18.7ms
5: learn: 0.6633421 total: 2.38ms remaining: 17.5ms
6: learn: 0.6587639 total: 2.73ms remaining: 16.8ms
7: learn: 0.6545091 total: 3.22ms remaining: 16.9ms
8: learn: 0.6492749 total: 3.65ms remaining: 16.6ms
9: learn: 0.6451569 total: 4.1ms remaining: 16.4ms
10: learn: 0.6404551 total: 4.47ms remaining: 15.8ms
11: learn: 0.6362056 total: 4.88ms remaining: 15.4ms
12: learn: 0.6319192 total: 5.23ms remaining: 14.9ms
13: learn: 0.6281266 total: 5.67ms remaining: 14.6ms
14: learn: 0.6229575 total: 6.06ms remaining: 14.1ms
15: learn: 0.6189410 total: 6.39ms remaining: 13.6ms
16: learn: 0.6150797 total: 6.65ms remaining: 12.9ms
17: learn: 0.6112747 total: 6.85ms remaining: 12.2ms
18: learn: 0.6075500 total: 7.29ms remaining: 11.9ms
19: learn: 0.6030214 total: 7.7ms remaining: 11.5ms
20: learn: 0.5989044 total: 8.16ms remaining: 11.3ms
21: learn: 0.5945766 total: 8.61ms remaining: 11ms
22: learn: 0.5899263 total: 9.02ms remaining: 10.6ms
23: learn: 0.5859453 total: 9.43ms remaining: 10.2ms
24: learn: 0.5817255 total: 9.97ms remaining: 9.97ms
25: learn: 0.5787281 total: 10.4ms remaining: 9.63ms
26: learn: 0.5750515 total: 10.9ms remaining: 9.28ms
27: learn: 0.5722523 total: 11.4ms remaining: 8.99ms
28: learn: 0.5688370 total: 11.9ms remaining: 8.65ms
29: learn: 0.5650730 total: 12.4ms remaining: 8.27ms
30: learn: 0.5614533 total: 12.8ms remaining: 7.84ms
31: learn: 0.5578750 total: 13.2ms remaining: 7.43ms
32: learn: 0.5542367 total: 13.9ms remaining: 7.17ms
33: learn: 0.5516665 total: 14.6ms remaining: 6.85ms
34: learn: 0.5487423 total: 14.9ms remaining: 6.4ms
35: learn: 0.5451976 total: 15.5ms remaining: 6.01ms
36: learn: 0.5415315 total: 15.9ms remaining: 5.58ms
37: learn: 0.5385923 total: 16.3ms remaining: 5.16ms
38: learn: 0.5357241 total: 16.8ms remaining: 4.74ms
39: learn: 0.5327391 total: 17.2ms remaining: 4.31ms
40: learn: 0.5294844 total: 17.6ms remaining: 3.86ms
41: learn: 0.5267391 total: 18ms remaining: 3.42ms
42: learn: 0.5243481 total: 18.5ms remaining: 3.01ms
43: learn: 0.5215191 total: 18.9ms remaining: 2.58ms
44: learn: 0.5183146 total: 19.5ms remaining: 2.16ms
45: learn: 0.5150006 total: 20.1ms remaining: 1.75ms
46: learn: 0.5117049 total: 20.6ms remaining: 1.31ms
47: learn: 0.5084576 total: 20.9ms remaining: 872us
48: learn: 0.5058160 total: 21.4ms remaining: 437us
49: learn: 0.5032421 total: 22ms remaining: 0us
0: learn: 0.6878694 total: 607us remaining: 29.8ms
1: learn: 0.6827595 total: 1.05ms remaining: 25.3ms
2: learn: 0.6778842 total: 1.42ms remaining: 22.3ms
3: learn: 0.6740388 total: 1.86ms remaining: 21.4ms
4: learn: 0.6691401 total: 2.29ms remaining: 20.6ms
5: learn: 0.6648926 total: 2.67ms remaining: 19.6ms
6: learn: 0.6611328 total: 3.06ms remaining: 18.8ms
7: learn: 0.6560550 total: 3.41ms remaining: 17.9ms
8: learn: 0.6511893 total: 3.83ms remaining: 17.4ms
9: learn: 0.6473611 total: 4.25ms remaining: 17ms
10: learn: 0.6425144 total: 4.66ms remaining: 16.5ms
11: learn: 0.6383460 total: 5.12ms remaining: 16.2ms
12: learn: 0.6332335 total: 5.5ms remaining: 15.7ms
13: learn: 0.6286910 total: 5.91ms remaining: 15.2ms
14: learn: 0.6242482 total: 6.33ms remaining: 14.8ms
15: learn: 0.6204319 total: 6.75ms remaining: 14.3ms
16: learn: 0.6162046 total: 7.14ms remaining: 13.9ms
17: learn: 0.6121904 total: 7.54ms remaining: 13.4ms
18: learn: 0.6077890 total: 7.92ms remaining: 12.9ms
19: learn: 0.6045111 total: 8.32ms remaining: 12.5ms
20: learn: 0.6007578 total: 8.73ms remaining: 12.1ms
21: learn: 0.5985328 total: 9.17ms remaining: 11.7ms
22: learn: 0.5943166 total: 9.58ms remaining: 11.2ms
23: learn: 0.5910600 total: 9.94ms remaining: 10.8ms
24: learn: 0.5876570 total: 10.4ms remaining: 10.4ms
25: learn: 0.5843096 total: 10.9ms remaining: 10ms
26: learn: 0.5808736 total: 11.3ms remaining: 9.61ms
27: learn: 0.5782591 total: 11.7ms remaining: 9.16ms
28: learn: 0.5752312 total: 12ms remaining: 8.71ms
29: learn: 0.5713676 total: 12.5ms remaining: 8.31ms
30: learn: 0.5680353 total: 12.9ms remaining: 7.93ms
31: learn: 0.5645367 total: 13.5ms remaining: 7.57ms
32: learn: 0.5611002 total: 14.1ms remaining: 7.25ms
33: learn: 0.5572774 total: 14.7ms remaining: 6.89ms
34: learn: 0.5541482 total: 15.1ms remaining: 6.49ms
35: learn: 0.5509010 total: 15.6ms remaining: 6.07ms
36: learn: 0.5473483 total: 16.1ms remaining: 5.67ms
37: learn: 0.5442582 total: 16.6ms remaining: 5.24ms
38: learn: 0.5409799 total: 17.1ms remaining: 4.83ms
39: learn: 0.5380030 total: 17.4ms remaining: 4.36ms
40: learn: 0.5347076 total: 17.9ms remaining: 3.94ms
41: learn: 0.5320555 total: 18.4ms remaining: 3.51ms
42: learn: 0.5289706 total: 19.1ms remaining: 3.1ms
43: learn: 0.5261645 total: 19.6ms remaining: 2.67ms
44: learn: 0.5233001 total: 20.1ms remaining: 2.23ms
45: learn: 0.5201561 total: 20.5ms remaining: 1.78ms
46: learn: 0.5175403 total: 21ms remaining: 1.34ms
47: learn: 0.5144315 total: 21.4ms remaining: 893us
48: learn: 0.5114985 total: 21.9ms remaining: 446us
49: learn: 0.5095739 total: 22.3ms remaining: 0us
0: learn: 0.6878111 total: 416us remaining: 20.4ms
1: learn: 0.6830677 total: 705us remaining: 16.9ms
2: learn: 0.6786716 total: 983us remaining: 15.4ms
3: learn: 0.6730396 total: 1.29ms remaining: 14.9ms
4: learn: 0.6684266 total: 1.41ms remaining: 12.7ms
5: learn: 0.6638953 total: 1.66ms remaining: 12.2ms
6: learn: 0.6589777 total: 1.99ms remaining: 12.3ms
7: learn: 0.6538375 total: 2.3ms remaining: 12.1ms
8: learn: 0.6500576 total: 2.61ms remaining: 11.9ms
9: learn: 0.6454226 total: 2.89ms remaining: 11.5ms
10: learn: 0.6414998 total: 3.15ms remaining: 11.2ms
11: learn: 0.6373951 total: 3.36ms remaining: 10.6ms
12: learn: 0.6336399 total: 3.66ms remaining: 10.4ms
13: learn: 0.6301762 total: 3.94ms remaining: 10.1ms
14: learn: 0.6265799 total: 4.57ms remaining: 10.7ms
15: learn: 0.6228602 total: 5.14ms remaining: 10.9ms
16: learn: 0.6182332 total: 5.4ms remaining: 10.5ms
17: learn: 0.6135086 total: 5.74ms remaining: 10.2ms
18: learn: 0.6104827 total: 6.14ms remaining: 10ms
19: learn: 0.6071668 total: 6.4ms remaining: 9.61ms
20: learn: 0.6035631 total: 6.85ms remaining: 9.46ms
21: learn: 0.5992836 total: 7.33ms remaining: 9.33ms
22: learn: 0.5954905 total: 7.69ms remaining: 9.03ms
23: learn: 0.5915550 total: 8.03ms remaining: 8.7ms
24: learn: 0.5876077 total: 8.48ms remaining: 8.48ms
25: learn: 0.5835114 total: 9.11ms remaining: 8.4ms
26: learn: 0.5796600 total: 9.56ms remaining: 8.14ms
27: learn: 0.5766405 total: 9.84ms remaining: 7.73ms
28: learn: 0.5731260 total: 10.2ms remaining: 7.36ms
29: learn: 0.5697091 total: 10.8ms remaining: 7.21ms
30: learn: 0.5658183 total: 11.1ms remaining: 6.83ms
31: learn: 0.5626938 total: 11.5ms remaining: 6.48ms
32: learn: 0.5596719 total: 12ms remaining: 6.18ms
33: learn: 0.5562376 total: 12.3ms remaining: 5.78ms
34: learn: 0.5526401 total: 12.9ms remaining: 5.53ms
35: learn: 0.5492243 total: 13.3ms remaining: 5.17ms
36: learn: 0.5457598 total: 13.6ms remaining: 4.78ms
37: learn: 0.5420782 total: 13.9ms remaining: 4.39ms
38: learn: 0.5390517 total: 14.3ms remaining: 4.03ms
39: learn: 0.5357773 total: 14.7ms remaining: 3.66ms
40: learn: 0.5328531 total: 15.1ms remaining: 3.31ms
41: learn: 0.5300062 total: 15.5ms remaining: 2.95ms
42: learn: 0.5272410 total: 15.9ms remaining: 2.58ms
43: learn: 0.5240909 total: 16.2ms remaining: 2.21ms
44: learn: 0.5214789 total: 16.6ms remaining: 1.85ms
45: learn: 0.5191970 total: 17.1ms remaining: 1.48ms
46: learn: 0.5161765 total: 17.4ms remaining: 1.11ms
47: learn: 0.5133697 total: 17.5ms remaining: 729us
48: learn: 0.5104980 total: 17.8ms remaining: 362us
49: learn: 0.5075510 total: 18.4ms remaining: 0us
0: learn: 0.6881984 total: 575us remaining: 28.2ms
1: learn: 0.6837389 total: 990us remaining: 23.8ms
2: learn: 0.6789850 total: 1.39ms remaining: 21.7ms
3: learn: 0.6740545 total: 2.19ms remaining: 25.2ms
4: learn: 0.6700245 total: 3.15ms remaining: 28.3ms
5: learn: 0.6664139 total: 3.96ms remaining: 29ms
6: learn: 0.6617050 total: 4.88ms remaining: 30ms
7: learn: 0.6579000 total: 5.83ms remaining: 30.6ms
8: learn: 0.6528585 total: 6.72ms remaining: 30.6ms
9: learn: 0.6487785 total: 7.64ms remaining: 30.6ms
10: learn: 0.6439531 total: 8.46ms remaining: 30ms
11: learn: 0.6395388 total: 9.34ms remaining: 29.6ms
12: learn: 0.6351148 total: 10.3ms remaining: 29.4ms
13: learn: 0.6311540 total: 11.2ms remaining: 28.8ms
14: learn: 0.6266968 total: 12ms remaining: 28.1ms
15: learn: 0.6225980 total: 12.9ms remaining: 27.5ms
16: learn: 0.6190533 total: 13.8ms remaining: 26.8ms
17: learn: 0.6146239 total: 14.7ms remaining: 26ms
18: learn: 0.6109725 total: 15.9ms remaining: 26ms
19: learn: 0.6069686 total: 16.7ms remaining: 25.1ms
20: learn: 0.6025956 total: 17.4ms remaining: 24.1ms
21: learn: 0.5986945 total: 18.2ms remaining: 23.1ms
22: learn: 0.5944782 total: 19ms remaining: 22.3ms
23: learn: 0.5917354 total: 20.3ms remaining: 22ms
24: learn: 0.5882377 total: 20.5ms remaining: 20.5ms
25: learn: 0.5847297 total: 21.3ms remaining: 19.6ms
26: learn: 0.5815059 total: 21.8ms remaining: 18.5ms
27: learn: 0.5781487 total: 22.3ms remaining: 17.5ms
28: learn: 0.5743941 total: 23.1ms remaining: 16.7ms
29: learn: 0.5714101 total: 23.9ms remaining: 15.9ms
30: learn: 0.5680936 total: 24.6ms remaining: 15.1ms
31: learn: 0.5645795 total: 25.3ms remaining: 14.2ms
32: learn: 0.5609279 total: 25.9ms remaining: 13.3ms
33: learn: 0.5577243 total: 26.6ms remaining: 12.5ms
34: learn: 0.5546932 total: 27.3ms remaining: 11.7ms
35: learn: 0.5516329 total: 27.9ms remaining: 10.8ms
36: learn: 0.5484561 total: 28.5ms remaining: 10ms
37: learn: 0.5458463 total: 29ms remaining: 9.16ms
38: learn: 0.5429156 total: 29.6ms remaining: 8.34ms
39: learn: 0.5401707 total: 30.1ms remaining: 7.52ms
40: learn: 0.5371278 total: 30.7ms remaining: 6.74ms
41: learn: 0.5348207 total: 31.2ms remaining: 5.93ms
42: learn: 0.5322859 total: 31.6ms remaining: 5.14ms
43: learn: 0.5295026 total: 32.2ms remaining: 4.39ms
44: learn: 0.5272640 total: 32.8ms remaining: 3.64ms
45: learn: 0.5245788 total: 32.9ms remaining: 2.86ms
46: learn: 0.5220263 total: 33.4ms remaining: 2.13ms
47: learn: 0.5199606 total: 33.9ms remaining: 1.41ms
48: learn: 0.5177430 total: 34.4ms remaining: 702us
49: learn: 0.5155567 total: 34.9ms remaining: 0us
0: learn: 0.6884902 total: 752us remaining: 36.9ms
1: learn: 0.6827814 total: 1.51ms remaining: 36.2ms
2: learn: 0.6775249 total: 1.88ms remaining: 29.5ms
3: learn: 0.6730922 total: 2.55ms remaining: 29.3ms
4: learn: 0.6678643 total: 3.31ms remaining: 29.8ms
5: learn: 0.6641156 total: 3.95ms remaining: 29ms
6: learn: 0.6591491 total: 4.58ms remaining: 28.2ms
7: learn: 0.6544223 total: 5.28ms remaining: 27.7ms
8: learn: 0.6493682 total: 6ms remaining: 27.3ms
9: learn: 0.6451104 total: 6.74ms remaining: 27ms
10: learn: 0.6405221 total: 7.38ms remaining: 26.2ms
11: learn: 0.6370647 total: 7.97ms remaining: 25.2ms
12: learn: 0.6327924 total: 8.52ms remaining: 24.3ms
13: learn: 0.6287428 total: 9.29ms remaining: 23.9ms
14: learn: 0.6240275 total: 10.1ms remaining: 23.7ms
15: learn: 0.6208828 total: 11.2ms remaining: 23.8ms
16: learn: 0.6173008 total: 12.3ms remaining: 23.8ms
17: learn: 0.6141311 total: 12.9ms remaining: 23ms
18: learn: 0.6099239 total: 13.6ms remaining: 22.2ms
19: learn: 0.6060360 total: 14.4ms remaining: 21.5ms
20: learn: 0.6018792 total: 14.9ms remaining: 20.6ms
21: learn: 0.5990134 total: 15.4ms remaining: 19.6ms
22: learn: 0.5950741 total: 16ms remaining: 18.8ms
23: learn: 0.5913950 total: 16.8ms remaining: 18.2ms
24: learn: 0.5878841 total: 17.5ms remaining: 17.5ms
25: learn: 0.5840340 total: 17.9ms remaining: 16.6ms
26: learn: 0.5802704 total: 18.6ms remaining: 15.8ms
27: learn: 0.5774147 total: 19.3ms remaining: 15.1ms
28: learn: 0.5737299 total: 19.8ms remaining: 14.4ms
29: learn: 0.5707954 total: 20ms remaining: 13.4ms
30: learn: 0.5676180 total: 20.6ms remaining: 12.7ms
31: learn: 0.5639358 total: 21.2ms remaining: 11.9ms
32: learn: 0.5602758 total: 21.8ms remaining: 11.2ms
33: learn: 0.5569481 total: 22.3ms remaining: 10.5ms
34: learn: 0.5540790 total: 22.9ms remaining: 9.79ms
35: learn: 0.5507536 total: 23.4ms remaining: 9.11ms
36: learn: 0.5476666 total: 23.9ms remaining: 8.4ms
37: learn: 0.5454448 total: 24.3ms remaining: 7.67ms
38: learn: 0.5422534 total: 24.9ms remaining: 7.01ms
39: learn: 0.5391597 total: 25.5ms remaining: 6.37ms
40: learn: 0.5361588 total: 25.9ms remaining: 5.69ms
41: learn: 0.5336828 total: 26.4ms remaining: 5.03ms
42: learn: 0.5301573 total: 26.8ms remaining: 4.36ms
43: learn: 0.5268429 total: 27.3ms remaining: 3.72ms
44: learn: 0.5243637 total: 27.6ms remaining: 3.07ms
45: learn: 0.5215132 total: 28ms remaining: 2.44ms
46: learn: 0.5184502 total: 28.5ms remaining: 1.82ms
47: learn: 0.5157361 total: 28.8ms remaining: 1.2ms
48: learn: 0.5133939 total: 29.2ms remaining: 596us
49: learn: 0.5110648 total: 29.8ms remaining: 0us
0: learn: 0.6882498 total: 608us remaining: 29.8ms
1: learn: 0.6828704 total: 1.06ms remaining: 25.4ms
2: learn: 0.6783092 total: 1.48ms remaining: 23.1ms
3: learn: 0.6728309 total: 1.91ms remaining: 21.9ms
4: learn: 0.6682349 total: 2.05ms remaining: 18.4ms
5: learn: 0.6635672 total: 2.43ms remaining: 17.8ms
6: learn: 0.6588318 total: 2.82ms remaining: 17.3ms
7: learn: 0.6534401 total: 3.17ms remaining: 16.6ms
8: learn: 0.6493226 total: 3.57ms remaining: 16.3ms
9: learn: 0.6449812 total: 4.07ms remaining: 16.3ms
10: learn: 0.6405017 total: 4.52ms remaining: 16ms
11: learn: 0.6369216 total: 4.86ms remaining: 15.4ms
12: learn: 0.6332908 total: 5.3ms remaining: 15.1ms
13: learn: 0.6289660 total: 5.73ms remaining: 14.7ms
14: learn: 0.6249958 total: 6.15ms remaining: 14.3ms
15: learn: 0.6212372 total: 6.3ms remaining: 13.4ms
16: learn: 0.6166556 total: 6.68ms remaining: 13ms
17: learn: 0.6131139 total: 7.04ms remaining: 12.5ms
18: learn: 0.6101849 total: 7.41ms remaining: 12.1ms
19: learn: 0.6060846 total: 7.81ms remaining: 11.7ms
20: learn: 0.6024470 total: 8.18ms remaining: 11.3ms
21: learn: 0.5990534 total: 8.54ms remaining: 10.9ms
22: learn: 0.5953439 total: 8.89ms remaining: 10.4ms
23: learn: 0.5917527 total: 9.36ms remaining: 10.1ms
24: learn: 0.5879160 total: 9.71ms remaining: 9.71ms
25: learn: 0.5843541 total: 10.1ms remaining: 9.3ms
26: learn: 0.5802603 total: 10.4ms remaining: 8.88ms
27: learn: 0.5771810 total: 10.8ms remaining: 8.52ms
28: learn: 0.5734008 total: 11.2ms remaining: 8.14ms
29: learn: 0.5700816 total: 11.7ms remaining: 7.77ms
30: learn: 0.5669234 total: 11.9ms remaining: 7.26ms
31: learn: 0.5643094 total: 12.2ms remaining: 6.85ms
32: learn: 0.5608718 total: 12.6ms remaining: 6.5ms
33: learn: 0.5573620 total: 13.1ms remaining: 6.17ms
34: learn: 0.5536805 total: 13.6ms remaining: 5.85ms
35: learn: 0.5506021 total: 14.1ms remaining: 5.46ms
36: learn: 0.5478504 total: 14.5ms remaining: 5.11ms
37: learn: 0.5445196 total: 15ms remaining: 4.73ms
38: learn: 0.5421972 total: 15.3ms remaining: 4.32ms
39: learn: 0.5397971 total: 15.7ms remaining: 3.93ms
40: learn: 0.5368651 total: 16.2ms remaining: 3.55ms
41: learn: 0.5341032 total: 16.6ms remaining: 3.17ms
42: learn: 0.5313339 total: 17ms remaining: 2.77ms
43: learn: 0.5282895 total: 17.5ms remaining: 2.38ms
44: learn: 0.5253477 total: 17.8ms remaining: 1.98ms
45: learn: 0.5226034 total: 18.3ms remaining: 1.59ms
46: learn: 0.5197743 total: 18.7ms remaining: 1.19ms
47: learn: 0.5174824 total: 19.1ms remaining: 797us
48: learn: 0.5148874 total: 19.6ms remaining: 399us
49: learn: 0.5125514 total: 19.9ms remaining: 0us
0: learn: 0.6884755 total: 403us remaining: 19.7ms
1: learn: 0.6840549 total: 743us remaining: 17.8ms
2: learn: 0.6800972 total: 1.02ms remaining: 16.1ms
3: learn: 0.6768483 total: 1.31ms remaining: 15.1ms
4: learn: 0.6715695 total: 1.6ms remaining: 14.4ms
5: learn: 0.6672446 total: 1.99ms remaining: 14.6ms
6: learn: 0.6625330 total: 2.5ms remaining: 15.4ms
7: learn: 0.6589198 total: 2.88ms remaining: 15.1ms
8: learn: 0.6546621 total: 3.39ms remaining: 15.5ms
9: learn: 0.6501226 total: 3.87ms remaining: 15.5ms
10: learn: 0.6460963 total: 4.29ms remaining: 15.2ms
11: learn: 0.6423398 total: 4.54ms remaining: 14.4ms
12: learn: 0.6379660 total: 5.04ms remaining: 14.3ms
13: learn: 0.6342444 total: 5.49ms remaining: 14.1ms
14: learn: 0.6305205 total: 5.79ms remaining: 13.5ms
15: learn: 0.6264874 total: 6.32ms remaining: 13.4ms
16: learn: 0.6230416 total: 6.88ms remaining: 13.4ms
17: learn: 0.6197055 total: 7.39ms remaining: 13.1ms
18: learn: 0.6157189 total: 7.75ms remaining: 12.7ms
19: learn: 0.6125868 total: 8.24ms remaining: 12.4ms
20: learn: 0.6083547 total: 8.77ms remaining: 12.1ms
21: learn: 0.6046788 total: 9.26ms remaining: 11.8ms
22: learn: 0.6015356 total: 9.65ms remaining: 11.3ms
23: learn: 0.5973653 total: 10.1ms remaining: 11ms
24: learn: 0.5941012 total: 10.5ms remaining: 10.5ms
25: learn: 0.5900899 total: 10.8ms remaining: 9.99ms
26: learn: 0.5864145 total: 11.6ms remaining: 9.88ms
27: learn: 0.5826043 total: 12.3ms remaining: 9.69ms
28: learn: 0.5791785 total: 12.9ms remaining: 9.32ms
29: learn: 0.5761813 total: 13.5ms remaining: 8.98ms
30: learn: 0.5732834 total: 14ms remaining: 8.57ms
31: learn: 0.5699500 total: 14.5ms remaining: 8.18ms
32: learn: 0.5664212 total: 14.9ms remaining: 7.67ms
33: learn: 0.5634228 total: 15.4ms remaining: 7.27ms
34: learn: 0.5609187 total: 15.9ms remaining: 6.82ms
35: learn: 0.5576450 total: 16.3ms remaining: 6.36ms
36: learn: 0.5546704 total: 16.8ms remaining: 5.92ms
37: learn: 0.5519747 total: 17.4ms remaining: 5.48ms
38: learn: 0.5486644 total: 17.9ms remaining: 5.05ms
39: learn: 0.5462319 total: 18.2ms remaining: 4.55ms
40: learn: 0.5433373 total: 18.6ms remaining: 4.07ms
41: learn: 0.5405130 total: 18.9ms remaining: 3.6ms
42: learn: 0.5383285 total: 19.5ms remaining: 3.17ms
43: learn: 0.5363352 total: 20ms remaining: 2.72ms
44: learn: 0.5333441 total: 20.5ms remaining: 2.27ms
45: learn: 0.5304898 total: 20.8ms remaining: 1.81ms
46: learn: 0.5277943 total: 21.3ms remaining: 1.36ms
47: learn: 0.5254893 total: 21.8ms remaining: 907us
48: learn: 0.5225329 total: 22.2ms remaining: 453us
49: learn: 0.5198716 total: 22.8ms remaining: 0us
0: learn: 0.6880341 total: 547us remaining: 26.8ms
1: learn: 0.6821741 total: 1.02ms remaining: 24.5ms
2: learn: 0.6772413 total: 1.41ms remaining: 22.1ms
3: learn: 0.6719167 total: 1.86ms remaining: 21.4ms
4: learn: 0.6673770 total: 2.16ms remaining: 19.5ms
5: learn: 0.6631005 total: 2.55ms remaining: 18.7ms
6: learn: 0.6583423 total: 2.94ms remaining: 18ms
7: learn: 0.6541579 total: 3.36ms remaining: 17.6ms
8: learn: 0.6500454 total: 3.76ms remaining: 17.1ms
9: learn: 0.6451953 total: 4.17ms remaining: 16.7ms
10: learn: 0.6408795 total: 4.53ms remaining: 16.1ms
11: learn: 0.6365962 total: 4.69ms remaining: 14.9ms
12: learn: 0.6326614 total: 5.2ms remaining: 14.8ms
13: learn: 0.6286050 total: 5.61ms remaining: 14.4ms
14: learn: 0.6244195 total: 6.03ms remaining: 14.1ms
15: learn: 0.6206224 total: 6.41ms remaining: 13.6ms
16: learn: 0.6166255 total: 6.79ms remaining: 13.2ms
17: learn: 0.6129454 total: 7.14ms remaining: 12.7ms
18: learn: 0.6091903 total: 7.51ms remaining: 12.2ms
19: learn: 0.6051157 total: 7.77ms remaining: 11.7ms
20: learn: 0.6008821 total: 8.17ms remaining: 11.3ms
21: learn: 0.5965324 total: 8.49ms remaining: 10.8ms
22: learn: 0.5923261 total: 8.87ms remaining: 10.4ms
23: learn: 0.5880464 total: 9.23ms remaining: 10ms
24: learn: 0.5839825 total: 9.54ms remaining: 9.54ms
25: learn: 0.5796767 total: 9.88ms remaining: 9.12ms
26: learn: 0.5762351 total: 10.3ms remaining: 8.75ms
27: learn: 0.5730085 total: 10.6ms remaining: 8.36ms
28: learn: 0.5698990 total: 11.1ms remaining: 8.07ms
29: learn: 0.5668733 total: 11.6ms remaining: 7.73ms
30: learn: 0.5631105 total: 11.9ms remaining: 7.32ms
31: learn: 0.5594105 total: 12.3ms remaining: 6.93ms
32: learn: 0.5556754 total: 12.7ms remaining: 6.55ms
33: learn: 0.5528355 total: 13.2ms remaining: 6.19ms
34: learn: 0.5497371 total: 13.6ms remaining: 5.82ms
35: learn: 0.5463105 total: 14ms remaining: 5.44ms
36: learn: 0.5429736 total: 14.6ms remaining: 5.13ms
37: learn: 0.5404733 total: 15ms remaining: 4.74ms
38: learn: 0.5372550 total: 15.4ms remaining: 4.35ms
39: learn: 0.5341701 total: 15.9ms remaining: 3.97ms
40: learn: 0.5312312 total: 16ms remaining: 3.52ms
41: learn: 0.5278372 total: 16.5ms remaining: 3.14ms
42: learn: 0.5251373 total: 17ms remaining: 2.77ms
43: learn: 0.5228357 total: 17.5ms remaining: 2.39ms
44: learn: 0.5200075 total: 18ms remaining: 2ms
45: learn: 0.5178263 total: 18.4ms remaining: 1.6ms
46: learn: 0.5150005 total: 18.7ms remaining: 1.2ms
47: learn: 0.5119695 total: 19.1ms remaining: 796us
48: learn: 0.5091177 total: 19.6ms remaining: 399us
49: learn: 0.5060475 total: 19.9ms remaining: 0us
0: learn: 0.6886303 total: 506us remaining: 24.8ms
1: learn: 0.6848097 total: 879us remaining: 21.1ms
2: learn: 0.6801248 total: 1.64ms remaining: 25.7ms
3: learn: 0.6770560 total: 2.02ms remaining: 23.3ms
4: learn: 0.6719868 total: 3.13ms remaining: 28.2ms
5: learn: 0.6677443 total: 3.43ms remaining: 25.2ms
6: learn: 0.6637682 total: 4.24ms remaining: 26.1ms
7: learn: 0.6595154 total: 5.58ms remaining: 29.3ms
8: learn: 0.6558830 total: 6.32ms remaining: 28.8ms
9: learn: 0.6517791 total: 7.6ms remaining: 30.4ms
10: learn: 0.6471647 total: 8.86ms remaining: 31.4ms
11: learn: 0.6430895 total: 9.68ms remaining: 30.7ms
12: learn: 0.6393340 total: 10.4ms remaining: 29.6ms
13: learn: 0.6354497 total: 11.3ms remaining: 29.1ms
14: learn: 0.6311215 total: 12.4ms remaining: 28.9ms
15: learn: 0.6275276 total: 13.6ms remaining: 28.9ms
16: learn: 0.6236488 total: 14.8ms remaining: 28.8ms
17: learn: 0.6202546 total: 15.9ms remaining: 28.3ms
18: learn: 0.6166884 total: 16.8ms remaining: 27.4ms
19: learn: 0.6129113 total: 18.1ms remaining: 27.2ms
20: learn: 0.6093491 total: 18.9ms remaining: 26.1ms
21: learn: 0.6065245 total: 19.4ms remaining: 24.7ms
22: learn: 0.6034042 total: 20ms remaining: 23.5ms
23: learn: 0.5997530 total: 20.6ms remaining: 22.3ms
24: learn: 0.5959880 total: 21.3ms remaining: 21.3ms
25: learn: 0.5922939 total: 22ms remaining: 20.3ms
26: learn: 0.5897792 total: 22.5ms remaining: 19.1ms
27: learn: 0.5861889 total: 23.4ms remaining: 18.4ms
28: learn: 0.5835273 total: 24.4ms remaining: 17.7ms
29: learn: 0.5808634 total: 25.1ms remaining: 16.7ms
30: learn: 0.5772724 total: 25.9ms remaining: 15.9ms
31: learn: 0.5740322 total: 26.6ms remaining: 15ms
32: learn: 0.5710547 total: 27.1ms remaining: 13.9ms
33: learn: 0.5677978 total: 27.6ms remaining: 13ms
34: learn: 0.5645430 total: 28.7ms remaining: 12.3ms
35: learn: 0.5610716 total: 29.7ms remaining: 11.6ms
36: learn: 0.5578719 total: 30.5ms remaining: 10.7ms
37: learn: 0.5552181 total: 31.1ms remaining: 9.81ms
38: learn: 0.5524750 total: 31.4ms remaining: 8.86ms
39: learn: 0.5500543 total: 31.9ms remaining: 7.98ms
40: learn: 0.5472859 total: 32.4ms remaining: 7.12ms
41: learn: 0.5445638 total: 33ms remaining: 6.29ms
42: learn: 0.5419454 total: 33.4ms remaining: 5.44ms
43: learn: 0.5391157 total: 33.9ms remaining: 4.63ms
44: learn: 0.5367238 total: 34.5ms remaining: 3.83ms
45: learn: 0.5340405 total: 35.1ms remaining: 3.05ms
46: learn: 0.5314801 total: 35.6ms remaining: 2.27ms
47: learn: 0.5288782 total: 36.1ms remaining: 1.5ms
48: learn: 0.5259124 total: 36.6ms remaining: 746us
49: learn: 0.5231423 total: 37.1ms remaining: 0us
0: learn: 0.6888083 total: 646us remaining: 31.7ms
1: learn: 0.6840100 total: 1.17ms remaining: 28ms
2: learn: 0.6794042 total: 1.54ms remaining: 24.2ms
3: learn: 0.6755347 total: 2.01ms remaining: 23.1ms
4: learn: 0.6710576 total: 2.44ms remaining: 22ms
5: learn: 0.6663095 total: 2.85ms remaining: 20.9ms
6: learn: 0.6626385 total: 3.24ms remaining: 19.9ms
7: learn: 0.6575897 total: 3.79ms remaining: 19.9ms
8: learn: 0.6527714 total: 4.26ms remaining: 19.4ms
9: learn: 0.6479300 total: 4.72ms remaining: 18.9ms
10: learn: 0.6447237 total: 5.23ms remaining: 18.5ms
11: learn: 0.6410750 total: 5.78ms remaining: 18.3ms
12: learn: 0.6368673 total: 6.23ms remaining: 17.7ms
13: learn: 0.6331436 total: 6.72ms remaining: 17.3ms
14: learn: 0.6287820 total: 7.17ms remaining: 16.7ms
15: learn: 0.6257715 total: 7.69ms remaining: 16.3ms
16: learn: 0.6217684 total: 8.13ms remaining: 15.8ms
17: learn: 0.6173308 total: 8.66ms remaining: 15.4ms
18: learn: 0.6138749 total: 9.17ms remaining: 15ms
19: learn: 0.6098700 total: 9.63ms remaining: 14.4ms
20: learn: 0.6058963 total: 10.2ms remaining: 14.1ms
21: learn: 0.6024329 total: 10.7ms remaining: 13.6ms
22: learn: 0.5992812 total: 11.1ms remaining: 13.1ms
23: learn: 0.5963980 total: 11.5ms remaining: 12.4ms
24: learn: 0.5932907 total: 11.9ms remaining: 11.9ms
25: learn: 0.5906091 total: 12.2ms remaining: 11.3ms
26: learn: 0.5863905 total: 12.7ms remaining: 10.8ms
27: learn: 0.5837556 total: 13.2ms remaining: 10.4ms
28: learn: 0.5809663 total: 13.8ms remaining: 9.96ms
29: learn: 0.5773555 total: 14.3ms remaining: 9.5ms
30: learn: 0.5738775 total: 14.7ms remaining: 9ms
31: learn: 0.5705333 total: 15.1ms remaining: 8.48ms
32: learn: 0.5675880 total: 15.6ms remaining: 8.04ms
33: learn: 0.5643922 total: 16.1ms remaining: 7.59ms
34: learn: 0.5619492 total: 16.5ms remaining: 7.07ms
35: learn: 0.5596123 total: 16.8ms remaining: 6.55ms
36: learn: 0.5563666 total: 17.1ms remaining: 6.02ms
37: learn: 0.5530458 total: 17.6ms remaining: 5.56ms
38: learn: 0.5505357 total: 17.9ms remaining: 5.04ms
39: learn: 0.5473357 total: 18.4ms remaining: 4.59ms
40: learn: 0.5448640 total: 18.8ms remaining: 4.13ms
41: learn: 0.5414981 total: 19.3ms remaining: 3.67ms
42: learn: 0.5392637 total: 19.7ms remaining: 3.21ms
43: learn: 0.5362985 total: 20.1ms remaining: 2.74ms
44: learn: 0.5339898 total: 20.4ms remaining: 2.27ms
45: learn: 0.5305500 total: 20.8ms remaining: 1.81ms
46: learn: 0.5278988 total: 21.2ms remaining: 1.35ms
47: learn: 0.5249098 total: 21.5ms remaining: 897us
48: learn: 0.5224657 total: 21.9ms remaining: 446us
49: learn: 0.5203313 total: 22.2ms remaining: 0us
0: learn: 0.6885484 total: 606us remaining: 29.7ms
1: learn: 0.6832990 total: 1.02ms remaining: 24.5ms
2: learn: 0.6784705 total: 1.37ms remaining: 21.5ms
3: learn: 0.6740281 total: 1.72ms remaining: 19.8ms
4: learn: 0.6689528 total: 2.1ms remaining: 18.9ms
5: learn: 0.6651681 total: 2.6ms remaining: 19.1ms
6: learn: 0.6602428 total: 3ms remaining: 18.4ms
7: learn: 0.6565745 total: 3.41ms remaining: 17.9ms
8: learn: 0.6526943 total: 3.78ms remaining: 17.2ms
9: learn: 0.6481739 total: 4.18ms remaining: 16.7ms
10: learn: 0.6445998 total: 4.52ms remaining: 16ms
11: learn: 0.6406037 total: 4.72ms remaining: 14.9ms
12: learn: 0.6369031 total: 5.14ms remaining: 14.6ms
13: learn: 0.6331369 total: 5.51ms remaining: 14.2ms
14: learn: 0.6293530 total: 5.95ms remaining: 13.9ms
15: learn: 0.6256101 total: 6.32ms remaining: 13.4ms
16: learn: 0.6223479 total: 6.75ms remaining: 13.1ms
17: learn: 0.6191322 total: 7.12ms remaining: 12.7ms
18: learn: 0.6159982 total: 7.48ms remaining: 12.2ms
19: learn: 0.6125919 total: 7.96ms remaining: 11.9ms
20: learn: 0.6087055 total: 8.45ms remaining: 11.7ms
21: learn: 0.6053173 total: 8.89ms remaining: 11.3ms
22: learn: 0.6016938 total: 9.3ms remaining: 10.9ms
23: learn: 0.5979304 total: 9.75ms remaining: 10.6ms
24: learn: 0.5951165 total: 10.1ms remaining: 10.1ms
25: learn: 0.5919794 total: 10.6ms remaining: 9.76ms
26: learn: 0.5887875 total: 11ms remaining: 9.35ms
27: learn: 0.5852175 total: 11.3ms remaining: 8.91ms
28: learn: 0.5813837 total: 11.5ms remaining: 8.36ms
29: learn: 0.5779479 total: 11.9ms remaining: 7.95ms
30: learn: 0.5747208 total: 12.3ms remaining: 7.57ms
31: learn: 0.5711797 total: 12.9ms remaining: 7.24ms
32: learn: 0.5692595 total: 13.4ms remaining: 6.89ms
33: learn: 0.5661201 total: 13.8ms remaining: 6.51ms
34: learn: 0.5634923 total: 14.4ms remaining: 6.17ms
35: learn: 0.5607615 total: 14.9ms remaining: 5.8ms
36: learn: 0.5580848 total: 15.4ms remaining: 5.39ms
37: learn: 0.5548941 total: 15.9ms remaining: 5.01ms
38: learn: 0.5520169 total: 16.3ms remaining: 4.59ms
39: learn: 0.5491247 total: 16.9ms remaining: 4.22ms
40: learn: 0.5471747 total: 17.6ms remaining: 3.86ms
41: learn: 0.5441979 total: 18.2ms remaining: 3.47ms
42: learn: 0.5408542 total: 18.7ms remaining: 3.04ms
43: learn: 0.5378777 total: 19.3ms remaining: 2.64ms
44: learn: 0.5352505 total: 19.8ms remaining: 2.2ms
45: learn: 0.5326579 total: 20.1ms remaining: 1.75ms
46: learn: 0.5301107 total: 20.5ms remaining: 1.31ms
47: learn: 0.5270970 total: 21ms remaining: 874us
48: learn: 0.5243750 total: 21.4ms remaining: 437us
49: learn: 0.5221046 total: 21.9ms remaining: 0us
0: learn: 0.6884063 total: 700us remaining: 34.3ms
1: learn: 0.6836852 total: 1.62ms remaining: 38.8ms
2: learn: 0.6790473 total: 1.79ms remaining: 28.1ms
3: learn: 0.6744778 total: 2.76ms remaining: 31.8ms
4: learn: 0.6704643 total: 4.36ms remaining: 39.2ms
5: learn: 0.6659800 total: 5.31ms remaining: 38.9ms
6: learn: 0.6620535 total: 7.33ms remaining: 45ms
7: learn: 0.6583088 total: 9.46ms remaining: 49.6ms
8: learn: 0.6545450 total: 11.6ms remaining: 53ms
9: learn: 0.6505076 total: 13.6ms remaining: 54.4ms
10: learn: 0.6461325 total: 15.5ms remaining: 55ms
11: learn: 0.6423199 total: 16.9ms remaining: 53.4ms
12: learn: 0.6383978 total: 19ms remaining: 54ms
13: learn: 0.6350323 total: 20.9ms remaining: 53.8ms
14: learn: 0.6308430 total: 22.7ms remaining: 53ms
15: learn: 0.6269783 total: 24.8ms remaining: 52.7ms
16: learn: 0.6229115 total: 26.7ms remaining: 51.8ms
17: learn: 0.6187152 total: 28.5ms remaining: 50.6ms
18: learn: 0.6157313 total: 30.9ms remaining: 50.5ms
19: learn: 0.6119853 total: 33ms remaining: 49.5ms
20: learn: 0.6076172 total: 34.8ms remaining: 48.1ms
21: learn: 0.6042139 total: 37.1ms remaining: 47.3ms
22: learn: 0.6002465 total: 39.3ms remaining: 46.1ms
23: learn: 0.5968348 total: 41ms remaining: 44.5ms
24: learn: 0.5937729 total: 42.5ms remaining: 42.5ms
25: learn: 0.5904123 total: 44.1ms remaining: 40.7ms
26: learn: 0.5879717 total: 45.5ms remaining: 38.8ms
27: learn: 0.5843604 total: 47.5ms remaining: 37.3ms
28: learn: 0.5809902 total: 49.4ms remaining: 35.8ms
29: learn: 0.5778176 total: 50.2ms remaining: 33.5ms
30: learn: 0.5746172 total: 50.8ms remaining: 31.2ms
31: learn: 0.5717418 total: 52ms remaining: 29.2ms
32: learn: 0.5683009 total: 53.8ms remaining: 27.7ms
33: learn: 0.5651867 total: 55.6ms remaining: 26.2ms
34: learn: 0.5622274 total: 57.6ms remaining: 24.7ms
35: learn: 0.5593118 total: 60ms remaining: 23.3ms
36: learn: 0.5561789 total: 61ms remaining: 21.4ms
37: learn: 0.5528329 total: 63.3ms remaining: 20ms
38: learn: 0.5501205 total: 65.1ms remaining: 18.3ms
39: learn: 0.5471151 total: 67.2ms remaining: 16.8ms
40: learn: 0.5442372 total: 69.3ms remaining: 15.2ms
41: learn: 0.5415876 total: 71.3ms remaining: 13.6ms
42: learn: 0.5386989 total: 72.6ms remaining: 11.8ms
43: learn: 0.5359933 total: 74.5ms remaining: 10.2ms
44: learn: 0.5330209 total: 75.3ms remaining: 8.37ms
45: learn: 0.5301928 total: 76.9ms remaining: 6.69ms
46: learn: 0.5281150 total: 78.8ms remaining: 5.03ms
47: learn: 0.5254504 total: 79.7ms remaining: 3.32ms
48: learn: 0.5232090 total: 81ms remaining: 1.65ms
49: learn: 0.5201480 total: 82.4ms remaining: 0us
0: learn: 0.6884317 total: 532us remaining: 26.1ms
1: learn: 0.6842219 total: 1.15ms remaining: 27.6ms
2: learn: 0.6788840 total: 1.6ms remaining: 25ms
3: learn: 0.6745675 total: 2.06ms remaining: 23.7ms
4: learn: 0.6701844 total: 2.23ms remaining: 20ms
5: learn: 0.6658574 total: 2.87ms remaining: 21ms
6: learn: 0.6613603 total: 3.43ms remaining: 21.1ms
7: learn: 0.6565105 total: 4.03ms remaining: 21.1ms
8: learn: 0.6527962 total: 4.6ms remaining: 21ms
9: learn: 0.6488140 total: 5.16ms remaining: 20.6ms
10: learn: 0.6442703 total: 5.76ms remaining: 20.4ms
11: learn: 0.6409121 total: 6.37ms remaining: 20.2ms
12: learn: 0.6361741 total: 6.75ms remaining: 19.2ms
13: learn: 0.6320954 total: 7.38ms remaining: 19ms
14: learn: 0.6277527 total: 8.08ms remaining: 18.9ms
15: learn: 0.6234568 total: 8.62ms remaining: 18.3ms
16: learn: 0.6197360 total: 9.21ms remaining: 17.9ms
17: learn: 0.6161622 total: 9.75ms remaining: 17.3ms
18: learn: 0.6126012 total: 10.3ms remaining: 16.8ms
19: learn: 0.6093254 total: 10.7ms remaining: 16.1ms
20: learn: 0.6050689 total: 11.4ms remaining: 15.7ms
21: learn: 0.6013927 total: 11.9ms remaining: 15.1ms
22: learn: 0.5975716 total: 12.5ms remaining: 14.7ms
23: learn: 0.5938500 total: 13.1ms remaining: 14.2ms
24: learn: 0.5904517 total: 13.7ms remaining: 13.7ms
25: learn: 0.5872580 total: 14.2ms remaining: 13.1ms
26: learn: 0.5841571 total: 14.6ms remaining: 12.4ms
27: learn: 0.5809397 total: 14.9ms remaining: 11.7ms
28: learn: 0.5776263 total: 15.4ms remaining: 11.2ms
29: learn: 0.5740388 total: 15.9ms remaining: 10.6ms
30: learn: 0.5708005 total: 16.2ms remaining: 9.94ms
31: learn: 0.5674383 total: 16.5ms remaining: 9.3ms
32: learn: 0.5643648 total: 16.9ms remaining: 8.7ms
33: learn: 0.5609892 total: 17.3ms remaining: 8.16ms
34: learn: 0.5574797 total: 17.8ms remaining: 7.64ms
35: learn: 0.5546357 total: 18.3ms remaining: 7.11ms
36: learn: 0.5519836 total: 18.7ms remaining: 6.58ms
37: learn: 0.5496245 total: 19ms remaining: 6.01ms
38: learn: 0.5462709 total: 19.3ms remaining: 5.44ms
39: learn: 0.5438816 total: 19.8ms remaining: 4.96ms
40: learn: 0.5412759 total: 20.1ms remaining: 4.41ms
41: learn: 0.5381534 total: 20.6ms remaining: 3.93ms
42: learn: 0.5350876 total: 21ms remaining: 3.41ms
43: learn: 0.5324341 total: 21.4ms remaining: 2.92ms
44: learn: 0.5297221 total: 21.8ms remaining: 2.42ms
45: learn: 0.5269718 total: 22.1ms remaining: 1.92ms
46: learn: 0.5241960 total: 22.3ms remaining: 1.43ms
47: learn: 0.5216740 total: 22.6ms remaining: 942us
48: learn: 0.5188038 total: 23ms remaining: 470us
49: learn: 0.5159962 total: 23.4ms remaining: 0us
0: learn: 0.6889045 total: 1.39ms remaining: 68.3ms
1: learn: 0.6837910 total: 3.27ms remaining: 78.6ms
2: learn: 0.6796625 total: 4.17ms remaining: 65.4ms
3: learn: 0.6755106 total: 4.88ms remaining: 56.1ms
4: learn: 0.6704930 total: 5.23ms remaining: 47.1ms
5: learn: 0.6659863 total: 6.64ms remaining: 48.7ms
6: learn: 0.6611236 total: 7.98ms remaining: 49ms
7: learn: 0.6570025 total: 10.1ms remaining: 52.9ms
8: learn: 0.6525141 total: 11.6ms remaining: 52.9ms
9: learn: 0.6488466 total: 13.7ms remaining: 54.7ms
10: learn: 0.6439622 total: 15.6ms remaining: 55.2ms
11: learn: 0.6399417 total: 17.5ms remaining: 55.3ms
12: learn: 0.6355915 total: 19.7ms remaining: 56.1ms
13: learn: 0.6311927 total: 22.2ms remaining: 57ms
14: learn: 0.6264151 total: 24.2ms remaining: 56.6ms
15: learn: 0.6224202 total: 26.3ms remaining: 55.8ms
16: learn: 0.6179388 total: 28.9ms remaining: 56ms
17: learn: 0.6142164 total: 30.9ms remaining: 55ms
18: learn: 0.6103310 total: 33.1ms remaining: 54.1ms
19: learn: 0.6061065 total: 35.6ms remaining: 53.4ms
20: learn: 0.6024065 total: 38.1ms remaining: 52.7ms
21: learn: 0.5988295 total: 40.5ms remaining: 51.5ms
22: learn: 0.5947404 total: 42.6ms remaining: 50ms
23: learn: 0.5927571 total: 45.1ms remaining: 48.9ms
24: learn: 0.5893932 total: 49.2ms remaining: 49.2ms
25: learn: 0.5859566 total: 53.1ms remaining: 49ms
26: learn: 0.5824083 total: 55.2ms remaining: 47ms
27: learn: 0.5785285 total: 57.4ms remaining: 45.1ms
28: learn: 0.5759217 total: 59.9ms remaining: 43.4ms
29: learn: 0.5725713 total: 61.2ms remaining: 40.8ms
30: learn: 0.5701297 total: 62.5ms remaining: 38.3ms
31: learn: 0.5669196 total: 64.9ms remaining: 36.5ms
32: learn: 0.5633647 total: 67ms remaining: 34.5ms
33: learn: 0.5600124 total: 68.7ms remaining: 32.3ms
34: learn: 0.5570166 total: 70.7ms remaining: 30.3ms
35: learn: 0.5536585 total: 72.7ms remaining: 28.3ms
36: learn: 0.5501919 total: 74.6ms remaining: 26.2ms
37: learn: 0.5476065 total: 76.6ms remaining: 24.2ms
38: learn: 0.5450105 total: 78.4ms remaining: 22.1ms
39: learn: 0.5427028 total: 80.1ms remaining: 20ms
40: learn: 0.5399803 total: 82.4ms remaining: 18.1ms
41: learn: 0.5374643 total: 83.3ms remaining: 15.9ms
42: learn: 0.5349718 total: 83.6ms remaining: 13.6ms
43: learn: 0.5319629 total: 85.1ms remaining: 11.6ms
44: learn: 0.5290022 total: 86.8ms remaining: 9.64ms
45: learn: 0.5263070 total: 89ms remaining: 7.74ms
46: learn: 0.5235474 total: 90.9ms remaining: 5.8ms
47: learn: 0.5206222 total: 91.9ms remaining: 3.83ms
48: learn: 0.5179628 total: 93.4ms remaining: 1.91ms
49: learn: 0.5158435 total: 94.9ms remaining: 0us
0: learn: 0.6881669 total: 464us remaining: 22.8ms
1: learn: 0.6833468 total: 833us remaining: 20ms
2: learn: 0.6791169 total: 1.19ms remaining: 18.6ms
3: learn: 0.6758376 total: 1.84ms remaining: 21.2ms
4: learn: 0.6712291 total: 2.57ms remaining: 23.1ms
5: learn: 0.6669118 total: 3.23ms remaining: 23.7ms
6: learn: 0.6632769 total: 3.93ms remaining: 24.2ms
7: learn: 0.6589892 total: 4.47ms remaining: 23.5ms
8: learn: 0.6545352 total: 4.91ms remaining: 22.4ms
9: learn: 0.6497433 total: 5.36ms remaining: 21.4ms
10: learn: 0.6459731 total: 5.86ms remaining: 20.8ms
11: learn: 0.6423070 total: 6.64ms remaining: 21ms
12: learn: 0.6378382 total: 7.08ms remaining: 20.2ms
13: learn: 0.6342830 total: 7.73ms remaining: 19.9ms
14: learn: 0.6305179 total: 8.7ms remaining: 20.3ms
15: learn: 0.6273236 total: 9.23ms remaining: 19.6ms
16: learn: 0.6233593 total: 9.75ms remaining: 18.9ms
17: learn: 0.6202737 total: 10.3ms remaining: 18.3ms
18: learn: 0.6164205 total: 10.7ms remaining: 17.5ms
19: learn: 0.6125395 total: 11.1ms remaining: 16.7ms
20: learn: 0.6093116 total: 11.5ms remaining: 15.8ms
21: learn: 0.6057181 total: 11.9ms remaining: 15.1ms
22: learn: 0.6021154 total: 12.3ms remaining: 14.5ms
23: learn: 0.5986431 total: 12.7ms remaining: 13.8ms
24: learn: 0.5948586 total: 13.1ms remaining: 13.1ms
25: learn: 0.5912135 total: 13.5ms remaining: 12.5ms
26: learn: 0.5876534 total: 13.9ms remaining: 11.9ms
27: learn: 0.5839998 total: 14.3ms remaining: 11.2ms
28: learn: 0.5802642 total: 14.7ms remaining: 10.6ms
29: learn: 0.5771353 total: 15.3ms remaining: 10.2ms
30: learn: 0.5739135 total: 15.7ms remaining: 9.65ms
31: learn: 0.5708365 total: 16.1ms remaining: 9.08ms
32: learn: 0.5672966 total: 16.5ms remaining: 8.47ms
33: learn: 0.5646367 total: 16.8ms remaining: 7.9ms
34: learn: 0.5611941 total: 17.1ms remaining: 7.35ms
35: learn: 0.5586881 total: 17.4ms remaining: 6.77ms
36: learn: 0.5552103 total: 17.8ms remaining: 6.26ms
37: learn: 0.5529219 total: 18.1ms remaining: 5.71ms
38: learn: 0.5496374 total: 18.4ms remaining: 5.17ms
39: learn: 0.5473225 total: 18.8ms remaining: 4.7ms
40: learn: 0.5442109 total: 19.3ms remaining: 4.24ms
41: learn: 0.5409920 total: 20ms remaining: 3.8ms
42: learn: 0.5386969 total: 20.3ms remaining: 3.31ms
43: learn: 0.5358866 total: 20.7ms remaining: 2.83ms
44: learn: 0.5330785 total: 21.1ms remaining: 2.35ms
45: learn: 0.5305521 total: 21.6ms remaining: 1.87ms
46: learn: 0.5278588 total: 22ms remaining: 1.4ms
47: learn: 0.5249340 total: 22.6ms remaining: 940us
48: learn: 0.5217724 total: 23ms remaining: 469us
49: learn: 0.5202045 total: 23.5ms remaining: 0us
0: learn: 0.6881313 total: 582us remaining: 28.5ms
1: learn: 0.6834396 total: 796us remaining: 19.1ms
2: learn: 0.6788678 total: 1.68ms remaining: 26.3ms
3: learn: 0.6745282 total: 2.65ms remaining: 30.5ms
4: learn: 0.6705313 total: 4.27ms remaining: 38.4ms
5: learn: 0.6664501 total: 5.62ms remaining: 41.2ms
6: learn: 0.6625251 total: 6.96ms remaining: 42.8ms
7: learn: 0.6589340 total: 8.45ms remaining: 44.4ms
8: learn: 0.6541913 total: 9.74ms remaining: 44.4ms
9: learn: 0.6498026 total: 10.8ms remaining: 43.2ms
10: learn: 0.6453986 total: 12.1ms remaining: 43ms
11: learn: 0.6421787 total: 13.4ms remaining: 42.4ms
12: learn: 0.6387500 total: 14.6ms remaining: 41.5ms
13: learn: 0.6352154 total: 15.6ms remaining: 40.2ms
14: learn: 0.6312877 total: 16.7ms remaining: 39.1ms
15: learn: 0.6279324 total: 18ms remaining: 38.3ms
16: learn: 0.6247067 total: 18.9ms remaining: 36.6ms
17: learn: 0.6210395 total: 20.1ms remaining: 35.7ms
18: learn: 0.6179746 total: 21.4ms remaining: 34.9ms
19: learn: 0.6143485 total: 22.2ms remaining: 33.3ms
20: learn: 0.6106524 total: 22.9ms remaining: 31.6ms
21: learn: 0.6066544 total: 24.2ms remaining: 30.8ms
22: learn: 0.6042317 total: 25.7ms remaining: 30.2ms
23: learn: 0.6005581 total: 27ms remaining: 29.2ms
24: learn: 0.5968835 total: 27.9ms remaining: 27.9ms
25: learn: 0.5931447 total: 29ms remaining: 26.7ms
26: learn: 0.5890090 total: 30.3ms remaining: 25.8ms
27: learn: 0.5856988 total: 31.3ms remaining: 24.6ms
28: learn: 0.5825885 total: 32.3ms remaining: 23.4ms
29: learn: 0.5794557 total: 33.3ms remaining: 22.2ms
30: learn: 0.5759834 total: 34.1ms remaining: 20.9ms
31: learn: 0.5738438 total: 35.2ms remaining: 19.8ms
32: learn: 0.5708319 total: 36.3ms remaining: 18.7ms
33: learn: 0.5679970 total: 37.2ms remaining: 17.5ms
34: learn: 0.5651570 total: 38.1ms remaining: 16.3ms
35: learn: 0.5631887 total: 39.2ms remaining: 15.2ms
36: learn: 0.5605275 total: 40.3ms remaining: 14.1ms
37: learn: 0.5577177 total: 41.5ms remaining: 13.1ms
38: learn: 0.5547583 total: 42.7ms remaining: 12ms
39: learn: 0.5524270 total: 44.3ms remaining: 11.1ms
40: learn: 0.5500806 total: 45.4ms remaining: 9.97ms
41: learn: 0.5475216 total: 46.9ms remaining: 8.94ms
42: learn: 0.5446629 total: 48.2ms remaining: 7.84ms
43: learn: 0.5419058 total: 49.3ms remaining: 6.72ms
44: learn: 0.5387193 total: 50.4ms remaining: 5.6ms
45: learn: 0.5362276 total: 51.7ms remaining: 4.49ms
46: learn: 0.5335108 total: 52.7ms remaining: 3.36ms
47: learn: 0.5313886 total: 53.6ms remaining: 2.23ms
48: learn: 0.5288603 total: 54.3ms remaining: 1.11ms
49: learn: 0.5261072 total: 55.3ms remaining: 0us
0: learn: 0.6882460 total: 494us remaining: 24.2ms
1: learn: 0.6843912 total: 903us remaining: 21.7ms
2: learn: 0.6800541 total: 1.32ms remaining: 20.8ms
3: learn: 0.6760891 total: 1.73ms remaining: 19.8ms
4: learn: 0.6712217 total: 2.19ms remaining: 19.7ms
5: learn: 0.6672614 total: 2.65ms remaining: 19.4ms
6: learn: 0.6625482 total: 3.1ms remaining: 19ms
7: learn: 0.6584332 total: 3.57ms remaining: 18.8ms
8: learn: 0.6542506 total: 3.98ms remaining: 18.1ms
9: learn: 0.6496036 total: 4.39ms remaining: 17.6ms
10: learn: 0.6458211 total: 4.76ms remaining: 16.9ms
11: learn: 0.6409358 total: 5.02ms remaining: 15.9ms
12: learn: 0.6366605 total: 5.49ms remaining: 15.6ms
13: learn: 0.6327545 total: 5.92ms remaining: 15.2ms
14: learn: 0.6285988 total: 6.28ms remaining: 14.7ms
15: learn: 0.6249008 total: 6.62ms remaining: 14.1ms
16: learn: 0.6202061 total: 7.04ms remaining: 13.7ms
17: learn: 0.6165730 total: 7.38ms remaining: 13.1ms
18: learn: 0.6143048 total: 7.8ms remaining: 12.7ms
19: learn: 0.6101026 total: 8.17ms remaining: 12.2ms
20: learn: 0.6062540 total: 8.57ms remaining: 11.8ms
21: learn: 0.6024226 total: 9.01ms remaining: 11.5ms
22: learn: 0.5992711 total: 9.41ms remaining: 11ms
23: learn: 0.5962842 total: 9.75ms remaining: 10.6ms
24: learn: 0.5930634 total: 10.1ms remaining: 10.1ms
25: learn: 0.5896212 total: 10.5ms remaining: 9.72ms
26: learn: 0.5862146 total: 10.9ms remaining: 9.31ms
27: learn: 0.5836004 total: 11.3ms remaining: 8.89ms
28: learn: 0.5806826 total: 11.7ms remaining: 8.51ms
29: learn: 0.5778009 total: 12.2ms remaining: 8.14ms
30: learn: 0.5747073 total: 12.6ms remaining: 7.73ms
31: learn: 0.5711173 total: 13ms remaining: 7.29ms
32: learn: 0.5676276 total: 13.4ms remaining: 6.9ms
33: learn: 0.5649525 total: 13.8ms remaining: 6.48ms
34: learn: 0.5614809 total: 14.1ms remaining: 6.06ms
35: learn: 0.5586365 total: 14.5ms remaining: 5.64ms
36: learn: 0.5555984 total: 15ms remaining: 5.28ms
37: learn: 0.5521355 total: 15.5ms remaining: 4.89ms
38: learn: 0.5491231 total: 16ms remaining: 4.52ms
39: learn: 0.5461905 total: 16.5ms remaining: 4.13ms
40: learn: 0.5435150 total: 17ms remaining: 3.73ms
41: learn: 0.5400794 total: 17.5ms remaining: 3.32ms
42: learn: 0.5369342 total: 18ms remaining: 2.92ms
43: learn: 0.5338750 total: 18.4ms remaining: 2.5ms
44: learn: 0.5307068 total: 18.8ms remaining: 2.09ms
45: learn: 0.5278970 total: 19.2ms remaining: 1.67ms
46: learn: 0.5253307 total: 19.6ms remaining: 1.25ms
47: learn: 0.5227516 total: 20ms remaining: 835us
48: learn: 0.5203989 total: 20.5ms remaining: 417us
49: learn: 0.5173267 total: 21ms remaining: 0us
0: learn: 0.6893152 total: 952us remaining: 46.7ms
1: learn: 0.6844835 total: 1.84ms remaining: 44.1ms
2: learn: 0.6800974 total: 2.55ms remaining: 40ms
3: learn: 0.6760208 total: 3.54ms remaining: 40.7ms
4: learn: 0.6719295 total: 5ms remaining: 45.1ms
5: learn: 0.6673758 total: 6.46ms remaining: 47.4ms
6: learn: 0.6621889 total: 8.23ms remaining: 50.5ms
7: learn: 0.6576188 total: 9.33ms remaining: 49ms
8: learn: 0.6534662 total: 11.5ms remaining: 52.4ms
9: learn: 0.6495321 total: 13.6ms remaining: 54.5ms
10: learn: 0.6456604 total: 15.1ms remaining: 53.6ms
11: learn: 0.6406508 total: 16.6ms remaining: 52.7ms
12: learn: 0.6362949 total: 18.3ms remaining: 52.1ms
13: learn: 0.6323097 total: 20.5ms remaining: 52.8ms
14: learn: 0.6280563 total: 21.6ms remaining: 50.4ms
15: learn: 0.6241475 total: 22.4ms remaining: 47.5ms
16: learn: 0.6197190 total: 23.8ms remaining: 46.2ms
17: learn: 0.6159291 total: 24.5ms remaining: 43.6ms
18: learn: 0.6123843 total: 25.9ms remaining: 42.2ms
19: learn: 0.6095165 total: 27.4ms remaining: 41.2ms
20: learn: 0.6066874 total: 28.8ms remaining: 39.8ms
21: learn: 0.6032681 total: 29.8ms remaining: 37.9ms
22: learn: 0.6000013 total: 31.5ms remaining: 37ms
23: learn: 0.5968636 total: 33ms remaining: 35.8ms
24: learn: 0.5936499 total: 34.7ms remaining: 34.7ms
25: learn: 0.5902258 total: 36.1ms remaining: 33.3ms
26: learn: 0.5872464 total: 37.8ms remaining: 32.2ms
27: learn: 0.5840496 total: 39ms remaining: 30.6ms
28: learn: 0.5803844 total: 40.6ms remaining: 29.4ms
29: learn: 0.5765966 total: 41.8ms remaining: 27.8ms
30: learn: 0.5729305 total: 43.1ms remaining: 26.4ms
31: learn: 0.5693852 total: 44.4ms remaining: 25ms
32: learn: 0.5662854 total: 45.5ms remaining: 23.4ms
33: learn: 0.5633348 total: 47ms remaining: 22.1ms
34: learn: 0.5603130 total: 48.4ms remaining: 20.7ms
35: learn: 0.5570380 total: 49.7ms remaining: 19.3ms
36: learn: 0.5535011 total: 51.1ms remaining: 18ms
37: learn: 0.5506970 total: 52.8ms remaining: 16.7ms
38: learn: 0.5475887 total: 54.1ms remaining: 15.3ms
39: learn: 0.5448545 total: 54.9ms remaining: 13.7ms
40: learn: 0.5416395 total: 55.7ms remaining: 12.2ms
41: learn: 0.5385460 total: 56.8ms remaining: 10.8ms
42: learn: 0.5358333 total: 58.3ms remaining: 9.49ms
43: learn: 0.5329987 total: 59ms remaining: 8.05ms
44: learn: 0.5306597 total: 59.6ms remaining: 6.63ms
45: learn: 0.5274482 total: 61.4ms remaining: 5.34ms
46: learn: 0.5249510 total: 63.2ms remaining: 4.03ms
47: learn: 0.5218486 total: 64.7ms remaining: 2.7ms
48: learn: 0.5192859 total: 66.2ms remaining: 1.35ms
49: learn: 0.5163437 total: 67.8ms remaining: 0us
0: learn: 0.6898834 total: 1.06ms remaining: 52.1ms
1: learn: 0.6842462 total: 1.84ms remaining: 44.3ms
2: learn: 0.6792858 total: 3.06ms remaining: 48ms
3: learn: 0.6746738 total: 4.41ms remaining: 50.7ms
4: learn: 0.6695928 total: 5.73ms remaining: 51.5ms
5: learn: 0.6661403 total: 7.44ms remaining: 54.6ms
6: learn: 0.6611720 total: 8.96ms remaining: 55.1ms
7: learn: 0.6566346 total: 9.51ms remaining: 49.9ms
8: learn: 0.6521007 total: 11.4ms remaining: 52.2ms
9: learn: 0.6483892 total: 12.9ms remaining: 51.7ms
10: learn: 0.6445378 total: 14.7ms remaining: 52.3ms
11: learn: 0.6402623 total: 16ms remaining: 50.6ms
12: learn: 0.6356433 total: 17.7ms remaining: 50.5ms
13: learn: 0.6320741 total: 19.7ms remaining: 50.6ms
14: learn: 0.6293125 total: 21.7ms remaining: 50.7ms
15: learn: 0.6252919 total: 23.9ms remaining: 50.8ms
16: learn: 0.6215804 total: 26ms remaining: 50.5ms
17: learn: 0.6181259 total: 28ms remaining: 49.7ms
18: learn: 0.6150490 total: 29.1ms remaining: 47.5ms
19: learn: 0.6107875 total: 30.4ms remaining: 45.6ms
20: learn: 0.6067928 total: 32ms remaining: 44.2ms
21: learn: 0.6026918 total: 33.9ms remaining: 43.1ms
22: learn: 0.5992985 total: 36.2ms remaining: 42.5ms
23: learn: 0.5966905 total: 37.4ms remaining: 40.5ms
24: learn: 0.5929736 total: 39.2ms remaining: 39.2ms
25: learn: 0.5903067 total: 41ms remaining: 37.8ms
26: learn: 0.5869536 total: 42.6ms remaining: 36.3ms
27: learn: 0.5834395 total: 44.8ms remaining: 35.2ms
28: learn: 0.5804730 total: 46.9ms remaining: 34ms
29: learn: 0.5774620 total: 48.9ms remaining: 32.6ms
30: learn: 0.5742536 total: 50.6ms remaining: 31ms
31: learn: 0.5709241 total: 51.8ms remaining: 29.1ms
32: learn: 0.5676292 total: 53.3ms remaining: 27.5ms
33: learn: 0.5644774 total: 55.5ms remaining: 26.1ms
34: learn: 0.5607723 total: 57.4ms remaining: 24.6ms
35: learn: 0.5575395 total: 59.2ms remaining: 23ms
36: learn: 0.5550180 total: 60.9ms remaining: 21.4ms
37: learn: 0.5519310 total: 62.6ms remaining: 19.8ms
38: learn: 0.5485639 total: 64.5ms remaining: 18.2ms
39: learn: 0.5453633 total: 66.3ms remaining: 16.6ms
40: learn: 0.5423500 total: 68.1ms remaining: 14.9ms
41: learn: 0.5392707 total: 70.2ms remaining: 13.4ms
42: learn: 0.5363255 total: 73.1ms remaining: 11.9ms
43: learn: 0.5335525 total: 75.3ms remaining: 10.3ms
44: learn: 0.5315166 total: 77.2ms remaining: 8.58ms
45: learn: 0.5284654 total: 79ms remaining: 6.87ms
46: learn: 0.5254547 total: 80.6ms remaining: 5.14ms
47: learn: 0.5226909 total: 81.4ms remaining: 3.39ms
48: learn: 0.5204454 total: 83.6ms remaining: 1.71ms
49: learn: 0.5174313 total: 85.7ms remaining: 0us
0: learn: 0.6896049 total: 565us remaining: 27.7ms
1: learn: 0.6849136 total: 1ms remaining: 24.1ms
2: learn: 0.6803576 total: 1.38ms remaining: 21.6ms
3: learn: 0.6768148 total: 1.74ms remaining: 20ms
4: learn: 0.6727358 total: 2.2ms remaining: 19.8ms
5: learn: 0.6684443 total: 2.84ms remaining: 20.9ms
6: learn: 0.6645143 total: 3.37ms remaining: 20.7ms
7: learn: 0.6608152 total: 3.92ms remaining: 20.6ms
8: learn: 0.6561977 total: 4.33ms remaining: 19.7ms
9: learn: 0.6516975 total: 5.02ms remaining: 20.1ms
10: learn: 0.6481278 total: 5.54ms remaining: 19.6ms
11: learn: 0.6440783 total: 6.18ms remaining: 19.6ms
12: learn: 0.6404162 total: 6.75ms remaining: 19.2ms
13: learn: 0.6370865 total: 7.22ms remaining: 18.6ms
14: learn: 0.6330448 total: 7.73ms remaining: 18ms
15: learn: 0.6293072 total: 8.2ms remaining: 17.4ms
16: learn: 0.6255331 total: 8.5ms remaining: 16.5ms
17: learn: 0.6212560 total: 8.99ms remaining: 16ms
18: learn: 0.6170654 total: 9.31ms remaining: 15.2ms
19: learn: 0.6131645 total: 9.74ms remaining: 14.6ms
20: learn: 0.6095121 total: 10.1ms remaining: 14ms
21: learn: 0.6071570 total: 10.5ms remaining: 13.3ms
22: learn: 0.6038082 total: 10.8ms remaining: 12.6ms
23: learn: 0.6003898 total: 11.1ms remaining: 12ms
24: learn: 0.5968834 total: 11.5ms remaining: 11.5ms
25: learn: 0.5930002 total: 11.9ms remaining: 10.9ms
26: learn: 0.5898503 total: 12.2ms remaining: 10.4ms
27: learn: 0.5866331 total: 12.6ms remaining: 9.9ms
28: learn: 0.5835247 total: 13ms remaining: 9.4ms
29: learn: 0.5802822 total: 13.4ms remaining: 8.91ms
30: learn: 0.5774329 total: 13.8ms remaining: 8.47ms
31: learn: 0.5738669 total: 14.2ms remaining: 7.98ms
32: learn: 0.5708731 total: 14.5ms remaining: 7.46ms
33: learn: 0.5672658 total: 14.8ms remaining: 6.95ms
34: learn: 0.5635881 total: 15.3ms remaining: 6.54ms
35: learn: 0.5611327 total: 15.5ms remaining: 6.04ms
36: learn: 0.5586970 total: 15.9ms remaining: 5.6ms
37: learn: 0.5557206 total: 16.2ms remaining: 5.12ms
38: learn: 0.5533724 total: 16.7ms remaining: 4.7ms
39: learn: 0.5506490 total: 16.8ms remaining: 4.21ms
40: learn: 0.5481978 total: 17.2ms remaining: 3.77ms
41: learn: 0.5449705 total: 17.8ms remaining: 3.38ms
42: learn: 0.5425293 total: 18.1ms remaining: 2.95ms
43: learn: 0.5401083 total: 18.6ms remaining: 2.54ms
44: learn: 0.5378922 total: 19.2ms remaining: 2.13ms
45: learn: 0.5347885 total: 19.7ms remaining: 1.72ms
46: learn: 0.5320211 total: 20.1ms remaining: 1.28ms
47: learn: 0.5299872 total: 20.4ms remaining: 851us
48: learn: 0.5269199 total: 20.9ms remaining: 426us
49: learn: 0.5241069 total: 21.3ms remaining: 0us
0: learn: 0.6881868 total: 610us remaining: 29.9ms
1: learn: 0.6829738 total: 1.24ms remaining: 29.7ms
2: learn: 0.6776030 total: 1.9ms remaining: 29.7ms
3: learn: 0.6738265 total: 2.62ms remaining: 30.1ms
4: learn: 0.6695172 total: 3.35ms remaining: 30.1ms
5: learn: 0.6648879 total: 4.14ms remaining: 30.4ms
6: learn: 0.6603464 total: 4.85ms remaining: 29.8ms
7: learn: 0.6554638 total: 5.56ms remaining: 29.2ms
8: learn: 0.6500920 total: 6.36ms remaining: 29ms
9: learn: 0.6452815 total: 6.96ms remaining: 27.8ms
10: learn: 0.6408534 total: 7.51ms remaining: 26.6ms
11: learn: 0.6366700 total: 8.23ms remaining: 26.1ms
12: learn: 0.6325433 total: 8.78ms remaining: 25ms
13: learn: 0.6274845 total: 9.3ms remaining: 23.9ms
14: learn: 0.6231921 total: 9.73ms remaining: 22.7ms
15: learn: 0.6185671 total: 10.3ms remaining: 21.9ms
16: learn: 0.6144366 total: 10.9ms remaining: 21.2ms
17: learn: 0.6104962 total: 11.6ms remaining: 20.6ms
18: learn: 0.6058526 total: 12.2ms remaining: 19.9ms
19: learn: 0.6011075 total: 12.7ms remaining: 19.1ms
20: learn: 0.5967877 total: 13.2ms remaining: 18.3ms
21: learn: 0.5921092 total: 13.7ms remaining: 17.4ms
22: learn: 0.5891360 total: 14.1ms remaining: 16.5ms
23: learn: 0.5852300 total: 14.5ms remaining: 15.7ms
24: learn: 0.5813717 total: 14.9ms remaining: 14.9ms
25: learn: 0.5776055 total: 15.3ms remaining: 14.1ms
26: learn: 0.5742867 total: 15.7ms remaining: 13.3ms
27: learn: 0.5700137 total: 16.1ms remaining: 12.7ms
28: learn: 0.5669218 total: 16.5ms remaining: 12ms
29: learn: 0.5633576 total: 16.9ms remaining: 11.3ms
30: learn: 0.5598367 total: 17.2ms remaining: 10.6ms
31: learn: 0.5569285 total: 17.7ms remaining: 9.93ms
32: learn: 0.5533664 total: 18.1ms remaining: 9.33ms
33: learn: 0.5500605 total: 18.6ms remaining: 8.73ms
34: learn: 0.5466335 total: 18.9ms remaining: 8.1ms
35: learn: 0.5439169 total: 19.2ms remaining: 7.48ms
36: learn: 0.5401360 total: 19.6ms remaining: 6.87ms
37: learn: 0.5374215 total: 19.9ms remaining: 6.29ms
38: learn: 0.5337420 total: 20.3ms remaining: 5.73ms
39: learn: 0.5306709 total: 20.8ms remaining: 5.2ms
40: learn: 0.5274138 total: 21.2ms remaining: 4.65ms
41: learn: 0.5238286 total: 21.5ms remaining: 4.1ms
42: learn: 0.5208717 total: 22ms remaining: 3.58ms
43: learn: 0.5179086 total: 22.3ms remaining: 3.04ms
44: learn: 0.5147624 total: 22.7ms remaining: 2.52ms
45: learn: 0.5118639 total: 23.2ms remaining: 2.02ms
46: learn: 0.5087510 total: 23.7ms remaining: 1.51ms
47: learn: 0.5062515 total: 24.2ms remaining: 1.01ms
48: learn: 0.5031440 total: 24.5ms remaining: 500us
49: learn: 0.4998412 total: 24.9ms remaining: 0us
0: learn: 0.6877711 total: 638us remaining: 31.3ms
1: learn: 0.6818905 total: 1.31ms remaining: 31.3ms
2: learn: 0.6769580 total: 1.89ms remaining: 29.6ms
3: learn: 0.6719819 total: 2.29ms remaining: 26.4ms
4: learn: 0.6675527 total: 2.46ms remaining: 22.1ms
5: learn: 0.6629202 total: 2.81ms remaining: 20.6ms
6: learn: 0.6587006 total: 3.24ms remaining: 19.9ms
7: learn: 0.6530943 total: 3.65ms remaining: 19.2ms
8: learn: 0.6487981 total: 4.16ms remaining: 18.9ms
9: learn: 0.6443788 total: 4.58ms remaining: 18.3ms
10: learn: 0.6410016 total: 5.07ms remaining: 18ms
11: learn: 0.6366268 total: 5.55ms remaining: 17.6ms
12: learn: 0.6316172 total: 5.79ms remaining: 16.5ms
13: learn: 0.6281314 total: 6.4ms remaining: 16.5ms
14: learn: 0.6239560 total: 6.87ms remaining: 16ms
15: learn: 0.6193631 total: 7.29ms remaining: 15.5ms
16: learn: 0.6150314 total: 7.8ms remaining: 15.1ms
17: learn: 0.6115652 total: 8.34ms remaining: 14.8ms
18: learn: 0.6084149 total: 8.91ms remaining: 14.5ms
19: learn: 0.6048026 total: 9.36ms remaining: 14ms
20: learn: 0.6008272 total: 9.81ms remaining: 13.5ms
21: learn: 0.5968209 total: 10.4ms remaining: 13.2ms
22: learn: 0.5930977 total: 10.9ms remaining: 12.8ms
23: learn: 0.5888537 total: 11.3ms remaining: 12.3ms
24: learn: 0.5851596 total: 11.8ms remaining: 11.8ms
25: learn: 0.5814760 total: 12.2ms remaining: 11.3ms
26: learn: 0.5787512 total: 12.6ms remaining: 10.7ms
27: learn: 0.5757786 total: 13.1ms remaining: 10.3ms
28: learn: 0.5730349 total: 13.6ms remaining: 9.82ms
29: learn: 0.5696539 total: 14.1ms remaining: 9.43ms
30: learn: 0.5661847 total: 14.6ms remaining: 8.93ms
31: learn: 0.5623431 total: 15ms remaining: 8.41ms
32: learn: 0.5595707 total: 15.4ms remaining: 7.92ms
33: learn: 0.5563657 total: 15.8ms remaining: 7.42ms
34: learn: 0.5532213 total: 16.1ms remaining: 6.91ms
35: learn: 0.5496803 total: 16.6ms remaining: 6.44ms
36: learn: 0.5464033 total: 17ms remaining: 5.96ms
37: learn: 0.5437436 total: 17.4ms remaining: 5.49ms
38: learn: 0.5404662 total: 17.6ms remaining: 4.95ms
39: learn: 0.5371853 total: 17.9ms remaining: 4.47ms
40: learn: 0.5340354 total: 18.3ms remaining: 4.01ms
41: learn: 0.5313063 total: 18.7ms remaining: 3.56ms
42: learn: 0.5287670 total: 19.2ms remaining: 3.12ms
43: learn: 0.5257274 total: 19.6ms remaining: 2.67ms
44: learn: 0.5226840 total: 20.1ms remaining: 2.23ms
45: learn: 0.5192317 total: 20.5ms remaining: 1.78ms
46: learn: 0.5162763 total: 20.7ms remaining: 1.32ms
47: learn: 0.5130371 total: 21.2ms remaining: 882us
48: learn: 0.5109865 total: 21.6ms remaining: 440us
49: learn: 0.5086913 total: 22.3ms remaining: 0us
0: learn: 0.6881719 total: 665us remaining: 32.6ms
1: learn: 0.6822875 total: 1.41ms remaining: 33.9ms
2: learn: 0.6772719 total: 2.07ms remaining: 32.5ms
3: learn: 0.6722092 total: 3.04ms remaining: 35ms
4: learn: 0.6668137 total: 3.89ms remaining: 35ms
5: learn: 0.6627716 total: 4.72ms remaining: 34.6ms
6: learn: 0.6577771 total: 5.54ms remaining: 34ms
7: learn: 0.6539072 total: 6.5ms remaining: 34.1ms
8: learn: 0.6498281 total: 7.41ms remaining: 33.8ms
9: learn: 0.6449070 total: 8.19ms remaining: 32.8ms
10: learn: 0.6409257 total: 8.86ms remaining: 31.4ms
11: learn: 0.6361254 total: 9.67ms remaining: 30.6ms
12: learn: 0.6330183 total: 10.8ms remaining: 30.8ms
13: learn: 0.6287459 total: 11.7ms remaining: 30.1ms
14: learn: 0.6242019 total: 13ms remaining: 30.2ms
15: learn: 0.6199916 total: 13.4ms remaining: 28.5ms
16: learn: 0.6158487 total: 14.4ms remaining: 27.9ms
17: learn: 0.6123479 total: 15.3ms remaining: 27.3ms
18: learn: 0.6079944 total: 16.4ms remaining: 26.7ms
19: learn: 0.6040598 total: 17.4ms remaining: 26ms
20: learn: 0.6006360 total: 18.1ms remaining: 25ms
21: learn: 0.5968636 total: 18.9ms remaining: 24.1ms
22: learn: 0.5938489 total: 20ms remaining: 23.4ms
23: learn: 0.5899764 total: 21.2ms remaining: 22.9ms
24: learn: 0.5861951 total: 22.1ms remaining: 22.1ms
25: learn: 0.5821266 total: 22.8ms remaining: 21.1ms
26: learn: 0.5785535 total: 23.7ms remaining: 20.2ms
27: learn: 0.5753401 total: 24.6ms remaining: 19.3ms
28: learn: 0.5713715 total: 25.3ms remaining: 18.3ms
29: learn: 0.5679069 total: 26ms remaining: 17.4ms
30: learn: 0.5639393 total: 26.7ms remaining: 16.4ms
31: learn: 0.5609833 total: 27.5ms remaining: 15.5ms
32: learn: 0.5575458 total: 28.2ms remaining: 14.5ms
33: learn: 0.5542170 total: 28.9ms remaining: 13.6ms
34: learn: 0.5514027 total: 29.8ms remaining: 12.8ms
35: learn: 0.5492300 total: 30.4ms remaining: 11.8ms
36: learn: 0.5457673 total: 31ms remaining: 10.9ms
37: learn: 0.5426376 total: 31.8ms remaining: 10.1ms
38: learn: 0.5399211 total: 32.8ms remaining: 9.24ms
39: learn: 0.5364066 total: 33.6ms remaining: 8.4ms
40: learn: 0.5339090 total: 34.4ms remaining: 7.54ms
41: learn: 0.5304076 total: 35.2ms remaining: 6.7ms
42: learn: 0.5277597 total: 36.2ms remaining: 5.89ms
43: learn: 0.5246306 total: 37ms remaining: 5.05ms
44: learn: 0.5211611 total: 37.8ms remaining: 4.2ms
45: learn: 0.5181130 total: 38.6ms remaining: 3.36ms
46: learn: 0.5151812 total: 39.4ms remaining: 2.51ms
47: learn: 0.5121221 total: 40.2ms remaining: 1.67ms
48: learn: 0.5090498 total: 41ms remaining: 837us
49: learn: 0.5066203 total: 41.7ms remaining: 0us
0: learn: 0.6885583 total: 678us remaining: 33.2ms
1: learn: 0.6834753 total: 1.24ms remaining: 29.8ms
2: learn: 0.6789470 total: 2.58ms remaining: 40.4ms
3: learn: 0.6755916 total: 4.21ms remaining: 48.4ms
4: learn: 0.6715284 total: 5.82ms remaining: 52.4ms
5: learn: 0.6673140 total: 6.7ms remaining: 49.2ms
6: learn: 0.6627801 total: 7.47ms remaining: 45.9ms
7: learn: 0.6575259 total: 8.77ms remaining: 46.1ms
8: learn: 0.6536172 total: 10.4ms remaining: 47.5ms
9: learn: 0.6497181 total: 11.8ms remaining: 47.1ms
10: learn: 0.6457477 total: 13.5ms remaining: 47.8ms
11: learn: 0.6413658 total: 14.3ms remaining: 45.4ms
12: learn: 0.6373549 total: 15.3ms remaining: 43.5ms
13: learn: 0.6336955 total: 16.1ms remaining: 41.3ms
14: learn: 0.6307611 total: 18ms remaining: 41.9ms
15: learn: 0.6264181 total: 19.8ms remaining: 42.2ms
16: learn: 0.6234381 total: 21.7ms remaining: 42.1ms
17: learn: 0.6204603 total: 23.1ms remaining: 41.1ms
18: learn: 0.6166111 total: 24.3ms remaining: 39.7ms
19: learn: 0.6133843 total: 26.1ms remaining: 39.1ms
20: learn: 0.6095574 total: 28.9ms remaining: 39.9ms
21: learn: 0.6058448 total: 30.3ms remaining: 38.5ms
22: learn: 0.6034212 total: 32.7ms remaining: 38.4ms
23: learn: 0.6010433 total: 34.6ms remaining: 37.4ms
24: learn: 0.5977298 total: 36.9ms remaining: 36.9ms
25: learn: 0.5951096 total: 39.3ms remaining: 36.3ms
26: learn: 0.5909807 total: 41.4ms remaining: 35.2ms
27: learn: 0.5869012 total: 43.1ms remaining: 33.9ms
28: learn: 0.5835725 total: 44.8ms remaining: 32.5ms
29: learn: 0.5798497 total: 46.4ms remaining: 30.9ms
30: learn: 0.5763549 total: 48.1ms remaining: 29.5ms
31: learn: 0.5730186 total: 50ms remaining: 28.1ms
32: learn: 0.5708685 total: 51.1ms remaining: 26.3ms
33: learn: 0.5670709 total: 52.4ms remaining: 24.6ms
34: learn: 0.5640302 total: 54.9ms remaining: 23.5ms
35: learn: 0.5606153 total: 56.5ms remaining: 22ms
36: learn: 0.5584798 total: 58ms remaining: 20.4ms
37: learn: 0.5551818 total: 60ms remaining: 18.9ms
38: learn: 0.5516796 total: 61.5ms remaining: 17.3ms
39: learn: 0.5484479 total: 63.3ms remaining: 15.8ms
40: learn: 0.5452117 total: 64.8ms remaining: 14.2ms
41: learn: 0.5417497 total: 66.1ms remaining: 12.6ms
42: learn: 0.5394314 total: 67.5ms remaining: 11ms
43: learn: 0.5371108 total: 69.1ms remaining: 9.42ms
44: learn: 0.5346540 total: 71.2ms remaining: 7.92ms
45: learn: 0.5318031 total: 72.6ms remaining: 6.32ms
46: learn: 0.5296798 total: 74.5ms remaining: 4.75ms
47: learn: 0.5272447 total: 76.5ms remaining: 3.19ms
48: learn: 0.5249427 total: 77.2ms remaining: 1.58ms
49: learn: 0.5225004 total: 77.5ms remaining: 0us
Evaluating param combo 4/16: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 6, 'boosted_cb_model__num_leaves': 16}
Duplicate found for this combo (hash=636eb05e979c6196fb2441a30772aa77). Removing old run: a1dc521c75e24062baf4b379706c7fd4
0: learn: 0.6882686 total: 1.59ms remaining: 77.9ms
1: learn: 0.6842287 total: 3.49ms remaining: 83.8ms
2: learn: 0.6795568 total: 6.48ms remaining: 102ms
3: learn: 0.6751320 total: 8.48ms remaining: 97.5ms
4: learn: 0.6706698 total: 10.2ms remaining: 92ms
5: learn: 0.6664309 total: 12.3ms remaining: 90.5ms
6: learn: 0.6621163 total: 14.7ms remaining: 90.4ms
7: learn: 0.6580139 total: 16.5ms remaining: 86.4ms
8: learn: 0.6528908 total: 19.9ms remaining: 90.7ms
9: learn: 0.6489797 total: 21.3ms remaining: 85.3ms
10: learn: 0.6451126 total: 24.3ms remaining: 86.1ms
11: learn: 0.6408115 total: 25.3ms remaining: 80ms
12: learn: 0.6371277 total: 28.5ms remaining: 81ms
13: learn: 0.6335652 total: 29.7ms remaining: 76.3ms
14: learn: 0.6295871 total: 33.2ms remaining: 77.5ms
15: learn: 0.6263508 total: 34.3ms remaining: 72.8ms
16: learn: 0.6230431 total: 36.5ms remaining: 70.8ms
17: learn: 0.6195287 total: 38.9ms remaining: 69.2ms
18: learn: 0.6158131 total: 41.2ms remaining: 67.3ms
19: learn: 0.6118211 total: 44.2ms remaining: 66.3ms
20: learn: 0.6079949 total: 44.9ms remaining: 62ms
21: learn: 0.6045961 total: 47.9ms remaining: 61ms
22: learn: 0.6018823 total: 50.7ms remaining: 59.5ms
23: learn: 0.5984668 total: 52.7ms remaining: 57.1ms
24: learn: 0.5950315 total: 53.8ms remaining: 53.8ms
25: learn: 0.5916049 total: 56.2ms remaining: 51.9ms
26: learn: 0.5879565 total: 58.2ms remaining: 49.6ms
27: learn: 0.5845201 total: 61.2ms remaining: 48.1ms
28: learn: 0.5810843 total: 62.5ms remaining: 45.2ms
29: learn: 0.5783354 total: 64.7ms remaining: 43.1ms
30: learn: 0.5757722 total: 66.9ms remaining: 41ms
31: learn: 0.5724279 total: 67.8ms remaining: 38.1ms
32: learn: 0.5683808 total: 70.1ms remaining: 36.1ms
33: learn: 0.5646839 total: 72.7ms remaining: 34.2ms
34: learn: 0.5617004 total: 75.6ms remaining: 32.4ms
35: learn: 0.5581773 total: 77ms remaining: 30ms
36: learn: 0.5554810 total: 80.2ms remaining: 28.2ms
37: learn: 0.5519617 total: 82.7ms remaining: 26.1ms
38: learn: 0.5486472 total: 84.8ms remaining: 23.9ms
39: learn: 0.5453312 total: 87.7ms remaining: 21.9ms
40: learn: 0.5429028 total: 88.7ms remaining: 19.5ms
41: learn: 0.5397943 total: 90.5ms remaining: 17.2ms
42: learn: 0.5365728 total: 92.4ms remaining: 15ms
43: learn: 0.5339146 total: 94.1ms remaining: 12.8ms
44: learn: 0.5313950 total: 97.2ms remaining: 10.8ms
45: learn: 0.5285409 total: 99.8ms remaining: 8.68ms
46: learn: 0.5255187 total: 102ms remaining: 6.53ms
47: learn: 0.5227771 total: 103ms remaining: 4.29ms
48: learn: 0.5197882 total: 105ms remaining: 2.15ms
49: learn: 0.5169313 total: 108ms remaining: 0us
0: learn: 0.6879001 total: 767us remaining: 37.6ms
1: learn: 0.6825664 total: 1.5ms remaining: 36ms
2: learn: 0.6778624 total: 1.94ms remaining: 30.4ms
3: learn: 0.6728604 total: 2.08ms remaining: 23.9ms
4: learn: 0.6685503 total: 3.05ms remaining: 27.5ms
5: learn: 0.6637032 total: 3.34ms remaining: 24.5ms
6: learn: 0.6594021 total: 4.92ms remaining: 30.2ms
7: learn: 0.6552368 total: 6.44ms remaining: 33.8ms
8: learn: 0.6509857 total: 8.28ms remaining: 37.7ms
9: learn: 0.6465138 total: 9.92ms remaining: 39.7ms
10: learn: 0.6427414 total: 11.5ms remaining: 40.7ms
11: learn: 0.6380331 total: 13.2ms remaining: 41.7ms
12: learn: 0.6340114 total: 14.7ms remaining: 41.9ms
13: learn: 0.6297944 total: 16.5ms remaining: 42.5ms
14: learn: 0.6258446 total: 17ms remaining: 39.7ms
15: learn: 0.6215928 total: 18.7ms remaining: 39.7ms
16: learn: 0.6177414 total: 20.5ms remaining: 39.8ms
17: learn: 0.6139682 total: 22.5ms remaining: 40ms
18: learn: 0.6104304 total: 23.5ms remaining: 38.4ms
19: learn: 0.6064870 total: 24.8ms remaining: 37.2ms
20: learn: 0.6031981 total: 25.6ms remaining: 35.4ms
21: learn: 0.5993083 total: 26.9ms remaining: 34.3ms
22: learn: 0.5955826 total: 28.5ms remaining: 33.5ms
23: learn: 0.5917941 total: 29.8ms remaining: 32.3ms
24: learn: 0.5885132 total: 31.7ms remaining: 31.7ms
25: learn: 0.5856755 total: 33.5ms remaining: 30.9ms
26: learn: 0.5819673 total: 34.4ms remaining: 29.3ms
27: learn: 0.5785210 total: 35.6ms remaining: 28ms
28: learn: 0.5748315 total: 37.4ms remaining: 27.1ms
29: learn: 0.5708227 total: 39ms remaining: 26ms
30: learn: 0.5676059 total: 39.5ms remaining: 24.2ms
31: learn: 0.5649729 total: 41.3ms remaining: 23.2ms
32: learn: 0.5616275 total: 43.5ms remaining: 22.4ms
33: learn: 0.5587318 total: 45.2ms remaining: 21.3ms
34: learn: 0.5551036 total: 46.5ms remaining: 19.9ms
35: learn: 0.5516173 total: 47.1ms remaining: 18.3ms
36: learn: 0.5482520 total: 49.4ms remaining: 17.4ms
37: learn: 0.5449372 total: 50.8ms remaining: 16ms
38: learn: 0.5416244 total: 52.1ms remaining: 14.7ms
39: learn: 0.5384832 total: 53.9ms remaining: 13.5ms
40: learn: 0.5356229 total: 54.3ms remaining: 11.9ms
41: learn: 0.5325318 total: 55.8ms remaining: 10.6ms
42: learn: 0.5291555 total: 57.7ms remaining: 9.39ms
43: learn: 0.5267413 total: 59.5ms remaining: 8.12ms
44: learn: 0.5239407 total: 61.5ms remaining: 6.84ms
45: learn: 0.5205549 total: 63.2ms remaining: 5.49ms
46: learn: 0.5179840 total: 64.5ms remaining: 4.12ms
47: learn: 0.5152208 total: 66.3ms remaining: 2.76ms
48: learn: 0.5124560 total: 68.1ms remaining: 1.39ms
49: learn: 0.5101575 total: 69.9ms remaining: 0us
0: learn: 0.6879038 total: 1.12ms remaining: 55ms
1: learn: 0.6833723 total: 2.73ms remaining: 65.5ms
2: learn: 0.6793155 total: 5.08ms remaining: 79.5ms
3: learn: 0.6755335 total: 6ms remaining: 69ms
4: learn: 0.6704722 total: 8.13ms remaining: 73.2ms
5: learn: 0.6661947 total: 11.7ms remaining: 85.8ms
6: learn: 0.6614249 total: 14.9ms remaining: 91.4ms
7: learn: 0.6571407 total: 16.2ms remaining: 85.1ms
8: learn: 0.6534196 total: 17.8ms remaining: 81ms
9: learn: 0.6486063 total: 20.5ms remaining: 82.1ms
10: learn: 0.6447700 total: 23.6ms remaining: 83.7ms
11: learn: 0.6408974 total: 25.5ms remaining: 80.8ms
12: learn: 0.6366198 total: 26.4ms remaining: 75.2ms
13: learn: 0.6325800 total: 29.5ms remaining: 75.8ms
14: learn: 0.6277010 total: 31.7ms remaining: 74ms
15: learn: 0.6239838 total: 35ms remaining: 74.4ms
16: learn: 0.6196373 total: 37.9ms remaining: 73.5ms
17: learn: 0.6153141 total: 40.1ms remaining: 71.3ms
18: learn: 0.6108362 total: 42.5ms remaining: 69.4ms
19: learn: 0.6073908 total: 43.4ms remaining: 65.1ms
20: learn: 0.6036408 total: 45.4ms remaining: 62.7ms
21: learn: 0.5996456 total: 47.7ms remaining: 60.8ms
22: learn: 0.5964015 total: 50.5ms remaining: 59.3ms
23: learn: 0.5925342 total: 51.7ms remaining: 56ms
24: learn: 0.5889169 total: 54.2ms remaining: 54.2ms
25: learn: 0.5853063 total: 56.6ms remaining: 52.2ms
26: learn: 0.5814669 total: 59.6ms remaining: 50.8ms
27: learn: 0.5781858 total: 60.9ms remaining: 47.8ms
28: learn: 0.5749039 total: 63.6ms remaining: 46ms
29: learn: 0.5712272 total: 65.4ms remaining: 43.6ms
30: learn: 0.5686921 total: 67.6ms remaining: 41.5ms
31: learn: 0.5653936 total: 70.3ms remaining: 39.6ms
32: learn: 0.5625110 total: 71.7ms remaining: 36.9ms
33: learn: 0.5589728 total: 74.2ms remaining: 34.9ms
34: learn: 0.5561277 total: 76.7ms remaining: 32.9ms
35: learn: 0.5527337 total: 79.2ms remaining: 30.8ms
36: learn: 0.5493706 total: 81.5ms remaining: 28.6ms
37: learn: 0.5462561 total: 83.8ms remaining: 26.5ms
38: learn: 0.5436280 total: 85.3ms remaining: 24.1ms
39: learn: 0.5413929 total: 86.2ms remaining: 21.5ms
40: learn: 0.5384565 total: 88.1ms remaining: 19.3ms
41: learn: 0.5354266 total: 90.5ms remaining: 17.2ms
42: learn: 0.5322718 total: 92.6ms remaining: 15.1ms
43: learn: 0.5294400 total: 95.2ms remaining: 13ms
44: learn: 0.5265466 total: 97.3ms remaining: 10.8ms
45: learn: 0.5235887 total: 99.7ms remaining: 8.67ms
46: learn: 0.5208130 total: 102ms remaining: 6.54ms
47: learn: 0.5181899 total: 105ms remaining: 4.37ms
48: learn: 0.5153383 total: 107ms remaining: 2.19ms
49: learn: 0.5125170 total: 109ms remaining: 0us
0: learn: 0.6877830 total: 603us remaining: 29.6ms
1: learn: 0.6834925 total: 1.57ms remaining: 37.8ms
2: learn: 0.6789587 total: 3.79ms remaining: 59.4ms
3: learn: 0.6742087 total: 4.76ms remaining: 54.7ms
4: learn: 0.6697901 total: 7.14ms remaining: 64.3ms
5: learn: 0.6651971 total: 9.17ms remaining: 67.2ms
6: learn: 0.6608724 total: 11.6ms remaining: 71.2ms
7: learn: 0.6570621 total: 14.1ms remaining: 74.3ms
8: learn: 0.6525786 total: 16.1ms remaining: 73.4ms
9: learn: 0.6485319 total: 19.1ms remaining: 76.5ms
10: learn: 0.6438388 total: 21.6ms remaining: 76.6ms
11: learn: 0.6399976 total: 22.7ms remaining: 71.9ms
12: learn: 0.6354262 total: 26.3ms remaining: 74.9ms
13: learn: 0.6318842 total: 27.8ms remaining: 71.6ms
14: learn: 0.6283602 total: 28.9ms remaining: 67.5ms
15: learn: 0.6239873 total: 30.5ms remaining: 64.8ms
16: learn: 0.6197663 total: 31.9ms remaining: 62ms
17: learn: 0.6160495 total: 34.1ms remaining: 60.6ms
18: learn: 0.6121943 total: 36.2ms remaining: 59.1ms
19: learn: 0.6087217 total: 38.4ms remaining: 57.5ms
20: learn: 0.6050074 total: 39.4ms remaining: 54.4ms
21: learn: 0.6011373 total: 40.9ms remaining: 52ms
22: learn: 0.5971935 total: 42.9ms remaining: 50.3ms
23: learn: 0.5927861 total: 44.1ms remaining: 47.8ms
24: learn: 0.5895579 total: 45.9ms remaining: 45.9ms
25: learn: 0.5859251 total: 46.9ms remaining: 43.3ms
26: learn: 0.5830791 total: 49.5ms remaining: 42.2ms
27: learn: 0.5796411 total: 51.1ms remaining: 40.2ms
28: learn: 0.5763626 total: 52.7ms remaining: 38.2ms
29: learn: 0.5729057 total: 53.9ms remaining: 35.9ms
30: learn: 0.5696363 total: 55.5ms remaining: 34ms
31: learn: 0.5660485 total: 56.8ms remaining: 31.9ms
32: learn: 0.5629348 total: 58.4ms remaining: 30.1ms
33: learn: 0.5591928 total: 60ms remaining: 28.2ms
34: learn: 0.5556857 total: 61.3ms remaining: 26.3ms
35: learn: 0.5529631 total: 62.2ms remaining: 24.2ms
36: learn: 0.5495135 total: 63.7ms remaining: 22.4ms
37: learn: 0.5466279 total: 65.1ms remaining: 20.6ms
38: learn: 0.5434318 total: 66.8ms remaining: 18.8ms
39: learn: 0.5405497 total: 68.1ms remaining: 17ms
40: learn: 0.5371825 total: 69.5ms remaining: 15.3ms
41: learn: 0.5341038 total: 70.6ms remaining: 13.4ms
42: learn: 0.5308408 total: 71.7ms remaining: 11.7ms
43: learn: 0.5280881 total: 73.3ms remaining: 9.99ms
44: learn: 0.5251138 total: 74.2ms remaining: 8.24ms
45: learn: 0.5217302 total: 75.7ms remaining: 6.58ms
46: learn: 0.5193920 total: 78ms remaining: 4.98ms
47: learn: 0.5161583 total: 79.3ms remaining: 3.3ms
48: learn: 0.5128156 total: 79.6ms remaining: 1.62ms
49: learn: 0.5101414 total: 81.2ms remaining: 0us
0: learn: 0.6885024 total: 748us remaining: 36.7ms
1: learn: 0.6846428 total: 1.91ms remaining: 45.9ms
2: learn: 0.6800668 total: 3.41ms remaining: 53.4ms
3: learn: 0.6756460 total: 4.67ms remaining: 53.7ms
4: learn: 0.6712975 total: 5.64ms remaining: 50.8ms
5: learn: 0.6676982 total: 7.21ms remaining: 52.9ms
6: learn: 0.6642036 total: 8.42ms remaining: 51.7ms
7: learn: 0.6601437 total: 9.43ms remaining: 49.5ms
8: learn: 0.6557718 total: 10.4ms remaining: 47.6ms
9: learn: 0.6518354 total: 11.9ms remaining: 47.7ms
10: learn: 0.6477087 total: 13.3ms remaining: 47.3ms
11: learn: 0.6435351 total: 14.1ms remaining: 44.5ms
12: learn: 0.6394047 total: 16.6ms remaining: 47.3ms
13: learn: 0.6358291 total: 18.9ms remaining: 48.6ms
14: learn: 0.6320091 total: 20.7ms remaining: 48.3ms
15: learn: 0.6279222 total: 22.1ms remaining: 47ms
16: learn: 0.6242789 total: 24ms remaining: 46.6ms
17: learn: 0.6204739 total: 24.6ms remaining: 43.8ms
18: learn: 0.6170662 total: 26.2ms remaining: 42.7ms
19: learn: 0.6138748 total: 27.8ms remaining: 41.7ms
20: learn: 0.6101235 total: 28.4ms remaining: 39.2ms
21: learn: 0.6064071 total: 30ms remaining: 38.2ms
22: learn: 0.6035068 total: 31.6ms remaining: 37.1ms
23: learn: 0.5998397 total: 33ms remaining: 35.8ms
24: learn: 0.5964036 total: 34.4ms remaining: 34.4ms
25: learn: 0.5933263 total: 35.7ms remaining: 32.9ms
26: learn: 0.5891559 total: 36.6ms remaining: 31.2ms
27: learn: 0.5862627 total: 38.4ms remaining: 30.2ms
28: learn: 0.5829233 total: 39.6ms remaining: 28.7ms
29: learn: 0.5799922 total: 40.8ms remaining: 27.2ms
30: learn: 0.5764535 total: 42.4ms remaining: 26ms
31: learn: 0.5732734 total: 43.8ms remaining: 24.6ms
32: learn: 0.5703912 total: 44.9ms remaining: 23.1ms
33: learn: 0.5677847 total: 46ms remaining: 21.6ms
34: learn: 0.5644089 total: 47.5ms remaining: 20.4ms
35: learn: 0.5612078 total: 48.7ms remaining: 19ms
36: learn: 0.5579158 total: 50ms remaining: 17.6ms
37: learn: 0.5548959 total: 51.2ms remaining: 16.2ms
38: learn: 0.5517562 total: 52.6ms remaining: 14.8ms
39: learn: 0.5485756 total: 54.1ms remaining: 13.5ms
40: learn: 0.5452331 total: 55.1ms remaining: 12.1ms
41: learn: 0.5428162 total: 56.4ms remaining: 10.7ms
42: learn: 0.5404160 total: 57.9ms remaining: 9.42ms
43: learn: 0.5380578 total: 58.6ms remaining: 8ms
44: learn: 0.5355467 total: 59.5ms remaining: 6.61ms
45: learn: 0.5328272 total: 60.6ms remaining: 5.27ms
46: learn: 0.5299021 total: 61.4ms remaining: 3.92ms
47: learn: 0.5268303 total: 62.4ms remaining: 2.6ms
48: learn: 0.5242317 total: 63.4ms remaining: 1.29ms
49: learn: 0.5215018 total: 64.4ms remaining: 0us
0: learn: 0.6884559 total: 686us remaining: 33.6ms
1: learn: 0.6843430 total: 1.75ms remaining: 42.1ms
2: learn: 0.6796675 total: 2.77ms remaining: 43.4ms
3: learn: 0.6752330 total: 3.87ms remaining: 44.5ms
4: learn: 0.6710458 total: 4.93ms remaining: 44.4ms
5: learn: 0.6662186 total: 5.87ms remaining: 43.1ms
6: learn: 0.6616622 total: 6.72ms remaining: 41.3ms
7: learn: 0.6570717 total: 7.71ms remaining: 40.5ms
8: learn: 0.6531693 total: 7.95ms remaining: 36.2ms
9: learn: 0.6481402 total: 8.86ms remaining: 35.4ms
10: learn: 0.6431442 total: 9.78ms remaining: 34.7ms
11: learn: 0.6392234 total: 10.9ms remaining: 34.5ms
12: learn: 0.6358571 total: 11.9ms remaining: 33.9ms
13: learn: 0.6329785 total: 12.8ms remaining: 33ms
14: learn: 0.6288396 total: 13.9ms remaining: 32.4ms
15: learn: 0.6247382 total: 14.9ms remaining: 31.6ms
16: learn: 0.6214486 total: 16ms remaining: 31ms
17: learn: 0.6172429 total: 16.8ms remaining: 29.8ms
18: learn: 0.6139519 total: 17.7ms remaining: 29ms
19: learn: 0.6099444 total: 18.6ms remaining: 27.9ms
20: learn: 0.6062216 total: 20.2ms remaining: 27.9ms
21: learn: 0.6022794 total: 21.4ms remaining: 27.3ms
22: learn: 0.5979682 total: 22.1ms remaining: 26ms
23: learn: 0.5944250 total: 22.8ms remaining: 24.7ms
24: learn: 0.5907559 total: 23.6ms remaining: 23.6ms
25: learn: 0.5870441 total: 24.3ms remaining: 22.4ms
26: learn: 0.5837875 total: 25.4ms remaining: 21.6ms
27: learn: 0.5807009 total: 26.2ms remaining: 20.6ms
28: learn: 0.5776645 total: 27ms remaining: 19.6ms
29: learn: 0.5749488 total: 28ms remaining: 18.7ms
30: learn: 0.5714051 total: 28.7ms remaining: 17.6ms
31: learn: 0.5682202 total: 29.5ms remaining: 16.6ms
32: learn: 0.5646589 total: 30.4ms remaining: 15.7ms
33: learn: 0.5612332 total: 31.2ms remaining: 14.7ms
34: learn: 0.5580240 total: 32ms remaining: 13.7ms
35: learn: 0.5552691 total: 32.5ms remaining: 12.6ms
36: learn: 0.5523508 total: 33ms remaining: 11.6ms
37: learn: 0.5496641 total: 33.6ms remaining: 10.6ms
38: learn: 0.5467321 total: 34.5ms remaining: 9.72ms
39: learn: 0.5439376 total: 35.3ms remaining: 8.83ms
40: learn: 0.5411322 total: 36.2ms remaining: 7.96ms
41: learn: 0.5381168 total: 36.7ms remaining: 6.99ms
42: learn: 0.5351125 total: 37.4ms remaining: 6.08ms
43: learn: 0.5321597 total: 38.1ms remaining: 5.2ms
44: learn: 0.5296471 total: 38.9ms remaining: 4.32ms
45: learn: 0.5268437 total: 39.7ms remaining: 3.45ms
46: learn: 0.5236156 total: 40.4ms remaining: 2.58ms
47: learn: 0.5209652 total: 41.2ms remaining: 1.71ms
48: learn: 0.5180780 total: 41.8ms remaining: 853us
49: learn: 0.5153886 total: 42.4ms remaining: 0us
0: learn: 0.6881859 total: 893us remaining: 43.8ms
1: learn: 0.6844111 total: 2.98ms remaining: 71.6ms
2: learn: 0.6799576 total: 5.42ms remaining: 84.9ms
3: learn: 0.6755406 total: 6.94ms remaining: 79.8ms
4: learn: 0.6713634 total: 9.12ms remaining: 82.1ms
5: learn: 0.6666994 total: 11ms remaining: 80.5ms
6: learn: 0.6625210 total: 13ms remaining: 79.7ms
7: learn: 0.6582164 total: 15.3ms remaining: 80.3ms
8: learn: 0.6542482 total: 17.7ms remaining: 80.5ms
9: learn: 0.6492810 total: 18.7ms remaining: 74.8ms
10: learn: 0.6452562 total: 19ms remaining: 67.3ms
11: learn: 0.6405148 total: 20.7ms remaining: 65.4ms
12: learn: 0.6367805 total: 22.3ms remaining: 63.5ms
13: learn: 0.6323872 total: 24.9ms remaining: 64ms
14: learn: 0.6287385 total: 27.6ms remaining: 64.4ms
15: learn: 0.6245734 total: 30.8ms remaining: 65.4ms
16: learn: 0.6206634 total: 33.6ms remaining: 65.2ms
17: learn: 0.6162322 total: 34.6ms remaining: 61.5ms
18: learn: 0.6130019 total: 37.6ms remaining: 61.3ms
19: learn: 0.6087847 total: 40.2ms remaining: 60.3ms
20: learn: 0.6049334 total: 42.8ms remaining: 59.1ms
21: learn: 0.6013334 total: 45.2ms remaining: 57.5ms
22: learn: 0.5976211 total: 47.8ms remaining: 56.1ms
23: learn: 0.5941389 total: 50.2ms remaining: 54.4ms
24: learn: 0.5904175 total: 52.8ms remaining: 52.8ms
25: learn: 0.5872989 total: 54.9ms remaining: 50.7ms
26: learn: 0.5843788 total: 56.7ms remaining: 48.3ms
27: learn: 0.5810017 total: 58.3ms remaining: 45.8ms
28: learn: 0.5770647 total: 60.2ms remaining: 43.6ms
29: learn: 0.5734684 total: 62.2ms remaining: 41.5ms
30: learn: 0.5696450 total: 63.5ms remaining: 38.9ms
31: learn: 0.5669904 total: 65.1ms remaining: 36.6ms
32: learn: 0.5633582 total: 66.7ms remaining: 34.4ms
33: learn: 0.5602036 total: 68.2ms remaining: 32.1ms
34: learn: 0.5574471 total: 69.9ms remaining: 29.9ms
35: learn: 0.5546641 total: 71ms remaining: 27.6ms
36: learn: 0.5520727 total: 72.3ms remaining: 25.4ms
37: learn: 0.5495315 total: 73.2ms remaining: 23.1ms
38: learn: 0.5463314 total: 74.2ms remaining: 20.9ms
39: learn: 0.5434985 total: 75.4ms remaining: 18.8ms
40: learn: 0.5406721 total: 76.5ms remaining: 16.8ms
41: learn: 0.5374358 total: 77.6ms remaining: 14.8ms
42: learn: 0.5343420 total: 79ms remaining: 12.9ms
43: learn: 0.5313562 total: 79.7ms remaining: 10.9ms
44: learn: 0.5281717 total: 80.7ms remaining: 8.97ms
45: learn: 0.5257638 total: 81.9ms remaining: 7.12ms
46: learn: 0.5230920 total: 82.9ms remaining: 5.29ms
47: learn: 0.5204169 total: 84ms remaining: 3.5ms
48: learn: 0.5177728 total: 85.1ms remaining: 1.74ms
49: learn: 0.5152570 total: 85.8ms remaining: 0us
0: learn: 0.6884949 total: 1.17ms remaining: 57.3ms
1: learn: 0.6839801 total: 2.88ms remaining: 69.3ms
2: learn: 0.6791613 total: 4.44ms remaining: 69.6ms
3: learn: 0.6742374 total: 6.5ms remaining: 74.8ms
4: learn: 0.6698433 total: 9.93ms remaining: 89.4ms
5: learn: 0.6657356 total: 13.3ms remaining: 97.7ms
6: learn: 0.6612373 total: 16.5ms remaining: 101ms
7: learn: 0.6568788 total: 18.4ms remaining: 96.4ms
8: learn: 0.6536179 total: 20ms remaining: 91.1ms
9: learn: 0.6493948 total: 20.8ms remaining: 83.2ms
10: learn: 0.6452865 total: 23ms remaining: 81.5ms
11: learn: 0.6416073 total: 24.7ms remaining: 78.3ms
12: learn: 0.6391135 total: 27.7ms remaining: 78.9ms
13: learn: 0.6346678 total: 30.6ms remaining: 78.6ms
14: learn: 0.6311541 total: 31.9ms remaining: 74.3ms
15: learn: 0.6275356 total: 34.7ms remaining: 73.8ms
16: learn: 0.6240274 total: 37.4ms remaining: 72.5ms
17: learn: 0.6205983 total: 39.2ms remaining: 69.6ms
18: learn: 0.6164443 total: 41.4ms remaining: 67.5ms
19: learn: 0.6122633 total: 42.1ms remaining: 63.2ms
20: learn: 0.6092096 total: 43.9ms remaining: 60.7ms
21: learn: 0.6057245 total: 46.8ms remaining: 59.5ms
22: learn: 0.6021422 total: 50ms remaining: 58.7ms
23: learn: 0.5981987 total: 52.2ms remaining: 56.5ms
24: learn: 0.5948994 total: 54.7ms remaining: 54.7ms
25: learn: 0.5921669 total: 57.1ms remaining: 52.7ms
26: learn: 0.5890569 total: 60.1ms remaining: 51.2ms
27: learn: 0.5853665 total: 62.4ms remaining: 49ms
28: learn: 0.5821117 total: 63.5ms remaining: 46ms
29: learn: 0.5789751 total: 66.6ms remaining: 44.4ms
30: learn: 0.5760359 total: 68.9ms remaining: 42.2ms
31: learn: 0.5726499 total: 70.8ms remaining: 39.8ms
32: learn: 0.5692102 total: 73.3ms remaining: 37.8ms
33: learn: 0.5659861 total: 75.4ms remaining: 35.5ms
34: learn: 0.5629366 total: 78.2ms remaining: 33.5ms
35: learn: 0.5603221 total: 80ms remaining: 31.1ms
36: learn: 0.5568870 total: 82.5ms remaining: 29ms
37: learn: 0.5537936 total: 84.9ms remaining: 26.8ms
38: learn: 0.5507915 total: 86.7ms remaining: 24.5ms
39: learn: 0.5481011 total: 88ms remaining: 22ms
40: learn: 0.5452670 total: 90.1ms remaining: 19.8ms
41: learn: 0.5417452 total: 91.6ms remaining: 17.5ms
42: learn: 0.5391062 total: 93.6ms remaining: 15.2ms
43: learn: 0.5367240 total: 95.3ms remaining: 13ms
44: learn: 0.5338743 total: 97.1ms remaining: 10.8ms
45: learn: 0.5310151 total: 98.9ms remaining: 8.6ms
46: learn: 0.5279937 total: 99.9ms remaining: 6.37ms
47: learn: 0.5251938 total: 101ms remaining: 4.22ms
48: learn: 0.5222575 total: 102ms remaining: 2.08ms
49: learn: 0.5198283 total: 104ms remaining: 0us
0: learn: 0.6878969 total: 708us remaining: 34.7ms
1: learn: 0.6834371 total: 1.71ms remaining: 41.1ms
2: learn: 0.6793659 total: 2.82ms remaining: 44.1ms
3: learn: 0.6746757 total: 3.55ms remaining: 40.9ms
4: learn: 0.6702315 total: 4.17ms remaining: 37.5ms
5: learn: 0.6660132 total: 5.16ms remaining: 37.8ms
6: learn: 0.6620074 total: 5.97ms remaining: 36.7ms
7: learn: 0.6577334 total: 6.72ms remaining: 35.3ms
8: learn: 0.6533813 total: 7.43ms remaining: 33.9ms
9: learn: 0.6487205 total: 8.21ms remaining: 32.8ms
10: learn: 0.6447954 total: 9ms remaining: 31.9ms
11: learn: 0.6401955 total: 9.67ms remaining: 30.6ms
12: learn: 0.6355459 total: 10.2ms remaining: 28.9ms
13: learn: 0.6317789 total: 10.9ms remaining: 28.1ms
14: learn: 0.6274385 total: 11.6ms remaining: 27.1ms
15: learn: 0.6239107 total: 12.3ms remaining: 26.1ms
16: learn: 0.6199303 total: 12.9ms remaining: 25.1ms
17: learn: 0.6165104 total: 13.5ms remaining: 24ms
18: learn: 0.6121992 total: 13.9ms remaining: 22.7ms
19: learn: 0.6090521 total: 14.5ms remaining: 21.8ms
20: learn: 0.6049247 total: 15.5ms remaining: 21.4ms
21: learn: 0.6009591 total: 16.2ms remaining: 20.6ms
22: learn: 0.5972106 total: 16.9ms remaining: 19.8ms
23: learn: 0.5934511 total: 17.1ms remaining: 18.5ms
24: learn: 0.5896801 total: 17.8ms remaining: 17.8ms
25: learn: 0.5857917 total: 18.3ms remaining: 16.9ms
26: learn: 0.5824441 total: 19ms remaining: 16.2ms
27: learn: 0.5782137 total: 19.7ms remaining: 15.5ms
28: learn: 0.5745073 total: 20.5ms remaining: 14.8ms
29: learn: 0.5714868 total: 21.2ms remaining: 14.2ms
30: learn: 0.5678615 total: 21.9ms remaining: 13.4ms
31: learn: 0.5640808 total: 22.4ms remaining: 12.6ms
32: learn: 0.5609747 total: 22.9ms remaining: 11.8ms
33: learn: 0.5572637 total: 23.4ms remaining: 11ms
34: learn: 0.5541527 total: 23.7ms remaining: 10.2ms
35: learn: 0.5513270 total: 24.3ms remaining: 9.47ms
36: learn: 0.5478448 total: 24.9ms remaining: 8.76ms
37: learn: 0.5444886 total: 25.3ms remaining: 7.99ms
38: learn: 0.5418822 total: 25.8ms remaining: 7.26ms
39: learn: 0.5388862 total: 26.6ms remaining: 6.64ms
40: learn: 0.5363399 total: 27.1ms remaining: 5.95ms
41: learn: 0.5334605 total: 28.2ms remaining: 5.36ms
42: learn: 0.5303391 total: 28.6ms remaining: 4.66ms
43: learn: 0.5271799 total: 29.2ms remaining: 3.98ms
44: learn: 0.5242581 total: 29.7ms remaining: 3.3ms
45: learn: 0.5213147 total: 30.2ms remaining: 2.63ms
46: learn: 0.5184782 total: 30.9ms remaining: 1.97ms
47: learn: 0.5158614 total: 31.5ms remaining: 1.31ms
48: learn: 0.5129389 total: 32.2ms remaining: 656us
49: learn: 0.5103840 total: 32.5ms remaining: 0us
0: learn: 0.6886303 total: 825us remaining: 40.4ms
1: learn: 0.6848532 total: 1.45ms remaining: 34.9ms
2: learn: 0.6807654 total: 2.12ms remaining: 33.2ms
3: learn: 0.6757109 total: 4.02ms remaining: 46.2ms
4: learn: 0.6712357 total: 6.43ms remaining: 57.9ms
5: learn: 0.6677912 total: 8.7ms remaining: 63.8ms
6: learn: 0.6633335 total: 10.5ms remaining: 64.5ms
7: learn: 0.6595120 total: 11.1ms remaining: 58.5ms
8: learn: 0.6547988 total: 11.7ms remaining: 53.2ms
9: learn: 0.6508939 total: 12.8ms remaining: 51.1ms
10: learn: 0.6474754 total: 14.5ms remaining: 51.6ms
11: learn: 0.6435513 total: 16.5ms remaining: 52.3ms
12: learn: 0.6399169 total: 18.5ms remaining: 52.8ms
13: learn: 0.6360485 total: 20.6ms remaining: 53ms
14: learn: 0.6320116 total: 21.3ms remaining: 49.7ms
15: learn: 0.6283337 total: 23.6ms remaining: 50.2ms
16: learn: 0.6250711 total: 25.9ms remaining: 50.3ms
17: learn: 0.6215353 total: 28.6ms remaining: 50.9ms
18: learn: 0.6181452 total: 30.5ms remaining: 49.8ms
19: learn: 0.6144406 total: 32.5ms remaining: 48.7ms
20: learn: 0.6107661 total: 34.4ms remaining: 47.6ms
21: learn: 0.6074419 total: 36.3ms remaining: 46.2ms
22: learn: 0.6040458 total: 38.3ms remaining: 45ms
23: learn: 0.6009696 total: 40.6ms remaining: 44ms
24: learn: 0.5974595 total: 42.8ms remaining: 42.8ms
25: learn: 0.5941549 total: 44.8ms remaining: 41.3ms
26: learn: 0.5910260 total: 46.4ms remaining: 39.6ms
27: learn: 0.5879450 total: 47.6ms remaining: 37.4ms
28: learn: 0.5848512 total: 49.6ms remaining: 35.9ms
29: learn: 0.5820150 total: 51.6ms remaining: 34.4ms
30: learn: 0.5785911 total: 53.5ms remaining: 32.8ms
31: learn: 0.5753528 total: 54.9ms remaining: 30.9ms
32: learn: 0.5725451 total: 57.2ms remaining: 29.5ms
33: learn: 0.5700475 total: 58.5ms remaining: 27.5ms
34: learn: 0.5667296 total: 60.8ms remaining: 26.1ms
35: learn: 0.5638499 total: 62.3ms remaining: 24.2ms
36: learn: 0.5609647 total: 63.7ms remaining: 22.4ms
37: learn: 0.5576302 total: 64.6ms remaining: 20.4ms
38: learn: 0.5545089 total: 66.3ms remaining: 18.7ms
39: learn: 0.5522755 total: 68.3ms remaining: 17.1ms
40: learn: 0.5490363 total: 70.3ms remaining: 15.4ms
41: learn: 0.5461510 total: 70.9ms remaining: 13.5ms
42: learn: 0.5430897 total: 71.3ms remaining: 11.6ms
43: learn: 0.5398943 total: 72.6ms remaining: 9.89ms
44: learn: 0.5372769 total: 74.6ms remaining: 8.29ms
45: learn: 0.5347099 total: 76.9ms remaining: 6.68ms
46: learn: 0.5322609 total: 78.4ms remaining: 5ms
47: learn: 0.5299848 total: 80.4ms remaining: 3.35ms
48: learn: 0.5275044 total: 83.8ms remaining: 1.71ms
49: learn: 0.5247466 total: 85.7ms remaining: 0us
0: learn: 0.6890516 total: 618us remaining: 30.3ms
1: learn: 0.6843684 total: 1.28ms remaining: 30.8ms
2: learn: 0.6798558 total: 3.25ms remaining: 50.9ms
3: learn: 0.6749312 total: 5.38ms remaining: 61.9ms
4: learn: 0.6708149 total: 8.43ms remaining: 75.9ms
5: learn: 0.6666183 total: 10.8ms remaining: 79.2ms
6: learn: 0.6614496 total: 12.9ms remaining: 79ms
7: learn: 0.6573563 total: 14.7ms remaining: 77.2ms
8: learn: 0.6532401 total: 16.5ms remaining: 75ms
9: learn: 0.6500008 total: 18.4ms remaining: 73.6ms
10: learn: 0.6454487 total: 20.2ms remaining: 71.5ms
11: learn: 0.6410700 total: 21.7ms remaining: 68.7ms
12: learn: 0.6370755 total: 24ms remaining: 68.3ms
13: learn: 0.6327961 total: 25.5ms remaining: 65.6ms
14: learn: 0.6294922 total: 27ms remaining: 63.1ms
15: learn: 0.6260024 total: 28.8ms remaining: 61.3ms
16: learn: 0.6225606 total: 30.8ms remaining: 59.7ms
17: learn: 0.6194160 total: 32.3ms remaining: 57.4ms
18: learn: 0.6160197 total: 34.3ms remaining: 56ms
19: learn: 0.6121442 total: 36.3ms remaining: 54.5ms
20: learn: 0.6080268 total: 38ms remaining: 52.5ms
21: learn: 0.6045500 total: 39.5ms remaining: 50.3ms
22: learn: 0.6005688 total: 41.3ms remaining: 48.5ms
23: learn: 0.5973170 total: 42.8ms remaining: 46.4ms
24: learn: 0.5939270 total: 45.1ms remaining: 45.1ms
25: learn: 0.5907113 total: 46.6ms remaining: 43ms
26: learn: 0.5874044 total: 48.3ms remaining: 41.1ms
27: learn: 0.5838297 total: 49.8ms remaining: 39.1ms
28: learn: 0.5804910 total: 51.3ms remaining: 37.2ms
29: learn: 0.5767455 total: 52.5ms remaining: 35ms
30: learn: 0.5729896 total: 53.8ms remaining: 33ms
31: learn: 0.5699794 total: 56.5ms remaining: 31.8ms
32: learn: 0.5664476 total: 58.1ms remaining: 30ms
33: learn: 0.5630630 total: 59.6ms remaining: 28ms
34: learn: 0.5606261 total: 61.2ms remaining: 26.2ms
35: learn: 0.5575230 total: 62.4ms remaining: 24.3ms
36: learn: 0.5543894 total: 63.5ms remaining: 22.3ms
37: learn: 0.5513296 total: 64.8ms remaining: 20.5ms
38: learn: 0.5481648 total: 66.1ms remaining: 18.7ms
39: learn: 0.5451467 total: 67.1ms remaining: 16.8ms
40: learn: 0.5421398 total: 68.4ms remaining: 15ms
41: learn: 0.5394561 total: 69.5ms remaining: 13.2ms
42: learn: 0.5365911 total: 70.5ms remaining: 11.5ms
43: learn: 0.5336141 total: 71.6ms remaining: 9.77ms
44: learn: 0.5307000 total: 72.9ms remaining: 8.1ms
45: learn: 0.5278690 total: 73.9ms remaining: 6.43ms
46: learn: 0.5250755 total: 74.9ms remaining: 4.78ms
47: learn: 0.5225611 total: 75.8ms remaining: 3.16ms
48: learn: 0.5201245 total: 77ms remaining: 1.57ms
49: learn: 0.5169272 total: 78ms remaining: 0us
0: learn: 0.6884397 total: 770us remaining: 37.7ms
1: learn: 0.6835109 total: 1.63ms remaining: 39ms
2: learn: 0.6795496 total: 2.26ms remaining: 35.4ms
3: learn: 0.6751464 total: 3.46ms remaining: 39.8ms
4: learn: 0.6712628 total: 4.67ms remaining: 42.1ms
5: learn: 0.6671028 total: 5.89ms remaining: 43.2ms
6: learn: 0.6631048 total: 7.3ms remaining: 44.8ms
7: learn: 0.6589732 total: 8.4ms remaining: 44.1ms
8: learn: 0.6543210 total: 9.5ms remaining: 43.3ms
9: learn: 0.6503692 total: 10.8ms remaining: 43.2ms
10: learn: 0.6460389 total: 11.3ms remaining: 39.9ms
11: learn: 0.6423343 total: 12.6ms remaining: 39.9ms
12: learn: 0.6382826 total: 13.6ms remaining: 38.7ms
13: learn: 0.6341497 total: 14.7ms remaining: 37.7ms
14: learn: 0.6306207 total: 16ms remaining: 37.3ms
15: learn: 0.6267048 total: 17.3ms remaining: 36.8ms
16: learn: 0.6235354 total: 18.5ms remaining: 35.9ms
17: learn: 0.6202240 total: 19.5ms remaining: 34.6ms
18: learn: 0.6164425 total: 20.4ms remaining: 33.3ms
19: learn: 0.6124860 total: 21.3ms remaining: 32ms
20: learn: 0.6090163 total: 22.6ms remaining: 31.2ms
21: learn: 0.6057270 total: 23.8ms remaining: 30.2ms
22: learn: 0.6017883 total: 24.8ms remaining: 29.1ms
23: learn: 0.5982530 total: 25.7ms remaining: 27.8ms
24: learn: 0.5953937 total: 27ms remaining: 27ms
25: learn: 0.5918257 total: 27.5ms remaining: 25.4ms
26: learn: 0.5884519 total: 28ms remaining: 23.8ms
27: learn: 0.5852720 total: 29ms remaining: 22.8ms
28: learn: 0.5822245 total: 30.1ms remaining: 21.8ms
29: learn: 0.5795334 total: 31.2ms remaining: 20.8ms
30: learn: 0.5760984 total: 32ms remaining: 19.6ms
31: learn: 0.5726693 total: 33.1ms remaining: 18.6ms
32: learn: 0.5692743 total: 34ms remaining: 17.5ms
33: learn: 0.5666976 total: 34.7ms remaining: 16.3ms
34: learn: 0.5634716 total: 35.5ms remaining: 15.2ms
35: learn: 0.5606266 total: 36.3ms remaining: 14.1ms
36: learn: 0.5577354 total: 37.4ms remaining: 13.1ms
37: learn: 0.5545374 total: 38.4ms remaining: 12.1ms
38: learn: 0.5516278 total: 39.2ms remaining: 11.1ms
39: learn: 0.5482702 total: 39.9ms remaining: 9.98ms
40: learn: 0.5451829 total: 40.4ms remaining: 8.87ms
41: learn: 0.5421858 total: 41.3ms remaining: 7.87ms
42: learn: 0.5394945 total: 41.8ms remaining: 6.8ms
43: learn: 0.5367497 total: 42.6ms remaining: 5.82ms
44: learn: 0.5341104 total: 43.4ms remaining: 4.82ms
45: learn: 0.5318895 total: 44.2ms remaining: 3.84ms
46: learn: 0.5292087 total: 44.9ms remaining: 2.87ms
47: learn: 0.5268572 total: 45.7ms remaining: 1.91ms
48: learn: 0.5241924 total: 46.5ms remaining: 949us
49: learn: 0.5215148 total: 47.2ms remaining: 0us
0: learn: 0.6884211 total: 624us remaining: 30.6ms
1: learn: 0.6839877 total: 1.78ms remaining: 42.8ms
2: learn: 0.6791304 total: 3.02ms remaining: 47.3ms
3: learn: 0.6745372 total: 4.2ms remaining: 48.3ms
4: learn: 0.6705564 total: 5.66ms remaining: 51ms
5: learn: 0.6657961 total: 6.95ms remaining: 50.9ms
6: learn: 0.6618196 total: 8ms remaining: 49.1ms
7: learn: 0.6577615 total: 9.38ms remaining: 49.2ms
8: learn: 0.6540484 total: 10.9ms remaining: 49.5ms
9: learn: 0.6503615 total: 12ms remaining: 48.1ms
10: learn: 0.6460645 total: 12.5ms remaining: 44.3ms
11: learn: 0.6420317 total: 13.8ms remaining: 43.6ms
12: learn: 0.6381292 total: 15ms remaining: 42.8ms
13: learn: 0.6339102 total: 16ms remaining: 41.2ms
14: learn: 0.6301877 total: 17.5ms remaining: 40.8ms
15: learn: 0.6264915 total: 18.9ms remaining: 40.3ms
16: learn: 0.6233760 total: 20.2ms remaining: 39.3ms
17: learn: 0.6198275 total: 21.6ms remaining: 38.5ms
18: learn: 0.6163971 total: 23ms remaining: 37.6ms
19: learn: 0.6123553 total: 24.3ms remaining: 36.4ms
20: learn: 0.6089223 total: 25.6ms remaining: 35.4ms
21: learn: 0.6059369 total: 27.8ms remaining: 35.4ms
22: learn: 0.6028116 total: 29.2ms remaining: 34.3ms
23: learn: 0.5990138 total: 30.5ms remaining: 33ms
24: learn: 0.5957435 total: 31.8ms remaining: 31.8ms
25: learn: 0.5925306 total: 33ms remaining: 30.5ms
26: learn: 0.5892411 total: 34.1ms remaining: 29.1ms
27: learn: 0.5857035 total: 35.6ms remaining: 28ms
28: learn: 0.5820093 total: 36.8ms remaining: 26.6ms
29: learn: 0.5790695 total: 37.8ms remaining: 25.2ms
30: learn: 0.5761151 total: 38.9ms remaining: 23.8ms
31: learn: 0.5730415 total: 40.2ms remaining: 22.6ms
32: learn: 0.5701165 total: 41.4ms remaining: 21.3ms
33: learn: 0.5666366 total: 42.9ms remaining: 20.2ms
34: learn: 0.5632009 total: 44ms remaining: 18.9ms
35: learn: 0.5600635 total: 45.3ms remaining: 17.6ms
36: learn: 0.5569608 total: 45.6ms remaining: 16ms
37: learn: 0.5540607 total: 46.8ms remaining: 14.8ms
38: learn: 0.5510633 total: 47.3ms remaining: 13.3ms
39: learn: 0.5483487 total: 49.2ms remaining: 12.3ms
40: learn: 0.5452854 total: 50.7ms remaining: 11.1ms
41: learn: 0.5423161 total: 52ms remaining: 9.91ms
42: learn: 0.5392697 total: 53.3ms remaining: 8.67ms
43: learn: 0.5359676 total: 54.3ms remaining: 7.41ms
44: learn: 0.5334258 total: 55.5ms remaining: 6.17ms
45: learn: 0.5303586 total: 56.6ms remaining: 4.92ms
46: learn: 0.5272792 total: 57.7ms remaining: 3.68ms
47: learn: 0.5248664 total: 58.6ms remaining: 2.44ms
48: learn: 0.5221360 total: 59.8ms remaining: 1.22ms
49: learn: 0.5195198 total: 60.3ms remaining: 0us
0: learn: 0.6878325 total: 641us remaining: 31.4ms
1: learn: 0.6829833 total: 1.04ms remaining: 25ms
2: learn: 0.6783131 total: 1.54ms remaining: 24.1ms
3: learn: 0.6737727 total: 1.93ms remaining: 22.2ms
4: learn: 0.6691293 total: 2.59ms remaining: 23.3ms
5: learn: 0.6650105 total: 3.62ms remaining: 26.6ms
6: learn: 0.6608788 total: 4.38ms remaining: 26.9ms
7: learn: 0.6565204 total: 5.24ms remaining: 27.5ms
8: learn: 0.6524720 total: 6.19ms remaining: 28.2ms
9: learn: 0.6474983 total: 7ms remaining: 28ms
10: learn: 0.6435726 total: 7.99ms remaining: 28.3ms
11: learn: 0.6388268 total: 8.78ms remaining: 27.8ms
12: learn: 0.6351269 total: 9.32ms remaining: 26.5ms
13: learn: 0.6311666 total: 10.3ms remaining: 26.6ms
14: learn: 0.6277222 total: 11.1ms remaining: 25.9ms
15: learn: 0.6240896 total: 11.5ms remaining: 24.5ms
16: learn: 0.6204197 total: 12.3ms remaining: 23.9ms
17: learn: 0.6168908 total: 13.1ms remaining: 23.3ms
18: learn: 0.6124770 total: 13.3ms remaining: 21.8ms
19: learn: 0.6086259 total: 14ms remaining: 21ms
20: learn: 0.6052431 total: 14.7ms remaining: 20.3ms
21: learn: 0.6019313 total: 15.9ms remaining: 20.2ms
22: learn: 0.5985107 total: 16.7ms remaining: 19.6ms
23: learn: 0.5946564 total: 17.4ms remaining: 18.8ms
24: learn: 0.5909908 total: 18.4ms remaining: 18.4ms
25: learn: 0.5875362 total: 18.7ms remaining: 17.3ms
26: learn: 0.5837100 total: 19.5ms remaining: 16.6ms
27: learn: 0.5800796 total: 20.3ms remaining: 15.9ms
28: learn: 0.5767052 total: 20.5ms remaining: 14.9ms
29: learn: 0.5738579 total: 21.8ms remaining: 14.5ms
30: learn: 0.5707203 total: 22.5ms remaining: 13.8ms
31: learn: 0.5674669 total: 22.7ms remaining: 12.8ms
32: learn: 0.5643868 total: 23.4ms remaining: 12.1ms
33: learn: 0.5607419 total: 23.9ms remaining: 11.2ms
34: learn: 0.5576943 total: 24ms remaining: 10.3ms
35: learn: 0.5545097 total: 24.7ms remaining: 9.6ms
36: learn: 0.5512279 total: 25.3ms remaining: 8.9ms
37: learn: 0.5483413 total: 26.1ms remaining: 8.24ms
38: learn: 0.5457237 total: 26.8ms remaining: 7.55ms
39: learn: 0.5426478 total: 27.6ms remaining: 6.9ms
40: learn: 0.5400138 total: 28.4ms remaining: 6.22ms
41: learn: 0.5371179 total: 29.1ms remaining: 5.54ms
42: learn: 0.5343056 total: 29.8ms remaining: 4.85ms
43: learn: 0.5319724 total: 30.2ms remaining: 4.12ms
44: learn: 0.5292683 total: 30.8ms remaining: 3.42ms
45: learn: 0.5263663 total: 31.4ms remaining: 2.73ms
46: learn: 0.5237196 total: 31.8ms remaining: 2.03ms
47: learn: 0.5209193 total: 32.7ms remaining: 1.36ms
48: learn: 0.5182532 total: 33.4ms remaining: 682us
49: learn: 0.5157383 total: 34.1ms remaining: 0us
0: learn: 0.6882585 total: 1.07ms remaining: 52.6ms
1: learn: 0.6839777 total: 1.69ms remaining: 40.6ms
2: learn: 0.6792552 total: 4.01ms remaining: 62.9ms
3: learn: 0.6748659 total: 5.25ms remaining: 60.4ms
4: learn: 0.6706315 total: 7.47ms remaining: 67.2ms
5: learn: 0.6666245 total: 10.6ms remaining: 77.8ms
6: learn: 0.6626422 total: 13.2ms remaining: 81.1ms
7: learn: 0.6586781 total: 15.9ms remaining: 83.3ms
8: learn: 0.6552025 total: 19.3ms remaining: 87.9ms
9: learn: 0.6506840 total: 20.8ms remaining: 83.2ms
10: learn: 0.6460304 total: 21.6ms remaining: 76.4ms
11: learn: 0.6419958 total: 22.9ms remaining: 72.4ms
12: learn: 0.6382144 total: 25.5ms remaining: 72.5ms
13: learn: 0.6336491 total: 26.8ms remaining: 68.8ms
14: learn: 0.6292397 total: 29.4ms remaining: 68.6ms
15: learn: 0.6255244 total: 31.8ms remaining: 67.6ms
16: learn: 0.6219032 total: 34.9ms remaining: 67.8ms
17: learn: 0.6189971 total: 36ms remaining: 64ms
18: learn: 0.6153793 total: 39.3ms remaining: 64.2ms
19: learn: 0.6114960 total: 42.3ms remaining: 63.4ms
20: learn: 0.6075407 total: 45.6ms remaining: 62.9ms
21: learn: 0.6039678 total: 48.3ms remaining: 61.5ms
22: learn: 0.6005275 total: 51.2ms remaining: 60.1ms
23: learn: 0.5969838 total: 54.6ms remaining: 59.1ms
24: learn: 0.5940994 total: 57.2ms remaining: 57.2ms
25: learn: 0.5903183 total: 58.3ms remaining: 53.8ms
26: learn: 0.5865176 total: 59ms remaining: 50.3ms
27: learn: 0.5827540 total: 61.6ms remaining: 48.4ms
28: learn: 0.5802146 total: 63.9ms remaining: 46.3ms
29: learn: 0.5768232 total: 65.1ms remaining: 43.4ms
30: learn: 0.5730392 total: 67ms remaining: 41.1ms
31: learn: 0.5706433 total: 69.6ms remaining: 39.1ms
32: learn: 0.5670191 total: 72.3ms remaining: 37.2ms
33: learn: 0.5640741 total: 74.1ms remaining: 34.9ms
34: learn: 0.5603547 total: 76.2ms remaining: 32.7ms
35: learn: 0.5572766 total: 78.6ms remaining: 30.5ms
36: learn: 0.5551564 total: 80.6ms remaining: 28.3ms
37: learn: 0.5523044 total: 82ms remaining: 25.9ms
38: learn: 0.5491651 total: 83.7ms remaining: 23.6ms
39: learn: 0.5457140 total: 84.6ms remaining: 21.1ms
40: learn: 0.5429322 total: 86.4ms remaining: 19ms
41: learn: 0.5395395 total: 88.2ms remaining: 16.8ms
42: learn: 0.5373249 total: 89.6ms remaining: 14.6ms
43: learn: 0.5341033 total: 91ms remaining: 12.4ms
44: learn: 0.5311522 total: 92.4ms remaining: 10.3ms
45: learn: 0.5283215 total: 94.1ms remaining: 8.18ms
46: learn: 0.5254838 total: 96ms remaining: 6.13ms
47: learn: 0.5232056 total: 97.6ms remaining: 4.07ms
48: learn: 0.5205159 total: 99.1ms remaining: 2.02ms
49: learn: 0.5177927 total: 101ms remaining: 0us
0: learn: 0.6881440 total: 705us remaining: 34.6ms
1: learn: 0.6838226 total: 1.66ms remaining: 39.9ms
2: learn: 0.6798522 total: 2.88ms remaining: 45.2ms
3: learn: 0.6753612 total: 3.84ms remaining: 44.1ms
4: learn: 0.6718949 total: 5.17ms remaining: 46.5ms
5: learn: 0.6680771 total: 6.26ms remaining: 45.9ms
6: learn: 0.6634730 total: 7.28ms remaining: 44.7ms
7: learn: 0.6597522 total: 8.52ms remaining: 44.7ms
8: learn: 0.6563034 total: 9.71ms remaining: 44.2ms
9: learn: 0.6521967 total: 10.1ms remaining: 40.3ms
10: learn: 0.6481657 total: 11.3ms remaining: 40ms
11: learn: 0.6448143 total: 12.4ms remaining: 39.4ms
12: learn: 0.6420479 total: 13.1ms remaining: 37.2ms
13: learn: 0.6381273 total: 14.1ms remaining: 36.3ms
14: learn: 0.6348032 total: 15.2ms remaining: 35.6ms
15: learn: 0.6309156 total: 17.1ms remaining: 36.4ms
16: learn: 0.6273113 total: 18.5ms remaining: 35.9ms
17: learn: 0.6236722 total: 19.5ms remaining: 34.6ms
18: learn: 0.6193329 total: 20.6ms remaining: 33.6ms
19: learn: 0.6151982 total: 21.7ms remaining: 32.5ms
20: learn: 0.6114231 total: 22.8ms remaining: 31.5ms
21: learn: 0.6080377 total: 23.9ms remaining: 30.4ms
22: learn: 0.6050294 total: 24.9ms remaining: 29.3ms
23: learn: 0.6013056 total: 26.2ms remaining: 28.4ms
24: learn: 0.5975738 total: 27.3ms remaining: 27.3ms
25: learn: 0.5942470 total: 28.3ms remaining: 26.1ms
26: learn: 0.5909084 total: 29.4ms remaining: 25.1ms
27: learn: 0.5876613 total: 30.5ms remaining: 24ms
28: learn: 0.5846476 total: 31.6ms remaining: 22.9ms
29: learn: 0.5813644 total: 32.9ms remaining: 21.9ms
30: learn: 0.5778006 total: 33.8ms remaining: 20.7ms
31: learn: 0.5740806 total: 34.7ms remaining: 19.5ms
32: learn: 0.5706020 total: 35.3ms remaining: 18.2ms
33: learn: 0.5675473 total: 36.2ms remaining: 17ms
34: learn: 0.5644498 total: 37.1ms remaining: 15.9ms
35: learn: 0.5610447 total: 38.3ms remaining: 14.9ms
36: learn: 0.5578353 total: 39.1ms remaining: 13.7ms
37: learn: 0.5551180 total: 39.9ms remaining: 12.6ms
38: learn: 0.5521754 total: 40.6ms remaining: 11.5ms
39: learn: 0.5491918 total: 41.4ms remaining: 10.4ms
40: learn: 0.5464784 total: 42.2ms remaining: 9.27ms
41: learn: 0.5434380 total: 43.2ms remaining: 8.23ms
42: learn: 0.5409694 total: 44.2ms remaining: 7.19ms
43: learn: 0.5386926 total: 44.9ms remaining: 6.12ms
44: learn: 0.5358634 total: 45.6ms remaining: 5.07ms
45: learn: 0.5334981 total: 45.8ms remaining: 3.99ms
46: learn: 0.5305617 total: 46.5ms remaining: 2.97ms
47: learn: 0.5279681 total: 47.2ms remaining: 1.97ms
48: learn: 0.5252795 total: 47.9ms remaining: 976us
49: learn: 0.5226056 total: 48.7ms remaining: 0us
0: learn: 0.6880598 total: 779us remaining: 38.2ms
1: learn: 0.6833246 total: 1.67ms remaining: 40ms
2: learn: 0.6789769 total: 2.73ms remaining: 42.7ms
3: learn: 0.6739423 total: 3.49ms remaining: 40.1ms
4: learn: 0.6690233 total: 4.13ms remaining: 37.2ms
5: learn: 0.6655579 total: 4.98ms remaining: 36.5ms
6: learn: 0.6615555 total: 5.79ms remaining: 35.6ms
7: learn: 0.6581583 total: 6.6ms remaining: 34.7ms
8: learn: 0.6542419 total: 7.43ms remaining: 33.8ms
9: learn: 0.6497969 total: 8.02ms remaining: 32.1ms
10: learn: 0.6456478 total: 8.22ms remaining: 29.1ms
11: learn: 0.6415730 total: 8.63ms remaining: 27.3ms
12: learn: 0.6372634 total: 9.42ms remaining: 26.8ms
13: learn: 0.6326632 total: 10.1ms remaining: 25.9ms
14: learn: 0.6293084 total: 10.9ms remaining: 25.3ms
15: learn: 0.6257282 total: 11.6ms remaining: 24.7ms
16: learn: 0.6216776 total: 12.4ms remaining: 24ms
17: learn: 0.6188229 total: 12.9ms remaining: 23ms
18: learn: 0.6149747 total: 13.8ms remaining: 22.5ms
19: learn: 0.6112811 total: 14ms remaining: 21ms
20: learn: 0.6074915 total: 14.8ms remaining: 20.4ms
21: learn: 0.6044135 total: 15.7ms remaining: 20ms
22: learn: 0.6009953 total: 16.6ms remaining: 19.5ms
23: learn: 0.5974793 total: 17.4ms remaining: 18.9ms
24: learn: 0.5937905 total: 18.3ms remaining: 18.3ms
25: learn: 0.5902730 total: 19.2ms remaining: 17.7ms
26: learn: 0.5875519 total: 20ms remaining: 17ms
27: learn: 0.5841550 total: 20.9ms remaining: 16.4ms
28: learn: 0.5808950 total: 21.7ms remaining: 15.7ms
29: learn: 0.5772600 total: 22.5ms remaining: 15ms
30: learn: 0.5745535 total: 23.4ms remaining: 14.3ms
31: learn: 0.5712423 total: 24ms remaining: 13.5ms
32: learn: 0.5676332 total: 24.8ms remaining: 12.8ms
33: learn: 0.5646369 total: 25.6ms remaining: 12.1ms
34: learn: 0.5617155 total: 26.4ms remaining: 11.3ms
35: learn: 0.5585733 total: 27ms remaining: 10.5ms
36: learn: 0.5559330 total: 27.3ms remaining: 9.61ms
37: learn: 0.5529272 total: 28.2ms remaining: 8.89ms
38: learn: 0.5503624 total: 28.4ms remaining: 8.01ms
39: learn: 0.5471022 total: 29.2ms remaining: 7.3ms
40: learn: 0.5445004 total: 30.1ms remaining: 6.6ms
41: learn: 0.5418501 total: 30.9ms remaining: 5.88ms
42: learn: 0.5393487 total: 31.7ms remaining: 5.16ms
43: learn: 0.5365438 total: 32.5ms remaining: 4.44ms
44: learn: 0.5334766 total: 33.5ms remaining: 3.72ms
45: learn: 0.5308641 total: 34.2ms remaining: 2.97ms
46: learn: 0.5284121 total: 35ms remaining: 2.23ms
47: learn: 0.5257992 total: 35.4ms remaining: 1.47ms
48: learn: 0.5233899 total: 36.1ms remaining: 737us
49: learn: 0.5203597 total: 37ms remaining: 0us
0: learn: 0.6882715 total: 478us remaining: 23.4ms
1: learn: 0.6837467 total: 982us remaining: 23.6ms
2: learn: 0.6783988 total: 1.66ms remaining: 26ms
3: learn: 0.6736412 total: 2.39ms remaining: 27.5ms
4: learn: 0.6688160 total: 3.45ms remaining: 31ms
5: learn: 0.6636849 total: 4.43ms remaining: 32.5ms
6: learn: 0.6595088 total: 5.38ms remaining: 33.1ms
7: learn: 0.6554600 total: 6.36ms remaining: 33.4ms
8: learn: 0.6513102 total: 7.26ms remaining: 33.1ms
9: learn: 0.6474276 total: 7.9ms remaining: 31.6ms
10: learn: 0.6433167 total: 8.48ms remaining: 30.1ms
11: learn: 0.6393555 total: 9.19ms remaining: 29.1ms
12: learn: 0.6353104 total: 9.94ms remaining: 28.3ms
13: learn: 0.6323059 total: 10.7ms remaining: 27.5ms
14: learn: 0.6292336 total: 11.6ms remaining: 27ms
15: learn: 0.6254054 total: 12.2ms remaining: 25.9ms
16: learn: 0.6217592 total: 13.2ms remaining: 25.5ms
17: learn: 0.6182798 total: 13.6ms remaining: 24.1ms
18: learn: 0.6145356 total: 14.1ms remaining: 23ms
19: learn: 0.6111740 total: 14.8ms remaining: 22.2ms
20: learn: 0.6075515 total: 15.6ms remaining: 21.5ms
21: learn: 0.6036147 total: 16.4ms remaining: 20.9ms
22: learn: 0.5997521 total: 17ms remaining: 20ms
23: learn: 0.5954807 total: 17.7ms remaining: 19.1ms
24: learn: 0.5914498 total: 18.4ms remaining: 18.4ms
25: learn: 0.5877992 total: 18.8ms remaining: 17.4ms
26: learn: 0.5840369 total: 19.4ms remaining: 16.5ms
27: learn: 0.5807457 total: 20.1ms remaining: 15.8ms
28: learn: 0.5773241 total: 20.7ms remaining: 15ms
29: learn: 0.5752621 total: 21.2ms remaining: 14.1ms
30: learn: 0.5711845 total: 22ms remaining: 13.5ms
31: learn: 0.5677277 total: 22.4ms remaining: 12.6ms
32: learn: 0.5642957 total: 22.9ms remaining: 11.8ms
33: learn: 0.5610649 total: 23.7ms remaining: 11.1ms
34: learn: 0.5579598 total: 24.1ms remaining: 10.3ms
35: learn: 0.5543794 total: 24.6ms remaining: 9.59ms
36: learn: 0.5516154 total: 25.2ms remaining: 8.84ms
37: learn: 0.5487977 total: 25.4ms remaining: 8.02ms
38: learn: 0.5459438 total: 26.1ms remaining: 7.35ms
39: learn: 0.5435490 total: 26.9ms remaining: 6.73ms
40: learn: 0.5410065 total: 27.4ms remaining: 6.01ms
41: learn: 0.5381863 total: 28.1ms remaining: 5.35ms
42: learn: 0.5346479 total: 28.6ms remaining: 4.66ms
43: learn: 0.5323395 total: 29.3ms remaining: 4ms
44: learn: 0.5297288 total: 29.9ms remaining: 3.32ms
45: learn: 0.5279816 total: 30.4ms remaining: 2.65ms
46: learn: 0.5253830 total: 31ms remaining: 1.98ms
47: learn: 0.5228773 total: 31.3ms remaining: 1.3ms
48: learn: 0.5200328 total: 32.1ms remaining: 655us
49: learn: 0.5170826 total: 32.9ms remaining: 0us
0: learn: 0.6895544 total: 1.18ms remaining: 57.8ms
1: learn: 0.6849260 total: 2.02ms remaining: 48.6ms
2: learn: 0.6810258 total: 3.65ms remaining: 57.2ms
3: learn: 0.6765089 total: 4.97ms remaining: 57.2ms
4: learn: 0.6731004 total: 6.37ms remaining: 57.3ms
5: learn: 0.6686184 total: 7.59ms remaining: 55.7ms
6: learn: 0.6644403 total: 8.96ms remaining: 55.1ms
7: learn: 0.6598710 total: 10.8ms remaining: 56.8ms
8: learn: 0.6550665 total: 13.1ms remaining: 59.5ms
9: learn: 0.6515600 total: 15.1ms remaining: 60.3ms
10: learn: 0.6484847 total: 16.5ms remaining: 58.5ms
11: learn: 0.6441061 total: 18.7ms remaining: 59.2ms
12: learn: 0.6395647 total: 19.7ms remaining: 56.1ms
13: learn: 0.6362526 total: 21.3ms remaining: 54.6ms
14: learn: 0.6329626 total: 22.4ms remaining: 52.3ms
15: learn: 0.6290173 total: 23.7ms remaining: 50.4ms
16: learn: 0.6262281 total: 24.6ms remaining: 47.8ms
17: learn: 0.6227024 total: 26.8ms remaining: 47.6ms
18: learn: 0.6188889 total: 27.4ms remaining: 44.7ms
19: learn: 0.6152467 total: 28.8ms remaining: 43.2ms
20: learn: 0.6111927 total: 30ms remaining: 41.4ms
21: learn: 0.6078590 total: 31.2ms remaining: 39.7ms
22: learn: 0.6043231 total: 32.9ms remaining: 38.6ms
23: learn: 0.6004283 total: 34.2ms remaining: 37.1ms
24: learn: 0.5967828 total: 35.4ms remaining: 35.4ms
25: learn: 0.5934005 total: 36.6ms remaining: 33.8ms
26: learn: 0.5897854 total: 37.7ms remaining: 32.1ms
27: learn: 0.5867783 total: 38.9ms remaining: 30.6ms
28: learn: 0.5835423 total: 39.8ms remaining: 28.8ms
29: learn: 0.5799524 total: 40.7ms remaining: 27.1ms
30: learn: 0.5767129 total: 41.7ms remaining: 25.6ms
31: learn: 0.5734765 total: 42.7ms remaining: 24ms
32: learn: 0.5697662 total: 43.5ms remaining: 22.4ms
33: learn: 0.5660582 total: 44.4ms remaining: 20.9ms
34: learn: 0.5634641 total: 44.9ms remaining: 19.3ms
35: learn: 0.5603953 total: 45.2ms remaining: 17.6ms
36: learn: 0.5577197 total: 46.2ms remaining: 16.2ms
37: learn: 0.5547286 total: 46.5ms remaining: 14.7ms
38: learn: 0.5513552 total: 47.2ms remaining: 13.3ms
39: learn: 0.5487145 total: 48.1ms remaining: 12ms
40: learn: 0.5458332 total: 48.7ms remaining: 10.7ms
41: learn: 0.5430226 total: 49ms remaining: 9.34ms
42: learn: 0.5405773 total: 50ms remaining: 8.13ms
43: learn: 0.5376341 total: 50.8ms remaining: 6.93ms
44: learn: 0.5347205 total: 51.3ms remaining: 5.7ms
45: learn: 0.5318596 total: 52.1ms remaining: 4.53ms
46: learn: 0.5296991 total: 53ms remaining: 3.38ms
47: learn: 0.5265906 total: 53.7ms remaining: 2.24ms
48: learn: 0.5243975 total: 54.5ms remaining: 1.11ms
49: learn: 0.5220615 total: 55.4ms remaining: 0us
0: learn: 0.6899173 total: 1.09ms remaining: 53.3ms
1: learn: 0.6859439 total: 2.87ms remaining: 68.8ms
2: learn: 0.6820029 total: 4.59ms remaining: 72ms
3: learn: 0.6775952 total: 7.32ms remaining: 84.2ms
4: learn: 0.6734936 total: 9.7ms remaining: 87.3ms
5: learn: 0.6694175 total: 11.3ms remaining: 83.1ms
6: learn: 0.6650204 total: 13.8ms remaining: 85ms
7: learn: 0.6604569 total: 15.9ms remaining: 83.2ms
8: learn: 0.6566835 total: 18.5ms remaining: 84.4ms
9: learn: 0.6522852 total: 21.7ms remaining: 87ms
10: learn: 0.6482039 total: 25.3ms remaining: 89.5ms
11: learn: 0.6452449 total: 27.9ms remaining: 88.5ms
12: learn: 0.6413751 total: 30.4ms remaining: 86.5ms
13: learn: 0.6373760 total: 32.6ms remaining: 83.9ms
14: learn: 0.6325962 total: 34.3ms remaining: 79.9ms
15: learn: 0.6287772 total: 36.2ms remaining: 77ms
16: learn: 0.6249406 total: 37.6ms remaining: 73ms
17: learn: 0.6213689 total: 39.9ms remaining: 71ms
18: learn: 0.6178202 total: 41.6ms remaining: 67.8ms
19: learn: 0.6138921 total: 43.7ms remaining: 65.5ms
20: learn: 0.6102038 total: 45.4ms remaining: 62.8ms
21: learn: 0.6067466 total: 47.3ms remaining: 60.2ms
22: learn: 0.6034256 total: 49.3ms remaining: 57.9ms
23: learn: 0.5995496 total: 51.4ms remaining: 55.6ms
24: learn: 0.5958312 total: 53.2ms remaining: 53.2ms
25: learn: 0.5922630 total: 54.8ms remaining: 50.5ms
26: learn: 0.5891180 total: 56.9ms remaining: 48.5ms
27: learn: 0.5855249 total: 58.2ms remaining: 45.8ms
28: learn: 0.5822194 total: 60.2ms remaining: 43.6ms
29: learn: 0.5783025 total: 61.8ms remaining: 41.2ms
30: learn: 0.5749866 total: 62.5ms remaining: 38.3ms
31: learn: 0.5717586 total: 63.9ms remaining: 36ms
32: learn: 0.5679180 total: 65.2ms remaining: 33.6ms
33: learn: 0.5647130 total: 66.7ms remaining: 31.4ms
34: learn: 0.5615301 total: 67.7ms remaining: 29ms
35: learn: 0.5582356 total: 68.5ms remaining: 26.6ms
36: learn: 0.5552371 total: 69ms remaining: 24.2ms
37: learn: 0.5522339 total: 70.1ms remaining: 22.1ms
38: learn: 0.5490700 total: 71.3ms remaining: 20.1ms
39: learn: 0.5460870 total: 72.4ms remaining: 18.1ms
40: learn: 0.5430329 total: 73.5ms remaining: 16.1ms
41: learn: 0.5394535 total: 74.5ms remaining: 14.2ms
42: learn: 0.5364412 total: 75.5ms remaining: 12.3ms
43: learn: 0.5343403 total: 76.4ms remaining: 10.4ms
44: learn: 0.5311697 total: 77.3ms remaining: 8.59ms
45: learn: 0.5285576 total: 78.3ms remaining: 6.81ms
46: learn: 0.5254773 total: 78.7ms remaining: 5.02ms
47: learn: 0.5228290 total: 79.5ms remaining: 3.31ms
48: learn: 0.5197728 total: 81.2ms remaining: 1.66ms
49: learn: 0.5170935 total: 82.3ms remaining: 0us
0: learn: 0.6895196 total: 1.07ms remaining: 52.4ms
1: learn: 0.6848229 total: 2.61ms remaining: 62.6ms
2: learn: 0.6798775 total: 4.29ms remaining: 67.2ms
3: learn: 0.6752787 total: 5.55ms remaining: 63.9ms
4: learn: 0.6702490 total: 6.99ms remaining: 62.9ms
5: learn: 0.6655144 total: 8.28ms remaining: 60.7ms
6: learn: 0.6609475 total: 9.38ms remaining: 57.6ms
7: learn: 0.6563440 total: 10.8ms remaining: 56.7ms
8: learn: 0.6525135 total: 12.6ms remaining: 57.3ms
9: learn: 0.6486859 total: 14ms remaining: 55.9ms
10: learn: 0.6448139 total: 15.3ms remaining: 54.1ms
11: learn: 0.6411210 total: 16.1ms remaining: 51ms
12: learn: 0.6382069 total: 17.9ms remaining: 50.8ms
13: learn: 0.6339793 total: 19.3ms remaining: 49.6ms
14: learn: 0.6297542 total: 20.5ms remaining: 47.9ms
15: learn: 0.6262754 total: 22.1ms remaining: 47ms
16: learn: 0.6232733 total: 23.5ms remaining: 45.6ms
17: learn: 0.6193642 total: 24.8ms remaining: 44.1ms
18: learn: 0.6150939 total: 25.9ms remaining: 42.3ms
19: learn: 0.6115523 total: 26.3ms remaining: 39.4ms
20: learn: 0.6078337 total: 27.4ms remaining: 37.8ms
21: learn: 0.6044395 total: 28.8ms remaining: 36.7ms
22: learn: 0.6008396 total: 30.4ms remaining: 35.6ms
23: learn: 0.5972830 total: 31.3ms remaining: 33.9ms
24: learn: 0.5934103 total: 32.5ms remaining: 32.5ms
25: learn: 0.5901748 total: 33.8ms remaining: 31.2ms
26: learn: 0.5868766 total: 35.2ms remaining: 30ms
27: learn: 0.5835131 total: 36.6ms remaining: 28.8ms
28: learn: 0.5805723 total: 38ms remaining: 27.5ms
29: learn: 0.5773479 total: 39.2ms remaining: 26.1ms
30: learn: 0.5736389 total: 40.1ms remaining: 24.6ms
31: learn: 0.5699806 total: 41.3ms remaining: 23.2ms
32: learn: 0.5668918 total: 42.6ms remaining: 21.9ms
33: learn: 0.5633195 total: 43.9ms remaining: 20.7ms
34: learn: 0.5598419 total: 45ms remaining: 19.3ms
35: learn: 0.5573127 total: 46.4ms remaining: 18.1ms
36: learn: 0.5542739 total: 47.4ms remaining: 16.7ms
37: learn: 0.5519820 total: 48.5ms remaining: 15.3ms
38: learn: 0.5491087 total: 49.9ms remaining: 14.1ms
39: learn: 0.5465576 total: 51.1ms remaining: 12.8ms
40: learn: 0.5436731 total: 52.3ms remaining: 11.5ms
41: learn: 0.5409845 total: 53.4ms remaining: 10.2ms
42: learn: 0.5383135 total: 54.4ms remaining: 8.86ms
43: learn: 0.5355018 total: 55.6ms remaining: 7.58ms
44: learn: 0.5333852 total: 56.8ms remaining: 6.31ms
45: learn: 0.5305767 total: 57.9ms remaining: 5.04ms
46: learn: 0.5275610 total: 58.8ms remaining: 3.76ms
47: learn: 0.5247445 total: 59.8ms remaining: 2.49ms
48: learn: 0.5223355 total: 60.6ms remaining: 1.24ms
49: learn: 0.5195292 total: 61.7ms remaining: 0us
0: learn: 0.6881645 total: 2.17ms remaining: 106ms
1: learn: 0.6828074 total: 5.25ms remaining: 126ms
2: learn: 0.6783972 total: 7.85ms remaining: 123ms
3: learn: 0.6738964 total: 10.7ms remaining: 123ms
4: learn: 0.6689173 total: 12.5ms remaining: 112ms
5: learn: 0.6650825 total: 15.7ms remaining: 115ms
6: learn: 0.6601666 total: 19.6ms remaining: 121ms
7: learn: 0.6554972 total: 22.4ms remaining: 118ms
8: learn: 0.6505983 total: 25ms remaining: 114ms
9: learn: 0.6462642 total: 28.4ms remaining: 114ms
10: learn: 0.6414765 total: 32.2ms remaining: 114ms
11: learn: 0.6373932 total: 34.7ms remaining: 110ms
12: learn: 0.6321228 total: 35.8ms remaining: 102ms
13: learn: 0.6283485 total: 38.4ms remaining: 98.7ms
14: learn: 0.6242799 total: 39.5ms remaining: 92.2ms
15: learn: 0.6194836 total: 41.3ms remaining: 87.9ms
16: learn: 0.6155681 total: 42.9ms remaining: 83.3ms
17: learn: 0.6117237 total: 43.4ms remaining: 77.1ms
18: learn: 0.6075215 total: 45.6ms remaining: 74.3ms
19: learn: 0.6036330 total: 46.7ms remaining: 70ms
20: learn: 0.5997560 total: 48.9ms remaining: 67.5ms
21: learn: 0.5961921 total: 50.1ms remaining: 63.8ms
22: learn: 0.5921406 total: 52.5ms remaining: 61.6ms
23: learn: 0.5881204 total: 53.6ms remaining: 58.1ms
24: learn: 0.5842689 total: 55.3ms remaining: 55.3ms
25: learn: 0.5804399 total: 57.8ms remaining: 53.4ms
26: learn: 0.5770751 total: 59.7ms remaining: 50.9ms
27: learn: 0.5733741 total: 61ms remaining: 47.9ms
28: learn: 0.5696210 total: 63ms remaining: 45.6ms
29: learn: 0.5660444 total: 64ms remaining: 42.7ms
30: learn: 0.5626817 total: 66.3ms remaining: 40.6ms
31: learn: 0.5591196 total: 68.2ms remaining: 38.3ms
32: learn: 0.5557326 total: 70.7ms remaining: 36.4ms
33: learn: 0.5523177 total: 72.9ms remaining: 34.3ms
34: learn: 0.5493842 total: 75.7ms remaining: 32.4ms
35: learn: 0.5460369 total: 77.8ms remaining: 30.3ms
36: learn: 0.5436892 total: 79.5ms remaining: 27.9ms
37: learn: 0.5404256 total: 81.8ms remaining: 25.8ms
38: learn: 0.5370325 total: 84ms remaining: 23.7ms
39: learn: 0.5341095 total: 86ms remaining: 21.5ms
40: learn: 0.5307599 total: 88.7ms remaining: 19.5ms
41: learn: 0.5281249 total: 90.4ms remaining: 17.2ms
42: learn: 0.5247488 total: 92.5ms remaining: 15.1ms
43: learn: 0.5220971 total: 95ms remaining: 13ms
44: learn: 0.5193386 total: 95.6ms remaining: 10.6ms
45: learn: 0.5168324 total: 96.8ms remaining: 8.42ms
46: learn: 0.5137939 total: 99.5ms remaining: 6.35ms
47: learn: 0.5105831 total: 102ms remaining: 4.25ms
48: learn: 0.5081495 total: 104ms remaining: 2.12ms
49: learn: 0.5052566 total: 106ms remaining: 0us
0: learn: 0.6878932 total: 758us remaining: 37.1ms
1: learn: 0.6839973 total: 1.7ms remaining: 40.7ms
2: learn: 0.6793372 total: 2.85ms remaining: 44.7ms
3: learn: 0.6751816 total: 3.77ms remaining: 43.4ms
4: learn: 0.6706232 total: 4.88ms remaining: 44ms
5: learn: 0.6661899 total: 5.39ms remaining: 39.5ms
6: learn: 0.6619799 total: 6.41ms remaining: 39.4ms
7: learn: 0.6578849 total: 7.09ms remaining: 37.2ms
8: learn: 0.6532810 total: 8.17ms remaining: 37.2ms
9: learn: 0.6492916 total: 8.78ms remaining: 35.1ms
10: learn: 0.6452099 total: 9.91ms remaining: 35.1ms
11: learn: 0.6407272 total: 10.2ms remaining: 32.4ms
12: learn: 0.6364104 total: 11.1ms remaining: 31.6ms
13: learn: 0.6328575 total: 12.1ms remaining: 31.2ms
14: learn: 0.6296023 total: 13ms remaining: 30.4ms
15: learn: 0.6265584 total: 14ms remaining: 29.8ms
16: learn: 0.6233491 total: 14.7ms remaining: 28.5ms
17: learn: 0.6191226 total: 15.6ms remaining: 27.8ms
18: learn: 0.6151361 total: 16.6ms remaining: 27.2ms
19: learn: 0.6117795 total: 17.8ms remaining: 26.7ms
20: learn: 0.6081684 total: 18.7ms remaining: 25.8ms
21: learn: 0.6042276 total: 19.5ms remaining: 24.8ms
22: learn: 0.6006605 total: 20.6ms remaining: 24.2ms
23: learn: 0.5971764 total: 20.9ms remaining: 22.7ms
24: learn: 0.5942878 total: 22ms remaining: 22ms
25: learn: 0.5906416 total: 23ms remaining: 21.3ms
26: learn: 0.5872862 total: 24ms remaining: 20.5ms
27: learn: 0.5834617 total: 24.7ms remaining: 19.4ms
28: learn: 0.5802959 total: 25.2ms remaining: 18.2ms
29: learn: 0.5769130 total: 26.1ms remaining: 17.4ms
30: learn: 0.5747204 total: 26.9ms remaining: 16.5ms
31: learn: 0.5714667 total: 28ms remaining: 15.7ms
32: learn: 0.5685505 total: 29ms remaining: 14.9ms
33: learn: 0.5654209 total: 29.7ms remaining: 14ms
34: learn: 0.5624272 total: 30.7ms remaining: 13.2ms
35: learn: 0.5589781 total: 31.1ms remaining: 12.1ms
36: learn: 0.5554954 total: 32ms remaining: 11.2ms
37: learn: 0.5527426 total: 33ms remaining: 10.4ms
38: learn: 0.5494957 total: 33.5ms remaining: 9.44ms
39: learn: 0.5462821 total: 34.2ms remaining: 8.54ms
40: learn: 0.5433351 total: 34.9ms remaining: 7.67ms
41: learn: 0.5402277 total: 36ms remaining: 6.85ms
42: learn: 0.5368235 total: 36.5ms remaining: 5.94ms
43: learn: 0.5342293 total: 37.1ms remaining: 5.06ms
44: learn: 0.5311675 total: 38ms remaining: 4.22ms
45: learn: 0.5286138 total: 38.7ms remaining: 3.37ms
46: learn: 0.5257986 total: 39.3ms remaining: 2.51ms
47: learn: 0.5230951 total: 40.1ms remaining: 1.67ms
48: learn: 0.5208991 total: 40.9ms remaining: 834us
49: learn: 0.5188314 total: 41.7ms remaining: 0us
0: learn: 0.6880858 total: 1.43ms remaining: 70.2ms
1: learn: 0.6835325 total: 2.48ms remaining: 59.5ms
2: learn: 0.6790942 total: 4.98ms remaining: 78.1ms
3: learn: 0.6743935 total: 6.13ms remaining: 70.5ms
4: learn: 0.6698191 total: 8.08ms remaining: 72.7ms
5: learn: 0.6646577 total: 10.6ms remaining: 77.5ms
6: learn: 0.6594989 total: 13.1ms remaining: 80.4ms
7: learn: 0.6558044 total: 15.7ms remaining: 82.3ms
8: learn: 0.6515938 total: 18.6ms remaining: 84.9ms
9: learn: 0.6462522 total: 19.9ms remaining: 79.5ms
10: learn: 0.6425140 total: 23ms remaining: 81.6ms
11: learn: 0.6381312 total: 24ms remaining: 75.9ms
12: learn: 0.6347719 total: 25.9ms remaining: 73.8ms
13: learn: 0.6302025 total: 27.2ms remaining: 69.9ms
14: learn: 0.6261653 total: 29.5ms remaining: 68.8ms
15: learn: 0.6226721 total: 31.3ms remaining: 66.6ms
16: learn: 0.6187113 total: 34.1ms remaining: 66.1ms
17: learn: 0.6140191 total: 36.2ms remaining: 64.4ms
18: learn: 0.6096826 total: 38.1ms remaining: 62.2ms
19: learn: 0.6052420 total: 40.2ms remaining: 60.3ms
20: learn: 0.6017107 total: 42.8ms remaining: 59.1ms
21: learn: 0.5983776 total: 45.9ms remaining: 58.5ms
22: learn: 0.5951994 total: 48.4ms remaining: 56.8ms
23: learn: 0.5918906 total: 50.7ms remaining: 55ms
24: learn: 0.5884141 total: 51.6ms remaining: 51.6ms
25: learn: 0.5841399 total: 51.9ms remaining: 47.9ms
26: learn: 0.5803691 total: 54.1ms remaining: 46.1ms
27: learn: 0.5769200 total: 56.4ms remaining: 44.3ms
28: learn: 0.5736473 total: 58.3ms remaining: 42.2ms
29: learn: 0.5700408 total: 60.2ms remaining: 40.2ms
30: learn: 0.5662298 total: 62.2ms remaining: 38.1ms
31: learn: 0.5626713 total: 64.2ms remaining: 36.1ms
32: learn: 0.5589932 total: 65.6ms remaining: 33.8ms
33: learn: 0.5555780 total: 66.4ms remaining: 31.3ms
34: learn: 0.5522308 total: 66.8ms remaining: 28.6ms
35: learn: 0.5489336 total: 67.5ms remaining: 26.2ms
36: learn: 0.5463816 total: 70ms remaining: 24.6ms
37: learn: 0.5430801 total: 72.2ms remaining: 22.8ms
38: learn: 0.5403064 total: 73.5ms remaining: 20.7ms
39: learn: 0.5369235 total: 76ms remaining: 19ms
40: learn: 0.5339607 total: 76.8ms remaining: 16.9ms
41: learn: 0.5311299 total: 79.4ms remaining: 15.1ms
42: learn: 0.5281621 total: 80.8ms remaining: 13.1ms
43: learn: 0.5248973 total: 83ms remaining: 11.3ms
44: learn: 0.5222278 total: 86.1ms remaining: 9.57ms
45: learn: 0.5195237 total: 88.2ms remaining: 7.67ms
46: learn: 0.5169016 total: 90.6ms remaining: 5.78ms
47: learn: 0.5141868 total: 93.3ms remaining: 3.88ms
48: learn: 0.5115914 total: 95.9ms remaining: 1.96ms
49: learn: 0.5085887 total: 97.7ms remaining: 0us
0: learn: 0.6881413 total: 2.73ms remaining: 134ms
1: learn: 0.6838407 total: 5.43ms remaining: 130ms
2: learn: 0.6798131 total: 6.78ms remaining: 106ms
3: learn: 0.6758858 total: 8.57ms remaining: 98.6ms
4: learn: 0.6712487 total: 10.4ms remaining: 93.6ms
5: learn: 0.6667165 total: 12.1ms remaining: 89ms
6: learn: 0.6624754 total: 13ms remaining: 79.6ms
7: learn: 0.6584117 total: 14.3ms remaining: 75.2ms
8: learn: 0.6543664 total: 16.1ms remaining: 73.4ms
9: learn: 0.6500307 total: 17.7ms remaining: 70.8ms
10: learn: 0.6463957 total: 18.8ms remaining: 66.7ms
11: learn: 0.6433796 total: 21.5ms remaining: 68.1ms
12: learn: 0.6393141 total: 23.3ms remaining: 66.2ms
13: learn: 0.6355468 total: 24.9ms remaining: 64ms
14: learn: 0.6316320 total: 26.2ms remaining: 61.1ms
15: learn: 0.6285376 total: 27.6ms remaining: 58.7ms
16: learn: 0.6239545 total: 28.5ms remaining: 55.3ms
17: learn: 0.6199367 total: 30.2ms remaining: 53.7ms
18: learn: 0.6159201 total: 31.6ms remaining: 51.6ms
19: learn: 0.6118337 total: 33ms remaining: 49.5ms
20: learn: 0.6088449 total: 33.8ms remaining: 46.7ms
21: learn: 0.6054882 total: 35ms remaining: 44.5ms
22: learn: 0.6018578 total: 36.2ms remaining: 42.5ms
23: learn: 0.5985672 total: 37.1ms remaining: 40.2ms
24: learn: 0.5946907 total: 38.3ms remaining: 38.3ms
25: learn: 0.5905861 total: 39ms remaining: 36ms
26: learn: 0.5873582 total: 40.1ms remaining: 34.2ms
27: learn: 0.5835815 total: 41.1ms remaining: 32.3ms
28: learn: 0.5802048 total: 42.1ms remaining: 30.5ms
29: learn: 0.5766559 total: 43ms remaining: 28.7ms
30: learn: 0.5741789 total: 44ms remaining: 27ms
31: learn: 0.5708948 total: 44.9ms remaining: 25.2ms
32: learn: 0.5679576 total: 45.6ms remaining: 23.5ms
33: learn: 0.5653729 total: 46.4ms remaining: 21.8ms
34: learn: 0.5628168 total: 47.2ms remaining: 20.2ms
35: learn: 0.5597310 total: 47.8ms remaining: 18.6ms
36: learn: 0.5574384 total: 48.5ms remaining: 17.1ms
37: learn: 0.5552292 total: 49ms remaining: 15.5ms
38: learn: 0.5518550 total: 49.5ms remaining: 14ms
39: learn: 0.5488987 total: 50.2ms remaining: 12.6ms
40: learn: 0.5458771 total: 50.9ms remaining: 11.2ms
41: learn: 0.5425878 total: 51.5ms remaining: 9.8ms
42: learn: 0.5398097 total: 52.2ms remaining: 8.49ms
43: learn: 0.5371612 total: 52.8ms remaining: 7.2ms
44: learn: 0.5340660 total: 53.5ms remaining: 5.95ms
45: learn: 0.5317800 total: 54.2ms remaining: 4.71ms
46: learn: 0.5293149 total: 54.9ms remaining: 3.5ms
47: learn: 0.5266531 total: 55.6ms remaining: 2.32ms
48: learn: 0.5238752 total: 56.2ms remaining: 1.15ms
49: learn: 0.5207962 total: 56.7ms remaining: 0us
Evaluating param combo 5/16: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.1, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 8}
Duplicate found for this combo (hash=9eef1bd532edbf812482fcf4794a0b6d). Removing old run: 5532b56cc94f4d5f8abcdffa62979125
0: learn: 0.6462693 total: 279us remaining: 13.7ms
1: learn: 0.6006126 total: 545us remaining: 13.1ms
2: learn: 0.5631732 total: 780us remaining: 12.2ms
3: learn: 0.5318762 total: 1.18ms remaining: 13.6ms
4: learn: 0.5054558 total: 1.58ms remaining: 14.2ms
5: learn: 0.4799320 total: 2.08ms remaining: 15.3ms
6: learn: 0.4585994 total: 3.2ms remaining: 19.6ms
7: learn: 0.4417764 total: 3.87ms remaining: 20.3ms
8: learn: 0.4225440 total: 4.4ms remaining: 20.1ms
9: learn: 0.4084612 total: 4.99ms remaining: 19.9ms
10: learn: 0.3962466 total: 5.58ms remaining: 19.8ms
11: learn: 0.3845676 total: 6.09ms remaining: 19.3ms
12: learn: 0.3717729 total: 6.36ms remaining: 18.1ms
13: learn: 0.3609883 total: 6.68ms remaining: 17.2ms
14: learn: 0.3500067 total: 6.87ms remaining: 16ms
15: learn: 0.3412218 total: 7.17ms remaining: 15.2ms
16: learn: 0.3315580 total: 7.44ms remaining: 14.4ms
17: learn: 0.3252685 total: 7.98ms remaining: 14.2ms
18: learn: 0.3195212 total: 8.34ms remaining: 13.6ms
19: learn: 0.3143782 total: 9.09ms remaining: 13.6ms
20: learn: 0.3089230 total: 9.82ms remaining: 13.6ms
21: learn: 0.3037625 total: 10.5ms remaining: 13.3ms
22: learn: 0.2990881 total: 10.8ms remaining: 12.7ms
23: learn: 0.2939303 total: 11.4ms remaining: 12.3ms
24: learn: 0.2891699 total: 11.6ms remaining: 11.6ms
25: learn: 0.2840286 total: 12.4ms remaining: 11.5ms
26: learn: 0.2792264 total: 13ms remaining: 11.1ms
27: learn: 0.2746170 total: 13.5ms remaining: 10.6ms
28: learn: 0.2711557 total: 14.1ms remaining: 10.2ms
29: learn: 0.2693929 total: 14.5ms remaining: 9.65ms
30: learn: 0.2660210 total: 15.1ms remaining: 9.26ms
31: learn: 0.2638487 total: 15.3ms remaining: 8.63ms
32: learn: 0.2615085 total: 16ms remaining: 8.23ms
33: learn: 0.2594956 total: 16.6ms remaining: 7.8ms
34: learn: 0.2566762 total: 17.2ms remaining: 7.38ms
35: learn: 0.2542834 total: 17.5ms remaining: 6.82ms
36: learn: 0.2522301 total: 18ms remaining: 6.31ms
37: learn: 0.2495766 total: 18.4ms remaining: 5.82ms
38: learn: 0.2473101 total: 18.8ms remaining: 5.31ms
39: learn: 0.2450895 total: 19.5ms remaining: 4.87ms
40: learn: 0.2426112 total: 20ms remaining: 4.38ms
41: learn: 0.2396711 total: 20.4ms remaining: 3.89ms
42: learn: 0.2378629 total: 20.7ms remaining: 3.38ms
43: learn: 0.2352862 total: 21.1ms remaining: 2.87ms
44: learn: 0.2334733 total: 21.5ms remaining: 2.38ms
45: learn: 0.2314837 total: 22ms remaining: 1.91ms
46: learn: 0.2290257 total: 22.6ms remaining: 1.44ms
47: learn: 0.2272964 total: 22.9ms remaining: 952us
48: learn: 0.2253947 total: 23.2ms remaining: 474us
49: learn: 0.2237654 total: 23.7ms remaining: 0us
0: learn: 0.6437160 total: 378us remaining: 18.6ms
1: learn: 0.5949760 total: 678us remaining: 16.3ms
2: learn: 0.5565073 total: 886us remaining: 13.9ms
3: learn: 0.5265837 total: 1.18ms remaining: 13.6ms
4: learn: 0.5005287 total: 1.41ms remaining: 12.7ms
5: learn: 0.4720428 total: 1.92ms remaining: 14.1ms
6: learn: 0.4484482 total: 2.33ms remaining: 14.3ms
7: learn: 0.4293736 total: 2.84ms remaining: 14.9ms
8: learn: 0.4148507 total: 3.35ms remaining: 15.3ms
9: learn: 0.3977165 total: 4.05ms remaining: 16.2ms
10: learn: 0.3825212 total: 4.55ms remaining: 16.1ms
11: learn: 0.3667856 total: 5.04ms remaining: 15.9ms
12: learn: 0.3547366 total: 5.71ms remaining: 16.2ms
13: learn: 0.3424929 total: 6.43ms remaining: 16.5ms
14: learn: 0.3349308 total: 7.09ms remaining: 16.5ms
15: learn: 0.3224127 total: 7.66ms remaining: 16.3ms
16: learn: 0.3178131 total: 8.17ms remaining: 15.9ms
17: learn: 0.3131939 total: 8.69ms remaining: 15.4ms
18: learn: 0.3045147 total: 9.4ms remaining: 15.3ms
19: learn: 0.2970139 total: 10.2ms remaining: 15.2ms
20: learn: 0.2918113 total: 11.1ms remaining: 15.3ms
21: learn: 0.2854668 total: 11.9ms remaining: 15.1ms
22: learn: 0.2779652 total: 12.5ms remaining: 14.6ms
23: learn: 0.2734233 total: 13ms remaining: 14.1ms
24: learn: 0.2685745 total: 14ms remaining: 14ms
25: learn: 0.2629943 total: 14.8ms remaining: 13.7ms
26: learn: 0.2604321 total: 15.6ms remaining: 13.2ms
27: learn: 0.2566107 total: 16.2ms remaining: 12.7ms
28: learn: 0.2531121 total: 16.9ms remaining: 12.2ms
29: learn: 0.2515028 total: 17.4ms remaining: 11.6ms
30: learn: 0.2482660 total: 18.2ms remaining: 11.1ms
31: learn: 0.2458159 total: 18.9ms remaining: 10.6ms
32: learn: 0.2416483 total: 19.7ms remaining: 10.1ms
33: learn: 0.2386672 total: 20.5ms remaining: 9.64ms
34: learn: 0.2359889 total: 21.2ms remaining: 9.1ms
35: learn: 0.2338575 total: 22ms remaining: 8.55ms
36: learn: 0.2298220 total: 23.1ms remaining: 8.11ms
37: learn: 0.2283967 total: 24.3ms remaining: 7.66ms
38: learn: 0.2263530 total: 25.3ms remaining: 7.14ms
39: learn: 0.2246581 total: 26.1ms remaining: 6.52ms
40: learn: 0.2223378 total: 27ms remaining: 5.92ms
41: learn: 0.2207038 total: 27.9ms remaining: 5.31ms
42: learn: 0.2197856 total: 28.5ms remaining: 4.64ms
43: learn: 0.2188898 total: 29.2ms remaining: 3.98ms
44: learn: 0.2176194 total: 29.8ms remaining: 3.31ms
45: learn: 0.2163312 total: 30.5ms remaining: 2.65ms
46: learn: 0.2154455 total: 31.2ms remaining: 1.99ms
47: learn: 0.2139407 total: 32ms remaining: 1.33ms
48: learn: 0.2120411 total: 32.7ms remaining: 666us
49: learn: 0.2106386 total: 33.3ms remaining: 0us
0: learn: 0.6454260 total: 671us remaining: 32.9ms
1: learn: 0.6019562 total: 1.37ms remaining: 32.8ms
2: learn: 0.5626070 total: 1.73ms remaining: 27.1ms
3: learn: 0.5307213 total: 2.38ms remaining: 27.3ms
4: learn: 0.5032371 total: 3.32ms remaining: 29.9ms
5: learn: 0.4776693 total: 4.34ms remaining: 31.8ms
6: learn: 0.4566797 total: 5.3ms remaining: 32.6ms
7: learn: 0.4389420 total: 6.26ms remaining: 32.9ms
8: learn: 0.4181396 total: 7.04ms remaining: 32.1ms
9: learn: 0.4027384 total: 7.82ms remaining: 31.3ms
10: learn: 0.3900032 total: 8.56ms remaining: 30.4ms
11: learn: 0.3777932 total: 9.65ms remaining: 30.5ms
12: learn: 0.3636268 total: 10.5ms remaining: 29.9ms
13: learn: 0.3525592 total: 11.2ms remaining: 28.9ms
14: learn: 0.3415734 total: 12.1ms remaining: 28.2ms
15: learn: 0.3322054 total: 13.2ms remaining: 28.1ms
16: learn: 0.3233730 total: 14.5ms remaining: 28.2ms
17: learn: 0.3167153 total: 15.8ms remaining: 28.1ms
18: learn: 0.3106400 total: 16.8ms remaining: 27.5ms
19: learn: 0.3023905 total: 18ms remaining: 27.1ms
20: learn: 0.2963086 total: 19.2ms remaining: 26.5ms
21: learn: 0.2904163 total: 19.9ms remaining: 25.4ms
22: learn: 0.2852205 total: 20.8ms remaining: 24.4ms
23: learn: 0.2798857 total: 21.7ms remaining: 23.5ms
24: learn: 0.2744258 total: 22.7ms remaining: 22.7ms
25: learn: 0.2718590 total: 24.2ms remaining: 22.3ms
26: learn: 0.2677934 total: 25.6ms remaining: 21.8ms
27: learn: 0.2637715 total: 27.1ms remaining: 21.3ms
28: learn: 0.2593053 total: 28.4ms remaining: 20.6ms
29: learn: 0.2574366 total: 29.5ms remaining: 19.7ms
30: learn: 0.2539525 total: 30.9ms remaining: 18.9ms
31: learn: 0.2512715 total: 31.7ms remaining: 17.8ms
32: learn: 0.2481380 total: 33ms remaining: 17ms
33: learn: 0.2458802 total: 34.2ms remaining: 16.1ms
34: learn: 0.2430230 total: 35.5ms remaining: 15.2ms
35: learn: 0.2402213 total: 36.7ms remaining: 14.3ms
36: learn: 0.2382592 total: 38ms remaining: 13.4ms
37: learn: 0.2366223 total: 39.2ms remaining: 12.4ms
38: learn: 0.2346630 total: 40.1ms remaining: 11.3ms
39: learn: 0.2316478 total: 41.4ms remaining: 10.4ms
40: learn: 0.2295176 total: 42.4ms remaining: 9.3ms
41: learn: 0.2275613 total: 43.4ms remaining: 8.27ms
42: learn: 0.2261304 total: 44.5ms remaining: 7.24ms
43: learn: 0.2230182 total: 45.6ms remaining: 6.22ms
44: learn: 0.2213852 total: 47.2ms remaining: 5.25ms
45: learn: 0.2198024 total: 48.7ms remaining: 4.23ms
46: learn: 0.2183045 total: 49.5ms remaining: 3.16ms
47: learn: 0.2166827 total: 50.2ms remaining: 2.09ms
48: learn: 0.2136031 total: 51.9ms remaining: 1.06ms
49: learn: 0.2121566 total: 52.8ms remaining: 0us
0: learn: 0.6470966 total: 452us remaining: 22.2ms
1: learn: 0.6001838 total: 923us remaining: 22.2ms
2: learn: 0.5604623 total: 1.58ms remaining: 24.7ms
3: learn: 0.5261755 total: 2.89ms remaining: 33.3ms
4: learn: 0.4992383 total: 3.73ms remaining: 33.5ms
5: learn: 0.4721898 total: 4.78ms remaining: 35ms
6: learn: 0.4498006 total: 5.98ms remaining: 36.8ms
7: learn: 0.4314508 total: 6.79ms remaining: 35.7ms
8: learn: 0.4101723 total: 7.84ms remaining: 35.7ms
9: learn: 0.3935443 total: 9.1ms remaining: 36.4ms
10: learn: 0.3798147 total: 10.1ms remaining: 35.9ms
11: learn: 0.3665670 total: 11.4ms remaining: 36.2ms
12: learn: 0.3536267 total: 12.8ms remaining: 36.4ms
13: learn: 0.3420766 total: 13.9ms remaining: 35.7ms
14: learn: 0.3326825 total: 14.8ms remaining: 34.6ms
15: learn: 0.3240086 total: 16ms remaining: 34ms
16: learn: 0.3134403 total: 16.9ms remaining: 32.9ms
17: learn: 0.3067898 total: 18.3ms remaining: 32.5ms
18: learn: 0.3003032 total: 19.7ms remaining: 32.1ms
19: learn: 0.2951168 total: 20.8ms remaining: 31.2ms
20: learn: 0.2888849 total: 22.3ms remaining: 30.8ms
21: learn: 0.2831076 total: 23.6ms remaining: 30ms
22: learn: 0.2763535 total: 24.7ms remaining: 28.9ms
23: learn: 0.2698591 total: 26.3ms remaining: 28.5ms
24: learn: 0.2647141 total: 27.7ms remaining: 27.7ms
25: learn: 0.2590451 total: 28.4ms remaining: 26.2ms
26: learn: 0.2557403 total: 29.2ms remaining: 24.9ms
27: learn: 0.2526011 total: 30.1ms remaining: 23.7ms
28: learn: 0.2495110 total: 30.8ms remaining: 22.3ms
29: learn: 0.2446900 total: 31.5ms remaining: 21ms
30: learn: 0.2404218 total: 31.9ms remaining: 19.6ms
31: learn: 0.2370704 total: 33.4ms remaining: 18.8ms
32: learn: 0.2333679 total: 35ms remaining: 18ms
33: learn: 0.2298418 total: 36.4ms remaining: 17.1ms
34: learn: 0.2271491 total: 37.6ms remaining: 16.1ms
35: learn: 0.2245757 total: 38.9ms remaining: 15.1ms
36: learn: 0.2214767 total: 40.2ms remaining: 14.1ms
37: learn: 0.2191640 total: 41.3ms remaining: 13ms
38: learn: 0.2178533 total: 42.5ms remaining: 12ms
39: learn: 0.2163079 total: 43.5ms remaining: 10.9ms
40: learn: 0.2149998 total: 45ms remaining: 9.88ms
41: learn: 0.2135240 total: 46.7ms remaining: 8.89ms
42: learn: 0.2118976 total: 48.5ms remaining: 7.9ms
43: learn: 0.2106812 total: 49.9ms remaining: 6.81ms
44: learn: 0.2087706 total: 51.3ms remaining: 5.7ms
45: learn: 0.2077590 total: 53.1ms remaining: 4.61ms
46: learn: 0.2062417 total: 54.4ms remaining: 3.47ms
47: learn: 0.2041230 total: 55.5ms remaining: 2.31ms
48: learn: 0.2029428 total: 56.5ms remaining: 1.15ms
49: learn: 0.2010316 total: 57.8ms remaining: 0us
0: learn: 0.6466390 total: 372us remaining: 18.2ms
1: learn: 0.6043306 total: 743us remaining: 17.8ms
2: learn: 0.5698679 total: 977us remaining: 15.3ms
3: learn: 0.5400060 total: 1.28ms remaining: 14.7ms
4: learn: 0.5139000 total: 1.49ms remaining: 13.4ms
5: learn: 0.4909190 total: 1.84ms remaining: 13.5ms
6: learn: 0.4663019 total: 2.26ms remaining: 13.9ms
7: learn: 0.4496656 total: 2.94ms remaining: 15.5ms
8: learn: 0.4302446 total: 3.47ms remaining: 15.8ms
9: learn: 0.4155090 total: 4.23ms remaining: 16.9ms
10: learn: 0.4046576 total: 4.85ms remaining: 17.2ms
11: learn: 0.3902395 total: 5.52ms remaining: 17.5ms
12: learn: 0.3781937 total: 6.31ms remaining: 18ms
13: learn: 0.3684001 total: 7.33ms remaining: 18.9ms
14: learn: 0.3594224 total: 8.53ms remaining: 19.9ms
15: learn: 0.3506654 total: 9.05ms remaining: 19.2ms
16: learn: 0.3427953 total: 9.35ms remaining: 18.2ms
17: learn: 0.3375828 total: 9.63ms remaining: 17.1ms
18: learn: 0.3304561 total: 10.4ms remaining: 16.9ms
19: learn: 0.3248426 total: 11.4ms remaining: 17.1ms
20: learn: 0.3187507 total: 12.6ms remaining: 17.4ms
21: learn: 0.3109731 total: 13.8ms remaining: 17.6ms
22: learn: 0.3052623 total: 14.6ms remaining: 17.1ms
23: learn: 0.3002449 total: 15.6ms remaining: 16.9ms
24: learn: 0.2967801 total: 16.7ms remaining: 16.7ms
25: learn: 0.2923089 total: 17.7ms remaining: 16.3ms
26: learn: 0.2868579 total: 19ms remaining: 16.2ms
27: learn: 0.2841698 total: 20ms remaining: 15.7ms
28: learn: 0.2810342 total: 20.9ms remaining: 15.1ms
29: learn: 0.2780382 total: 21.8ms remaining: 14.6ms
30: learn: 0.2758407 total: 22.8ms remaining: 14ms
31: learn: 0.2730709 total: 24ms remaining: 13.5ms
32: learn: 0.2704504 total: 25.2ms remaining: 13ms
33: learn: 0.2684170 total: 26.3ms remaining: 12.4ms
34: learn: 0.2646933 total: 27.5ms remaining: 11.8ms
35: learn: 0.2624324 total: 29.2ms remaining: 11.4ms
36: learn: 0.2604280 total: 30.4ms remaining: 10.7ms
37: learn: 0.2579343 total: 31.2ms remaining: 9.85ms
38: learn: 0.2550934 total: 31.6ms remaining: 8.91ms
39: learn: 0.2526229 total: 32.6ms remaining: 8.14ms
40: learn: 0.2500359 total: 33.6ms remaining: 7.38ms
41: learn: 0.2471391 total: 34.4ms remaining: 6.55ms
42: learn: 0.2454036 total: 34.7ms remaining: 5.65ms
43: learn: 0.2433846 total: 35.2ms remaining: 4.8ms
44: learn: 0.2406459 total: 36.4ms remaining: 4.04ms
45: learn: 0.2380629 total: 37.9ms remaining: 3.3ms
46: learn: 0.2358969 total: 39.6ms remaining: 2.52ms
47: learn: 0.2344105 total: 40.9ms remaining: 1.7ms
48: learn: 0.2324208 total: 42.2ms remaining: 860us
49: learn: 0.2305732 total: 43.6ms remaining: 0us
0: learn: 0.6501714 total: 307us remaining: 15.1ms
1: learn: 0.6066480 total: 522us remaining: 12.6ms
2: learn: 0.5657020 total: 690us remaining: 10.8ms
3: learn: 0.5314888 total: 934us remaining: 10.8ms
4: learn: 0.5033352 total: 1.4ms remaining: 12.6ms
5: learn: 0.4746589 total: 1.96ms remaining: 14.3ms
6: learn: 0.4545285 total: 2.74ms remaining: 16.8ms
7: learn: 0.4381047 total: 3.19ms remaining: 16.7ms
8: learn: 0.4177237 total: 3.48ms remaining: 15.9ms
9: learn: 0.4023522 total: 3.95ms remaining: 15.8ms
10: learn: 0.3914068 total: 4.24ms remaining: 15ms
11: learn: 0.3787263 total: 4.55ms remaining: 14.4ms
12: learn: 0.3650055 total: 5.1ms remaining: 14.5ms
13: learn: 0.3522909 total: 5.7ms remaining: 14.7ms
14: learn: 0.3408388 total: 5.94ms remaining: 13.9ms
15: learn: 0.3301578 total: 6.2ms remaining: 13.2ms
16: learn: 0.3207327 total: 6.61ms remaining: 12.8ms
17: learn: 0.3122013 total: 7.02ms remaining: 12.5ms
18: learn: 0.3065739 total: 7.36ms remaining: 12ms
19: learn: 0.2986789 total: 7.87ms remaining: 11.8ms
20: learn: 0.2920784 total: 8.17ms remaining: 11.3ms
21: learn: 0.2861408 total: 8.48ms remaining: 10.8ms
22: learn: 0.2806232 total: 9ms remaining: 10.6ms
23: learn: 0.2757454 total: 9.37ms remaining: 10.2ms
24: learn: 0.2715914 total: 9.63ms remaining: 9.63ms
25: learn: 0.2662000 total: 9.93ms remaining: 9.16ms
26: learn: 0.2620222 total: 10.7ms remaining: 9.09ms
27: learn: 0.2586619 total: 11.3ms remaining: 8.89ms
28: learn: 0.2559411 total: 11.6ms remaining: 8.38ms
29: learn: 0.2521874 total: 11.9ms remaining: 7.92ms
30: learn: 0.2487332 total: 12.4ms remaining: 7.61ms
31: learn: 0.2455035 total: 12.7ms remaining: 7.15ms
32: learn: 0.2428433 total: 13ms remaining: 6.69ms
33: learn: 0.2410762 total: 13.1ms remaining: 6.18ms
34: learn: 0.2382084 total: 13.5ms remaining: 5.79ms
35: learn: 0.2366788 total: 13.8ms remaining: 5.37ms
36: learn: 0.2347484 total: 14ms remaining: 4.93ms
37: learn: 0.2324811 total: 14.5ms remaining: 4.57ms
38: learn: 0.2302696 total: 14.9ms remaining: 4.21ms
39: learn: 0.2282705 total: 15.2ms remaining: 3.8ms
40: learn: 0.2261110 total: 15.6ms remaining: 3.42ms
41: learn: 0.2238719 total: 16ms remaining: 3.06ms
42: learn: 0.2215777 total: 16.4ms remaining: 2.67ms
43: learn: 0.2196766 total: 16.9ms remaining: 2.31ms
44: learn: 0.2185631 total: 17.4ms remaining: 1.93ms
45: learn: 0.2166483 total: 17.9ms remaining: 1.55ms
46: learn: 0.2139387 total: 18.3ms remaining: 1.17ms
47: learn: 0.2129512 total: 18.5ms remaining: 771us
48: learn: 0.2119949 total: 18.8ms remaining: 383us
49: learn: 0.2109265 total: 19.1ms remaining: 0us
0: learn: 0.6481110 total: 402us remaining: 19.7ms
1: learn: 0.6015437 total: 1.19ms remaining: 28.7ms
2: learn: 0.5637141 total: 2.3ms remaining: 36ms
3: learn: 0.5285975 total: 3.48ms remaining: 40.1ms
4: learn: 0.5021036 total: 4.29ms remaining: 38.6ms
5: learn: 0.4762132 total: 4.9ms remaining: 35.9ms
6: learn: 0.4571360 total: 5.71ms remaining: 35ms
7: learn: 0.4392327 total: 7.54ms remaining: 39.6ms
8: learn: 0.4235303 total: 8.14ms remaining: 37.1ms
9: learn: 0.4067796 total: 9.19ms remaining: 36.8ms
10: learn: 0.3956489 total: 10.1ms remaining: 35.8ms
11: learn: 0.3834309 total: 11.7ms remaining: 36.9ms
12: learn: 0.3706993 total: 12.7ms remaining: 36.1ms
13: learn: 0.3593553 total: 13.7ms remaining: 35.2ms
14: learn: 0.3495868 total: 14.5ms remaining: 33.8ms
15: learn: 0.3380214 total: 16ms remaining: 33.9ms
16: learn: 0.3303325 total: 17.1ms remaining: 33.1ms
17: learn: 0.3239962 total: 18.1ms remaining: 32.3ms
18: learn: 0.3173096 total: 19.2ms remaining: 31.4ms
19: learn: 0.3130535 total: 20.4ms remaining: 30.6ms
20: learn: 0.3053856 total: 22.7ms remaining: 31.3ms
21: learn: 0.2967920 total: 23.8ms remaining: 30.3ms
22: learn: 0.2911773 total: 25.1ms remaining: 29.4ms
23: learn: 0.2846182 total: 26.5ms remaining: 28.7ms
24: learn: 0.2808455 total: 27.9ms remaining: 27.9ms
25: learn: 0.2780712 total: 29.1ms remaining: 26.9ms
26: learn: 0.2744681 total: 31ms remaining: 26.4ms
27: learn: 0.2696076 total: 32.4ms remaining: 25.5ms
28: learn: 0.2644929 total: 33.4ms remaining: 24.2ms
29: learn: 0.2616771 total: 34.3ms remaining: 22.9ms
30: learn: 0.2582177 total: 35.8ms remaining: 21.9ms
31: learn: 0.2542861 total: 37.3ms remaining: 21ms
32: learn: 0.2512674 total: 39ms remaining: 20.1ms
33: learn: 0.2476332 total: 40.3ms remaining: 19ms
34: learn: 0.2439946 total: 41.8ms remaining: 17.9ms
35: learn: 0.2408321 total: 43.3ms remaining: 16.8ms
36: learn: 0.2375090 total: 45.6ms remaining: 16ms
37: learn: 0.2354509 total: 47.5ms remaining: 15ms
38: learn: 0.2341470 total: 49.2ms remaining: 13.9ms
39: learn: 0.2321929 total: 50.5ms remaining: 12.6ms
40: learn: 0.2300608 total: 52ms remaining: 11.4ms
41: learn: 0.2276641 total: 53.5ms remaining: 10.2ms
42: learn: 0.2262292 total: 54.5ms remaining: 8.87ms
43: learn: 0.2238001 total: 55.5ms remaining: 7.57ms
44: learn: 0.2225238 total: 56.5ms remaining: 6.27ms
45: learn: 0.2207839 total: 57.6ms remaining: 5.01ms
46: learn: 0.2192435 total: 58.4ms remaining: 3.73ms
47: learn: 0.2170573 total: 58.9ms remaining: 2.45ms
48: learn: 0.2151633 total: 59.7ms remaining: 1.22ms
49: learn: 0.2135362 total: 60.6ms remaining: 0us
0: learn: 0.6515506 total: 365us remaining: 17.9ms
1: learn: 0.6073513 total: 660us remaining: 15.8ms
2: learn: 0.5704869 total: 881us remaining: 13.8ms
3: learn: 0.5398446 total: 1.23ms remaining: 14.2ms
4: learn: 0.5111154 total: 1.76ms remaining: 15.8ms
5: learn: 0.4857031 total: 2.41ms remaining: 17.7ms
6: learn: 0.4649189 total: 3.18ms remaining: 19.5ms
7: learn: 0.4473799 total: 3.97ms remaining: 20.8ms
8: learn: 0.4283679 total: 4.67ms remaining: 21.3ms
9: learn: 0.4129013 total: 5.41ms remaining: 21.6ms
10: learn: 0.3981913 total: 6.05ms remaining: 21.5ms
11: learn: 0.3860850 total: 6.88ms remaining: 21.8ms
12: learn: 0.3747197 total: 7.77ms remaining: 22.1ms
13: learn: 0.3643286 total: 8.6ms remaining: 22.1ms
14: learn: 0.3560632 total: 9.06ms remaining: 21.1ms
15: learn: 0.3493369 total: 9.85ms remaining: 20.9ms
16: learn: 0.3405992 total: 10.6ms remaining: 20.5ms
17: learn: 0.3364019 total: 11.1ms remaining: 19.8ms
18: learn: 0.3284740 total: 11.9ms remaining: 19.4ms
19: learn: 0.3207455 total: 12.9ms remaining: 19.4ms
20: learn: 0.3136963 total: 13.9ms remaining: 19.1ms
21: learn: 0.3068343 total: 14.9ms remaining: 18.9ms
22: learn: 0.3015201 total: 15.4ms remaining: 18.1ms
23: learn: 0.2973018 total: 16.2ms remaining: 17.5ms
24: learn: 0.2925741 total: 17ms remaining: 17ms
25: learn: 0.2863699 total: 18.1ms remaining: 16.7ms
26: learn: 0.2828654 total: 19.2ms remaining: 16.4ms
27: learn: 0.2796819 total: 20.2ms remaining: 15.9ms
28: learn: 0.2773885 total: 20.9ms remaining: 15.1ms
29: learn: 0.2764725 total: 21.9ms remaining: 14.6ms
30: learn: 0.2727544 total: 22.7ms remaining: 13.9ms
31: learn: 0.2691427 total: 23.7ms remaining: 13.3ms
32: learn: 0.2657733 total: 24.7ms remaining: 12.7ms
33: learn: 0.2627910 total: 25.6ms remaining: 12ms
34: learn: 0.2593242 total: 26.5ms remaining: 11.4ms
35: learn: 0.2568494 total: 27.6ms remaining: 10.7ms
36: learn: 0.2522878 total: 28.7ms remaining: 10.1ms
37: learn: 0.2495150 total: 29.6ms remaining: 9.33ms
38: learn: 0.2464842 total: 30.5ms remaining: 8.61ms
39: learn: 0.2445408 total: 31.5ms remaining: 7.87ms
40: learn: 0.2418177 total: 32.7ms remaining: 7.19ms
41: learn: 0.2396222 total: 33.6ms remaining: 6.4ms
42: learn: 0.2366848 total: 34.7ms remaining: 5.65ms
43: learn: 0.2330100 total: 35.8ms remaining: 4.89ms
44: learn: 0.2310290 total: 37.1ms remaining: 4.12ms
45: learn: 0.2291962 total: 38.5ms remaining: 3.34ms
46: learn: 0.2279771 total: 39.6ms remaining: 2.53ms
47: learn: 0.2257751 total: 40.5ms remaining: 1.69ms
48: learn: 0.2228804 total: 41.5ms remaining: 846us
49: learn: 0.2201211 total: 42.5ms remaining: 0us
0: learn: 0.6455145 total: 313us remaining: 15.3ms
1: learn: 0.5988760 total: 537us remaining: 12.9ms
2: learn: 0.5594107 total: 698us remaining: 10.9ms
3: learn: 0.5244881 total: 937us remaining: 10.8ms
4: learn: 0.4973720 total: 1.1ms remaining: 9.93ms
5: learn: 0.4741560 total: 1.3ms remaining: 9.51ms
6: learn: 0.4507672 total: 1.48ms remaining: 9.09ms
7: learn: 0.4334544 total: 1.72ms remaining: 9.02ms
8: learn: 0.4127818 total: 1.92ms remaining: 8.75ms
9: learn: 0.3964144 total: 2.32ms remaining: 9.28ms
10: learn: 0.3815367 total: 2.51ms remaining: 8.91ms
11: learn: 0.3706824 total: 2.72ms remaining: 8.63ms
12: learn: 0.3603382 total: 2.92ms remaining: 8.3ms
13: learn: 0.3494307 total: 3.23ms remaining: 8.31ms
14: learn: 0.3417150 total: 3.42ms remaining: 7.99ms
15: learn: 0.3334102 total: 3.78ms remaining: 8.04ms
16: learn: 0.3233884 total: 4.21ms remaining: 8.18ms
17: learn: 0.3176727 total: 4.5ms remaining: 8.01ms
18: learn: 0.3103917 total: 5.03ms remaining: 8.2ms
19: learn: 0.3034642 total: 5.29ms remaining: 7.94ms
20: learn: 0.2980677 total: 5.97ms remaining: 8.25ms
21: learn: 0.2918482 total: 6.4ms remaining: 8.14ms
22: learn: 0.2854194 total: 7.27ms remaining: 8.53ms
23: learn: 0.2799811 total: 7.92ms remaining: 8.58ms
24: learn: 0.2749249 total: 8.53ms remaining: 8.53ms
25: learn: 0.2704717 total: 9.12ms remaining: 8.42ms
26: learn: 0.2662482 total: 9.77ms remaining: 8.32ms
27: learn: 0.2633140 total: 10.5ms remaining: 8.21ms
28: learn: 0.2597887 total: 11.5ms remaining: 8.29ms
29: learn: 0.2568451 total: 12.1ms remaining: 8.07ms
30: learn: 0.2544384 total: 12.4ms remaining: 7.58ms
31: learn: 0.2516443 total: 12.7ms remaining: 7.15ms
32: learn: 0.2467225 total: 13.2ms remaining: 6.79ms
33: learn: 0.2444546 total: 13.7ms remaining: 6.45ms
34: learn: 0.2425635 total: 14.2ms remaining: 6.07ms
35: learn: 0.2394869 total: 14.7ms remaining: 5.71ms
36: learn: 0.2373269 total: 15ms remaining: 5.27ms
37: learn: 0.2343314 total: 15.5ms remaining: 4.89ms
38: learn: 0.2317488 total: 15.9ms remaining: 4.49ms
39: learn: 0.2306135 total: 16.4ms remaining: 4.09ms
40: learn: 0.2289222 total: 17.1ms remaining: 3.75ms
41: learn: 0.2267190 total: 17.5ms remaining: 3.33ms
42: learn: 0.2254531 total: 18ms remaining: 2.94ms
43: learn: 0.2235827 total: 18.7ms remaining: 2.54ms
44: learn: 0.2210251 total: 19.1ms remaining: 2.13ms
45: learn: 0.2194663 total: 19.8ms remaining: 1.72ms
46: learn: 0.2180275 total: 20ms remaining: 1.28ms
47: learn: 0.2169634 total: 20.5ms remaining: 852us
48: learn: 0.2141994 total: 20.8ms remaining: 424us
49: learn: 0.2126256 total: 21.1ms remaining: 0us
0: learn: 0.6503257 total: 436us remaining: 21.4ms
1: learn: 0.6091405 total: 863us remaining: 20.7ms
2: learn: 0.5749262 total: 1.65ms remaining: 25.9ms
3: learn: 0.5438978 total: 2.73ms remaining: 31.4ms
4: learn: 0.5155700 total: 3.64ms remaining: 32.8ms
5: learn: 0.4927320 total: 4.75ms remaining: 34.8ms
6: learn: 0.4727667 total: 5.81ms remaining: 35.7ms
7: learn: 0.4557302 total: 6.53ms remaining: 34.3ms
8: learn: 0.4388609 total: 6.81ms remaining: 31ms
9: learn: 0.4247644 total: 6.95ms remaining: 27.8ms
10: learn: 0.4144140 total: 7.87ms remaining: 27.9ms
11: learn: 0.4019024 total: 9.21ms remaining: 29.2ms
12: learn: 0.3916899 total: 9.68ms remaining: 27.6ms
13: learn: 0.3815831 total: 9.97ms remaining: 25.6ms
14: learn: 0.3717996 total: 10.3ms remaining: 24.1ms
15: learn: 0.3647230 total: 11ms remaining: 23.3ms
16: learn: 0.3545912 total: 11.9ms remaining: 23.1ms
17: learn: 0.3502944 total: 12.8ms remaining: 22.7ms
18: learn: 0.3428250 total: 14ms remaining: 22.8ms
19: learn: 0.3374503 total: 14.9ms remaining: 22.4ms
20: learn: 0.3317534 total: 16.1ms remaining: 22.2ms
21: learn: 0.3248470 total: 17ms remaining: 21.6ms
22: learn: 0.3197077 total: 17.6ms remaining: 20.6ms
23: learn: 0.3149752 total: 18.6ms remaining: 20.2ms
24: learn: 0.3115804 total: 19.4ms remaining: 19.4ms
25: learn: 0.3073465 total: 19.8ms remaining: 18.3ms
26: learn: 0.3014933 total: 21.2ms remaining: 18ms
27: learn: 0.2982788 total: 22ms remaining: 17.3ms
28: learn: 0.2954439 total: 22.7ms remaining: 16.4ms
29: learn: 0.2917875 total: 23.6ms remaining: 15.8ms
30: learn: 0.2900107 total: 24.7ms remaining: 15.2ms
31: learn: 0.2864544 total: 25.8ms remaining: 14.5ms
32: learn: 0.2810382 total: 26.8ms remaining: 13.8ms
33: learn: 0.2787524 total: 27.7ms remaining: 13ms
34: learn: 0.2764095 total: 28.8ms remaining: 12.3ms
35: learn: 0.2748109 total: 29.9ms remaining: 11.6ms
36: learn: 0.2728280 total: 30.8ms remaining: 10.8ms
37: learn: 0.2703365 total: 31.4ms remaining: 9.92ms
38: learn: 0.2683979 total: 32.3ms remaining: 9.1ms
39: learn: 0.2667767 total: 33.6ms remaining: 8.39ms
40: learn: 0.2652805 total: 34.4ms remaining: 7.56ms
41: learn: 0.2621843 total: 35.2ms remaining: 6.71ms
42: learn: 0.2596001 total: 35.9ms remaining: 5.84ms
43: learn: 0.2571810 total: 36.7ms remaining: 5ms
44: learn: 0.2554186 total: 37.8ms remaining: 4.2ms
45: learn: 0.2533093 total: 38.5ms remaining: 3.35ms
46: learn: 0.2511507 total: 39.7ms remaining: 2.54ms
47: learn: 0.2495800 total: 40.8ms remaining: 1.7ms
48: learn: 0.2474001 total: 41.5ms remaining: 847us
49: learn: 0.2461070 total: 41.8ms remaining: 0us
0: learn: 0.6515750 total: 609us remaining: 29.8ms
1: learn: 0.6134885 total: 1.15ms remaining: 27.6ms
2: learn: 0.5837400 total: 1.72ms remaining: 26.9ms
3: learn: 0.5489154 total: 2.56ms remaining: 29.4ms
4: learn: 0.5169299 total: 3.53ms remaining: 31.8ms
5: learn: 0.4907425 total: 5.61ms remaining: 41.1ms
6: learn: 0.4713171 total: 6.71ms remaining: 41.2ms
7: learn: 0.4534210 total: 7.43ms remaining: 39ms
8: learn: 0.4367045 total: 8.56ms remaining: 39ms
9: learn: 0.4205350 total: 9.39ms remaining: 37.6ms
10: learn: 0.4056829 total: 10.4ms remaining: 36.8ms
11: learn: 0.3903177 total: 11.2ms remaining: 35.5ms
12: learn: 0.3766978 total: 12.2ms remaining: 34.7ms
13: learn: 0.3669745 total: 13.1ms remaining: 33.8ms
14: learn: 0.3548570 total: 14.1ms remaining: 32.8ms
15: learn: 0.3456593 total: 15.1ms remaining: 32.2ms
16: learn: 0.3378650 total: 15.9ms remaining: 30.9ms
17: learn: 0.3310731 total: 16.4ms remaining: 29.1ms
18: learn: 0.3233461 total: 17.2ms remaining: 28.1ms
19: learn: 0.3165295 total: 18.4ms remaining: 27.6ms
20: learn: 0.3094843 total: 19.5ms remaining: 27ms
21: learn: 0.3020178 total: 20.6ms remaining: 26.2ms
22: learn: 0.2965073 total: 21.5ms remaining: 25.3ms
23: learn: 0.2896890 total: 22.2ms remaining: 24ms
24: learn: 0.2844204 total: 23.4ms remaining: 23.4ms
25: learn: 0.2797083 total: 24.5ms remaining: 22.6ms
26: learn: 0.2751533 total: 25.8ms remaining: 22ms
27: learn: 0.2715553 total: 26.8ms remaining: 21.1ms
28: learn: 0.2691510 total: 27.6ms remaining: 20ms
29: learn: 0.2671634 total: 28.8ms remaining: 19.2ms
30: learn: 0.2650620 total: 29.4ms remaining: 18ms
31: learn: 0.2613732 total: 30ms remaining: 16.9ms
32: learn: 0.2592097 total: 30.5ms remaining: 15.7ms
33: learn: 0.2555304 total: 31.8ms remaining: 14.9ms
34: learn: 0.2532133 total: 33ms remaining: 14.2ms
35: learn: 0.2505760 total: 34.7ms remaining: 13.5ms
36: learn: 0.2488293 total: 35.7ms remaining: 12.5ms
37: learn: 0.2453197 total: 37ms remaining: 11.7ms
38: learn: 0.2419295 total: 38.4ms remaining: 10.8ms
39: learn: 0.2391228 total: 39.3ms remaining: 9.83ms
40: learn: 0.2363322 total: 40.3ms remaining: 8.84ms
41: learn: 0.2343441 total: 41.1ms remaining: 7.83ms
42: learn: 0.2325132 total: 42.2ms remaining: 6.87ms
43: learn: 0.2296161 total: 43.7ms remaining: 5.96ms
44: learn: 0.2287773 total: 44.6ms remaining: 4.96ms
45: learn: 0.2272871 total: 45.7ms remaining: 3.98ms
46: learn: 0.2260889 total: 46.5ms remaining: 2.97ms
47: learn: 0.2244358 total: 46.9ms remaining: 1.95ms
48: learn: 0.2231578 total: 47.9ms remaining: 977us
49: learn: 0.2215341 total: 49.3ms remaining: 0us
0: learn: 0.6495190 total: 365us remaining: 17.9ms
1: learn: 0.6095526 total: 660us remaining: 15.8ms
2: learn: 0.5719992 total: 929us remaining: 14.6ms
3: learn: 0.5425656 total: 1.73ms remaining: 19.9ms
4: learn: 0.5159659 total: 2.8ms remaining: 25.2ms
5: learn: 0.4917545 total: 3.44ms remaining: 25.3ms
6: learn: 0.4701324 total: 3.68ms remaining: 22.6ms
7: learn: 0.4524320 total: 4.61ms remaining: 24.2ms
8: learn: 0.4325252 total: 5.49ms remaining: 25ms
9: learn: 0.4165980 total: 6.8ms remaining: 27.2ms
10: learn: 0.4035100 total: 7.82ms remaining: 27.7ms
11: learn: 0.3907931 total: 8.84ms remaining: 28ms
12: learn: 0.3789276 total: 9.73ms remaining: 27.7ms
13: learn: 0.3688594 total: 10.7ms remaining: 27.5ms
14: learn: 0.3595050 total: 11.7ms remaining: 27.3ms
15: learn: 0.3510985 total: 12.8ms remaining: 27.3ms
16: learn: 0.3435509 total: 13.7ms remaining: 26.6ms
17: learn: 0.3372297 total: 14.8ms remaining: 26.3ms
18: learn: 0.3310716 total: 15.6ms remaining: 25.4ms
19: learn: 0.3258688 total: 16.3ms remaining: 24.4ms
20: learn: 0.3195430 total: 17ms remaining: 23.5ms
21: learn: 0.3125519 total: 18.1ms remaining: 23ms
22: learn: 0.3074617 total: 18.8ms remaining: 22.1ms
23: learn: 0.3031872 total: 19ms remaining: 20.5ms
24: learn: 0.2996652 total: 19.5ms remaining: 19.5ms
25: learn: 0.2955970 total: 20.5ms remaining: 18.9ms
26: learn: 0.2919958 total: 21.5ms remaining: 18.3ms
27: learn: 0.2880784 total: 21.9ms remaining: 17.2ms
28: learn: 0.2850178 total: 23.1ms remaining: 16.7ms
29: learn: 0.2831896 total: 23.6ms remaining: 15.7ms
30: learn: 0.2807488 total: 23.9ms remaining: 14.7ms
31: learn: 0.2781251 total: 24.2ms remaining: 13.6ms
32: learn: 0.2755400 total: 25.1ms remaining: 12.9ms
33: learn: 0.2735858 total: 25.8ms remaining: 12.2ms
34: learn: 0.2709428 total: 26.7ms remaining: 11.4ms
35: learn: 0.2680968 total: 27.8ms remaining: 10.8ms
36: learn: 0.2653391 total: 29ms remaining: 10.2ms
37: learn: 0.2622118 total: 29.7ms remaining: 9.39ms
38: learn: 0.2589166 total: 30.5ms remaining: 8.59ms
39: learn: 0.2560415 total: 31.4ms remaining: 7.85ms
40: learn: 0.2545774 total: 32ms remaining: 7.03ms
41: learn: 0.2521408 total: 32.6ms remaining: 6.2ms
42: learn: 0.2508771 total: 33ms remaining: 5.37ms
43: learn: 0.2500696 total: 33.7ms remaining: 4.6ms
44: learn: 0.2485904 total: 34.1ms remaining: 3.79ms
45: learn: 0.2465004 total: 35.3ms remaining: 3.07ms
46: learn: 0.2446957 total: 36.1ms remaining: 2.31ms
47: learn: 0.2427096 total: 36.8ms remaining: 1.53ms
48: learn: 0.2415083 total: 37.9ms remaining: 772us
49: learn: 0.2387823 total: 38.8ms remaining: 0us
0: learn: 0.6487726 total: 442us remaining: 21.7ms
1: learn: 0.6054752 total: 740us remaining: 17.8ms
2: learn: 0.5690516 total: 989us remaining: 15.5ms
3: learn: 0.5376892 total: 1.42ms remaining: 16.3ms
4: learn: 0.5107400 total: 1.86ms remaining: 16.8ms
5: learn: 0.4860125 total: 2.41ms remaining: 17.7ms
6: learn: 0.4650487 total: 3ms remaining: 18.4ms
7: learn: 0.4474529 total: 3.58ms remaining: 18.8ms
8: learn: 0.4286769 total: 4.14ms remaining: 18.9ms
9: learn: 0.4136039 total: 4.7ms remaining: 18.8ms
10: learn: 0.4019389 total: 5.15ms remaining: 18.3ms
11: learn: 0.3894000 total: 5.79ms remaining: 18.3ms
12: learn: 0.3778865 total: 6.42ms remaining: 18.3ms
13: learn: 0.3672801 total: 7.1ms remaining: 18.3ms
14: learn: 0.3576247 total: 7.65ms remaining: 17.9ms
15: learn: 0.3495637 total: 8.28ms remaining: 17.6ms
16: learn: 0.3412530 total: 8.95ms remaining: 17.4ms
17: learn: 0.3350939 total: 9.36ms remaining: 16.6ms
18: learn: 0.3290651 total: 9.92ms remaining: 16.2ms
19: learn: 0.3248385 total: 10.2ms remaining: 15.3ms
20: learn: 0.3197584 total: 10.7ms remaining: 14.8ms
21: learn: 0.3123746 total: 11.3ms remaining: 14.3ms
22: learn: 0.3067161 total: 11.9ms remaining: 14ms
23: learn: 0.3018604 total: 12.9ms remaining: 14ms
24: learn: 0.2972128 total: 13.5ms remaining: 13.5ms
25: learn: 0.2947712 total: 14.1ms remaining: 13ms
26: learn: 0.2912988 total: 14.5ms remaining: 12.3ms
27: learn: 0.2870765 total: 15.2ms remaining: 11.9ms
28: learn: 0.2831592 total: 15.8ms remaining: 11.4ms
29: learn: 0.2813999 total: 16.4ms remaining: 10.9ms
30: learn: 0.2782774 total: 17.4ms remaining: 10.6ms
31: learn: 0.2757420 total: 18ms remaining: 10.1ms
32: learn: 0.2733458 total: 18.9ms remaining: 9.76ms
33: learn: 0.2714703 total: 19.5ms remaining: 9.19ms
34: learn: 0.2692580 total: 20.1ms remaining: 8.63ms
35: learn: 0.2669193 total: 20.9ms remaining: 8.14ms
36: learn: 0.2654010 total: 21.6ms remaining: 7.6ms
37: learn: 0.2626552 total: 22.4ms remaining: 7.08ms
38: learn: 0.2591544 total: 23.5ms remaining: 6.63ms
39: learn: 0.2568921 total: 24.6ms remaining: 6.15ms
40: learn: 0.2539529 total: 26.2ms remaining: 5.75ms
41: learn: 0.2519482 total: 27.4ms remaining: 5.21ms
42: learn: 0.2496161 total: 28.1ms remaining: 4.57ms
43: learn: 0.2481991 total: 28.4ms remaining: 3.88ms
44: learn: 0.2469000 total: 29.2ms remaining: 3.24ms
45: learn: 0.2455710 total: 30.2ms remaining: 2.63ms
46: learn: 0.2415597 total: 31.3ms remaining: 2ms
47: learn: 0.2399522 total: 32.2ms remaining: 1.34ms
48: learn: 0.2386716 total: 33ms remaining: 673us
49: learn: 0.2371674 total: 34ms remaining: 0us
0: learn: 0.6470146 total: 375us remaining: 18.4ms
1: learn: 0.6064299 total: 742us remaining: 17.8ms
2: learn: 0.5691283 total: 1.08ms remaining: 16.9ms
3: learn: 0.5366569 total: 1.78ms remaining: 20.5ms
4: learn: 0.5098000 total: 2.24ms remaining: 20.2ms
5: learn: 0.4832216 total: 2.8ms remaining: 20.5ms
6: learn: 0.4596274 total: 3.29ms remaining: 20.2ms
7: learn: 0.4437963 total: 3.88ms remaining: 20.4ms
8: learn: 0.4236547 total: 4.3ms remaining: 19.6ms
9: learn: 0.4091465 total: 4.75ms remaining: 19ms
10: learn: 0.3961872 total: 5.17ms remaining: 18.3ms
11: learn: 0.3834046 total: 5.73ms remaining: 18.2ms
12: learn: 0.3710681 total: 6.2ms remaining: 17.6ms
13: learn: 0.3595058 total: 6.71ms remaining: 17.3ms
14: learn: 0.3509086 total: 7.17ms remaining: 16.7ms
15: learn: 0.3450153 total: 7.69ms remaining: 16.3ms
16: learn: 0.3371444 total: 8.15ms remaining: 15.8ms
17: learn: 0.3298510 total: 8.6ms remaining: 15.3ms
18: learn: 0.3232019 total: 8.85ms remaining: 14.4ms
19: learn: 0.3162856 total: 9.34ms remaining: 14ms
20: learn: 0.3111943 total: 9.81ms remaining: 13.5ms
21: learn: 0.3057793 total: 10.3ms remaining: 13.1ms
22: learn: 0.3007321 total: 10.7ms remaining: 12.5ms
23: learn: 0.2962390 total: 11ms remaining: 11.9ms
24: learn: 0.2927857 total: 11.3ms remaining: 11.3ms
25: learn: 0.2900541 total: 11.8ms remaining: 10.9ms
26: learn: 0.2865934 total: 12.2ms remaining: 10.4ms
27: learn: 0.2819302 total: 12.8ms remaining: 10ms
28: learn: 0.2778663 total: 13.2ms remaining: 9.59ms
29: learn: 0.2737055 total: 13.8ms remaining: 9.18ms
30: learn: 0.2722615 total: 14.1ms remaining: 8.67ms
31: learn: 0.2689709 total: 14.6ms remaining: 8.2ms
32: learn: 0.2664546 total: 15ms remaining: 7.73ms
33: learn: 0.2629107 total: 15.5ms remaining: 7.27ms
34: learn: 0.2603216 total: 16ms remaining: 6.84ms
35: learn: 0.2574981 total: 16.4ms remaining: 6.38ms
36: learn: 0.2557634 total: 16.7ms remaining: 5.87ms
37: learn: 0.2525677 total: 17.2ms remaining: 5.44ms
38: learn: 0.2487591 total: 17.6ms remaining: 4.97ms
39: learn: 0.2454105 total: 18.1ms remaining: 4.52ms
40: learn: 0.2435551 total: 18.6ms remaining: 4.08ms
41: learn: 0.2401939 total: 19.1ms remaining: 3.63ms
42: learn: 0.2387428 total: 19.4ms remaining: 3.15ms
43: learn: 0.2376608 total: 19.8ms remaining: 2.7ms
44: learn: 0.2358510 total: 20.3ms remaining: 2.25ms
45: learn: 0.2332892 total: 20.8ms remaining: 1.8ms
46: learn: 0.2316049 total: 21.1ms remaining: 1.34ms
47: learn: 0.2293167 total: 21.5ms remaining: 895us
48: learn: 0.2264321 total: 21.9ms remaining: 446us
49: learn: 0.2240784 total: 22.3ms remaining: 0us
0: learn: 0.6531507 total: 466us remaining: 22.8ms
1: learn: 0.6065162 total: 1.3ms remaining: 31.2ms
2: learn: 0.5670336 total: 1.53ms remaining: 23.9ms
3: learn: 0.5358549 total: 1.81ms remaining: 20.8ms
4: learn: 0.5074412 total: 2.03ms remaining: 18.3ms
5: learn: 0.4804679 total: 2.31ms remaining: 17ms
6: learn: 0.4657784 total: 2.56ms remaining: 15.7ms
7: learn: 0.4441830 total: 3.14ms remaining: 16.5ms
8: learn: 0.4246293 total: 3.63ms remaining: 16.6ms
9: learn: 0.4082806 total: 4.25ms remaining: 17ms
10: learn: 0.3971826 total: 4.79ms remaining: 17ms
11: learn: 0.3859011 total: 5.51ms remaining: 17.5ms
12: learn: 0.3732273 total: 6.29ms remaining: 17.9ms
13: learn: 0.3599254 total: 6.98ms remaining: 17.9ms
14: learn: 0.3520452 total: 7.51ms remaining: 17.5ms
15: learn: 0.3434055 total: 8.17ms remaining: 17.4ms
16: learn: 0.3346383 total: 8.8ms remaining: 17.1ms
17: learn: 0.3283062 total: 9.37ms remaining: 16.7ms
18: learn: 0.3212423 total: 9.92ms remaining: 16.2ms
19: learn: 0.3155269 total: 11.5ms remaining: 17.3ms
20: learn: 0.3091024 total: 12.7ms remaining: 17.6ms
21: learn: 0.3038824 total: 13.5ms remaining: 17.2ms
22: learn: 0.2960116 total: 14.1ms remaining: 16.5ms
23: learn: 0.2915019 total: 15.1ms remaining: 16.3ms
24: learn: 0.2851576 total: 15.7ms remaining: 15.7ms
25: learn: 0.2824845 total: 16.5ms remaining: 15.2ms
26: learn: 0.2779683 total: 17.1ms remaining: 14.6ms
27: learn: 0.2739306 total: 17.8ms remaining: 14ms
28: learn: 0.2711215 total: 18.4ms remaining: 13.4ms
29: learn: 0.2672378 total: 19.1ms remaining: 12.8ms
30: learn: 0.2642976 total: 20ms remaining: 12.2ms
31: learn: 0.2610812 total: 20.7ms remaining: 11.6ms
32: learn: 0.2570838 total: 21.3ms remaining: 11ms
33: learn: 0.2537967 total: 21.9ms remaining: 10.3ms
34: learn: 0.2509818 total: 22.5ms remaining: 9.63ms
35: learn: 0.2490697 total: 23ms remaining: 8.94ms
36: learn: 0.2463164 total: 23.4ms remaining: 8.23ms
37: learn: 0.2441294 total: 24.1ms remaining: 7.61ms
38: learn: 0.2423599 total: 24.8ms remaining: 7ms
39: learn: 0.2392679 total: 25.5ms remaining: 6.37ms
40: learn: 0.2373584 total: 26.2ms remaining: 5.74ms
41: learn: 0.2351607 total: 26.9ms remaining: 5.12ms
42: learn: 0.2329397 total: 27.5ms remaining: 4.47ms
43: learn: 0.2311672 total: 28.1ms remaining: 3.83ms
44: learn: 0.2291315 total: 28.7ms remaining: 3.18ms
45: learn: 0.2273520 total: 29.2ms remaining: 2.54ms
46: learn: 0.2248574 total: 29.7ms remaining: 1.9ms
47: learn: 0.2237799 total: 30.2ms remaining: 1.26ms
48: learn: 0.2229512 total: 30.7ms remaining: 626us
49: learn: 0.2207943 total: 31.3ms remaining: 0us
0: learn: 0.6532610 total: 310us remaining: 15.2ms
1: learn: 0.6102409 total: 602us remaining: 14.5ms
2: learn: 0.5726766 total: 844us remaining: 13.2ms
3: learn: 0.5399936 total: 1.08ms remaining: 12.4ms
4: learn: 0.5149233 total: 1.25ms remaining: 11.2ms
5: learn: 0.4883109 total: 1.76ms remaining: 12.9ms
6: learn: 0.4641478 total: 1.95ms remaining: 12ms
7: learn: 0.4460774 total: 2.74ms remaining: 14.4ms
8: learn: 0.4323097 total: 3.13ms remaining: 14.2ms
9: learn: 0.4157583 total: 3.77ms remaining: 15.1ms
10: learn: 0.4022919 total: 4.31ms remaining: 15.3ms
11: learn: 0.3898014 total: 4.9ms remaining: 15.5ms
12: learn: 0.3788049 total: 5.12ms remaining: 14.6ms
13: learn: 0.3683505 total: 5.61ms remaining: 14.4ms
14: learn: 0.3577059 total: 6.17ms remaining: 14.4ms
15: learn: 0.3493490 total: 6.87ms remaining: 14.6ms
16: learn: 0.3384982 total: 7.65ms remaining: 14.9ms
17: learn: 0.3329396 total: 8.13ms remaining: 14.4ms
18: learn: 0.3265991 total: 8.76ms remaining: 14.3ms
19: learn: 0.3205432 total: 9.57ms remaining: 14.4ms
20: learn: 0.3157322 total: 10.4ms remaining: 14.4ms
21: learn: 0.3109822 total: 11ms remaining: 14ms
22: learn: 0.3063239 total: 11.4ms remaining: 13.4ms
23: learn: 0.3018177 total: 11.9ms remaining: 12.9ms
24: learn: 0.2976308 total: 12.2ms remaining: 12.2ms
25: learn: 0.2931462 total: 12.9ms remaining: 11.9ms
26: learn: 0.2895543 total: 13.5ms remaining: 11.5ms
27: learn: 0.2858063 total: 14.4ms remaining: 11.3ms
28: learn: 0.2817349 total: 15ms remaining: 10.9ms
29: learn: 0.2783638 total: 15.3ms remaining: 10.2ms
30: learn: 0.2755228 total: 16ms remaining: 9.79ms
31: learn: 0.2726879 total: 16.5ms remaining: 9.29ms
32: learn: 0.2707994 total: 16.9ms remaining: 8.7ms
33: learn: 0.2671753 total: 17.7ms remaining: 8.34ms
34: learn: 0.2642467 total: 18.6ms remaining: 7.96ms
35: learn: 0.2618695 total: 19ms remaining: 7.4ms
36: learn: 0.2601981 total: 19.6ms remaining: 6.88ms
37: learn: 0.2580945 total: 20.2ms remaining: 6.38ms
38: learn: 0.2563573 total: 20.8ms remaining: 5.87ms
39: learn: 0.2534017 total: 21.4ms remaining: 5.35ms
40: learn: 0.2503710 total: 22ms remaining: 4.84ms
41: learn: 0.2467603 total: 22.6ms remaining: 4.29ms
42: learn: 0.2445181 total: 23.1ms remaining: 3.75ms
43: learn: 0.2436733 total: 23.6ms remaining: 3.22ms
44: learn: 0.2424590 total: 24.1ms remaining: 2.68ms
45: learn: 0.2413170 total: 24.7ms remaining: 2.15ms
46: learn: 0.2395461 total: 25.1ms remaining: 1.6ms
47: learn: 0.2376051 total: 25.5ms remaining: 1.06ms
48: learn: 0.2348047 total: 25.9ms remaining: 527us
49: learn: 0.2325867 total: 26.4ms remaining: 0us
0: learn: 0.6478370 total: 429us remaining: 21.1ms
1: learn: 0.6050464 total: 1.22ms remaining: 29.2ms
2: learn: 0.5715635 total: 2.4ms remaining: 37.7ms
3: learn: 0.5388645 total: 3.74ms remaining: 43.1ms
4: learn: 0.5127410 total: 4.45ms remaining: 40ms
5: learn: 0.4894968 total: 5.11ms remaining: 37.5ms
6: learn: 0.4690824 total: 5.75ms remaining: 35.3ms
7: learn: 0.4527488 total: 6.52ms remaining: 34.2ms
8: learn: 0.4367211 total: 7.08ms remaining: 32.3ms
9: learn: 0.4197791 total: 8.05ms remaining: 32.2ms
10: learn: 0.4076903 total: 8.88ms remaining: 31.5ms
11: learn: 0.3953699 total: 10.2ms remaining: 32.2ms
12: learn: 0.3835805 total: 11.6ms remaining: 33.1ms
13: learn: 0.3706580 total: 12.8ms remaining: 32.9ms
14: learn: 0.3632837 total: 13.6ms remaining: 31.8ms
15: learn: 0.3557629 total: 14.9ms remaining: 31.7ms
16: learn: 0.3465254 total: 16.4ms remaining: 31.8ms
17: learn: 0.3424635 total: 17.6ms remaining: 31.2ms
18: learn: 0.3340110 total: 19ms remaining: 31ms
19: learn: 0.3290275 total: 21.3ms remaining: 32ms
20: learn: 0.3221192 total: 21.9ms remaining: 30.2ms
21: learn: 0.3160905 total: 22.4ms remaining: 28.5ms
22: learn: 0.3106294 total: 23.1ms remaining: 27.1ms
23: learn: 0.3056046 total: 24.2ms remaining: 26.2ms
24: learn: 0.3024425 total: 25.2ms remaining: 25.2ms
25: learn: 0.2989832 total: 26.3ms remaining: 24.3ms
26: learn: 0.2944404 total: 27.4ms remaining: 23.3ms
27: learn: 0.2902619 total: 28.3ms remaining: 22.2ms
28: learn: 0.2872351 total: 29.2ms remaining: 21.1ms
29: learn: 0.2832049 total: 30.2ms remaining: 20.1ms
30: learn: 0.2811090 total: 30.9ms remaining: 18.9ms
31: learn: 0.2773095 total: 31.6ms remaining: 17.8ms
32: learn: 0.2748773 total: 32.3ms remaining: 16.6ms
33: learn: 0.2728913 total: 33.1ms remaining: 15.6ms
34: learn: 0.2704194 total: 34.2ms remaining: 14.6ms
35: learn: 0.2677007 total: 35.3ms remaining: 13.7ms
36: learn: 0.2654716 total: 36.3ms remaining: 12.7ms
37: learn: 0.2625555 total: 37.4ms remaining: 11.8ms
38: learn: 0.2588682 total: 38.3ms remaining: 10.8ms
39: learn: 0.2557431 total: 39.1ms remaining: 9.79ms
40: learn: 0.2541972 total: 39.8ms remaining: 8.74ms
41: learn: 0.2533046 total: 40.4ms remaining: 7.7ms
42: learn: 0.2508947 total: 41.2ms remaining: 6.7ms
43: learn: 0.2488413 total: 42ms remaining: 5.73ms
44: learn: 0.2460522 total: 42.9ms remaining: 4.77ms
45: learn: 0.2437101 total: 43.6ms remaining: 3.79ms
46: learn: 0.2420143 total: 44.2ms remaining: 2.82ms
47: learn: 0.2394717 total: 45ms remaining: 1.87ms
48: learn: 0.2377849 total: 45.6ms remaining: 931us
49: learn: 0.2356327 total: 46.3ms remaining: 0us
0: learn: 0.6489461 total: 657us remaining: 32.2ms
1: learn: 0.6064240 total: 1.32ms remaining: 31.7ms
2: learn: 0.5700715 total: 2ms remaining: 31.4ms
3: learn: 0.5400758 total: 2.67ms remaining: 30.7ms
4: learn: 0.5145103 total: 3.25ms remaining: 29.3ms
5: learn: 0.4866673 total: 3.9ms remaining: 28.6ms
6: learn: 0.4673804 total: 4.76ms remaining: 29.2ms
7: learn: 0.4481874 total: 5.4ms remaining: 28.4ms
8: learn: 0.4300384 total: 6.27ms remaining: 28.6ms
9: learn: 0.4134195 total: 6.81ms remaining: 27.2ms
10: learn: 0.3962502 total: 7.39ms remaining: 26.2ms
11: learn: 0.3811386 total: 7.83ms remaining: 24.8ms
12: learn: 0.3693335 total: 8.46ms remaining: 24.1ms
13: learn: 0.3598914 total: 9.19ms remaining: 23.6ms
14: learn: 0.3516374 total: 9.89ms remaining: 23.1ms
15: learn: 0.3406200 total: 10.7ms remaining: 22.8ms
16: learn: 0.3323188 total: 11.4ms remaining: 22ms
17: learn: 0.3250314 total: 11.7ms remaining: 20.8ms
18: learn: 0.3192270 total: 12.4ms remaining: 20.2ms
19: learn: 0.3128178 total: 13.1ms remaining: 19.6ms
20: learn: 0.3059293 total: 13.8ms remaining: 19ms
21: learn: 0.2987314 total: 14.4ms remaining: 18.3ms
22: learn: 0.2940187 total: 15ms remaining: 17.6ms
23: learn: 0.2900778 total: 15.6ms remaining: 16.9ms
24: learn: 0.2872265 total: 16.3ms remaining: 16.3ms
25: learn: 0.2842832 total: 17ms remaining: 15.7ms
26: learn: 0.2804576 total: 17.6ms remaining: 15ms
27: learn: 0.2770150 total: 18.2ms remaining: 14.3ms
28: learn: 0.2748851 total: 18.7ms remaining: 13.5ms
29: learn: 0.2718988 total: 19.3ms remaining: 12.8ms
30: learn: 0.2689076 total: 19.7ms remaining: 12.1ms
31: learn: 0.2655543 total: 20.6ms remaining: 11.6ms
32: learn: 0.2633803 total: 21.1ms remaining: 10.9ms
33: learn: 0.2607521 total: 21.6ms remaining: 10.2ms
34: learn: 0.2586816 total: 22.1ms remaining: 9.49ms
35: learn: 0.2561481 total: 22.7ms remaining: 8.82ms
36: learn: 0.2535555 total: 23.2ms remaining: 8.17ms
37: learn: 0.2498320 total: 23.9ms remaining: 7.54ms
38: learn: 0.2465524 total: 24.5ms remaining: 6.91ms
39: learn: 0.2453466 total: 25ms remaining: 6.25ms
40: learn: 0.2437222 total: 25.4ms remaining: 5.58ms
41: learn: 0.2416669 total: 26.1ms remaining: 4.97ms
42: learn: 0.2391285 total: 26.6ms remaining: 4.33ms
43: learn: 0.2365899 total: 27.2ms remaining: 3.71ms
44: learn: 0.2348016 total: 27.7ms remaining: 3.08ms
45: learn: 0.2321103 total: 28.2ms remaining: 2.46ms
46: learn: 0.2307285 total: 28.7ms remaining: 1.83ms
47: learn: 0.2292365 total: 29.2ms remaining: 1.22ms
48: learn: 0.2281501 total: 29.5ms remaining: 601us
49: learn: 0.2266474 total: 29.9ms remaining: 0us
0: learn: 0.6590402 total: 871us remaining: 42.7ms
1: learn: 0.6114009 total: 1.84ms remaining: 44.3ms
2: learn: 0.5810295 total: 2.84ms remaining: 44.5ms
3: learn: 0.5494286 total: 4.14ms remaining: 47.7ms
4: learn: 0.5218481 total: 4.99ms remaining: 44.9ms
5: learn: 0.4961409 total: 5.64ms remaining: 41.4ms
6: learn: 0.4825933 total: 7.32ms remaining: 45ms
7: learn: 0.4612439 total: 8.06ms remaining: 42.3ms
8: learn: 0.4405597 total: 8.78ms remaining: 40ms
9: learn: 0.4236230 total: 9.7ms remaining: 38.8ms
10: learn: 0.4086284 total: 10.5ms remaining: 37.1ms
11: learn: 0.3966478 total: 11.2ms remaining: 35.6ms
12: learn: 0.3845862 total: 12.7ms remaining: 36.2ms
13: learn: 0.3734868 total: 14ms remaining: 35.9ms
14: learn: 0.3612373 total: 15ms remaining: 35.1ms
15: learn: 0.3563945 total: 16.1ms remaining: 34.1ms
16: learn: 0.3520736 total: 16.8ms remaining: 32.6ms
17: learn: 0.3437252 total: 17.3ms remaining: 30.8ms
18: learn: 0.3366232 total: 18.6ms remaining: 30.3ms
19: learn: 0.3305014 total: 19.5ms remaining: 29.3ms
20: learn: 0.3237168 total: 20.3ms remaining: 28ms
21: learn: 0.3165086 total: 21.4ms remaining: 27.3ms
22: learn: 0.3115073 total: 22.7ms remaining: 26.6ms
23: learn: 0.3070963 total: 23.5ms remaining: 25.5ms
24: learn: 0.3018950 total: 24.8ms remaining: 24.8ms
25: learn: 0.2978404 total: 25.7ms remaining: 23.8ms
26: learn: 0.2944708 total: 26.5ms remaining: 22.6ms
27: learn: 0.2896176 total: 27.4ms remaining: 21.5ms
28: learn: 0.2862532 total: 28.9ms remaining: 20.9ms
29: learn: 0.2832470 total: 29.6ms remaining: 19.7ms
30: learn: 0.2798789 total: 29.9ms remaining: 18.3ms
31: learn: 0.2760300 total: 31ms remaining: 17.5ms
32: learn: 0.2725581 total: 31.9ms remaining: 16.4ms
33: learn: 0.2704228 total: 32.8ms remaining: 15.5ms
34: learn: 0.2677841 total: 33.3ms remaining: 14.3ms
35: learn: 0.2653151 total: 34.5ms remaining: 13.4ms
36: learn: 0.2633276 total: 35.5ms remaining: 12.5ms
37: learn: 0.2602114 total: 36.5ms remaining: 11.5ms
38: learn: 0.2575564 total: 37.5ms remaining: 10.6ms
39: learn: 0.2550264 total: 38.1ms remaining: 9.51ms
40: learn: 0.2531548 total: 38.8ms remaining: 8.52ms
41: learn: 0.2507302 total: 40ms remaining: 7.61ms
42: learn: 0.2494902 total: 40.9ms remaining: 6.66ms
43: learn: 0.2463801 total: 41.6ms remaining: 5.67ms
44: learn: 0.2447248 total: 42.7ms remaining: 4.75ms
45: learn: 0.2424518 total: 44ms remaining: 3.83ms
46: learn: 0.2411762 total: 44.9ms remaining: 2.87ms
47: learn: 0.2387374 total: 46ms remaining: 1.92ms
48: learn: 0.2362627 total: 46.9ms remaining: 957us
49: learn: 0.2354837 total: 47.7ms remaining: 0us
0: learn: 0.6577557 total: 724us remaining: 35.5ms
1: learn: 0.6115590 total: 1.42ms remaining: 34.1ms
2: learn: 0.5782661 total: 2.48ms remaining: 38.9ms
3: learn: 0.5504365 total: 3.88ms remaining: 44.6ms
4: learn: 0.5239138 total: 4.66ms remaining: 42ms
5: learn: 0.4960141 total: 5.75ms remaining: 42.1ms
6: learn: 0.4751299 total: 6.67ms remaining: 40.9ms
7: learn: 0.4522825 total: 7.62ms remaining: 40ms
8: learn: 0.4350004 total: 8.54ms remaining: 38.9ms
9: learn: 0.4172532 total: 9.66ms remaining: 38.7ms
10: learn: 0.4031272 total: 10.8ms remaining: 38.5ms
11: learn: 0.3894778 total: 11.9ms remaining: 37.7ms
12: learn: 0.3759268 total: 13.2ms remaining: 37.7ms
13: learn: 0.3646396 total: 14.2ms remaining: 36.4ms
14: learn: 0.3550212 total: 15.5ms remaining: 36.3ms
15: learn: 0.3459478 total: 17ms remaining: 36.2ms
16: learn: 0.3387418 total: 18.2ms remaining: 35.3ms
17: learn: 0.3317651 total: 19.7ms remaining: 35ms
18: learn: 0.3245328 total: 21ms remaining: 34.3ms
19: learn: 0.3186379 total: 21.9ms remaining: 32.8ms
20: learn: 0.3123720 total: 22.9ms remaining: 31.6ms
21: learn: 0.3066610 total: 23.7ms remaining: 30.2ms
22: learn: 0.3021609 total: 24.5ms remaining: 28.8ms
23: learn: 0.2981775 total: 25.7ms remaining: 27.9ms
24: learn: 0.2947302 total: 27.5ms remaining: 27.5ms
25: learn: 0.2894158 total: 29.2ms remaining: 26.9ms
26: learn: 0.2849985 total: 30.7ms remaining: 26.1ms
27: learn: 0.2816687 total: 32ms remaining: 25.1ms
28: learn: 0.2784744 total: 33.9ms remaining: 24.5ms
29: learn: 0.2753041 total: 35ms remaining: 23.4ms
30: learn: 0.2704793 total: 37ms remaining: 22.7ms
31: learn: 0.2655361 total: 38.5ms remaining: 21.7ms
32: learn: 0.2598502 total: 39.9ms remaining: 20.6ms
33: learn: 0.2568552 total: 40.9ms remaining: 19.2ms
34: learn: 0.2540340 total: 42.6ms remaining: 18.2ms
35: learn: 0.2503535 total: 44.2ms remaining: 17.2ms
36: learn: 0.2479424 total: 45.4ms remaining: 15.9ms
37: learn: 0.2453326 total: 46.6ms remaining: 14.7ms
38: learn: 0.2425282 total: 48.3ms remaining: 13.6ms
39: learn: 0.2398649 total: 49.6ms remaining: 12.4ms
40: learn: 0.2385556 total: 50.8ms remaining: 11.2ms
41: learn: 0.2365283 total: 52.9ms remaining: 10.1ms
42: learn: 0.2331381 total: 54.3ms remaining: 8.83ms
43: learn: 0.2300152 total: 55.5ms remaining: 7.57ms
44: learn: 0.2279245 total: 57.1ms remaining: 6.34ms
45: learn: 0.2263903 total: 57.9ms remaining: 5.03ms
46: learn: 0.2238257 total: 58.8ms remaining: 3.75ms
47: learn: 0.2216545 total: 59.9ms remaining: 2.5ms
48: learn: 0.2196501 total: 61.2ms remaining: 1.25ms
49: learn: 0.2168023 total: 62.4ms remaining: 0us
0: learn: 0.6583442 total: 660us remaining: 32.4ms
1: learn: 0.6160693 total: 1.17ms remaining: 28ms
2: learn: 0.5795190 total: 1.74ms remaining: 27.2ms
3: learn: 0.5466177 total: 2.68ms remaining: 30.8ms
4: learn: 0.5207080 total: 3.32ms remaining: 29.9ms
5: learn: 0.4950930 total: 4.04ms remaining: 29.6ms
6: learn: 0.4717440 total: 4.98ms remaining: 30.6ms
7: learn: 0.4536730 total: 5.88ms remaining: 30.9ms
8: learn: 0.4349721 total: 6.75ms remaining: 30.7ms
9: learn: 0.4202796 total: 8.06ms remaining: 32.3ms
10: learn: 0.4077376 total: 9.38ms remaining: 33.3ms
11: learn: 0.3960184 total: 10.8ms remaining: 34.1ms
12: learn: 0.3826414 total: 12ms remaining: 34ms
13: learn: 0.3722208 total: 13ms remaining: 33.3ms
14: learn: 0.3627836 total: 14.4ms remaining: 33.7ms
15: learn: 0.3526643 total: 15.4ms remaining: 32.8ms
16: learn: 0.3433313 total: 16.7ms remaining: 32.5ms
17: learn: 0.3337858 total: 18.1ms remaining: 32.1ms
18: learn: 0.3281039 total: 19.7ms remaining: 32.1ms
19: learn: 0.3204933 total: 21.7ms remaining: 32.5ms
20: learn: 0.3172523 total: 22.8ms remaining: 31.4ms
21: learn: 0.3112378 total: 24ms remaining: 30.6ms
22: learn: 0.3055845 total: 24.6ms remaining: 28.8ms
23: learn: 0.3017372 total: 24.9ms remaining: 27ms
24: learn: 0.2956202 total: 25.7ms remaining: 25.7ms
25: learn: 0.2904468 total: 26.8ms remaining: 24.8ms
26: learn: 0.2863467 total: 28.3ms remaining: 24.1ms
27: learn: 0.2831806 total: 29.9ms remaining: 23.5ms
28: learn: 0.2799353 total: 31.5ms remaining: 22.8ms
29: learn: 0.2769338 total: 32.9ms remaining: 22ms
30: learn: 0.2725319 total: 33.6ms remaining: 20.6ms
31: learn: 0.2690325 total: 34ms remaining: 19.1ms
32: learn: 0.2653722 total: 35.4ms remaining: 18.2ms
33: learn: 0.2626298 total: 37.1ms remaining: 17.5ms
34: learn: 0.2613544 total: 37.6ms remaining: 16.1ms
35: learn: 0.2590320 total: 38.3ms remaining: 14.9ms
36: learn: 0.2566611 total: 39.2ms remaining: 13.8ms
37: learn: 0.2535962 total: 39.9ms remaining: 12.6ms
38: learn: 0.2497372 total: 40.7ms remaining: 11.5ms
39: learn: 0.2470181 total: 41ms remaining: 10.2ms
40: learn: 0.2445742 total: 41.2ms remaining: 9.05ms
41: learn: 0.2430848 total: 42.4ms remaining: 8.08ms
42: learn: 0.2422891 total: 43.1ms remaining: 7.01ms
43: learn: 0.2397990 total: 43.5ms remaining: 5.93ms
44: learn: 0.2371706 total: 44.9ms remaining: 4.99ms
45: learn: 0.2352512 total: 46.2ms remaining: 4.01ms
46: learn: 0.2327897 total: 47.7ms remaining: 3.04ms
47: learn: 0.2308399 total: 49.3ms remaining: 2.05ms
48: learn: 0.2296719 total: 50.3ms remaining: 1.03ms
49: learn: 0.2277718 total: 51.6ms remaining: 0us
0: learn: 0.6475054 total: 1.6ms remaining: 78.3ms
1: learn: 0.6071370 total: 2.32ms remaining: 55.8ms
2: learn: 0.5688928 total: 2.97ms remaining: 46.5ms
3: learn: 0.5371729 total: 4.17ms remaining: 48ms
4: learn: 0.5086493 total: 5.99ms remaining: 53.9ms
5: learn: 0.4795564 total: 7.7ms remaining: 56.5ms
6: learn: 0.4579098 total: 8.87ms remaining: 54.5ms
7: learn: 0.4308290 total: 10.1ms remaining: 53ms
8: learn: 0.4093250 total: 11.1ms remaining: 50.5ms
9: learn: 0.3941826 total: 12ms remaining: 48.1ms
10: learn: 0.3800904 total: 13.2ms remaining: 46.9ms
11: learn: 0.3639103 total: 14.1ms remaining: 44.6ms
12: learn: 0.3496290 total: 15.3ms remaining: 43.4ms
13: learn: 0.3407169 total: 16.2ms remaining: 41.7ms
14: learn: 0.3297829 total: 17.4ms remaining: 40.7ms
15: learn: 0.3189842 total: 18.7ms remaining: 39.8ms
16: learn: 0.3114926 total: 19.4ms remaining: 37.7ms
17: learn: 0.3031443 total: 20ms remaining: 35.6ms
18: learn: 0.2955707 total: 20.3ms remaining: 33.1ms
19: learn: 0.2880411 total: 20.9ms remaining: 31.3ms
20: learn: 0.2805200 total: 21.8ms remaining: 30.1ms
21: learn: 0.2726320 total: 22.6ms remaining: 28.8ms
22: learn: 0.2675664 total: 23.2ms remaining: 27.2ms
23: learn: 0.2634753 total: 23.5ms remaining: 25.5ms
24: learn: 0.2596825 total: 23.9ms remaining: 23.9ms
25: learn: 0.2559484 total: 25ms remaining: 23.1ms
26: learn: 0.2515160 total: 25.7ms remaining: 21.9ms
27: learn: 0.2483297 total: 26.3ms remaining: 20.7ms
28: learn: 0.2462752 total: 27.1ms remaining: 19.7ms
29: learn: 0.2429713 total: 28.1ms remaining: 18.7ms
30: learn: 0.2395684 total: 28.6ms remaining: 17.5ms
31: learn: 0.2364600 total: 29.7ms remaining: 16.7ms
32: learn: 0.2339638 total: 30.4ms remaining: 15.7ms
33: learn: 0.2316042 total: 30.6ms remaining: 14.4ms
34: learn: 0.2292298 total: 31ms remaining: 13.3ms
35: learn: 0.2264514 total: 32ms remaining: 12.4ms
36: learn: 0.2223391 total: 33.1ms remaining: 11.6ms
37: learn: 0.2194596 total: 34.5ms remaining: 10.9ms
38: learn: 0.2165141 total: 35.5ms remaining: 10ms
39: learn: 0.2141335 total: 36.3ms remaining: 9.07ms
40: learn: 0.2121124 total: 37.4ms remaining: 8.21ms
41: learn: 0.2102779 total: 38.2ms remaining: 7.28ms
42: learn: 0.2082953 total: 38.6ms remaining: 6.29ms
43: learn: 0.2072916 total: 39.6ms remaining: 5.4ms
44: learn: 0.2049074 total: 40.3ms remaining: 4.48ms
45: learn: 0.2036420 total: 40.7ms remaining: 3.54ms
46: learn: 0.2014209 total: 41.9ms remaining: 2.67ms
47: learn: 0.1999690 total: 42.9ms remaining: 1.79ms
48: learn: 0.1990182 total: 43.6ms remaining: 890us
49: learn: 0.1973890 total: 45.1ms remaining: 0us
0: learn: 0.6426234 total: 378us remaining: 18.5ms
1: learn: 0.5966344 total: 653us remaining: 15.7ms
2: learn: 0.5610290 total: 883us remaining: 13.8ms
3: learn: 0.5301488 total: 1.2ms remaining: 13.8ms
4: learn: 0.5029961 total: 1.48ms remaining: 13.3ms
5: learn: 0.4793889 total: 1.87ms remaining: 13.7ms
6: learn: 0.4547454 total: 2.11ms remaining: 13ms
7: learn: 0.4372305 total: 2.4ms remaining: 12.6ms
8: learn: 0.4185281 total: 2.71ms remaining: 12.4ms
9: learn: 0.4020008 total: 3.17ms remaining: 12.7ms
10: learn: 0.3875013 total: 3.48ms remaining: 12.3ms
11: learn: 0.3742145 total: 3.81ms remaining: 12.1ms
12: learn: 0.3622586 total: 4.33ms remaining: 12.3ms
13: learn: 0.3518018 total: 4.79ms remaining: 12.3ms
14: learn: 0.3435048 total: 5.31ms remaining: 12.4ms
15: learn: 0.3326371 total: 5.66ms remaining: 12ms
16: learn: 0.3257278 total: 6.08ms remaining: 11.8ms
17: learn: 0.3167057 total: 6.53ms remaining: 11.6ms
18: learn: 0.3097649 total: 6.77ms remaining: 11ms
19: learn: 0.3042157 total: 7.23ms remaining: 10.8ms
20: learn: 0.2982697 total: 7.67ms remaining: 10.6ms
21: learn: 0.2921152 total: 8.11ms remaining: 10.3ms
22: learn: 0.2875303 total: 8.47ms remaining: 9.94ms
23: learn: 0.2834689 total: 8.87ms remaining: 9.61ms
24: learn: 0.2785533 total: 9.14ms remaining: 9.14ms
25: learn: 0.2762674 total: 9.55ms remaining: 8.82ms
26: learn: 0.2729498 total: 9.97ms remaining: 8.49ms
27: learn: 0.2685974 total: 10.3ms remaining: 8.12ms
28: learn: 0.2648926 total: 10.7ms remaining: 7.74ms
29: learn: 0.2629856 total: 11ms remaining: 7.33ms
30: learn: 0.2605379 total: 11.4ms remaining: 6.98ms
31: learn: 0.2583238 total: 11.6ms remaining: 6.53ms
32: learn: 0.2549441 total: 12ms remaining: 6.17ms
33: learn: 0.2528828 total: 12.4ms remaining: 5.82ms
34: learn: 0.2500136 total: 12.7ms remaining: 5.45ms
35: learn: 0.2472779 total: 13ms remaining: 5.07ms
36: learn: 0.2443514 total: 13.4ms remaining: 4.71ms
37: learn: 0.2422131 total: 13.7ms remaining: 4.34ms
38: learn: 0.2400465 total: 14.1ms remaining: 3.98ms
39: learn: 0.2378067 total: 14.5ms remaining: 3.62ms
40: learn: 0.2362807 total: 14.8ms remaining: 3.26ms
41: learn: 0.2342833 total: 15.2ms remaining: 2.89ms
42: learn: 0.2325844 total: 15.5ms remaining: 2.52ms
43: learn: 0.2309996 total: 15.9ms remaining: 2.17ms
44: learn: 0.2297499 total: 16.2ms remaining: 1.8ms
45: learn: 0.2277609 total: 16.6ms remaining: 1.45ms
46: learn: 0.2248966 total: 17ms remaining: 1.08ms
47: learn: 0.2233626 total: 17.3ms remaining: 720us
48: learn: 0.2210270 total: 17.6ms remaining: 360us
49: learn: 0.2200753 total: 17.9ms remaining: 0us
0: learn: 0.6457342 total: 1.09ms remaining: 53.5ms
1: learn: 0.6010041 total: 2.28ms remaining: 54.8ms
2: learn: 0.5607770 total: 3.25ms remaining: 50.9ms
3: learn: 0.5254774 total: 4.79ms remaining: 55.1ms
4: learn: 0.4970300 total: 6.01ms remaining: 54.1ms
5: learn: 0.4712240 total: 6.89ms remaining: 50.5ms
6: learn: 0.4472541 total: 8.31ms remaining: 51.1ms
7: learn: 0.4302047 total: 9.69ms remaining: 50.9ms
8: learn: 0.4096284 total: 10.7ms remaining: 48.7ms
9: learn: 0.3943730 total: 11.8ms remaining: 47.1ms
10: learn: 0.3825996 total: 12.7ms remaining: 45.1ms
11: learn: 0.3682381 total: 14.1ms remaining: 44.7ms
12: learn: 0.3542027 total: 15.5ms remaining: 44.1ms
13: learn: 0.3416179 total: 16.9ms remaining: 43.6ms
14: learn: 0.3315787 total: 17.9ms remaining: 41.8ms
15: learn: 0.3215391 total: 19.1ms remaining: 40.7ms
16: learn: 0.3128192 total: 20.5ms remaining: 39.9ms
17: learn: 0.3060670 total: 21.6ms remaining: 38.5ms
18: learn: 0.2996001 total: 22.7ms remaining: 37.1ms
19: learn: 0.2933228 total: 24.1ms remaining: 36.2ms
20: learn: 0.2865127 total: 25.7ms remaining: 35.5ms
21: learn: 0.2823357 total: 27ms remaining: 34.3ms
22: learn: 0.2769019 total: 28.4ms remaining: 33.3ms
23: learn: 0.2713227 total: 29.8ms remaining: 32.2ms
24: learn: 0.2655899 total: 31ms remaining: 31ms
25: learn: 0.2602218 total: 32.2ms remaining: 29.7ms
26: learn: 0.2562777 total: 33.6ms remaining: 28.6ms
27: learn: 0.2537223 total: 34.7ms remaining: 27.2ms
28: learn: 0.2511636 total: 35.2ms remaining: 25.5ms
29: learn: 0.2464586 total: 36.3ms remaining: 24.2ms
30: learn: 0.2430363 total: 37.4ms remaining: 22.9ms
31: learn: 0.2402076 total: 38.5ms remaining: 21.7ms
32: learn: 0.2372463 total: 39.6ms remaining: 20.4ms
33: learn: 0.2343819 total: 40.8ms remaining: 19.2ms
34: learn: 0.2322473 total: 41.9ms remaining: 18ms
35: learn: 0.2294492 total: 43ms remaining: 16.7ms
36: learn: 0.2274605 total: 43.9ms remaining: 15.4ms
37: learn: 0.2247957 total: 44.9ms remaining: 14.2ms
38: learn: 0.2220142 total: 45.8ms remaining: 12.9ms
39: learn: 0.2205209 total: 46.7ms remaining: 11.7ms
40: learn: 0.2186829 total: 47.6ms remaining: 10.4ms
41: learn: 0.2163716 total: 48.4ms remaining: 9.21ms
42: learn: 0.2141476 total: 49ms remaining: 7.98ms
43: learn: 0.2117608 total: 49.7ms remaining: 6.78ms
44: learn: 0.2093530 total: 50.5ms remaining: 5.62ms
45: learn: 0.2075321 total: 51.1ms remaining: 4.45ms
46: learn: 0.2060894 total: 52.1ms remaining: 3.32ms
47: learn: 0.2051089 total: 52.7ms remaining: 2.19ms
48: learn: 0.2037288 total: 53.3ms remaining: 1.09ms
49: learn: 0.2020560 total: 54.2ms remaining: 0us
0: learn: 0.6495311 total: 665us remaining: 32.6ms
1: learn: 0.6040737 total: 1.52ms remaining: 36.5ms
2: learn: 0.5672379 total: 2.18ms remaining: 34.2ms
3: learn: 0.5406197 total: 2.94ms remaining: 33.8ms
4: learn: 0.5151644 total: 3.7ms remaining: 33.3ms
5: learn: 0.4914903 total: 4.28ms remaining: 31.4ms
6: learn: 0.4692605 total: 5.04ms remaining: 31ms
7: learn: 0.4519017 total: 5.5ms remaining: 28.9ms
8: learn: 0.4380025 total: 6.08ms remaining: 27.7ms
9: learn: 0.4235189 total: 6.45ms remaining: 25.8ms
10: learn: 0.4113278 total: 6.99ms remaining: 24.8ms
11: learn: 0.3991034 total: 7.81ms remaining: 24.7ms
12: learn: 0.3858848 total: 8.35ms remaining: 23.8ms
13: learn: 0.3728550 total: 8.89ms remaining: 22.9ms
14: learn: 0.3636348 total: 9.37ms remaining: 21.9ms
15: learn: 0.3534055 total: 10.1ms remaining: 21.4ms
16: learn: 0.3436837 total: 10.7ms remaining: 20.8ms
17: learn: 0.3372644 total: 11.3ms remaining: 20.1ms
18: learn: 0.3313884 total: 11.9ms remaining: 19.4ms
19: learn: 0.3262682 total: 12.8ms remaining: 19.2ms
20: learn: 0.3172674 total: 13.4ms remaining: 18.5ms
21: learn: 0.3110035 total: 14.1ms remaining: 17.9ms
22: learn: 0.3032744 total: 14.7ms remaining: 17.3ms
23: learn: 0.2978424 total: 15.2ms remaining: 16.5ms
24: learn: 0.2923408 total: 15.7ms remaining: 15.7ms
25: learn: 0.2896119 total: 16.2ms remaining: 15ms
26: learn: 0.2859146 total: 16.7ms remaining: 14.3ms
27: learn: 0.2821476 total: 17.3ms remaining: 13.6ms
28: learn: 0.2781989 total: 17.8ms remaining: 12.9ms
29: learn: 0.2739802 total: 18.4ms remaining: 12.2ms
30: learn: 0.2710510 total: 19ms remaining: 11.6ms
31: learn: 0.2675356 total: 19.4ms remaining: 10.9ms
32: learn: 0.2641957 total: 19.9ms remaining: 10.2ms
33: learn: 0.2593319 total: 20.2ms remaining: 9.52ms
34: learn: 0.2557795 total: 20.7ms remaining: 8.88ms
35: learn: 0.2535475 total: 21.3ms remaining: 8.28ms
36: learn: 0.2505609 total: 21.9ms remaining: 7.68ms
37: learn: 0.2480422 total: 22.4ms remaining: 7.07ms
38: learn: 0.2455669 total: 22.9ms remaining: 6.47ms
39: learn: 0.2439872 total: 23.5ms remaining: 5.89ms
40: learn: 0.2422787 total: 23.9ms remaining: 5.25ms
41: learn: 0.2409530 total: 24.4ms remaining: 4.65ms
42: learn: 0.2383699 total: 24.9ms remaining: 4.06ms
43: learn: 0.2357869 total: 25.4ms remaining: 3.47ms
44: learn: 0.2337662 total: 26.1ms remaining: 2.9ms
45: learn: 0.2326745 total: 26.5ms remaining: 2.3ms
46: learn: 0.2309611 total: 27ms remaining: 1.72ms
47: learn: 0.2293255 total: 27.5ms remaining: 1.14ms
48: learn: 0.2276355 total: 27.9ms remaining: 569us
49: learn: 0.2256338 total: 28.3ms remaining: 0us
Evaluating param combo 6/16: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.1, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 16}
Duplicate found for this combo (hash=62dee38dd8a5646163858f2fe7ae5a00). Removing old run: 739adf0d65fc40beab24427215476e9e
0: learn: 0.6462693 total: 534us remaining: 26.2ms
1: learn: 0.6006126 total: 1.11ms remaining: 26.8ms
2: learn: 0.5631732 total: 2.17ms remaining: 34.1ms
3: learn: 0.5318762 total: 2.97ms remaining: 34.2ms
4: learn: 0.5054558 total: 3.47ms remaining: 31.2ms
5: learn: 0.4799320 total: 4.37ms remaining: 32ms
6: learn: 0.4585994 total: 5.02ms remaining: 30.9ms
7: learn: 0.4417764 total: 5.78ms remaining: 30.4ms
8: learn: 0.4225440 total: 6.55ms remaining: 29.9ms
9: learn: 0.4084612 total: 7.35ms remaining: 29.4ms
10: learn: 0.3962466 total: 7.95ms remaining: 28.2ms
11: learn: 0.3845676 total: 8.95ms remaining: 28.3ms
12: learn: 0.3717729 total: 9.47ms remaining: 27ms
13: learn: 0.3609883 total: 10.3ms remaining: 26.4ms
14: learn: 0.3500067 total: 10.6ms remaining: 24.7ms
15: learn: 0.3412218 total: 11.3ms remaining: 24ms
16: learn: 0.3315580 total: 11.8ms remaining: 22.9ms
17: learn: 0.3252685 total: 12.4ms remaining: 22ms
18: learn: 0.3195212 total: 13.3ms remaining: 21.6ms
19: learn: 0.3143782 total: 14ms remaining: 21ms
20: learn: 0.3089230 total: 14.6ms remaining: 20.1ms
21: learn: 0.3037625 total: 15.3ms remaining: 19.4ms
22: learn: 0.2990881 total: 15.9ms remaining: 18.7ms
23: learn: 0.2939303 total: 16.6ms remaining: 18ms
24: learn: 0.2891699 total: 17.1ms remaining: 17.1ms
25: learn: 0.2840286 total: 17.7ms remaining: 16.3ms
26: learn: 0.2792264 total: 18.4ms remaining: 15.7ms
27: learn: 0.2746170 total: 19.1ms remaining: 15ms
28: learn: 0.2711557 total: 19.6ms remaining: 14.2ms
29: learn: 0.2693929 total: 20.1ms remaining: 13.4ms
30: learn: 0.2660210 total: 20.7ms remaining: 12.7ms
31: learn: 0.2638487 total: 21ms remaining: 11.8ms
32: learn: 0.2615085 total: 21.8ms remaining: 11.2ms
33: learn: 0.2594956 total: 22.3ms remaining: 10.5ms
34: learn: 0.2566762 total: 22.9ms remaining: 9.8ms
35: learn: 0.2542834 total: 23.5ms remaining: 9.13ms
36: learn: 0.2522301 total: 24ms remaining: 8.44ms
37: learn: 0.2495766 total: 24.5ms remaining: 7.72ms
38: learn: 0.2473101 total: 25ms remaining: 7.06ms
39: learn: 0.2450895 total: 25.7ms remaining: 6.42ms
40: learn: 0.2426112 total: 26.2ms remaining: 5.76ms
41: learn: 0.2396711 total: 26.7ms remaining: 5.09ms
42: learn: 0.2378629 total: 27.3ms remaining: 4.45ms
43: learn: 0.2352862 total: 28.1ms remaining: 3.83ms
44: learn: 0.2334733 total: 28.8ms remaining: 3.19ms
45: learn: 0.2314837 total: 29.2ms remaining: 2.54ms
46: learn: 0.2290257 total: 29.7ms remaining: 1.9ms
47: learn: 0.2272964 total: 30.3ms remaining: 1.26ms
48: learn: 0.2253947 total: 30.8ms remaining: 628us
49: learn: 0.2237654 total: 31.4ms remaining: 0us
0: learn: 0.6437160 total: 344us remaining: 16.9ms
1: learn: 0.5949760 total: 667us remaining: 16ms
2: learn: 0.5565073 total: 1.03ms remaining: 16.2ms
3: learn: 0.5265837 total: 1.54ms remaining: 17.7ms
4: learn: 0.5005287 total: 2.01ms remaining: 18.1ms
5: learn: 0.4720428 total: 2.56ms remaining: 18.8ms
6: learn: 0.4484482 total: 3.11ms remaining: 19.1ms
7: learn: 0.4293736 total: 3.81ms remaining: 20ms
8: learn: 0.4148507 total: 4.35ms remaining: 19.8ms
9: learn: 0.3977165 total: 5.07ms remaining: 20.3ms
10: learn: 0.3825212 total: 5.63ms remaining: 20ms
11: learn: 0.3667856 total: 6.01ms remaining: 19ms
12: learn: 0.3547366 total: 6.61ms remaining: 18.8ms
13: learn: 0.3424929 total: 7.17ms remaining: 18.4ms
14: learn: 0.3349308 total: 7.91ms remaining: 18.5ms
15: learn: 0.3224127 total: 8.49ms remaining: 18ms
16: learn: 0.3178131 total: 8.94ms remaining: 17.4ms
17: learn: 0.3131939 total: 9.34ms remaining: 16.6ms
18: learn: 0.3045147 total: 9.88ms remaining: 16.1ms
19: learn: 0.2970139 total: 10.4ms remaining: 15.6ms
20: learn: 0.2918113 total: 11.1ms remaining: 15.3ms
21: learn: 0.2854668 total: 11.7ms remaining: 14.9ms
22: learn: 0.2779652 total: 12.2ms remaining: 14.3ms
23: learn: 0.2734233 total: 12.7ms remaining: 13.8ms
24: learn: 0.2685745 total: 13.4ms remaining: 13.4ms
25: learn: 0.2629943 total: 14ms remaining: 12.9ms
26: learn: 0.2604321 total: 14.6ms remaining: 12.5ms
27: learn: 0.2566107 total: 15.2ms remaining: 11.9ms
28: learn: 0.2531121 total: 15.8ms remaining: 11.4ms
29: learn: 0.2515028 total: 16.2ms remaining: 10.8ms
30: learn: 0.2482660 total: 16.8ms remaining: 10.3ms
31: learn: 0.2458159 total: 17.6ms remaining: 9.91ms
32: learn: 0.2416483 total: 18.3ms remaining: 9.44ms
33: learn: 0.2386672 total: 18.9ms remaining: 8.92ms
34: learn: 0.2359889 total: 19.5ms remaining: 8.36ms
35: learn: 0.2338575 total: 20.1ms remaining: 7.8ms
36: learn: 0.2298220 total: 20.8ms remaining: 7.32ms
37: learn: 0.2283967 total: 21.3ms remaining: 6.74ms
38: learn: 0.2263530 total: 21.8ms remaining: 6.15ms
39: learn: 0.2246581 total: 22.3ms remaining: 5.58ms
40: learn: 0.2223378 total: 23ms remaining: 5.04ms
41: learn: 0.2207038 total: 23.4ms remaining: 4.46ms
42: learn: 0.2197856 total: 23.8ms remaining: 3.87ms
43: learn: 0.2188898 total: 24.2ms remaining: 3.29ms
44: learn: 0.2176194 total: 24.5ms remaining: 2.72ms
45: learn: 0.2163312 total: 25ms remaining: 2.17ms
46: learn: 0.2154455 total: 25.5ms remaining: 1.63ms
47: learn: 0.2139407 total: 26ms remaining: 1.08ms
48: learn: 0.2120411 total: 26.5ms remaining: 541us
49: learn: 0.2106386 total: 26.9ms remaining: 0us
0: learn: 0.6454260 total: 349us remaining: 17.1ms
1: learn: 0.6019562 total: 616us remaining: 14.8ms
2: learn: 0.5626070 total: 843us remaining: 13.2ms
3: learn: 0.5307213 total: 1.13ms remaining: 13ms
4: learn: 0.5032371 total: 1.36ms remaining: 12.2ms
5: learn: 0.4776693 total: 1.6ms remaining: 11.8ms
6: learn: 0.4566797 total: 1.82ms remaining: 11.2ms
7: learn: 0.4389420 total: 2.08ms remaining: 10.9ms
8: learn: 0.4181396 total: 2.27ms remaining: 10.4ms
9: learn: 0.4027384 total: 2.51ms remaining: 10ms
10: learn: 0.3900032 total: 2.7ms remaining: 9.58ms
11: learn: 0.3777932 total: 2.98ms remaining: 9.43ms
12: learn: 0.3636268 total: 3.23ms remaining: 9.19ms
13: learn: 0.3525592 total: 3.53ms remaining: 9.07ms
14: learn: 0.3415734 total: 3.78ms remaining: 8.81ms
15: learn: 0.3322054 total: 4.11ms remaining: 8.74ms
16: learn: 0.3233730 total: 4.37ms remaining: 8.49ms
17: learn: 0.3167153 total: 4.59ms remaining: 8.17ms
18: learn: 0.3106400 total: 4.84ms remaining: 7.89ms
19: learn: 0.3023905 total: 5.1ms remaining: 7.64ms
20: learn: 0.2963086 total: 5.44ms remaining: 7.51ms
21: learn: 0.2904163 total: 5.7ms remaining: 7.26ms
22: learn: 0.2852205 total: 6.04ms remaining: 7.09ms
23: learn: 0.2798857 total: 6.33ms remaining: 6.85ms
24: learn: 0.2744258 total: 6.54ms remaining: 6.54ms
25: learn: 0.2718590 total: 6.83ms remaining: 6.31ms
26: learn: 0.2677934 total: 7.13ms remaining: 6.07ms
27: learn: 0.2637715 total: 7.45ms remaining: 5.85ms
28: learn: 0.2593053 total: 7.8ms remaining: 5.65ms
29: learn: 0.2574366 total: 8.04ms remaining: 5.36ms
30: learn: 0.2539525 total: 8.35ms remaining: 5.12ms
31: learn: 0.2512715 total: 8.5ms remaining: 4.78ms
32: learn: 0.2481380 total: 8.89ms remaining: 4.58ms
33: learn: 0.2458802 total: 9.23ms remaining: 4.34ms
34: learn: 0.2430230 total: 9.56ms remaining: 4.1ms
35: learn: 0.2402213 total: 9.85ms remaining: 3.83ms
36: learn: 0.2382592 total: 10.2ms remaining: 3.59ms
37: learn: 0.2366223 total: 10.5ms remaining: 3.33ms
38: learn: 0.2346630 total: 10.8ms remaining: 3.05ms
39: learn: 0.2316478 total: 11.2ms remaining: 2.79ms
40: learn: 0.2295176 total: 11.5ms remaining: 2.52ms
41: learn: 0.2275613 total: 11.7ms remaining: 2.23ms
42: learn: 0.2261304 total: 12ms remaining: 1.95ms
43: learn: 0.2230182 total: 12.3ms remaining: 1.67ms
44: learn: 0.2213852 total: 12.6ms remaining: 1.4ms
45: learn: 0.2198024 total: 13ms remaining: 1.13ms
46: learn: 0.2183045 total: 13.3ms remaining: 846us
47: learn: 0.2166827 total: 13.6ms remaining: 566us
48: learn: 0.2136031 total: 14ms remaining: 285us
49: learn: 0.2121566 total: 14.3ms remaining: 0us
0: learn: 0.6470966 total: 505us remaining: 24.8ms
1: learn: 0.6001838 total: 1.14ms remaining: 27.4ms
2: learn: 0.5604623 total: 1.55ms remaining: 24.4ms
3: learn: 0.5261755 total: 2.06ms remaining: 23.7ms
4: learn: 0.4992383 total: 2.4ms remaining: 21.6ms
5: learn: 0.4721898 total: 2.78ms remaining: 20.4ms
6: learn: 0.4498006 total: 3.33ms remaining: 20.5ms
7: learn: 0.4314508 total: 3.9ms remaining: 20.5ms
8: learn: 0.4101723 total: 4.32ms remaining: 19.7ms
9: learn: 0.3935443 total: 4.77ms remaining: 19.1ms
10: learn: 0.3798147 total: 5.22ms remaining: 18.5ms
11: learn: 0.3665670 total: 5.76ms remaining: 18.3ms
12: learn: 0.3536267 total: 6.29ms remaining: 17.9ms
13: learn: 0.3420766 total: 6.75ms remaining: 17.3ms
14: learn: 0.3326825 total: 7.12ms remaining: 16.6ms
15: learn: 0.3240086 total: 7.49ms remaining: 15.9ms
16: learn: 0.3134403 total: 7.99ms remaining: 15.5ms
17: learn: 0.3067898 total: 8.36ms remaining: 14.9ms
18: learn: 0.3003032 total: 8.81ms remaining: 14.4ms
19: learn: 0.2951168 total: 9.26ms remaining: 13.9ms
20: learn: 0.2888849 total: 9.73ms remaining: 13.4ms
21: learn: 0.2831076 total: 10.1ms remaining: 12.8ms
22: learn: 0.2763535 total: 10.6ms remaining: 12.4ms
23: learn: 0.2698591 total: 11.1ms remaining: 12.1ms
24: learn: 0.2647141 total: 11.8ms remaining: 11.8ms
25: learn: 0.2590451 total: 12.2ms remaining: 11.3ms
26: learn: 0.2557403 total: 12.7ms remaining: 10.8ms
27: learn: 0.2526011 total: 13.2ms remaining: 10.4ms
28: learn: 0.2495110 total: 13.5ms remaining: 9.8ms
29: learn: 0.2446900 total: 14ms remaining: 9.33ms
30: learn: 0.2404218 total: 14.5ms remaining: 8.9ms
31: learn: 0.2370704 total: 14.9ms remaining: 8.4ms
32: learn: 0.2333679 total: 15.4ms remaining: 7.92ms
33: learn: 0.2298418 total: 15.9ms remaining: 7.47ms
34: learn: 0.2271491 total: 16.4ms remaining: 7.03ms
35: learn: 0.2245757 total: 16.9ms remaining: 6.58ms
36: learn: 0.2214767 total: 17.3ms remaining: 6.08ms
37: learn: 0.2191640 total: 17.9ms remaining: 5.64ms
38: learn: 0.2178533 total: 18.3ms remaining: 5.17ms
39: learn: 0.2163079 total: 18.7ms remaining: 4.68ms
40: learn: 0.2149998 total: 19.2ms remaining: 4.21ms
41: learn: 0.2135240 total: 19.7ms remaining: 3.75ms
42: learn: 0.2118976 total: 20.1ms remaining: 3.27ms
43: learn: 0.2106812 total: 20.6ms remaining: 2.8ms
44: learn: 0.2087706 total: 21.1ms remaining: 2.34ms
45: learn: 0.2077590 total: 21.4ms remaining: 1.86ms
46: learn: 0.2062417 total: 21.7ms remaining: 1.39ms
47: learn: 0.2041230 total: 22.1ms remaining: 918us
48: learn: 0.2029428 total: 22.4ms remaining: 456us
49: learn: 0.2010316 total: 22.8ms remaining: 0us
0: learn: 0.6466390 total: 318us remaining: 15.6ms
1: learn: 0.6043306 total: 558us remaining: 13.4ms
2: learn: 0.5698679 total: 789us remaining: 12.4ms
3: learn: 0.5400060 total: 1.09ms remaining: 12.5ms
4: learn: 0.5139000 total: 1.34ms remaining: 12.1ms
5: learn: 0.4909190 total: 1.65ms remaining: 12.1ms
6: learn: 0.4663019 total: 1.89ms remaining: 11.6ms
7: learn: 0.4496656 total: 2.18ms remaining: 11.5ms
8: learn: 0.4302446 total: 2.4ms remaining: 10.9ms
9: learn: 0.4155090 total: 2.75ms remaining: 11ms
10: learn: 0.4046576 total: 2.99ms remaining: 10.6ms
11: learn: 0.3902395 total: 3.24ms remaining: 10.3ms
12: learn: 0.3781937 total: 3.57ms remaining: 10.2ms
13: learn: 0.3684001 total: 3.89ms remaining: 9.99ms
14: learn: 0.3594224 total: 4.23ms remaining: 9.88ms
15: learn: 0.3506654 total: 4.42ms remaining: 9.4ms
16: learn: 0.3427953 total: 4.67ms remaining: 9.06ms
17: learn: 0.3375828 total: 4.97ms remaining: 8.85ms
18: learn: 0.3304561 total: 5.3ms remaining: 8.64ms
19: learn: 0.3248426 total: 5.62ms remaining: 8.42ms
20: learn: 0.3187507 total: 6.06ms remaining: 8.37ms
21: learn: 0.3109731 total: 6.38ms remaining: 8.13ms
22: learn: 0.3052623 total: 6.59ms remaining: 7.74ms
23: learn: 0.3002449 total: 6.89ms remaining: 7.47ms
24: learn: 0.2967801 total: 7.22ms remaining: 7.22ms
25: learn: 0.2923089 total: 7.51ms remaining: 6.93ms
26: learn: 0.2868579 total: 7.88ms remaining: 6.71ms
27: learn: 0.2841698 total: 8.17ms remaining: 6.42ms
28: learn: 0.2810342 total: 8.43ms remaining: 6.1ms
29: learn: 0.2780382 total: 8.7ms remaining: 5.8ms
30: learn: 0.2758407 total: 8.91ms remaining: 5.46ms
31: learn: 0.2730709 total: 9.19ms remaining: 5.17ms
32: learn: 0.2704504 total: 9.47ms remaining: 4.88ms
33: learn: 0.2684170 total: 9.73ms remaining: 4.58ms
34: learn: 0.2646933 total: 10ms remaining: 4.29ms
35: learn: 0.2624324 total: 10.3ms remaining: 4.01ms
36: learn: 0.2604280 total: 10.6ms remaining: 3.72ms
37: learn: 0.2579343 total: 10.9ms remaining: 3.44ms
38: learn: 0.2550934 total: 11.2ms remaining: 3.16ms
39: learn: 0.2526229 total: 11.5ms remaining: 2.86ms
40: learn: 0.2500359 total: 11.7ms remaining: 2.57ms
41: learn: 0.2471391 total: 11.9ms remaining: 2.27ms
42: learn: 0.2454036 total: 12.2ms remaining: 1.99ms
43: learn: 0.2433846 total: 12.5ms remaining: 1.7ms
44: learn: 0.2406459 total: 12.8ms remaining: 1.42ms
45: learn: 0.2380629 total: 13.1ms remaining: 1.14ms
46: learn: 0.2358969 total: 13.3ms remaining: 850us
47: learn: 0.2344105 total: 13.6ms remaining: 567us
48: learn: 0.2324208 total: 13.9ms remaining: 283us
49: learn: 0.2305732 total: 14.2ms remaining: 0us
0: learn: 0.6501714 total: 292us remaining: 14.3ms
1: learn: 0.6066480 total: 553us remaining: 13.3ms
2: learn: 0.5657020 total: 800us remaining: 12.5ms
3: learn: 0.5314888 total: 1.07ms remaining: 12.3ms
4: learn: 0.5033352 total: 1.39ms remaining: 12.5ms
5: learn: 0.4746589 total: 1.89ms remaining: 13.8ms
6: learn: 0.4545285 total: 2.3ms remaining: 14.1ms
7: learn: 0.4381047 total: 2.69ms remaining: 14.1ms
8: learn: 0.4177237 total: 3.11ms remaining: 14.2ms
9: learn: 0.4023522 total: 3.49ms remaining: 14ms
10: learn: 0.3914068 total: 3.79ms remaining: 13.4ms
11: learn: 0.3787263 total: 4.4ms remaining: 13.9ms
12: learn: 0.3650055 total: 4.87ms remaining: 13.9ms
13: learn: 0.3522909 total: 5.32ms remaining: 13.7ms
14: learn: 0.3408388 total: 5.71ms remaining: 13.3ms
15: learn: 0.3301578 total: 6.23ms remaining: 13.2ms
16: learn: 0.3207327 total: 6.74ms remaining: 13.1ms
17: learn: 0.3122013 total: 7.21ms remaining: 12.8ms
18: learn: 0.3065739 total: 7.99ms remaining: 13ms
19: learn: 0.2986789 total: 8.48ms remaining: 12.7ms
20: learn: 0.2920784 total: 8.91ms remaining: 12.3ms
21: learn: 0.2861408 total: 9.37ms remaining: 11.9ms
22: learn: 0.2806232 total: 9.72ms remaining: 11.4ms
23: learn: 0.2757454 total: 10.1ms remaining: 11ms
24: learn: 0.2715914 total: 10.5ms remaining: 10.5ms
25: learn: 0.2662000 total: 10.8ms remaining: 9.98ms
26: learn: 0.2620222 total: 11.2ms remaining: 9.54ms
27: learn: 0.2586619 total: 11.5ms remaining: 9.05ms
28: learn: 0.2559411 total: 12ms remaining: 8.68ms
29: learn: 0.2521874 total: 13.3ms remaining: 8.86ms
30: learn: 0.2487332 total: 14ms remaining: 8.57ms
31: learn: 0.2455035 total: 14.4ms remaining: 8.12ms
32: learn: 0.2428433 total: 14.9ms remaining: 7.68ms
33: learn: 0.2410762 total: 15.2ms remaining: 7.14ms
34: learn: 0.2382084 total: 15.6ms remaining: 6.67ms
35: learn: 0.2366788 total: 16ms remaining: 6.21ms
36: learn: 0.2347484 total: 16.4ms remaining: 5.78ms
37: learn: 0.2324811 total: 16.8ms remaining: 5.32ms
38: learn: 0.2302696 total: 17.2ms remaining: 4.86ms
39: learn: 0.2282705 total: 17.7ms remaining: 4.43ms
40: learn: 0.2261110 total: 18.1ms remaining: 3.98ms
41: learn: 0.2238719 total: 18.4ms remaining: 3.51ms
42: learn: 0.2215777 total: 18.8ms remaining: 3.07ms
43: learn: 0.2196766 total: 19.4ms remaining: 2.64ms
44: learn: 0.2185631 total: 19.6ms remaining: 2.18ms
45: learn: 0.2166483 total: 20ms remaining: 1.74ms
46: learn: 0.2139387 total: 20.5ms remaining: 1.3ms
47: learn: 0.2129512 total: 20.7ms remaining: 863us
48: learn: 0.2119949 total: 21ms remaining: 427us
49: learn: 0.2109265 total: 21.2ms remaining: 0us
0: learn: 0.6481110 total: 291us remaining: 14.3ms
1: learn: 0.6015437 total: 534us remaining: 12.8ms
2: learn: 0.5637141 total: 711us remaining: 11.1ms
3: learn: 0.5285975 total: 988us remaining: 11.4ms
4: learn: 0.5021036 total: 1.23ms remaining: 11.1ms
5: learn: 0.4762132 total: 1.51ms remaining: 11.1ms
6: learn: 0.4571360 total: 1.78ms remaining: 11ms
7: learn: 0.4392327 total: 2.1ms remaining: 11ms
8: learn: 0.4235303 total: 2.26ms remaining: 10.3ms
9: learn: 0.4067796 total: 2.58ms remaining: 10.3ms
10: learn: 0.3956489 total: 2.77ms remaining: 9.81ms
11: learn: 0.3834309 total: 3.05ms remaining: 9.67ms
12: learn: 0.3706993 total: 3.34ms remaining: 9.52ms
13: learn: 0.3593553 total: 3.64ms remaining: 9.37ms
14: learn: 0.3495868 total: 4.02ms remaining: 9.38ms
15: learn: 0.3380214 total: 4.34ms remaining: 9.23ms
16: learn: 0.3303325 total: 4.71ms remaining: 9.15ms
17: learn: 0.3239962 total: 4.96ms remaining: 8.82ms
18: learn: 0.3173096 total: 5.25ms remaining: 8.56ms
19: learn: 0.3130535 total: 5.56ms remaining: 8.34ms
20: learn: 0.3053856 total: 5.93ms remaining: 8.19ms
21: learn: 0.2967920 total: 6.23ms remaining: 7.93ms
22: learn: 0.2911773 total: 6.57ms remaining: 7.71ms
23: learn: 0.2846182 total: 6.8ms remaining: 7.37ms
24: learn: 0.2808455 total: 7.06ms remaining: 7.06ms
25: learn: 0.2780712 total: 7.37ms remaining: 6.8ms
26: learn: 0.2744681 total: 7.68ms remaining: 6.54ms
27: learn: 0.2696076 total: 7.96ms remaining: 6.25ms
28: learn: 0.2644929 total: 8.3ms remaining: 6.01ms
29: learn: 0.2616771 total: 8.54ms remaining: 5.69ms
30: learn: 0.2582177 total: 8.88ms remaining: 5.44ms
31: learn: 0.2542861 total: 9.22ms remaining: 5.18ms
32: learn: 0.2512674 total: 9.51ms remaining: 4.9ms
33: learn: 0.2476332 total: 9.84ms remaining: 4.63ms
34: learn: 0.2439946 total: 10.1ms remaining: 4.35ms
35: learn: 0.2408321 total: 10.4ms remaining: 4.05ms
36: learn: 0.2375090 total: 10.8ms remaining: 3.78ms
37: learn: 0.2354509 total: 11ms remaining: 3.48ms
38: learn: 0.2341470 total: 11.4ms remaining: 3.21ms
39: learn: 0.2321929 total: 11.7ms remaining: 2.91ms
40: learn: 0.2300608 total: 12ms remaining: 2.64ms
41: learn: 0.2276641 total: 12.4ms remaining: 2.36ms
42: learn: 0.2262292 total: 12.7ms remaining: 2.06ms
43: learn: 0.2238001 total: 12.9ms remaining: 1.76ms
44: learn: 0.2225238 total: 13.2ms remaining: 1.46ms
45: learn: 0.2207839 total: 13.6ms remaining: 1.18ms
46: learn: 0.2192435 total: 13.9ms remaining: 887us
47: learn: 0.2170573 total: 14.2ms remaining: 592us
48: learn: 0.2151633 total: 14.5ms remaining: 295us
49: learn: 0.2135362 total: 14.8ms remaining: 0us
0: learn: 0.6515506 total: 383us remaining: 18.8ms
1: learn: 0.6073513 total: 641us remaining: 15.4ms
2: learn: 0.5704869 total: 827us remaining: 13ms
3: learn: 0.5398446 total: 1.13ms remaining: 13ms
4: learn: 0.5111154 total: 1.35ms remaining: 12.1ms
5: learn: 0.4857031 total: 1.6ms remaining: 11.7ms
6: learn: 0.4649189 total: 1.84ms remaining: 11.3ms
7: learn: 0.4473799 total: 2.09ms remaining: 11ms
8: learn: 0.4283679 total: 2.28ms remaining: 10.4ms
9: learn: 0.4129013 total: 2.52ms remaining: 10.1ms
10: learn: 0.3981913 total: 2.74ms remaining: 9.71ms
11: learn: 0.3860850 total: 3.04ms remaining: 9.62ms
12: learn: 0.3747197 total: 3.3ms remaining: 9.38ms
13: learn: 0.3643286 total: 3.61ms remaining: 9.29ms
14: learn: 0.3560632 total: 3.78ms remaining: 8.82ms
15: learn: 0.3493369 total: 4.06ms remaining: 8.62ms
16: learn: 0.3405992 total: 4.35ms remaining: 8.45ms
17: learn: 0.3364019 total: 4.56ms remaining: 8.11ms
18: learn: 0.3284740 total: 4.82ms remaining: 7.86ms
19: learn: 0.3207455 total: 5.12ms remaining: 7.69ms
20: learn: 0.3136963 total: 5.42ms remaining: 7.48ms
21: learn: 0.3068343 total: 5.71ms remaining: 7.26ms
22: learn: 0.3015201 total: 5.9ms remaining: 6.92ms
23: learn: 0.2973018 total: 6.12ms remaining: 6.63ms
24: learn: 0.2925741 total: 6.4ms remaining: 6.4ms
25: learn: 0.2863699 total: 6.68ms remaining: 6.16ms
26: learn: 0.2828654 total: 6.97ms remaining: 5.94ms
27: learn: 0.2796819 total: 7.22ms remaining: 5.67ms
28: learn: 0.2773885 total: 7.45ms remaining: 5.39ms
29: learn: 0.2764725 total: 7.71ms remaining: 5.14ms
30: learn: 0.2727544 total: 7.95ms remaining: 4.88ms
31: learn: 0.2691427 total: 8.25ms remaining: 4.64ms
32: learn: 0.2657733 total: 8.55ms remaining: 4.4ms
33: learn: 0.2627910 total: 8.82ms remaining: 4.15ms
34: learn: 0.2593242 total: 9.1ms remaining: 3.9ms
35: learn: 0.2568494 total: 9.38ms remaining: 3.65ms
36: learn: 0.2522878 total: 9.66ms remaining: 3.39ms
37: learn: 0.2495150 total: 9.92ms remaining: 3.13ms
38: learn: 0.2464842 total: 10.2ms remaining: 2.88ms
39: learn: 0.2445408 total: 10.5ms remaining: 2.63ms
40: learn: 0.2418177 total: 10.8ms remaining: 2.38ms
41: learn: 0.2396222 total: 11.1ms remaining: 2.12ms
42: learn: 0.2366848 total: 11.4ms remaining: 1.86ms
43: learn: 0.2330100 total: 11.7ms remaining: 1.6ms
44: learn: 0.2310290 total: 12ms remaining: 1.33ms
45: learn: 0.2291962 total: 12.3ms remaining: 1.07ms
46: learn: 0.2279771 total: 12.7ms remaining: 808us
47: learn: 0.2257751 total: 13ms remaining: 540us
48: learn: 0.2228804 total: 13.3ms remaining: 271us
49: learn: 0.2201211 total: 13.6ms remaining: 0us
0: learn: 0.6455145 total: 347us remaining: 17ms
1: learn: 0.5988760 total: 589us remaining: 14.1ms
2: learn: 0.5594107 total: 761us remaining: 11.9ms
3: learn: 0.5244881 total: 1ms remaining: 11.5ms
4: learn: 0.4973720 total: 1.19ms remaining: 10.7ms
5: learn: 0.4741560 total: 1.39ms remaining: 10.2ms
6: learn: 0.4507672 total: 1.57ms remaining: 9.64ms
7: learn: 0.4334544 total: 1.75ms remaining: 9.22ms
8: learn: 0.4127818 total: 1.91ms remaining: 8.69ms
9: learn: 0.3964144 total: 2.12ms remaining: 8.49ms
10: learn: 0.3815367 total: 2.33ms remaining: 8.26ms
11: learn: 0.3706824 total: 2.55ms remaining: 8.09ms
12: learn: 0.3603382 total: 2.88ms remaining: 8.19ms
13: learn: 0.3494307 total: 3.25ms remaining: 8.36ms
14: learn: 0.3417150 total: 3.56ms remaining: 8.31ms
15: learn: 0.3334102 total: 3.91ms remaining: 8.31ms
16: learn: 0.3233884 total: 4.23ms remaining: 8.2ms
17: learn: 0.3176727 total: 4.49ms remaining: 7.98ms
18: learn: 0.3103917 total: 4.9ms remaining: 7.99ms
19: learn: 0.3034642 total: 5.24ms remaining: 7.87ms
20: learn: 0.2980677 total: 5.64ms remaining: 7.79ms
21: learn: 0.2918482 total: 5.98ms remaining: 7.61ms
22: learn: 0.2854194 total: 6.16ms remaining: 7.23ms
23: learn: 0.2799811 total: 6.39ms remaining: 6.92ms
24: learn: 0.2749249 total: 6.62ms remaining: 6.62ms
25: learn: 0.2704717 total: 6.84ms remaining: 6.31ms
26: learn: 0.2662482 total: 7.13ms remaining: 6.07ms
27: learn: 0.2633140 total: 7.43ms remaining: 5.84ms
28: learn: 0.2597887 total: 7.79ms remaining: 5.64ms
29: learn: 0.2568451 total: 8.07ms remaining: 5.38ms
30: learn: 0.2544384 total: 8.23ms remaining: 5.04ms
31: learn: 0.2516443 total: 8.5ms remaining: 4.78ms
32: learn: 0.2467225 total: 8.7ms remaining: 4.48ms
33: learn: 0.2444546 total: 9.05ms remaining: 4.26ms
34: learn: 0.2425635 total: 9.44ms remaining: 4.04ms
35: learn: 0.2394869 total: 9.65ms remaining: 3.75ms
36: learn: 0.2373269 total: 10.1ms remaining: 3.53ms
37: learn: 0.2343314 total: 10.5ms remaining: 3.3ms
38: learn: 0.2317488 total: 10.8ms remaining: 3.05ms
39: learn: 0.2306135 total: 11.1ms remaining: 2.78ms
40: learn: 0.2289222 total: 11.4ms remaining: 2.5ms
41: learn: 0.2267190 total: 11.8ms remaining: 2.24ms
42: learn: 0.2254531 total: 12ms remaining: 1.96ms
43: learn: 0.2235827 total: 12.3ms remaining: 1.67ms
44: learn: 0.2210251 total: 12.5ms remaining: 1.39ms
45: learn: 0.2194663 total: 12.8ms remaining: 1.11ms
46: learn: 0.2180275 total: 13.1ms remaining: 837us
47: learn: 0.2169634 total: 13.3ms remaining: 555us
48: learn: 0.2141994 total: 13.5ms remaining: 276us
49: learn: 0.2126256 total: 13.9ms remaining: 0us
0: learn: 0.6503257 total: 367us remaining: 18ms
1: learn: 0.6091405 total: 840us remaining: 20.2ms
2: learn: 0.5749262 total: 1.08ms remaining: 16.9ms
3: learn: 0.5438978 total: 1.32ms remaining: 15.2ms
4: learn: 0.5155700 total: 1.54ms remaining: 13.9ms
5: learn: 0.4927320 total: 1.82ms remaining: 13.4ms
6: learn: 0.4727667 total: 2.02ms remaining: 12.4ms
7: learn: 0.4557302 total: 2.46ms remaining: 12.9ms
8: learn: 0.4388609 total: 2.77ms remaining: 12.6ms
9: learn: 0.4247644 total: 2.98ms remaining: 11.9ms
10: learn: 0.4144140 total: 3.27ms remaining: 11.6ms
11: learn: 0.4019024 total: 3.6ms remaining: 11.4ms
12: learn: 0.3916899 total: 3.86ms remaining: 11ms
13: learn: 0.3815831 total: 4.2ms remaining: 10.8ms
14: learn: 0.3717996 total: 4.52ms remaining: 10.5ms
15: learn: 0.3647230 total: 4.8ms remaining: 10.2ms
16: learn: 0.3545912 total: 5.18ms remaining: 10.1ms
17: learn: 0.3502944 total: 5.42ms remaining: 9.64ms
18: learn: 0.3428250 total: 5.73ms remaining: 9.35ms
19: learn: 0.3374503 total: 6.01ms remaining: 9.02ms
20: learn: 0.3317534 total: 6.33ms remaining: 8.74ms
21: learn: 0.3248470 total: 6.57ms remaining: 8.36ms
22: learn: 0.3197077 total: 6.79ms remaining: 7.97ms
23: learn: 0.3149752 total: 7.1ms remaining: 7.69ms
24: learn: 0.3115804 total: 7.37ms remaining: 7.37ms
25: learn: 0.3073465 total: 7.66ms remaining: 7.07ms
26: learn: 0.3014933 total: 7.94ms remaining: 6.76ms
27: learn: 0.2982788 total: 8.23ms remaining: 6.47ms
28: learn: 0.2954439 total: 8.53ms remaining: 6.18ms
29: learn: 0.2917875 total: 8.82ms remaining: 5.88ms
30: learn: 0.2900107 total: 8.98ms remaining: 5.5ms
31: learn: 0.2864544 total: 9.25ms remaining: 5.2ms
32: learn: 0.2810382 total: 9.52ms remaining: 4.9ms
33: learn: 0.2787524 total: 9.91ms remaining: 4.66ms
34: learn: 0.2764095 total: 10.3ms remaining: 4.42ms
35: learn: 0.2748109 total: 10.6ms remaining: 4.12ms
36: learn: 0.2728280 total: 10.9ms remaining: 3.82ms
37: learn: 0.2703365 total: 11.2ms remaining: 3.52ms
38: learn: 0.2683979 total: 11.4ms remaining: 3.22ms
39: learn: 0.2667767 total: 11.6ms remaining: 2.9ms
40: learn: 0.2652805 total: 11.9ms remaining: 2.62ms
41: learn: 0.2621843 total: 12.2ms remaining: 2.33ms
42: learn: 0.2596001 total: 12.4ms remaining: 2.02ms
43: learn: 0.2571810 total: 12.7ms remaining: 1.73ms
44: learn: 0.2554186 total: 12.9ms remaining: 1.44ms
45: learn: 0.2533093 total: 13.2ms remaining: 1.14ms
46: learn: 0.2511507 total: 13.4ms remaining: 854us
47: learn: 0.2495800 total: 13.6ms remaining: 566us
48: learn: 0.2474001 total: 13.9ms remaining: 283us
49: learn: 0.2461070 total: 14.1ms remaining: 0us
0: learn: 0.6515750 total: 416us remaining: 20.4ms
1: learn: 0.6134885 total: 985us remaining: 23.7ms
2: learn: 0.5837400 total: 1.44ms remaining: 22.5ms
3: learn: 0.5489154 total: 2.03ms remaining: 23.3ms
4: learn: 0.5169299 total: 2.75ms remaining: 24.8ms
5: learn: 0.4907425 total: 3.23ms remaining: 23.7ms
6: learn: 0.4713171 total: 3.54ms remaining: 21.7ms
7: learn: 0.4534210 total: 3.9ms remaining: 20.5ms
8: learn: 0.4367045 total: 4.41ms remaining: 20.1ms
9: learn: 0.4205350 total: 4.8ms remaining: 19.2ms
10: learn: 0.4056829 total: 5.2ms remaining: 18.4ms
11: learn: 0.3903177 total: 5.67ms remaining: 18ms
12: learn: 0.3766978 total: 6.1ms remaining: 17.4ms
13: learn: 0.3669745 total: 6.63ms remaining: 17.1ms
14: learn: 0.3548570 total: 7.07ms remaining: 16.5ms
15: learn: 0.3456593 total: 7.64ms remaining: 16.2ms
16: learn: 0.3378650 total: 8.21ms remaining: 15.9ms
17: learn: 0.3310731 total: 8.64ms remaining: 15.4ms
18: learn: 0.3233461 total: 8.94ms remaining: 14.6ms
19: learn: 0.3165295 total: 9.19ms remaining: 13.8ms
20: learn: 0.3094843 total: 9.62ms remaining: 13.3ms
21: learn: 0.3020178 total: 9.9ms remaining: 12.6ms
22: learn: 0.2965073 total: 10.3ms remaining: 12.1ms
23: learn: 0.2896890 total: 10.7ms remaining: 11.6ms
24: learn: 0.2844204 total: 11.3ms remaining: 11.3ms
25: learn: 0.2797083 total: 11.9ms remaining: 10.9ms
26: learn: 0.2751533 total: 12.2ms remaining: 10.4ms
27: learn: 0.2715553 total: 12.8ms remaining: 10ms
28: learn: 0.2691510 total: 13.1ms remaining: 9.45ms
29: learn: 0.2671634 total: 13.5ms remaining: 8.98ms
30: learn: 0.2650620 total: 13.7ms remaining: 8.41ms
31: learn: 0.2613732 total: 14.1ms remaining: 7.92ms
32: learn: 0.2592097 total: 14.4ms remaining: 7.4ms
33: learn: 0.2555304 total: 14.7ms remaining: 6.92ms
34: learn: 0.2532133 total: 15ms remaining: 6.41ms
35: learn: 0.2505760 total: 15.3ms remaining: 5.95ms
36: learn: 0.2488293 total: 15.7ms remaining: 5.52ms
37: learn: 0.2453197 total: 15.9ms remaining: 5.02ms
38: learn: 0.2419295 total: 16.3ms remaining: 4.61ms
39: learn: 0.2391228 total: 16.8ms remaining: 4.19ms
40: learn: 0.2363322 total: 17.2ms remaining: 3.77ms
41: learn: 0.2343441 total: 17.6ms remaining: 3.36ms
42: learn: 0.2325132 total: 18ms remaining: 2.94ms
43: learn: 0.2296161 total: 18.4ms remaining: 2.51ms
44: learn: 0.2287773 total: 18.8ms remaining: 2.08ms
45: learn: 0.2272871 total: 19ms remaining: 1.66ms
46: learn: 0.2260889 total: 19.3ms remaining: 1.23ms
47: learn: 0.2244358 total: 19.7ms remaining: 820us
48: learn: 0.2231578 total: 20ms remaining: 408us
49: learn: 0.2215341 total: 20.3ms remaining: 0us
0: learn: 0.6495190 total: 342us remaining: 16.8ms
1: learn: 0.6095526 total: 559us remaining: 13.4ms
2: learn: 0.5719992 total: 891us remaining: 14ms
3: learn: 0.5425656 total: 1.2ms remaining: 13.8ms
4: learn: 0.5159659 total: 1.43ms remaining: 12.9ms
5: learn: 0.4917545 total: 1.71ms remaining: 12.5ms
6: learn: 0.4701324 total: 2.02ms remaining: 12.4ms
7: learn: 0.4524320 total: 2.28ms remaining: 12ms
8: learn: 0.4325252 total: 2.51ms remaining: 11.5ms
9: learn: 0.4165980 total: 2.85ms remaining: 11.4ms
10: learn: 0.4035100 total: 3.04ms remaining: 10.8ms
11: learn: 0.3907931 total: 3.29ms remaining: 10.4ms
12: learn: 0.3789276 total: 3.5ms remaining: 9.95ms
13: learn: 0.3688594 total: 3.8ms remaining: 9.77ms
14: learn: 0.3595050 total: 4.08ms remaining: 9.53ms
15: learn: 0.3510985 total: 4.39ms remaining: 9.32ms
16: learn: 0.3435509 total: 4.7ms remaining: 9.12ms
17: learn: 0.3372297 total: 4.95ms remaining: 8.81ms
18: learn: 0.3310716 total: 5.18ms remaining: 8.45ms
19: learn: 0.3258688 total: 5.47ms remaining: 8.21ms
20: learn: 0.3195430 total: 5.81ms remaining: 8.02ms
21: learn: 0.3125519 total: 6.09ms remaining: 7.75ms
22: learn: 0.3074617 total: 6.27ms remaining: 7.36ms
23: learn: 0.3031872 total: 6.42ms remaining: 6.95ms
24: learn: 0.2996652 total: 6.68ms remaining: 6.68ms
25: learn: 0.2955970 total: 7.01ms remaining: 6.47ms
26: learn: 0.2919958 total: 7.29ms remaining: 6.21ms
27: learn: 0.2880784 total: 7.51ms remaining: 5.9ms
28: learn: 0.2850178 total: 7.78ms remaining: 5.63ms
29: learn: 0.2831896 total: 8.06ms remaining: 5.37ms
30: learn: 0.2807488 total: 8.33ms remaining: 5.11ms
31: learn: 0.2781251 total: 8.45ms remaining: 4.75ms
32: learn: 0.2755400 total: 8.73ms remaining: 4.5ms
33: learn: 0.2735858 total: 8.97ms remaining: 4.22ms
34: learn: 0.2709428 total: 9.18ms remaining: 3.94ms
35: learn: 0.2680968 total: 9.51ms remaining: 3.7ms
36: learn: 0.2653391 total: 9.83ms remaining: 3.45ms
37: learn: 0.2622118 total: 10.2ms remaining: 3.21ms
38: learn: 0.2589166 total: 10.4ms remaining: 2.94ms
39: learn: 0.2560415 total: 10.7ms remaining: 2.68ms
40: learn: 0.2545774 total: 11ms remaining: 2.41ms
41: learn: 0.2521408 total: 11.3ms remaining: 2.15ms
42: learn: 0.2508771 total: 11.5ms remaining: 1.87ms
43: learn: 0.2500696 total: 11.7ms remaining: 1.59ms
44: learn: 0.2485904 total: 11.9ms remaining: 1.32ms
45: learn: 0.2465004 total: 12.2ms remaining: 1.06ms
46: learn: 0.2446957 total: 12.4ms remaining: 792us
47: learn: 0.2427096 total: 12.7ms remaining: 528us
48: learn: 0.2415083 total: 13ms remaining: 264us
49: learn: 0.2387823 total: 13.2ms remaining: 0us
0: learn: 0.6487726 total: 497us remaining: 24.4ms
1: learn: 0.6054752 total: 1.07ms remaining: 25.7ms
2: learn: 0.5690516 total: 1.45ms remaining: 22.7ms
3: learn: 0.5376892 total: 1.93ms remaining: 22.2ms
4: learn: 0.5107400 total: 2.25ms remaining: 20.3ms
5: learn: 0.4860125 total: 2.47ms remaining: 18.1ms
6: learn: 0.4650487 total: 3.03ms remaining: 18.6ms
7: learn: 0.4474529 total: 3.6ms remaining: 18.9ms
8: learn: 0.4286769 total: 3.96ms remaining: 18.1ms
9: learn: 0.4136039 total: 4.5ms remaining: 18ms
10: learn: 0.4019389 total: 4.84ms remaining: 17.1ms
11: learn: 0.3894000 total: 5.26ms remaining: 16.7ms
12: learn: 0.3778865 total: 5.71ms remaining: 16.3ms
13: learn: 0.3672801 total: 6.16ms remaining: 15.8ms
14: learn: 0.3576247 total: 6.6ms remaining: 15.4ms
15: learn: 0.3495637 total: 7.18ms remaining: 15.3ms
16: learn: 0.3412530 total: 7.84ms remaining: 15.2ms
17: learn: 0.3350939 total: 8.26ms remaining: 14.7ms
18: learn: 0.3290651 total: 8.83ms remaining: 14.4ms
19: learn: 0.3248385 total: 9.55ms remaining: 14.3ms
20: learn: 0.3197584 total: 10.4ms remaining: 14.4ms
21: learn: 0.3123746 total: 10.9ms remaining: 13.9ms
22: learn: 0.3067161 total: 11.4ms remaining: 13.4ms
23: learn: 0.3018604 total: 11.9ms remaining: 12.9ms
24: learn: 0.2972128 total: 12.2ms remaining: 12.2ms
25: learn: 0.2947712 total: 12.4ms remaining: 11.4ms
26: learn: 0.2912988 total: 13ms remaining: 11.1ms
27: learn: 0.2870765 total: 13.4ms remaining: 10.5ms
28: learn: 0.2831592 total: 14ms remaining: 10.1ms
29: learn: 0.2813999 total: 14.2ms remaining: 9.5ms
30: learn: 0.2782774 total: 14.8ms remaining: 9.05ms
31: learn: 0.2757420 total: 15.1ms remaining: 8.5ms
32: learn: 0.2733458 total: 15.9ms remaining: 8.18ms
33: learn: 0.2714703 total: 16.1ms remaining: 7.59ms
34: learn: 0.2692580 total: 16.4ms remaining: 7.04ms
35: learn: 0.2669193 total: 17.2ms remaining: 6.67ms
36: learn: 0.2654010 total: 17.7ms remaining: 6.21ms
37: learn: 0.2626552 total: 18ms remaining: 5.69ms
38: learn: 0.2591544 total: 18.2ms remaining: 5.14ms
39: learn: 0.2568921 total: 18.5ms remaining: 4.63ms
40: learn: 0.2539529 total: 19ms remaining: 4.17ms
41: learn: 0.2519482 total: 19.3ms remaining: 3.68ms
42: learn: 0.2496161 total: 19.8ms remaining: 3.22ms
43: learn: 0.2481991 total: 20.2ms remaining: 2.76ms
44: learn: 0.2469000 total: 20.8ms remaining: 2.31ms
45: learn: 0.2455710 total: 21.3ms remaining: 1.85ms
46: learn: 0.2415597 total: 21.8ms remaining: 1.39ms
47: learn: 0.2399522 total: 22.2ms remaining: 926us
48: learn: 0.2386716 total: 22.6ms remaining: 461us
49: learn: 0.2371674 total: 23ms remaining: 0us
0: learn: 0.6470146 total: 321us remaining: 15.7ms
1: learn: 0.6064299 total: 570us remaining: 13.7ms
2: learn: 0.5691283 total: 764us remaining: 12ms
3: learn: 0.5366569 total: 1.02ms remaining: 11.7ms
4: learn: 0.5098000 total: 1.21ms remaining: 10.9ms
5: learn: 0.4832216 total: 1.42ms remaining: 10.4ms
6: learn: 0.4596274 total: 1.62ms remaining: 9.98ms
7: learn: 0.4437963 total: 1.89ms remaining: 9.91ms
8: learn: 0.4236547 total: 2.12ms remaining: 9.66ms
9: learn: 0.4091465 total: 2.38ms remaining: 9.51ms
10: learn: 0.3961872 total: 2.58ms remaining: 9.15ms
11: learn: 0.3834046 total: 2.79ms remaining: 8.85ms
12: learn: 0.3710681 total: 3.06ms remaining: 8.7ms
13: learn: 0.3595058 total: 3.29ms remaining: 8.46ms
14: learn: 0.3509086 total: 3.61ms remaining: 8.43ms
15: learn: 0.3450153 total: 3.93ms remaining: 8.36ms
16: learn: 0.3371444 total: 4.24ms remaining: 8.23ms
17: learn: 0.3298510 total: 4.58ms remaining: 8.13ms
18: learn: 0.3232019 total: 4.7ms remaining: 7.66ms
19: learn: 0.3162856 total: 5.07ms remaining: 7.61ms
20: learn: 0.3111943 total: 5.33ms remaining: 7.36ms
21: learn: 0.3057793 total: 5.64ms remaining: 7.18ms
22: learn: 0.3007321 total: 5.91ms remaining: 6.94ms
23: learn: 0.2962390 total: 6.18ms remaining: 6.69ms
24: learn: 0.2927857 total: 6.37ms remaining: 6.37ms
25: learn: 0.2900541 total: 6.77ms remaining: 6.25ms
26: learn: 0.2865934 total: 7.03ms remaining: 5.99ms
27: learn: 0.2819302 total: 7.38ms remaining: 5.8ms
28: learn: 0.2778663 total: 7.7ms remaining: 5.57ms
29: learn: 0.2737055 total: 7.99ms remaining: 5.33ms
30: learn: 0.2722615 total: 8.21ms remaining: 5.03ms
31: learn: 0.2689709 total: 8.54ms remaining: 4.8ms
32: learn: 0.2664546 total: 8.82ms remaining: 4.54ms
33: learn: 0.2629107 total: 9.15ms remaining: 4.3ms
34: learn: 0.2603216 total: 9.41ms remaining: 4.03ms
35: learn: 0.2574981 total: 9.7ms remaining: 3.77ms
36: learn: 0.2557634 total: 9.83ms remaining: 3.45ms
37: learn: 0.2525677 total: 10.2ms remaining: 3.21ms
38: learn: 0.2487591 total: 10.5ms remaining: 2.95ms
39: learn: 0.2454105 total: 10.7ms remaining: 2.68ms
40: learn: 0.2435551 total: 11ms remaining: 2.42ms
41: learn: 0.2401939 total: 11.3ms remaining: 2.16ms
42: learn: 0.2387428 total: 11.6ms remaining: 1.88ms
43: learn: 0.2376608 total: 11.9ms remaining: 1.63ms
44: learn: 0.2358510 total: 12.4ms remaining: 1.37ms
45: learn: 0.2332892 total: 12.7ms remaining: 1.11ms
46: learn: 0.2316049 total: 13ms remaining: 830us
47: learn: 0.2293167 total: 13.4ms remaining: 556us
48: learn: 0.2264321 total: 13.6ms remaining: 278us
49: learn: 0.2240784 total: 13.9ms remaining: 0us
0: learn: 0.6531507 total: 381us remaining: 18.7ms
1: learn: 0.6065162 total: 1.41ms remaining: 33.7ms
2: learn: 0.5670336 total: 2.13ms remaining: 33.3ms
3: learn: 0.5358549 total: 3.06ms remaining: 35.2ms
4: learn: 0.5074412 total: 3.81ms remaining: 34.3ms
5: learn: 0.4804679 total: 4.36ms remaining: 32ms
6: learn: 0.4657784 total: 4.68ms remaining: 28.8ms
7: learn: 0.4441830 total: 5.43ms remaining: 28.5ms
8: learn: 0.4246293 total: 6.25ms remaining: 28.5ms
9: learn: 0.4082806 total: 7.13ms remaining: 28.5ms
10: learn: 0.3971826 total: 7.87ms remaining: 27.9ms
11: learn: 0.3859011 total: 8.96ms remaining: 28.4ms
12: learn: 0.3732273 total: 10ms remaining: 28.6ms
13: learn: 0.3599254 total: 11.2ms remaining: 28.8ms
14: learn: 0.3520452 total: 11.9ms remaining: 27.8ms
15: learn: 0.3434055 total: 13.1ms remaining: 27.8ms
16: learn: 0.3346383 total: 14ms remaining: 27.1ms
17: learn: 0.3283062 total: 14.7ms remaining: 26.1ms
18: learn: 0.3212423 total: 15.5ms remaining: 25.3ms
19: learn: 0.3155269 total: 16.2ms remaining: 24.4ms
20: learn: 0.3091024 total: 17.3ms remaining: 23.9ms
21: learn: 0.3038824 total: 18ms remaining: 22.9ms
22: learn: 0.2960116 total: 18.2ms remaining: 21.4ms
23: learn: 0.2915019 total: 19.3ms remaining: 20.9ms
24: learn: 0.2851576 total: 19.9ms remaining: 19.9ms
25: learn: 0.2824845 total: 20.8ms remaining: 19.2ms
26: learn: 0.2779683 total: 21.8ms remaining: 18.5ms
27: learn: 0.2739306 total: 22.6ms remaining: 17.7ms
28: learn: 0.2711215 total: 23.7ms remaining: 17.1ms
29: learn: 0.2672378 total: 24.8ms remaining: 16.5ms
30: learn: 0.2642976 total: 25.5ms remaining: 15.7ms
31: learn: 0.2610812 total: 26.6ms remaining: 15ms
32: learn: 0.2570838 total: 27.6ms remaining: 14.2ms
33: learn: 0.2537967 total: 28.7ms remaining: 13.5ms
34: learn: 0.2509818 total: 29.7ms remaining: 12.7ms
35: learn: 0.2490697 total: 30.7ms remaining: 11.9ms
36: learn: 0.2463164 total: 31.3ms remaining: 11ms
37: learn: 0.2441294 total: 31.9ms remaining: 10.1ms
38: learn: 0.2423599 total: 32.2ms remaining: 9.07ms
39: learn: 0.2392679 total: 33ms remaining: 8.26ms
40: learn: 0.2373584 total: 34.2ms remaining: 7.5ms
41: learn: 0.2351607 total: 35ms remaining: 6.67ms
42: learn: 0.2329397 total: 35.9ms remaining: 5.84ms
43: learn: 0.2311672 total: 36.9ms remaining: 5.03ms
44: learn: 0.2291315 total: 37.9ms remaining: 4.22ms
45: learn: 0.2273520 total: 38.8ms remaining: 3.37ms
46: learn: 0.2248574 total: 39.9ms remaining: 2.54ms
47: learn: 0.2237799 total: 40.4ms remaining: 1.68ms
48: learn: 0.2229512 total: 40.9ms remaining: 835us
49: learn: 0.2207943 total: 41.6ms remaining: 0us
0: learn: 0.6532610 total: 533us remaining: 26.1ms
1: learn: 0.6102409 total: 958us remaining: 23ms
2: learn: 0.5726766 total: 1.26ms remaining: 19.8ms
3: learn: 0.5399936 total: 1.62ms remaining: 18.7ms
4: learn: 0.5149233 total: 1.98ms remaining: 17.8ms
5: learn: 0.4883109 total: 2.32ms remaining: 17ms
6: learn: 0.4641478 total: 2.65ms remaining: 16.3ms
7: learn: 0.4460774 total: 3.09ms remaining: 16.2ms
8: learn: 0.4323097 total: 3.31ms remaining: 15.1ms
9: learn: 0.4157583 total: 3.71ms remaining: 14.8ms
10: learn: 0.4022919 total: 4.03ms remaining: 14.3ms
11: learn: 0.3898014 total: 4.43ms remaining: 14ms
12: learn: 0.3788049 total: 4.8ms remaining: 13.7ms
13: learn: 0.3683505 total: 5.16ms remaining: 13.3ms
14: learn: 0.3577059 total: 5.5ms remaining: 12.8ms
15: learn: 0.3493490 total: 5.87ms remaining: 12.5ms
16: learn: 0.3384982 total: 6.43ms remaining: 12.5ms
17: learn: 0.3329396 total: 6.76ms remaining: 12ms
18: learn: 0.3265991 total: 7.14ms remaining: 11.6ms
19: learn: 0.3205432 total: 7.72ms remaining: 11.6ms
20: learn: 0.3157322 total: 8.21ms remaining: 11.3ms
21: learn: 0.3109822 total: 8.67ms remaining: 11ms
22: learn: 0.3063239 total: 9.11ms remaining: 10.7ms
23: learn: 0.3018177 total: 9.48ms remaining: 10.3ms
24: learn: 0.2976308 total: 9.87ms remaining: 9.87ms
25: learn: 0.2931462 total: 10.3ms remaining: 9.48ms
26: learn: 0.2895543 total: 10.8ms remaining: 9.21ms
27: learn: 0.2858063 total: 11.3ms remaining: 8.88ms
28: learn: 0.2817349 total: 11.6ms remaining: 8.39ms
29: learn: 0.2783638 total: 12.1ms remaining: 8.04ms
30: learn: 0.2755228 total: 12.5ms remaining: 7.68ms
31: learn: 0.2726879 total: 12.9ms remaining: 7.27ms
32: learn: 0.2707994 total: 13.3ms remaining: 6.83ms
33: learn: 0.2671753 total: 13.6ms remaining: 6.42ms
34: learn: 0.2642467 total: 14ms remaining: 6.01ms
35: learn: 0.2618695 total: 14.4ms remaining: 5.61ms
36: learn: 0.2601981 total: 14.7ms remaining: 5.17ms
37: learn: 0.2580945 total: 15.1ms remaining: 4.77ms
38: learn: 0.2563573 total: 15.5ms remaining: 4.37ms
39: learn: 0.2534017 total: 15.9ms remaining: 3.99ms
40: learn: 0.2503710 total: 16.3ms remaining: 3.57ms
41: learn: 0.2467603 total: 16.6ms remaining: 3.16ms
42: learn: 0.2445181 total: 17ms remaining: 2.76ms
43: learn: 0.2436733 total: 17.4ms remaining: 2.37ms
44: learn: 0.2424590 total: 17.8ms remaining: 1.98ms
45: learn: 0.2413170 total: 18.4ms remaining: 1.6ms
46: learn: 0.2395461 total: 18.7ms remaining: 1.19ms
47: learn: 0.2376051 total: 19.1ms remaining: 793us
48: learn: 0.2348047 total: 19.4ms remaining: 395us
49: learn: 0.2325867 total: 20ms remaining: 0us
0: learn: 0.6478370 total: 406us remaining: 19.9ms
1: learn: 0.6050464 total: 757us remaining: 18.2ms
2: learn: 0.5715635 total: 1.05ms remaining: 16.5ms
3: learn: 0.5388645 total: 1.41ms remaining: 16.3ms
4: learn: 0.5127410 total: 1.64ms remaining: 14.7ms
5: learn: 0.4894968 total: 1.97ms remaining: 14.4ms
6: learn: 0.4690824 total: 2.27ms remaining: 14ms
7: learn: 0.4527488 total: 2.65ms remaining: 13.9ms
8: learn: 0.4367211 total: 2.83ms remaining: 12.9ms
9: learn: 0.4197791 total: 3.18ms remaining: 12.7ms
10: learn: 0.4076903 total: 3.55ms remaining: 12.6ms
11: learn: 0.3953699 total: 3.94ms remaining: 12.5ms
12: learn: 0.3835805 total: 4.4ms remaining: 12.5ms
13: learn: 0.3706580 total: 4.71ms remaining: 12.1ms
14: learn: 0.3632837 total: 5.19ms remaining: 12.1ms
15: learn: 0.3557629 total: 5.65ms remaining: 12ms
16: learn: 0.3465254 total: 5.98ms remaining: 11.6ms
17: learn: 0.3424635 total: 6.31ms remaining: 11.2ms
18: learn: 0.3340110 total: 6.7ms remaining: 10.9ms
19: learn: 0.3290275 total: 7.08ms remaining: 10.6ms
20: learn: 0.3221192 total: 7.52ms remaining: 10.4ms
21: learn: 0.3160905 total: 7.95ms remaining: 10.1ms
22: learn: 0.3106294 total: 8.18ms remaining: 9.6ms
23: learn: 0.3056046 total: 8.49ms remaining: 9.2ms
24: learn: 0.3024425 total: 8.86ms remaining: 8.86ms
25: learn: 0.2989832 total: 9.31ms remaining: 8.59ms
26: learn: 0.2944404 total: 9.74ms remaining: 8.29ms
27: learn: 0.2902619 total: 10.1ms remaining: 7.92ms
28: learn: 0.2872351 total: 10.4ms remaining: 7.56ms
29: learn: 0.2832049 total: 10.8ms remaining: 7.23ms
30: learn: 0.2811090 total: 11.1ms remaining: 6.82ms
31: learn: 0.2773095 total: 11.5ms remaining: 6.47ms
32: learn: 0.2748773 total: 11.8ms remaining: 6.09ms
33: learn: 0.2728913 total: 12.3ms remaining: 5.77ms
34: learn: 0.2704194 total: 12.7ms remaining: 5.42ms
35: learn: 0.2677007 total: 13.1ms remaining: 5.08ms
36: learn: 0.2654716 total: 13.5ms remaining: 4.74ms
37: learn: 0.2625555 total: 13.8ms remaining: 4.37ms
38: learn: 0.2588682 total: 14.3ms remaining: 4.02ms
39: learn: 0.2557431 total: 14.5ms remaining: 3.62ms
40: learn: 0.2541972 total: 14.8ms remaining: 3.25ms
41: learn: 0.2533046 total: 15.1ms remaining: 2.88ms
42: learn: 0.2508947 total: 15.5ms remaining: 2.52ms
43: learn: 0.2488413 total: 15.9ms remaining: 2.17ms
44: learn: 0.2460522 total: 16.4ms remaining: 1.82ms
45: learn: 0.2437101 total: 16.8ms remaining: 1.46ms
46: learn: 0.2420143 total: 17.3ms remaining: 1.1ms
47: learn: 0.2394717 total: 17.7ms remaining: 737us
48: learn: 0.2377849 total: 18.1ms remaining: 368us
49: learn: 0.2356327 total: 18.5ms remaining: 0us
0: learn: 0.6489461 total: 300us remaining: 14.7ms
1: learn: 0.6064240 total: 543us remaining: 13ms
2: learn: 0.5700715 total: 799us remaining: 12.5ms
3: learn: 0.5400758 total: 1.03ms remaining: 11.9ms
4: learn: 0.5145103 total: 1.25ms remaining: 11.3ms
5: learn: 0.4866673 total: 1.47ms remaining: 10.8ms
6: learn: 0.4673804 total: 1.72ms remaining: 10.6ms
7: learn: 0.4481874 total: 2ms remaining: 10.5ms
8: learn: 0.4300384 total: 2.25ms remaining: 10.2ms
9: learn: 0.4134195 total: 2.4ms remaining: 9.62ms
10: learn: 0.3962502 total: 2.67ms remaining: 9.47ms
11: learn: 0.3811386 total: 2.85ms remaining: 9.03ms
12: learn: 0.3693335 total: 3.25ms remaining: 9.25ms
13: learn: 0.3598914 total: 3.6ms remaining: 9.24ms
14: learn: 0.3516374 total: 4.02ms remaining: 9.38ms
15: learn: 0.3406200 total: 4.34ms remaining: 9.21ms
16: learn: 0.3323188 total: 4.72ms remaining: 9.15ms
17: learn: 0.3250314 total: 4.99ms remaining: 8.88ms
18: learn: 0.3192270 total: 5.57ms remaining: 9.09ms
19: learn: 0.3128178 total: 6.06ms remaining: 9.1ms
20: learn: 0.3059293 total: 6.47ms remaining: 8.93ms
21: learn: 0.2987314 total: 6.89ms remaining: 8.77ms
22: learn: 0.2940187 total: 7.2ms remaining: 8.45ms
23: learn: 0.2900778 total: 7.52ms remaining: 8.15ms
24: learn: 0.2872265 total: 7.86ms remaining: 7.86ms
25: learn: 0.2842832 total: 8.2ms remaining: 7.57ms
26: learn: 0.2804576 total: 8.56ms remaining: 7.29ms
27: learn: 0.2770150 total: 8.83ms remaining: 6.94ms
28: learn: 0.2748851 total: 9.01ms remaining: 6.52ms
29: learn: 0.2718988 total: 9.7ms remaining: 6.47ms
30: learn: 0.2689076 total: 9.86ms remaining: 6.04ms
31: learn: 0.2655543 total: 10.3ms remaining: 5.82ms
32: learn: 0.2633803 total: 10.7ms remaining: 5.5ms
33: learn: 0.2607521 total: 11ms remaining: 5.18ms
34: learn: 0.2586816 total: 11.3ms remaining: 4.83ms
35: learn: 0.2561481 total: 11.9ms remaining: 4.63ms
36: learn: 0.2535555 total: 12.2ms remaining: 4.27ms
37: learn: 0.2498320 total: 12.4ms remaining: 3.93ms
38: learn: 0.2465524 total: 12.6ms remaining: 3.56ms
39: learn: 0.2453466 total: 12.9ms remaining: 3.24ms
40: learn: 0.2437222 total: 13.2ms remaining: 2.9ms
41: learn: 0.2416669 total: 13.5ms remaining: 2.57ms
42: learn: 0.2391285 total: 13.7ms remaining: 2.24ms
43: learn: 0.2365899 total: 14.3ms remaining: 1.94ms
44: learn: 0.2348016 total: 14.7ms remaining: 1.64ms
45: learn: 0.2321103 total: 15.2ms remaining: 1.32ms
46: learn: 0.2307285 total: 15.6ms remaining: 993us
47: learn: 0.2292365 total: 15.8ms remaining: 658us
48: learn: 0.2281501 total: 16.1ms remaining: 327us
49: learn: 0.2266474 total: 16.3ms remaining: 0us
0: learn: 0.6590402 total: 402us remaining: 19.7ms
1: learn: 0.6114009 total: 699us remaining: 16.8ms
2: learn: 0.5810295 total: 971us remaining: 15.2ms
3: learn: 0.5494286 total: 1.33ms remaining: 15.3ms
4: learn: 0.5218481 total: 1.67ms remaining: 15ms
5: learn: 0.4961409 total: 2ms remaining: 14.7ms
6: learn: 0.4825933 total: 2.35ms remaining: 14.4ms
7: learn: 0.4612439 total: 2.62ms remaining: 13.8ms
8: learn: 0.4405597 total: 2.94ms remaining: 13.4ms
9: learn: 0.4236230 total: 3.17ms remaining: 12.7ms
10: learn: 0.4086284 total: 3.41ms remaining: 12.1ms
11: learn: 0.3966478 total: 3.76ms remaining: 11.9ms
12: learn: 0.3845862 total: 4.1ms remaining: 11.7ms
13: learn: 0.3734868 total: 4.43ms remaining: 11.4ms
14: learn: 0.3612373 total: 4.77ms remaining: 11.1ms
15: learn: 0.3563945 total: 5.1ms remaining: 10.8ms
16: learn: 0.3520736 total: 5.34ms remaining: 10.4ms
17: learn: 0.3437252 total: 5.7ms remaining: 10.1ms
18: learn: 0.3366232 total: 6.12ms remaining: 9.98ms
19: learn: 0.3305014 total: 6.54ms remaining: 9.81ms
20: learn: 0.3237168 total: 6.87ms remaining: 9.48ms
21: learn: 0.3165086 total: 7.16ms remaining: 9.11ms
22: learn: 0.3115073 total: 7.46ms remaining: 8.76ms
23: learn: 0.3070963 total: 7.8ms remaining: 8.45ms
24: learn: 0.3018950 total: 8.13ms remaining: 8.13ms
25: learn: 0.2978404 total: 8.55ms remaining: 7.89ms
26: learn: 0.2944708 total: 8.88ms remaining: 7.57ms
27: learn: 0.2896176 total: 9.22ms remaining: 7.24ms
28: learn: 0.2862532 total: 9.51ms remaining: 6.88ms
29: learn: 0.2832470 total: 9.77ms remaining: 6.51ms
30: learn: 0.2798789 total: 10ms remaining: 6.16ms
31: learn: 0.2760300 total: 10.3ms remaining: 5.81ms
32: learn: 0.2725581 total: 10.5ms remaining: 5.43ms
33: learn: 0.2704228 total: 10.8ms remaining: 5.11ms
34: learn: 0.2677841 total: 11.1ms remaining: 4.76ms
35: learn: 0.2653151 total: 11.4ms remaining: 4.42ms
36: learn: 0.2633276 total: 11.7ms remaining: 4.1ms
37: learn: 0.2602114 total: 11.9ms remaining: 3.77ms
38: learn: 0.2575564 total: 12.3ms remaining: 3.46ms
39: learn: 0.2550264 total: 12.6ms remaining: 3.15ms
40: learn: 0.2531548 total: 12.9ms remaining: 2.83ms
41: learn: 0.2507302 total: 13.2ms remaining: 2.52ms
42: learn: 0.2494902 total: 13.7ms remaining: 2.23ms
43: learn: 0.2463801 total: 14ms remaining: 1.91ms
44: learn: 0.2447248 total: 14.4ms remaining: 1.6ms
45: learn: 0.2424518 total: 14.7ms remaining: 1.28ms
46: learn: 0.2411762 total: 15ms remaining: 954us
47: learn: 0.2387374 total: 15.2ms remaining: 635us
48: learn: 0.2362627 total: 15.6ms remaining: 317us
49: learn: 0.2354837 total: 15.8ms remaining: 0us
0: learn: 0.6577557 total: 434us remaining: 21.3ms
1: learn: 0.6115590 total: 730us remaining: 17.5ms
2: learn: 0.5782661 total: 1.09ms remaining: 17ms
3: learn: 0.5504365 total: 1.38ms remaining: 15.9ms
4: learn: 0.5239138 total: 1.62ms remaining: 14.6ms
5: learn: 0.4960141 total: 1.88ms remaining: 13.8ms
6: learn: 0.4751299 total: 2.2ms remaining: 13.5ms
7: learn: 0.4522825 total: 2.43ms remaining: 12.8ms
8: learn: 0.4350004 total: 2.69ms remaining: 12.3ms
9: learn: 0.4172532 total: 2.92ms remaining: 11.7ms
10: learn: 0.4031272 total: 3.24ms remaining: 11.5ms
11: learn: 0.3894778 total: 3.49ms remaining: 11.1ms
12: learn: 0.3759268 total: 3.82ms remaining: 10.9ms
13: learn: 0.3646396 total: 4.07ms remaining: 10.5ms
14: learn: 0.3550212 total: 4.37ms remaining: 10.2ms
15: learn: 0.3459478 total: 4.67ms remaining: 9.91ms
16: learn: 0.3387418 total: 4.91ms remaining: 9.54ms
17: learn: 0.3317651 total: 5.14ms remaining: 9.15ms
18: learn: 0.3245328 total: 5.44ms remaining: 8.87ms
19: learn: 0.3186379 total: 5.7ms remaining: 8.55ms
20: learn: 0.3123720 total: 6ms remaining: 8.28ms
21: learn: 0.3066610 total: 6.26ms remaining: 7.97ms
22: learn: 0.3021609 total: 6.44ms remaining: 7.56ms
23: learn: 0.2981775 total: 6.7ms remaining: 7.25ms
24: learn: 0.2947302 total: 7ms remaining: 7ms
25: learn: 0.2894158 total: 7.29ms remaining: 6.73ms
26: learn: 0.2849985 total: 7.56ms remaining: 6.44ms
27: learn: 0.2816687 total: 7.83ms remaining: 6.15ms
28: learn: 0.2784744 total: 8.2ms remaining: 5.93ms
29: learn: 0.2753041 total: 8.49ms remaining: 5.66ms
30: learn: 0.2704793 total: 8.88ms remaining: 5.44ms
31: learn: 0.2655361 total: 9.24ms remaining: 5.2ms
32: learn: 0.2598502 total: 9.55ms remaining: 4.92ms
33: learn: 0.2568552 total: 9.88ms remaining: 4.65ms
34: learn: 0.2540340 total: 10.2ms remaining: 4.36ms
35: learn: 0.2503535 total: 10.5ms remaining: 4.09ms
36: learn: 0.2479424 total: 10.8ms remaining: 3.79ms
37: learn: 0.2453326 total: 11.1ms remaining: 3.51ms
38: learn: 0.2425282 total: 11.5ms remaining: 3.24ms
39: learn: 0.2398649 total: 11.8ms remaining: 2.96ms
40: learn: 0.2385556 total: 12.2ms remaining: 2.67ms
41: learn: 0.2365283 total: 12.6ms remaining: 2.41ms
42: learn: 0.2331381 total: 12.9ms remaining: 2.11ms
43: learn: 0.2300152 total: 13.3ms remaining: 1.81ms
44: learn: 0.2279245 total: 13.6ms remaining: 1.52ms
45: learn: 0.2263903 total: 13.9ms remaining: 1.21ms
46: learn: 0.2238257 total: 14.2ms remaining: 908us
47: learn: 0.2216545 total: 14.6ms remaining: 607us
48: learn: 0.2196501 total: 14.9ms remaining: 304us
49: learn: 0.2168023 total: 15.3ms remaining: 0us
0: learn: 0.6583442 total: 1.05ms remaining: 51.6ms
1: learn: 0.6160693 total: 2.02ms remaining: 48.5ms
2: learn: 0.5795190 total: 2.71ms remaining: 42.5ms
3: learn: 0.5466177 total: 3.81ms remaining: 43.8ms
4: learn: 0.5207080 total: 4.71ms remaining: 42.4ms
5: learn: 0.4950930 total: 5.79ms remaining: 42.4ms
6: learn: 0.4717440 total: 6.56ms remaining: 40.3ms
7: learn: 0.4536730 total: 7.25ms remaining: 38.1ms
8: learn: 0.4349721 total: 7.79ms remaining: 35.5ms
9: learn: 0.4202796 total: 8.94ms remaining: 35.8ms
10: learn: 0.4077376 total: 9.79ms remaining: 34.7ms
11: learn: 0.3960184 total: 10.8ms remaining: 34.2ms
12: learn: 0.3826414 total: 11.7ms remaining: 33.2ms
13: learn: 0.3722208 total: 12.5ms remaining: 32.2ms
14: learn: 0.3627836 total: 13.9ms remaining: 32.5ms
15: learn: 0.3526643 total: 15ms remaining: 31.9ms
16: learn: 0.3433313 total: 16ms remaining: 31.1ms
17: learn: 0.3337858 total: 16.8ms remaining: 29.9ms
18: learn: 0.3281039 total: 17.7ms remaining: 28.9ms
19: learn: 0.3204933 total: 18.8ms remaining: 28.2ms
20: learn: 0.3172523 total: 19.8ms remaining: 27.3ms
21: learn: 0.3112378 total: 20.7ms remaining: 26.4ms
22: learn: 0.3055845 total: 21.4ms remaining: 25.2ms
23: learn: 0.3017372 total: 22.4ms remaining: 24.3ms
24: learn: 0.2956202 total: 23.6ms remaining: 23.6ms
25: learn: 0.2904468 total: 24.5ms remaining: 22.6ms
26: learn: 0.2863467 total: 25.5ms remaining: 21.7ms
27: learn: 0.2831806 total: 26.7ms remaining: 21ms
28: learn: 0.2799353 total: 27.8ms remaining: 20.1ms
29: learn: 0.2769338 total: 28.9ms remaining: 19.3ms
30: learn: 0.2725319 total: 29.9ms remaining: 18.3ms
31: learn: 0.2690325 total: 30.9ms remaining: 17.4ms
32: learn: 0.2653722 total: 31.9ms remaining: 16.4ms
33: learn: 0.2626298 total: 32.8ms remaining: 15.4ms
34: learn: 0.2613544 total: 34.1ms remaining: 14.6ms
35: learn: 0.2590320 total: 35.1ms remaining: 13.6ms
36: learn: 0.2566611 total: 36.1ms remaining: 12.7ms
37: learn: 0.2535962 total: 37.3ms remaining: 11.8ms
38: learn: 0.2497372 total: 38.4ms remaining: 10.8ms
39: learn: 0.2470181 total: 39.2ms remaining: 9.79ms
40: learn: 0.2445742 total: 40ms remaining: 8.78ms
41: learn: 0.2430848 total: 40.8ms remaining: 7.78ms
42: learn: 0.2422891 total: 41.5ms remaining: 6.76ms
43: learn: 0.2397990 total: 42ms remaining: 5.72ms
44: learn: 0.2371706 total: 42.6ms remaining: 4.74ms
45: learn: 0.2352512 total: 43.7ms remaining: 3.8ms
46: learn: 0.2327897 total: 44.6ms remaining: 2.85ms
47: learn: 0.2308399 total: 45.6ms remaining: 1.9ms
48: learn: 0.2296719 total: 46.5ms remaining: 948us
49: learn: 0.2277718 total: 47.5ms remaining: 0us
0: learn: 0.6475054 total: 687us remaining: 33.7ms
1: learn: 0.6071370 total: 1.49ms remaining: 35.8ms
2: learn: 0.5688928 total: 1.84ms remaining: 28.9ms
3: learn: 0.5371729 total: 2.35ms remaining: 27ms
4: learn: 0.5086493 total: 3.12ms remaining: 28ms
5: learn: 0.4795564 total: 4.05ms remaining: 29.7ms
6: learn: 0.4579098 total: 5.01ms remaining: 30.8ms
7: learn: 0.4308290 total: 5.7ms remaining: 29.9ms
8: learn: 0.4093250 total: 6.54ms remaining: 29.8ms
9: learn: 0.3941826 total: 7.27ms remaining: 29.1ms
10: learn: 0.3800904 total: 8.16ms remaining: 28.9ms
11: learn: 0.3639103 total: 8.81ms remaining: 27.9ms
12: learn: 0.3496290 total: 9.61ms remaining: 27.3ms
13: learn: 0.3407169 total: 10.2ms remaining: 26.2ms
14: learn: 0.3297829 total: 11ms remaining: 25.7ms
15: learn: 0.3189842 total: 11.7ms remaining: 25ms
16: learn: 0.3114926 total: 12.5ms remaining: 24.3ms
17: learn: 0.3031443 total: 13.3ms remaining: 23.6ms
18: learn: 0.2955707 total: 14.1ms remaining: 22.9ms
19: learn: 0.2880411 total: 14.7ms remaining: 22.1ms
20: learn: 0.2805200 total: 15.4ms remaining: 21.3ms
21: learn: 0.2726320 total: 16.1ms remaining: 20.5ms
22: learn: 0.2675664 total: 16.6ms remaining: 19.5ms
23: learn: 0.2634753 total: 17.5ms remaining: 18.9ms
24: learn: 0.2596825 total: 18.2ms remaining: 18.2ms
25: learn: 0.2559484 total: 19.1ms remaining: 17.6ms
26: learn: 0.2515160 total: 19.7ms remaining: 16.8ms
27: learn: 0.2483297 total: 20.4ms remaining: 16ms
28: learn: 0.2462752 total: 20.9ms remaining: 15.2ms
29: learn: 0.2429713 total: 21.7ms remaining: 14.5ms
30: learn: 0.2395684 total: 22.1ms remaining: 13.5ms
31: learn: 0.2364600 total: 22.7ms remaining: 12.8ms
32: learn: 0.2339638 total: 23.2ms remaining: 12ms
33: learn: 0.2316042 total: 23.7ms remaining: 11.2ms
34: learn: 0.2292298 total: 24ms remaining: 10.3ms
35: learn: 0.2264514 total: 24.3ms remaining: 9.43ms
36: learn: 0.2223391 total: 25ms remaining: 8.79ms
37: learn: 0.2194596 total: 25.9ms remaining: 8.19ms
38: learn: 0.2165141 total: 26.7ms remaining: 7.53ms
39: learn: 0.2141335 total: 27.5ms remaining: 6.88ms
40: learn: 0.2121124 total: 28.2ms remaining: 6.2ms
41: learn: 0.2102779 total: 28.9ms remaining: 5.51ms
42: learn: 0.2082953 total: 29.6ms remaining: 4.83ms
43: learn: 0.2072916 total: 30.1ms remaining: 4.11ms
44: learn: 0.2049074 total: 30.5ms remaining: 3.39ms
45: learn: 0.2036420 total: 31.2ms remaining: 2.71ms
46: learn: 0.2014209 total: 31.8ms remaining: 2.03ms
47: learn: 0.1999690 total: 32.4ms remaining: 1.35ms
48: learn: 0.1990182 total: 33.1ms remaining: 675us
49: learn: 0.1973890 total: 34ms remaining: 0us
0: learn: 0.6426234 total: 288us remaining: 14.2ms
1: learn: 0.5966344 total: 843us remaining: 20.2ms
2: learn: 0.5610290 total: 1.46ms remaining: 22.8ms
3: learn: 0.5301488 total: 2.24ms remaining: 25.7ms
4: learn: 0.5029961 total: 2.64ms remaining: 23.8ms
5: learn: 0.4793889 total: 3.2ms remaining: 23.5ms
6: learn: 0.4547454 total: 3.76ms remaining: 23.1ms
7: learn: 0.4372305 total: 4.52ms remaining: 23.7ms
8: learn: 0.4185281 total: 5.05ms remaining: 23ms
9: learn: 0.4020008 total: 5.94ms remaining: 23.8ms
10: learn: 0.3875013 total: 6.47ms remaining: 22.9ms
11: learn: 0.3742145 total: 6.83ms remaining: 21.6ms
12: learn: 0.3622586 total: 7.43ms remaining: 21.1ms
13: learn: 0.3518018 total: 7.94ms remaining: 20.4ms
14: learn: 0.3435048 total: 8.62ms remaining: 20.1ms
15: learn: 0.3326371 total: 9.03ms remaining: 19.2ms
16: learn: 0.3257278 total: 9.74ms remaining: 18.9ms
17: learn: 0.3167057 total: 10.4ms remaining: 18.4ms
18: learn: 0.3097649 total: 10.8ms remaining: 17.6ms
19: learn: 0.3042157 total: 11.6ms remaining: 17.4ms
20: learn: 0.2982697 total: 12.1ms remaining: 16.8ms
21: learn: 0.2921152 total: 12.8ms remaining: 16.3ms
22: learn: 0.2875303 total: 13.3ms remaining: 15.6ms
23: learn: 0.2834689 total: 13.7ms remaining: 14.8ms
24: learn: 0.2785533 total: 14.2ms remaining: 14.2ms
25: learn: 0.2762674 total: 14.6ms remaining: 13.5ms
26: learn: 0.2729498 total: 15.2ms remaining: 13ms
27: learn: 0.2685974 total: 16.1ms remaining: 12.6ms
28: learn: 0.2648926 total: 16.7ms remaining: 12.1ms
29: learn: 0.2629856 total: 17.2ms remaining: 11.5ms
30: learn: 0.2605379 total: 17.9ms remaining: 11ms
31: learn: 0.2583238 total: 18.1ms remaining: 10.2ms
32: learn: 0.2549441 total: 18.5ms remaining: 9.52ms
33: learn: 0.2528828 total: 19ms remaining: 8.95ms
34: learn: 0.2500136 total: 19.6ms remaining: 8.4ms
35: learn: 0.2472779 total: 20.1ms remaining: 7.83ms
36: learn: 0.2443514 total: 20.7ms remaining: 7.29ms
37: learn: 0.2422131 total: 21.3ms remaining: 6.72ms
38: learn: 0.2400465 total: 21.9ms remaining: 6.18ms
39: learn: 0.2378067 total: 22.5ms remaining: 5.61ms
40: learn: 0.2362807 total: 22.9ms remaining: 5.03ms
41: learn: 0.2342833 total: 23.5ms remaining: 4.47ms
42: learn: 0.2325844 total: 24ms remaining: 3.9ms
43: learn: 0.2309996 total: 24.5ms remaining: 3.34ms
44: learn: 0.2297499 total: 24.9ms remaining: 2.77ms
45: learn: 0.2277609 total: 25.5ms remaining: 2.22ms
46: learn: 0.2248966 total: 26ms remaining: 1.66ms
47: learn: 0.2233626 total: 26.5ms remaining: 1.1ms
48: learn: 0.2210270 total: 27ms remaining: 550us
49: learn: 0.2200753 total: 27.5ms remaining: 0us
0: learn: 0.6457342 total: 704us remaining: 34.5ms
1: learn: 0.6010041 total: 1.39ms remaining: 33.5ms
2: learn: 0.5607770 total: 1.9ms remaining: 29.8ms
3: learn: 0.5254774 total: 2.6ms remaining: 29.9ms
4: learn: 0.4970300 total: 3.23ms remaining: 29.1ms
5: learn: 0.4712240 total: 3.59ms remaining: 26.4ms
6: learn: 0.4472541 total: 4.07ms remaining: 25ms
7: learn: 0.4302047 total: 4.47ms remaining: 23.5ms
8: learn: 0.4096284 total: 4.86ms remaining: 22.2ms
9: learn: 0.3943730 total: 5.45ms remaining: 21.8ms
10: learn: 0.3825996 total: 5.99ms remaining: 21.2ms
11: learn: 0.3682381 total: 6.72ms remaining: 21.3ms
12: learn: 0.3542027 total: 7.23ms remaining: 20.6ms
13: learn: 0.3416179 total: 7.83ms remaining: 20.1ms
14: learn: 0.3315787 total: 8.33ms remaining: 19.4ms
15: learn: 0.3215391 total: 8.9ms remaining: 18.9ms
16: learn: 0.3128192 total: 9.54ms remaining: 18.5ms
17: learn: 0.3060670 total: 9.97ms remaining: 17.7ms
18: learn: 0.2996001 total: 10.5ms remaining: 17.1ms
19: learn: 0.2933228 total: 11ms remaining: 16.5ms
20: learn: 0.2865127 total: 11.4ms remaining: 15.7ms
21: learn: 0.2823357 total: 11.9ms remaining: 15.1ms
22: learn: 0.2769019 total: 12.5ms remaining: 14.6ms
23: learn: 0.2713227 total: 13ms remaining: 14ms
24: learn: 0.2655899 total: 13.4ms remaining: 13.4ms
25: learn: 0.2602218 total: 13.9ms remaining: 12.9ms
26: learn: 0.2562777 total: 14.3ms remaining: 12.2ms
27: learn: 0.2537223 total: 14.8ms remaining: 11.6ms
28: learn: 0.2511636 total: 15.1ms remaining: 10.9ms
29: learn: 0.2464586 total: 15.6ms remaining: 10.4ms
30: learn: 0.2430363 total: 16.1ms remaining: 9.89ms
31: learn: 0.2402076 total: 16.6ms remaining: 9.33ms
32: learn: 0.2372463 total: 17.1ms remaining: 8.81ms
33: learn: 0.2343819 total: 17.6ms remaining: 8.3ms
34: learn: 0.2322473 total: 18ms remaining: 7.73ms
35: learn: 0.2294492 total: 18.6ms remaining: 7.25ms
36: learn: 0.2274605 total: 19ms remaining: 6.67ms
37: learn: 0.2247957 total: 19.4ms remaining: 6.13ms
38: learn: 0.2220142 total: 19.9ms remaining: 5.6ms
39: learn: 0.2205209 total: 20.3ms remaining: 5.07ms
40: learn: 0.2186829 total: 20.7ms remaining: 4.55ms
41: learn: 0.2163716 total: 21.2ms remaining: 4.04ms
42: learn: 0.2141476 total: 21.5ms remaining: 3.51ms
43: learn: 0.2117608 total: 22ms remaining: 3ms
44: learn: 0.2093530 total: 22.5ms remaining: 2.5ms
45: learn: 0.2075321 total: 22.8ms remaining: 1.98ms
46: learn: 0.2060894 total: 23.3ms remaining: 1.49ms
47: learn: 0.2051089 total: 23.6ms remaining: 985us
48: learn: 0.2037288 total: 24.1ms remaining: 492us
49: learn: 0.2020560 total: 24.6ms remaining: 0us
0: learn: 0.6495311 total: 434us remaining: 21.3ms
1: learn: 0.6040737 total: 738us remaining: 17.7ms
2: learn: 0.5672379 total: 1.03ms remaining: 16.2ms
3: learn: 0.5406197 total: 1.42ms remaining: 16.3ms
4: learn: 0.5151644 total: 1.74ms remaining: 15.6ms
5: learn: 0.4914903 total: 1.96ms remaining: 14.4ms
6: learn: 0.4692605 total: 2.23ms remaining: 13.7ms
7: learn: 0.4519017 total: 2.53ms remaining: 13.3ms
8: learn: 0.4380025 total: 2.82ms remaining: 12.8ms
9: learn: 0.4235189 total: 3.05ms remaining: 12.2ms
10: learn: 0.4113278 total: 3.29ms remaining: 11.7ms
11: learn: 0.3991034 total: 3.62ms remaining: 11.5ms
12: learn: 0.3858848 total: 3.92ms remaining: 11.2ms
13: learn: 0.3728550 total: 4.19ms remaining: 10.8ms
14: learn: 0.3636348 total: 4.44ms remaining: 10.4ms
15: learn: 0.3534055 total: 4.77ms remaining: 10.1ms
16: learn: 0.3436837 total: 5.05ms remaining: 9.81ms
17: learn: 0.3372644 total: 5.3ms remaining: 9.42ms
18: learn: 0.3313884 total: 5.53ms remaining: 9.02ms
19: learn: 0.3262682 total: 5.8ms remaining: 8.7ms
20: learn: 0.3172674 total: 6.09ms remaining: 8.41ms
21: learn: 0.3110035 total: 6.39ms remaining: 8.14ms
22: learn: 0.3032744 total: 6.66ms remaining: 7.82ms
23: learn: 0.2978424 total: 6.87ms remaining: 7.44ms
24: learn: 0.2923408 total: 7.12ms remaining: 7.12ms
25: learn: 0.2896119 total: 7.38ms remaining: 6.82ms
26: learn: 0.2859146 total: 7.62ms remaining: 6.49ms
27: learn: 0.2821476 total: 7.91ms remaining: 6.21ms
28: learn: 0.2781989 total: 8.2ms remaining: 5.94ms
29: learn: 0.2739802 total: 8.47ms remaining: 5.65ms
30: learn: 0.2710510 total: 8.75ms remaining: 5.37ms
31: learn: 0.2675356 total: 9.01ms remaining: 5.07ms
32: learn: 0.2641957 total: 9.22ms remaining: 4.75ms
33: learn: 0.2593319 total: 9.43ms remaining: 4.44ms
34: learn: 0.2557795 total: 9.71ms remaining: 4.16ms
35: learn: 0.2535475 total: 9.96ms remaining: 3.87ms
36: learn: 0.2505609 total: 10.3ms remaining: 3.6ms
37: learn: 0.2480422 total: 10.5ms remaining: 3.33ms
38: learn: 0.2455669 total: 10.8ms remaining: 3.04ms
39: learn: 0.2439872 total: 11ms remaining: 2.76ms
40: learn: 0.2422787 total: 11.2ms remaining: 2.46ms
41: learn: 0.2409530 total: 11.5ms remaining: 2.18ms
42: learn: 0.2383699 total: 11.7ms remaining: 1.91ms
43: learn: 0.2357869 total: 12ms remaining: 1.64ms
44: learn: 0.2337662 total: 12.3ms remaining: 1.37ms
45: learn: 0.2326745 total: 12.5ms remaining: 1.09ms
46: learn: 0.2309611 total: 12.8ms remaining: 817us
47: learn: 0.2293255 total: 13.1ms remaining: 545us
48: learn: 0.2276355 total: 13.4ms remaining: 273us
49: learn: 0.2256338 total: 13.7ms remaining: 0us
Evaluating param combo 7/16: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.1, 'boosted_cb_model__max_depth': 6, 'boosted_cb_model__num_leaves': 8}
Duplicate found for this combo (hash=fd87acd1ab479998ff679253ed4cbee0). Removing old run: e44770b5fa044c6597e290af4a1977d8
0: learn: 0.6460229 total: 1.72ms remaining: 84.2ms
1: learn: 0.5986942 total: 3.22ms remaining: 77.4ms
2: learn: 0.5698636 total: 5.63ms remaining: 88.3ms
3: learn: 0.5334742 total: 7.71ms remaining: 88.7ms
4: learn: 0.5066109 total: 9.77ms remaining: 87.9ms
5: learn: 0.4843498 total: 11.8ms remaining: 86.4ms
6: learn: 0.4615797 total: 13.2ms remaining: 81.1ms
7: learn: 0.4449571 total: 14.2ms remaining: 74.8ms
8: learn: 0.4229021 total: 16ms remaining: 73.1ms
9: learn: 0.4101460 total: 17.6ms remaining: 70.5ms
10: learn: 0.3974463 total: 19.2ms remaining: 68.2ms
11: learn: 0.3876826 total: 20.8ms remaining: 65.8ms
12: learn: 0.3757080 total: 22.8ms remaining: 64.8ms
13: learn: 0.3620377 total: 24.1ms remaining: 62ms
14: learn: 0.3517450 total: 25.6ms remaining: 59.7ms
15: learn: 0.3423458 total: 26.8ms remaining: 57ms
16: learn: 0.3313032 total: 28.2ms remaining: 54.8ms
17: learn: 0.3212979 total: 30.1ms remaining: 53.4ms
18: learn: 0.3162087 total: 31.5ms remaining: 51.4ms
19: learn: 0.3091345 total: 33ms remaining: 49.4ms
20: learn: 0.3015900 total: 34.9ms remaining: 48.2ms
21: learn: 0.2984053 total: 36.6ms remaining: 46.6ms
22: learn: 0.2936107 total: 38.1ms remaining: 44.8ms
23: learn: 0.2878470 total: 39.6ms remaining: 42.9ms
24: learn: 0.2836974 total: 40.6ms remaining: 40.6ms
25: learn: 0.2780346 total: 42.2ms remaining: 38.9ms
26: learn: 0.2743370 total: 43.1ms remaining: 36.7ms
27: learn: 0.2683212 total: 44.5ms remaining: 34.9ms
28: learn: 0.2662361 total: 45ms remaining: 32.6ms
29: learn: 0.2639247 total: 46.3ms remaining: 30.9ms
30: learn: 0.2606274 total: 48.3ms remaining: 29.6ms
31: learn: 0.2570408 total: 50.3ms remaining: 28.3ms
32: learn: 0.2553281 total: 51.5ms remaining: 26.5ms
33: learn: 0.2520468 total: 52.2ms remaining: 24.6ms
34: learn: 0.2495426 total: 53.1ms remaining: 22.8ms
35: learn: 0.2475286 total: 53.9ms remaining: 20.9ms
36: learn: 0.2457805 total: 54.5ms remaining: 19.1ms
37: learn: 0.2425179 total: 55.3ms remaining: 17.5ms
38: learn: 0.2409787 total: 55.9ms remaining: 15.8ms
39: learn: 0.2392860 total: 57.4ms remaining: 14.4ms
40: learn: 0.2368910 total: 59.1ms remaining: 13ms
41: learn: 0.2336958 total: 60ms remaining: 11.4ms
42: learn: 0.2302500 total: 61ms remaining: 9.93ms
43: learn: 0.2295681 total: 61.6ms remaining: 8.4ms
44: learn: 0.2256848 total: 63ms remaining: 7ms
45: learn: 0.2228299 total: 63.7ms remaining: 5.54ms
46: learn: 0.2209182 total: 64.6ms remaining: 4.13ms
47: learn: 0.2189152 total: 65.6ms remaining: 2.73ms
48: learn: 0.2158229 total: 66.5ms remaining: 1.36ms
49: learn: 0.2140103 total: 67.7ms remaining: 0us
0: learn: 0.6434696 total: 462us remaining: 22.6ms
1: learn: 0.5908671 total: 788us remaining: 18.9ms
2: learn: 0.5540161 total: 1.12ms remaining: 17.5ms
3: learn: 0.5260698 total: 1.62ms remaining: 18.6ms
4: learn: 0.4972882 total: 2.24ms remaining: 20.2ms
5: learn: 0.4743007 total: 3ms remaining: 22ms
6: learn: 0.4529556 total: 3.68ms remaining: 22.6ms
7: learn: 0.4365312 total: 4.09ms remaining: 21.5ms
8: learn: 0.4267163 total: 4.67ms remaining: 21.3ms
9: learn: 0.4125750 total: 5.42ms remaining: 21.7ms
10: learn: 0.3975618 total: 6.2ms remaining: 22ms
11: learn: 0.3840776 total: 7.13ms remaining: 22.6ms
12: learn: 0.3664514 total: 7.66ms remaining: 21.8ms
13: learn: 0.3545502 total: 8.37ms remaining: 21.5ms
14: learn: 0.3431669 total: 8.9ms remaining: 20.8ms
15: learn: 0.3336461 total: 9.38ms remaining: 19.9ms
16: learn: 0.3257986 total: 10ms remaining: 19.4ms
17: learn: 0.3165228 total: 10.8ms remaining: 19.2ms
18: learn: 0.3073494 total: 11.6ms remaining: 19ms
19: learn: 0.3013560 total: 12.4ms remaining: 18.6ms
20: learn: 0.2931992 total: 13ms remaining: 18ms
21: learn: 0.2889907 total: 13.7ms remaining: 17.5ms
22: learn: 0.2834065 total: 14.3ms remaining: 16.7ms
23: learn: 0.2772113 total: 14.8ms remaining: 16ms
24: learn: 0.2703179 total: 15.5ms remaining: 15.5ms
25: learn: 0.2673409 total: 16.2ms remaining: 14.9ms
26: learn: 0.2622606 total: 16.8ms remaining: 14.3ms
27: learn: 0.2564959 total: 17.5ms remaining: 13.7ms
28: learn: 0.2518273 total: 18.2ms remaining: 13.2ms
29: learn: 0.2477439 total: 18.7ms remaining: 12.5ms
30: learn: 0.2441075 total: 19.2ms remaining: 11.7ms
31: learn: 0.2419642 total: 19.6ms remaining: 11ms
32: learn: 0.2381178 total: 20.1ms remaining: 10.3ms
33: learn: 0.2347685 total: 20.5ms remaining: 9.66ms
34: learn: 0.2324366 total: 21ms remaining: 9ms
35: learn: 0.2309254 total: 21.5ms remaining: 8.38ms
36: learn: 0.2282484 total: 22.1ms remaining: 7.76ms
37: learn: 0.2263869 total: 22.6ms remaining: 7.14ms
38: learn: 0.2241798 total: 23.2ms remaining: 6.53ms
39: learn: 0.2218812 total: 23.7ms remaining: 5.93ms
40: learn: 0.2187091 total: 24.2ms remaining: 5.32ms
41: learn: 0.2167929 total: 24.8ms remaining: 4.72ms
42: learn: 0.2157566 total: 25.2ms remaining: 4.1ms
43: learn: 0.2137873 total: 25.8ms remaining: 3.51ms
44: learn: 0.2117076 total: 26.2ms remaining: 2.92ms
45: learn: 0.2088020 total: 26.8ms remaining: 2.33ms
46: learn: 0.2060638 total: 27.2ms remaining: 1.74ms
47: learn: 0.2018728 total: 27.7ms remaining: 1.16ms
48: learn: 0.2001262 total: 28.2ms remaining: 575us
49: learn: 0.1994772 total: 28.7ms remaining: 0us
0: learn: 0.6422518 total: 837us remaining: 41ms
1: learn: 0.5993870 total: 1.66ms remaining: 39.9ms
2: learn: 0.5628584 total: 2.94ms remaining: 46.1ms
3: learn: 0.5386216 total: 4.13ms remaining: 47.5ms
4: learn: 0.5066031 total: 4.99ms remaining: 44.9ms
5: learn: 0.4820305 total: 6.16ms remaining: 45.2ms
6: learn: 0.4639540 total: 7.31ms remaining: 44.9ms
7: learn: 0.4407244 total: 8.35ms remaining: 43.8ms
8: learn: 0.4206859 total: 9.75ms remaining: 44.4ms
9: learn: 0.4032046 total: 10.8ms remaining: 43.3ms
10: learn: 0.3882634 total: 12ms remaining: 42.7ms
11: learn: 0.3760172 total: 13.1ms remaining: 41.4ms
12: learn: 0.3631583 total: 14.8ms remaining: 42ms
13: learn: 0.3521457 total: 15.5ms remaining: 39.9ms
14: learn: 0.3431454 total: 16.8ms remaining: 39.2ms
15: learn: 0.3315988 total: 18.5ms remaining: 39.4ms
16: learn: 0.3202798 total: 20.5ms remaining: 39.7ms
17: learn: 0.3134526 total: 21.3ms remaining: 37.9ms
18: learn: 0.3069279 total: 22.8ms remaining: 37.3ms
19: learn: 0.3004042 total: 23.6ms remaining: 35.4ms
20: learn: 0.2938632 total: 24.6ms remaining: 34ms
21: learn: 0.2883339 total: 26.3ms remaining: 33.4ms
22: learn: 0.2812017 total: 26.9ms remaining: 31.6ms
23: learn: 0.2786116 total: 27.7ms remaining: 30.1ms
24: learn: 0.2723876 total: 29.5ms remaining: 29.5ms
25: learn: 0.2681037 total: 31.3ms remaining: 28.9ms
26: learn: 0.2635183 total: 32.3ms remaining: 27.5ms
27: learn: 0.2580335 total: 33.3ms remaining: 26.1ms
28: learn: 0.2537586 total: 34.5ms remaining: 25ms
29: learn: 0.2492840 total: 35.4ms remaining: 23.6ms
30: learn: 0.2446996 total: 35.7ms remaining: 21.9ms
31: learn: 0.2426673 total: 37.2ms remaining: 20.9ms
32: learn: 0.2403391 total: 38.4ms remaining: 19.8ms
33: learn: 0.2370377 total: 39.8ms remaining: 18.7ms
34: learn: 0.2346893 total: 41.2ms remaining: 17.7ms
35: learn: 0.2319904 total: 42.8ms remaining: 16.7ms
36: learn: 0.2293336 total: 44ms remaining: 15.5ms
37: learn: 0.2261521 total: 45.4ms remaining: 14.3ms
38: learn: 0.2221190 total: 47.2ms remaining: 13.3ms
39: learn: 0.2200094 total: 48.1ms remaining: 12ms
40: learn: 0.2169681 total: 48.9ms remaining: 10.7ms
41: learn: 0.2148130 total: 50ms remaining: 9.53ms
42: learn: 0.2138348 total: 51.5ms remaining: 8.38ms
43: learn: 0.2114684 total: 52.8ms remaining: 7.2ms
44: learn: 0.2087580 total: 54ms remaining: 6ms
45: learn: 0.2072098 total: 55.1ms remaining: 4.79ms
46: learn: 0.2049209 total: 56.4ms remaining: 3.6ms
47: learn: 0.2036974 total: 57.6ms remaining: 2.4ms
48: learn: 0.2021657 total: 58.6ms remaining: 1.2ms
49: learn: 0.1997194 total: 59.6ms remaining: 0us
0: learn: 0.6416822 total: 433us remaining: 21.3ms
1: learn: 0.6019648 total: 723us remaining: 17.4ms
2: learn: 0.5687979 total: 1ms remaining: 15.7ms
3: learn: 0.5302771 total: 1.27ms remaining: 14.6ms
4: learn: 0.5040872 total: 1.38ms remaining: 12.4ms
5: learn: 0.4812169 total: 1.64ms remaining: 12ms
6: learn: 0.4567565 total: 2.16ms remaining: 13.3ms
7: learn: 0.4352898 total: 2.67ms remaining: 14ms
8: learn: 0.4165105 total: 3.05ms remaining: 13.9ms
9: learn: 0.3972385 total: 3.48ms remaining: 13.9ms
10: learn: 0.3820132 total: 3.86ms remaining: 13.7ms
11: learn: 0.3688061 total: 4.12ms remaining: 13ms
12: learn: 0.3540180 total: 4.27ms remaining: 12.1ms
13: learn: 0.3438991 total: 4.71ms remaining: 12.1ms
14: learn: 0.3371155 total: 5.16ms remaining: 12ms
15: learn: 0.3263958 total: 5.58ms remaining: 11.9ms
16: learn: 0.3163591 total: 5.92ms remaining: 11.5ms
17: learn: 0.3080109 total: 6.4ms remaining: 11.4ms
18: learn: 0.3011187 total: 6.8ms remaining: 11.1ms
19: learn: 0.2948551 total: 7.17ms remaining: 10.8ms
20: learn: 0.2862073 total: 7.62ms remaining: 10.5ms
21: learn: 0.2797604 total: 8.35ms remaining: 10.6ms
22: learn: 0.2738126 total: 8.89ms remaining: 10.4ms
23: learn: 0.2686730 total: 9.27ms remaining: 10ms
24: learn: 0.2626725 total: 9.73ms remaining: 9.73ms
25: learn: 0.2583266 total: 10.2ms remaining: 9.45ms
26: learn: 0.2549457 total: 10.6ms remaining: 8.99ms
27: learn: 0.2516413 total: 10.9ms remaining: 8.56ms
28: learn: 0.2460561 total: 11.4ms remaining: 8.26ms
29: learn: 0.2425478 total: 11.9ms remaining: 7.91ms
30: learn: 0.2389663 total: 12.3ms remaining: 7.53ms
31: learn: 0.2359938 total: 12.7ms remaining: 7.15ms
32: learn: 0.2322823 total: 13.1ms remaining: 6.76ms
33: learn: 0.2297660 total: 13.6ms remaining: 6.38ms
34: learn: 0.2275496 total: 14.1ms remaining: 6.06ms
35: learn: 0.2255628 total: 14.5ms remaining: 5.63ms
36: learn: 0.2230388 total: 14.9ms remaining: 5.24ms
37: learn: 0.2198693 total: 15.4ms remaining: 4.86ms
38: learn: 0.2175725 total: 15.7ms remaining: 4.44ms
39: learn: 0.2148144 total: 16.3ms remaining: 4.08ms
40: learn: 0.2123082 total: 16.7ms remaining: 3.66ms
41: learn: 0.2110802 total: 17.1ms remaining: 3.25ms
42: learn: 0.2096018 total: 17.5ms remaining: 2.85ms
43: learn: 0.2060006 total: 18.3ms remaining: 2.5ms
44: learn: 0.2032244 total: 19.1ms remaining: 2.12ms
45: learn: 0.2007813 total: 19.8ms remaining: 1.73ms
46: learn: 0.1999685 total: 20.5ms remaining: 1.31ms
47: learn: 0.1972742 total: 20.9ms remaining: 870us
48: learn: 0.1961341 total: 21.4ms remaining: 436us
49: learn: 0.1941166 total: 21.7ms remaining: 0us
0: learn: 0.6454307 total: 531us remaining: 26.1ms
1: learn: 0.6039107 total: 1ms remaining: 24ms
2: learn: 0.5680897 total: 1.36ms remaining: 21.3ms
3: learn: 0.5412015 total: 1.82ms remaining: 20.9ms
4: learn: 0.5182422 total: 2.25ms remaining: 20.2ms
5: learn: 0.4899378 total: 2.67ms remaining: 19.6ms
6: learn: 0.4712526 total: 3.06ms remaining: 18.8ms
7: learn: 0.4534745 total: 3.53ms remaining: 18.5ms
8: learn: 0.4335757 total: 3.88ms remaining: 17.7ms
9: learn: 0.4179533 total: 4.24ms remaining: 17ms
10: learn: 0.4047999 total: 4.65ms remaining: 16.5ms
11: learn: 0.3926710 total: 5.04ms remaining: 16ms
12: learn: 0.3830573 total: 5.42ms remaining: 15.4ms
13: learn: 0.3704344 total: 5.84ms remaining: 15ms
14: learn: 0.3596217 total: 6.25ms remaining: 14.6ms
15: learn: 0.3512870 total: 6.74ms remaining: 14.3ms
16: learn: 0.3442628 total: 7.29ms remaining: 14.2ms
17: learn: 0.3356590 total: 7.84ms remaining: 13.9ms
18: learn: 0.3259936 total: 8.48ms remaining: 13.8ms
19: learn: 0.3201304 total: 8.67ms remaining: 13ms
20: learn: 0.3140818 total: 9.13ms remaining: 12.6ms
21: learn: 0.3092937 total: 9.78ms remaining: 12.4ms
22: learn: 0.3023720 total: 10.3ms remaining: 12.1ms
23: learn: 0.2960557 total: 10.8ms remaining: 11.8ms
24: learn: 0.2915371 total: 11.3ms remaining: 11.3ms
25: learn: 0.2871922 total: 11.8ms remaining: 10.9ms
26: learn: 0.2829314 total: 12.3ms remaining: 10.5ms
27: learn: 0.2767304 total: 12.8ms remaining: 10ms
28: learn: 0.2737164 total: 12.9ms remaining: 9.36ms
29: learn: 0.2700901 total: 13.4ms remaining: 8.94ms
30: learn: 0.2675257 total: 13.8ms remaining: 8.46ms
31: learn: 0.2635270 total: 14.3ms remaining: 8.02ms
32: learn: 0.2598304 total: 14.8ms remaining: 7.62ms
33: learn: 0.2557064 total: 15.3ms remaining: 7.18ms
34: learn: 0.2518669 total: 15.7ms remaining: 6.75ms
35: learn: 0.2491863 total: 16.1ms remaining: 6.27ms
36: learn: 0.2459948 total: 16.6ms remaining: 5.83ms
37: learn: 0.2429728 total: 17.1ms remaining: 5.39ms
38: learn: 0.2407984 total: 17.6ms remaining: 4.96ms
39: learn: 0.2391175 total: 18.1ms remaining: 4.54ms
40: learn: 0.2358972 total: 18.6ms remaining: 4.08ms
41: learn: 0.2344892 total: 18.8ms remaining: 3.58ms
42: learn: 0.2323901 total: 19.1ms remaining: 3.11ms
43: learn: 0.2306705 total: 19.6ms remaining: 2.67ms
44: learn: 0.2291525 total: 20ms remaining: 2.22ms
45: learn: 0.2285173 total: 20.3ms remaining: 1.77ms
46: learn: 0.2262623 total: 20.8ms remaining: 1.33ms
47: learn: 0.2230465 total: 21.3ms remaining: 886us
48: learn: 0.2225959 total: 21.8ms remaining: 445us
49: learn: 0.2210729 total: 22.3ms remaining: 0us
0: learn: 0.6482460 total: 577us remaining: 28.3ms
1: learn: 0.5994620 total: 951us remaining: 22.8ms
2: learn: 0.5597606 total: 1.24ms remaining: 19.5ms
3: learn: 0.5297838 total: 1.66ms remaining: 19.1ms
4: learn: 0.4998076 total: 1.99ms remaining: 17.9ms
5: learn: 0.4730549 total: 2.32ms remaining: 17ms
6: learn: 0.4488273 total: 2.75ms remaining: 16.9ms
7: learn: 0.4308397 total: 3.12ms remaining: 16.4ms
8: learn: 0.4166495 total: 3.51ms remaining: 16ms
9: learn: 0.3991651 total: 3.89ms remaining: 15.6ms
10: learn: 0.3836696 total: 4.3ms remaining: 15.2ms
11: learn: 0.3709195 total: 4.62ms remaining: 14.6ms
12: learn: 0.3607502 total: 4.95ms remaining: 14.1ms
13: learn: 0.3534224 total: 5.34ms remaining: 13.7ms
14: learn: 0.3415889 total: 5.73ms remaining: 13.4ms
15: learn: 0.3315429 total: 6.14ms remaining: 13ms
16: learn: 0.3227477 total: 6.55ms remaining: 12.7ms
17: learn: 0.3143145 total: 6.95ms remaining: 12.4ms
18: learn: 0.3076283 total: 7.37ms remaining: 12ms
19: learn: 0.3030449 total: 7.76ms remaining: 11.6ms
20: learn: 0.2978045 total: 8.12ms remaining: 11.2ms
21: learn: 0.2912842 total: 8.48ms remaining: 10.8ms
22: learn: 0.2878362 total: 8.86ms remaining: 10.4ms
23: learn: 0.2841128 total: 9.2ms remaining: 9.97ms
24: learn: 0.2788583 total: 9.54ms remaining: 9.54ms
25: learn: 0.2749961 total: 9.89ms remaining: 9.13ms
26: learn: 0.2695994 total: 10.2ms remaining: 8.71ms
27: learn: 0.2649796 total: 10.6ms remaining: 8.35ms
28: learn: 0.2618379 total: 11.1ms remaining: 8ms
29: learn: 0.2589920 total: 11.5ms remaining: 7.67ms
30: learn: 0.2569354 total: 11.8ms remaining: 7.25ms
31: learn: 0.2524786 total: 12.2ms remaining: 6.89ms
32: learn: 0.2489823 total: 12.7ms remaining: 6.54ms
33: learn: 0.2444734 total: 13.2ms remaining: 6.19ms
34: learn: 0.2422778 total: 13.6ms remaining: 5.83ms
35: learn: 0.2410345 total: 13.9ms remaining: 5.42ms
36: learn: 0.2357798 total: 14.3ms remaining: 5.03ms
37: learn: 0.2320050 total: 14.7ms remaining: 4.63ms
38: learn: 0.2296439 total: 15.1ms remaining: 4.25ms
39: learn: 0.2275001 total: 15.4ms remaining: 3.85ms
40: learn: 0.2265651 total: 15.7ms remaining: 3.46ms
41: learn: 0.2250516 total: 16.1ms remaining: 3.07ms
42: learn: 0.2221955 total: 16.5ms remaining: 2.68ms
43: learn: 0.2200962 total: 16.9ms remaining: 2.31ms
44: learn: 0.2183577 total: 17.3ms remaining: 1.93ms
45: learn: 0.2171256 total: 17.5ms remaining: 1.52ms
46: learn: 0.2154213 total: 17.9ms remaining: 1.14ms
47: learn: 0.2144414 total: 18.2ms remaining: 758us
48: learn: 0.2114539 total: 18.6ms remaining: 379us
49: learn: 0.2087620 total: 19ms remaining: 0us
0: learn: 0.6459251 total: 856us remaining: 42ms
1: learn: 0.5967732 total: 1.4ms remaining: 33.5ms
2: learn: 0.5604266 total: 1.83ms remaining: 28.7ms
3: learn: 0.5301914 total: 2.27ms remaining: 26.1ms
4: learn: 0.5001033 total: 2.54ms remaining: 22.9ms
5: learn: 0.4757285 total: 3.01ms remaining: 22.1ms
6: learn: 0.4534027 total: 3.47ms remaining: 21.3ms
7: learn: 0.4332717 total: 4.38ms remaining: 23ms
8: learn: 0.4169403 total: 5.32ms remaining: 24.2ms
9: learn: 0.4024786 total: 5.68ms remaining: 22.7ms
10: learn: 0.3870025 total: 6.04ms remaining: 21.4ms
11: learn: 0.3708718 total: 6.34ms remaining: 20.1ms
12: learn: 0.3621849 total: 6.69ms remaining: 19.1ms
13: learn: 0.3487949 total: 7.08ms remaining: 18.2ms
14: learn: 0.3388521 total: 7.58ms remaining: 17.7ms
15: learn: 0.3304012 total: 7.71ms remaining: 16.4ms
16: learn: 0.3191993 total: 8.1ms remaining: 15.7ms
17: learn: 0.3131216 total: 8.59ms remaining: 15.3ms
18: learn: 0.3060119 total: 9.35ms remaining: 15.3ms
19: learn: 0.2994195 total: 9.82ms remaining: 14.7ms
20: learn: 0.2932420 total: 10.2ms remaining: 14.1ms
21: learn: 0.2870467 total: 10.7ms remaining: 13.6ms
22: learn: 0.2822750 total: 11.4ms remaining: 13.3ms
23: learn: 0.2740107 total: 11.7ms remaining: 12.6ms
24: learn: 0.2716019 total: 12ms remaining: 12ms
25: learn: 0.2674096 total: 12.3ms remaining: 11.3ms
26: learn: 0.2645899 total: 12.7ms remaining: 10.8ms
27: learn: 0.2602843 total: 13.2ms remaining: 10.4ms
28: learn: 0.2526958 total: 13.5ms remaining: 9.8ms
29: learn: 0.2493028 total: 13.9ms remaining: 9.3ms
30: learn: 0.2472036 total: 14.8ms remaining: 9.08ms
31: learn: 0.2430176 total: 15.3ms remaining: 8.61ms
32: learn: 0.2391329 total: 15.7ms remaining: 8.11ms
33: learn: 0.2358302 total: 16.3ms remaining: 7.66ms
34: learn: 0.2310165 total: 16.9ms remaining: 7.22ms
35: learn: 0.2277757 total: 17.2ms remaining: 6.67ms
36: learn: 0.2253298 total: 17.7ms remaining: 6.21ms
37: learn: 0.2233005 total: 18.2ms remaining: 5.76ms
38: learn: 0.2212937 total: 18.8ms remaining: 5.29ms
39: learn: 0.2194811 total: 19.1ms remaining: 4.77ms
40: learn: 0.2147043 total: 19.4ms remaining: 4.25ms
41: learn: 0.2138031 total: 19.8ms remaining: 3.77ms
42: learn: 0.2100221 total: 20.3ms remaining: 3.31ms
43: learn: 0.2084343 total: 21ms remaining: 2.86ms
44: learn: 0.2054505 total: 21.4ms remaining: 2.38ms
45: learn: 0.2016074 total: 21.9ms remaining: 1.91ms
46: learn: 0.1987326 total: 22.3ms remaining: 1.42ms
47: learn: 0.1961880 total: 22.6ms remaining: 941us
48: learn: 0.1951000 total: 22.9ms remaining: 467us
49: learn: 0.1916939 total: 23.4ms remaining: 0us
0: learn: 0.6481220 total: 688us remaining: 33.8ms
1: learn: 0.6111293 total: 1.32ms remaining: 31.7ms
2: learn: 0.5802804 total: 1.96ms remaining: 30.7ms
3: learn: 0.5439726 total: 2.7ms remaining: 31ms
4: learn: 0.5119527 total: 3.48ms remaining: 31.3ms
5: learn: 0.4901590 total: 4.39ms remaining: 32.2ms
6: learn: 0.4674184 total: 5.27ms remaining: 32.4ms
7: learn: 0.4472015 total: 6.2ms remaining: 32.5ms
8: learn: 0.4320357 total: 6.93ms remaining: 31.6ms
9: learn: 0.4137385 total: 7.8ms remaining: 31.2ms
10: learn: 0.4027291 total: 8.37ms remaining: 29.7ms
11: learn: 0.3875597 total: 8.79ms remaining: 27.8ms
12: learn: 0.3749398 total: 9.4ms remaining: 26.7ms
13: learn: 0.3680215 total: 9.98ms remaining: 25.7ms
14: learn: 0.3587988 total: 10.8ms remaining: 25.2ms
15: learn: 0.3490724 total: 11.5ms remaining: 24.4ms
16: learn: 0.3430159 total: 12.5ms remaining: 24.2ms
17: learn: 0.3346041 total: 13.2ms remaining: 23.4ms
18: learn: 0.3270005 total: 13.9ms remaining: 22.8ms
19: learn: 0.3193122 total: 14.6ms remaining: 21.9ms
20: learn: 0.3131098 total: 15.4ms remaining: 21.3ms
21: learn: 0.3077639 total: 16.1ms remaining: 20.5ms
22: learn: 0.3023597 total: 16.9ms remaining: 19.8ms
23: learn: 0.2988230 total: 17.7ms remaining: 19.1ms
24: learn: 0.2948105 total: 18.4ms remaining: 18.4ms
25: learn: 0.2903789 total: 19ms remaining: 17.6ms
26: learn: 0.2834294 total: 19.7ms remaining: 16.8ms
27: learn: 0.2779859 total: 20.3ms remaining: 15.9ms
28: learn: 0.2730893 total: 20.8ms remaining: 15.1ms
29: learn: 0.2678348 total: 21.7ms remaining: 14.5ms
30: learn: 0.2641220 total: 22.3ms remaining: 13.7ms
31: learn: 0.2609596 total: 22.9ms remaining: 12.9ms
32: learn: 0.2563068 total: 23.5ms remaining: 12.1ms
33: learn: 0.2530053 total: 24.1ms remaining: 11.3ms
34: learn: 0.2495580 total: 24.7ms remaining: 10.6ms
35: learn: 0.2460362 total: 25.1ms remaining: 9.78ms
36: learn: 0.2422195 total: 25.7ms remaining: 9.03ms
37: learn: 0.2397035 total: 26.3ms remaining: 8.3ms
38: learn: 0.2375111 total: 26.9ms remaining: 7.58ms
39: learn: 0.2351878 total: 27.3ms remaining: 6.83ms
40: learn: 0.2322549 total: 27.7ms remaining: 6.07ms
41: learn: 0.2296954 total: 28.1ms remaining: 5.35ms
42: learn: 0.2271196 total: 28.6ms remaining: 4.66ms
43: learn: 0.2242304 total: 29.2ms remaining: 3.98ms
44: learn: 0.2222700 total: 29.7ms remaining: 3.3ms
45: learn: 0.2202307 total: 30.2ms remaining: 2.63ms
46: learn: 0.2180234 total: 30.7ms remaining: 1.96ms
47: learn: 0.2162309 total: 31.2ms remaining: 1.3ms
48: learn: 0.2143964 total: 31.6ms remaining: 645us
49: learn: 0.2133614 total: 32.2ms remaining: 0us
0: learn: 0.6438231 total: 528us remaining: 25.9ms
1: learn: 0.5942339 total: 996us remaining: 23.9ms
2: learn: 0.5577436 total: 1.38ms remaining: 21.7ms
3: learn: 0.5227298 total: 1.84ms remaining: 21.1ms
4: learn: 0.4957887 total: 2.18ms remaining: 19.6ms
5: learn: 0.4699463 total: 2.65ms remaining: 19.5ms
6: learn: 0.4473918 total: 3.07ms remaining: 18.9ms
7: learn: 0.4301896 total: 3.39ms remaining: 17.8ms
8: learn: 0.4101801 total: 3.81ms remaining: 17.4ms
9: learn: 0.3951413 total: 4.23ms remaining: 16.9ms
10: learn: 0.3797073 total: 4.7ms remaining: 16.7ms
11: learn: 0.3670122 total: 5.12ms remaining: 16.2ms
12: learn: 0.3548199 total: 5.59ms remaining: 15.9ms
13: learn: 0.3447084 total: 6.04ms remaining: 15.5ms
14: learn: 0.3357773 total: 6.49ms remaining: 15.2ms
15: learn: 0.3263992 total: 6.64ms remaining: 14.1ms
16: learn: 0.3192555 total: 7.07ms remaining: 13.7ms
17: learn: 0.3126032 total: 7.45ms remaining: 13.2ms
18: learn: 0.3073360 total: 7.92ms remaining: 12.9ms
19: learn: 0.2996768 total: 8.38ms remaining: 12.6ms
20: learn: 0.2937950 total: 8.92ms remaining: 12.3ms
21: learn: 0.2894520 total: 9.22ms remaining: 11.7ms
22: learn: 0.2850523 total: 9.62ms remaining: 11.3ms
23: learn: 0.2786365 total: 10.1ms remaining: 10.9ms
24: learn: 0.2739428 total: 10.5ms remaining: 10.5ms
25: learn: 0.2704767 total: 10.9ms remaining: 10.1ms
26: learn: 0.2672330 total: 11.4ms remaining: 9.72ms
27: learn: 0.2617352 total: 11.8ms remaining: 9.27ms
28: learn: 0.2572261 total: 12.1ms remaining: 8.79ms
29: learn: 0.2552006 total: 12.6ms remaining: 8.39ms
30: learn: 0.2514638 total: 12.9ms remaining: 7.91ms
31: learn: 0.2491248 total: 13.4ms remaining: 7.55ms
32: learn: 0.2460170 total: 14ms remaining: 7.21ms
33: learn: 0.2432369 total: 14.5ms remaining: 6.81ms
34: learn: 0.2395334 total: 15.1ms remaining: 6.46ms
35: learn: 0.2375372 total: 15.5ms remaining: 6.02ms
36: learn: 0.2354697 total: 15.8ms remaining: 5.56ms
37: learn: 0.2335946 total: 16.2ms remaining: 5.13ms
38: learn: 0.2303347 total: 16.7ms remaining: 4.71ms
39: learn: 0.2283544 total: 17.1ms remaining: 4.28ms
40: learn: 0.2269470 total: 17.6ms remaining: 3.86ms
41: learn: 0.2246784 total: 18.2ms remaining: 3.46ms
42: learn: 0.2233697 total: 18.8ms remaining: 3.06ms
43: learn: 0.2206222 total: 19.5ms remaining: 2.65ms
44: learn: 0.2184053 total: 20ms remaining: 2.22ms
45: learn: 0.2162331 total: 20.4ms remaining: 1.78ms
46: learn: 0.2135035 total: 21ms remaining: 1.34ms
47: learn: 0.2124312 total: 21.4ms remaining: 892us
48: learn: 0.2112126 total: 21.9ms remaining: 447us
49: learn: 0.2093780 total: 22.4ms remaining: 0us
0: learn: 0.6495864 total: 539us remaining: 26.4ms
1: learn: 0.6173131 total: 1.33ms remaining: 32ms
2: learn: 0.5809182 total: 1.91ms remaining: 29.9ms
3: learn: 0.5592879 total: 2.57ms remaining: 29.5ms
4: learn: 0.5333497 total: 2.81ms remaining: 25.3ms
5: learn: 0.5100357 total: 3.17ms remaining: 23.3ms
6: learn: 0.4871572 total: 3.6ms remaining: 22.1ms
7: learn: 0.4656432 total: 4.1ms remaining: 21.5ms
8: learn: 0.4488647 total: 4.59ms remaining: 20.9ms
9: learn: 0.4359401 total: 5.1ms remaining: 20.4ms
10: learn: 0.4213552 total: 5.67ms remaining: 20.1ms
11: learn: 0.4112009 total: 6.11ms remaining: 19.3ms
12: learn: 0.3990163 total: 6.3ms remaining: 17.9ms
13: learn: 0.3899301 total: 6.69ms remaining: 17.2ms
14: learn: 0.3778913 total: 7.13ms remaining: 16.6ms
15: learn: 0.3704319 total: 7.54ms remaining: 16ms
16: learn: 0.3637520 total: 7.88ms remaining: 15.3ms
17: learn: 0.3549960 total: 8.3ms remaining: 14.8ms
18: learn: 0.3469922 total: 8.68ms remaining: 14.2ms
19: learn: 0.3421919 total: 9.07ms remaining: 13.6ms
20: learn: 0.3359789 total: 9.46ms remaining: 13.1ms
21: learn: 0.3289882 total: 9.82ms remaining: 12.5ms
22: learn: 0.3247038 total: 10.2ms remaining: 12ms
23: learn: 0.3173687 total: 10.6ms remaining: 11.5ms
24: learn: 0.3117996 total: 11ms remaining: 11ms
25: learn: 0.3062690 total: 11.4ms remaining: 10.6ms
26: learn: 0.3010276 total: 11.8ms remaining: 10.1ms
27: learn: 0.2966909 total: 12.3ms remaining: 9.63ms
28: learn: 0.2916840 total: 12.7ms remaining: 9.19ms
29: learn: 0.2884667 total: 13ms remaining: 8.68ms
30: learn: 0.2841525 total: 13.4ms remaining: 8.2ms
31: learn: 0.2803788 total: 13.8ms remaining: 7.75ms
32: learn: 0.2780639 total: 14.1ms remaining: 7.28ms
33: learn: 0.2750141 total: 14.5ms remaining: 6.83ms
34: learn: 0.2714735 total: 15ms remaining: 6.41ms
35: learn: 0.2683093 total: 15.4ms remaining: 6ms
36: learn: 0.2664962 total: 15.8ms remaining: 5.55ms
37: learn: 0.2626528 total: 16.2ms remaining: 5.12ms
38: learn: 0.2595677 total: 16.7ms remaining: 4.71ms
39: learn: 0.2553350 total: 17.2ms remaining: 4.3ms
40: learn: 0.2529060 total: 17.6ms remaining: 3.87ms
41: learn: 0.2497261 total: 18ms remaining: 3.44ms
42: learn: 0.2471071 total: 18.6ms remaining: 3.03ms
43: learn: 0.2460101 total: 19ms remaining: 2.59ms
44: learn: 0.2438296 total: 19.7ms remaining: 2.19ms
45: learn: 0.2411975 total: 20.3ms remaining: 1.76ms
46: learn: 0.2397885 total: 21ms remaining: 1.34ms
47: learn: 0.2384377 total: 21.6ms remaining: 898us
48: learn: 0.2360721 total: 22.1ms remaining: 451us
49: learn: 0.2334278 total: 22.7ms remaining: 0us
0: learn: 0.6511846 total: 857us remaining: 42ms
1: learn: 0.6076570 total: 1.44ms remaining: 34.6ms
2: learn: 0.5723418 total: 1.93ms remaining: 30.3ms
3: learn: 0.5438088 total: 2.53ms remaining: 29.1ms
4: learn: 0.5133112 total: 3.09ms remaining: 27.8ms
5: learn: 0.4856231 total: 3.67ms remaining: 26.9ms
6: learn: 0.4656300 total: 4.2ms remaining: 25.8ms
7: learn: 0.4409271 total: 4.72ms remaining: 24.8ms
8: learn: 0.4197159 total: 5.18ms remaining: 23.6ms
9: learn: 0.3996955 total: 5.78ms remaining: 23.1ms
10: learn: 0.3883977 total: 6.44ms remaining: 22.8ms
11: learn: 0.3761112 total: 7.17ms remaining: 22.7ms
12: learn: 0.3651722 total: 7.68ms remaining: 21.9ms
13: learn: 0.3541517 total: 8.17ms remaining: 21ms
14: learn: 0.3505325 total: 8.7ms remaining: 20.3ms
15: learn: 0.3446431 total: 8.93ms remaining: 19ms
16: learn: 0.3309643 total: 9.49ms remaining: 18.4ms
17: learn: 0.3234425 total: 9.94ms remaining: 17.7ms
18: learn: 0.3167732 total: 10.4ms remaining: 17ms
19: learn: 0.3116347 total: 10.8ms remaining: 16.2ms
20: learn: 0.3032964 total: 11.4ms remaining: 15.7ms
21: learn: 0.2968374 total: 11.9ms remaining: 15.1ms
22: learn: 0.2927937 total: 12.5ms remaining: 14.7ms
23: learn: 0.2860013 total: 13.1ms remaining: 14.2ms
24: learn: 0.2835729 total: 13.5ms remaining: 13.5ms
25: learn: 0.2788450 total: 13.9ms remaining: 12.8ms
26: learn: 0.2749840 total: 14.3ms remaining: 12.1ms
27: learn: 0.2704904 total: 14.6ms remaining: 11.5ms
28: learn: 0.2651415 total: 15ms remaining: 10.9ms
29: learn: 0.2608183 total: 15.4ms remaining: 10.3ms
30: learn: 0.2566741 total: 15.8ms remaining: 9.71ms
31: learn: 0.2537151 total: 16.3ms remaining: 9.15ms
32: learn: 0.2517625 total: 16.8ms remaining: 8.67ms
33: learn: 0.2481143 total: 17.4ms remaining: 8.19ms
34: learn: 0.2460842 total: 17.9ms remaining: 7.67ms
35: learn: 0.2418761 total: 18.3ms remaining: 7.13ms
36: learn: 0.2385708 total: 18.7ms remaining: 6.58ms
37: learn: 0.2351499 total: 19.1ms remaining: 6.02ms
38: learn: 0.2325407 total: 19.5ms remaining: 5.5ms
39: learn: 0.2308443 total: 19.8ms remaining: 4.96ms
40: learn: 0.2286408 total: 20.2ms remaining: 4.44ms
41: learn: 0.2263965 total: 20.6ms remaining: 3.92ms
42: learn: 0.2249888 total: 20.9ms remaining: 3.41ms
43: learn: 0.2228905 total: 21.3ms remaining: 2.9ms
44: learn: 0.2200580 total: 21.7ms remaining: 2.4ms
45: learn: 0.2176117 total: 22ms remaining: 1.92ms
46: learn: 0.2158570 total: 22.5ms remaining: 1.44ms
47: learn: 0.2132175 total: 23ms remaining: 957us
48: learn: 0.2113158 total: 23.4ms remaining: 477us
49: learn: 0.2085489 total: 23.8ms remaining: 0us
0: learn: 0.6488410 total: 483us remaining: 23.7ms
1: learn: 0.6041830 total: 792us remaining: 19ms
2: learn: 0.5666197 total: 1.25ms remaining: 19.6ms
3: learn: 0.5371348 total: 1.65ms remaining: 18.9ms
4: learn: 0.5069919 total: 1.99ms remaining: 17.9ms
5: learn: 0.4873744 total: 2.61ms remaining: 19.1ms
6: learn: 0.4635549 total: 3.2ms remaining: 19.7ms
7: learn: 0.4477490 total: 3.64ms remaining: 19.1ms
8: learn: 0.4331475 total: 3.99ms remaining: 18.2ms
9: learn: 0.4162288 total: 4.61ms remaining: 18.4ms
10: learn: 0.4022050 total: 4.98ms remaining: 17.7ms
11: learn: 0.3883253 total: 5.12ms remaining: 16.2ms
12: learn: 0.3768763 total: 5.44ms remaining: 15.5ms
13: learn: 0.3671179 total: 5.88ms remaining: 15.1ms
14: learn: 0.3574952 total: 6.35ms remaining: 14.8ms
15: learn: 0.3486668 total: 6.7ms remaining: 14.2ms
16: learn: 0.3413200 total: 7.25ms remaining: 14.1ms
17: learn: 0.3349934 total: 7.6ms remaining: 13.5ms
18: learn: 0.3267828 total: 8.18ms remaining: 13.3ms
19: learn: 0.3197310 total: 8.52ms remaining: 12.8ms
20: learn: 0.3149930 total: 8.77ms remaining: 12.1ms
21: learn: 0.3076532 total: 9.08ms remaining: 11.6ms
22: learn: 0.3028168 total: 9.62ms remaining: 11.3ms
23: learn: 0.2967363 total: 10.1ms remaining: 10.9ms
24: learn: 0.2906921 total: 10.4ms remaining: 10.4ms
25: learn: 0.2859462 total: 11ms remaining: 10.2ms
26: learn: 0.2822333 total: 11.3ms remaining: 9.67ms
27: learn: 0.2779236 total: 11.6ms remaining: 9.14ms
28: learn: 0.2729841 total: 11.9ms remaining: 8.63ms
29: learn: 0.2699343 total: 12.2ms remaining: 8.16ms
30: learn: 0.2659386 total: 12.7ms remaining: 7.77ms
31: learn: 0.2629158 total: 13.2ms remaining: 7.45ms
32: learn: 0.2604045 total: 13.6ms remaining: 7.01ms
33: learn: 0.2582302 total: 14.2ms remaining: 6.69ms
34: learn: 0.2554016 total: 14.7ms remaining: 6.29ms
35: learn: 0.2539357 total: 15.1ms remaining: 5.85ms
36: learn: 0.2511286 total: 15.5ms remaining: 5.46ms
37: learn: 0.2493651 total: 16ms remaining: 5.04ms
38: learn: 0.2467166 total: 16.4ms remaining: 4.64ms
39: learn: 0.2437269 total: 16.7ms remaining: 4.18ms
40: learn: 0.2427853 total: 17.2ms remaining: 3.79ms
41: learn: 0.2427569 total: 17.3ms remaining: 3.3ms
42: learn: 0.2406668 total: 17.6ms remaining: 2.87ms
43: learn: 0.2381068 total: 18ms remaining: 2.45ms
44: learn: 0.2354987 total: 18.4ms remaining: 2.04ms
45: learn: 0.2324478 total: 18.9ms remaining: 1.64ms
46: learn: 0.2301235 total: 19.2ms remaining: 1.23ms
47: learn: 0.2284376 total: 19.8ms remaining: 824us
48: learn: 0.2268030 total: 20.1ms remaining: 411us
49: learn: 0.2256024 total: 20.5ms remaining: 0us
0: learn: 0.6474641 total: 491us remaining: 24.1ms
1: learn: 0.6077883 total: 846us remaining: 20.3ms
2: learn: 0.5742449 total: 966us remaining: 15.1ms
3: learn: 0.5421671 total: 1.35ms remaining: 15.6ms
4: learn: 0.5167581 total: 1.83ms remaining: 16.5ms
5: learn: 0.4928078 total: 2.46ms remaining: 18.1ms
6: learn: 0.4707335 total: 2.83ms remaining: 17.4ms
7: learn: 0.4500471 total: 3.15ms remaining: 16.5ms
8: learn: 0.4322172 total: 3.47ms remaining: 15.8ms
9: learn: 0.4154537 total: 3.77ms remaining: 15.1ms
10: learn: 0.4026877 total: 3.97ms remaining: 14.1ms
11: learn: 0.3917246 total: 4.25ms remaining: 13.5ms
12: learn: 0.3834524 total: 4.53ms remaining: 12.9ms
13: learn: 0.3745413 total: 4.85ms remaining: 12.5ms
14: learn: 0.3678016 total: 5.17ms remaining: 12.1ms
15: learn: 0.3580243 total: 5.56ms remaining: 11.8ms
16: learn: 0.3506033 total: 5.75ms remaining: 11.2ms
17: learn: 0.3415945 total: 6.08ms remaining: 10.8ms
18: learn: 0.3352856 total: 6.41ms remaining: 10.5ms
19: learn: 0.3292979 total: 6.8ms remaining: 10.2ms
20: learn: 0.3232338 total: 7.14ms remaining: 9.86ms
21: learn: 0.3176816 total: 7.5ms remaining: 9.55ms
22: learn: 0.3105225 total: 7.81ms remaining: 9.17ms
23: learn: 0.3060575 total: 8.14ms remaining: 8.82ms
24: learn: 0.3022308 total: 8.48ms remaining: 8.48ms
25: learn: 0.2980852 total: 8.77ms remaining: 8.1ms
26: learn: 0.2926775 total: 9.06ms remaining: 7.72ms
27: learn: 0.2897220 total: 9.36ms remaining: 7.35ms
28: learn: 0.2849458 total: 9.67ms remaining: 7ms
29: learn: 0.2816172 total: 10ms remaining: 6.67ms
30: learn: 0.2802763 total: 10.3ms remaining: 6.29ms
31: learn: 0.2776989 total: 10.6ms remaining: 5.95ms
32: learn: 0.2734024 total: 11.1ms remaining: 5.71ms
33: learn: 0.2700002 total: 11.5ms remaining: 5.43ms
34: learn: 0.2674873 total: 11.9ms remaining: 5.12ms
35: learn: 0.2658062 total: 12.4ms remaining: 4.8ms
36: learn: 0.2625090 total: 12.8ms remaining: 4.48ms
37: learn: 0.2614340 total: 13.2ms remaining: 4.16ms
38: learn: 0.2592705 total: 13.6ms remaining: 3.83ms
39: learn: 0.2550580 total: 14ms remaining: 3.51ms
40: learn: 0.2526475 total: 14.5ms remaining: 3.18ms
41: learn: 0.2498844 total: 14.9ms remaining: 2.83ms
42: learn: 0.2485077 total: 15.3ms remaining: 2.48ms
43: learn: 0.2473002 total: 15.6ms remaining: 2.13ms
44: learn: 0.2447949 total: 16ms remaining: 1.78ms
45: learn: 0.2438473 total: 16.4ms remaining: 1.42ms
46: learn: 0.2420054 total: 16.8ms remaining: 1.07ms
47: learn: 0.2399732 total: 17.3ms remaining: 719us
48: learn: 0.2368189 total: 17.7ms remaining: 361us
49: learn: 0.2339327 total: 18.1ms remaining: 0us
0: learn: 0.6477647 total: 707us remaining: 34.7ms
1: learn: 0.6127518 total: 1.09ms remaining: 26.3ms
2: learn: 0.5886244 total: 1.56ms remaining: 24.4ms
3: learn: 0.5523313 total: 1.94ms remaining: 22.3ms
4: learn: 0.5216426 total: 2.43ms remaining: 21.9ms
5: learn: 0.4985728 total: 2.6ms remaining: 19.1ms
6: learn: 0.4714201 total: 2.98ms remaining: 18.3ms
7: learn: 0.4519625 total: 3.37ms remaining: 17.7ms
8: learn: 0.4321295 total: 3.81ms remaining: 17.4ms
9: learn: 0.4141023 total: 4.21ms remaining: 16.8ms
10: learn: 0.4001725 total: 4.59ms remaining: 16.3ms
11: learn: 0.3879811 total: 5ms remaining: 15.8ms
12: learn: 0.3739023 total: 5.32ms remaining: 15.2ms
13: learn: 0.3611474 total: 5.63ms remaining: 14.5ms
14: learn: 0.3500715 total: 5.9ms remaining: 13.8ms
15: learn: 0.3390426 total: 6.2ms remaining: 13.2ms
16: learn: 0.3304780 total: 6.51ms remaining: 12.6ms
17: learn: 0.3243833 total: 6.82ms remaining: 12.1ms
18: learn: 0.3174226 total: 7.14ms remaining: 11.7ms
19: learn: 0.3090807 total: 7.49ms remaining: 11.2ms
20: learn: 0.3010340 total: 7.8ms remaining: 10.8ms
21: learn: 0.2940049 total: 8.12ms remaining: 10.3ms
22: learn: 0.2890414 total: 8.43ms remaining: 9.9ms
23: learn: 0.2838094 total: 8.72ms remaining: 9.45ms
24: learn: 0.2793484 total: 9.05ms remaining: 9.05ms
25: learn: 0.2745319 total: 9.4ms remaining: 8.67ms
26: learn: 0.2711734 total: 9.7ms remaining: 8.26ms
27: learn: 0.2679744 total: 10ms remaining: 7.85ms
28: learn: 0.2612836 total: 10.5ms remaining: 7.61ms
29: learn: 0.2591571 total: 10.8ms remaining: 7.2ms
30: learn: 0.2573990 total: 11.3ms remaining: 6.9ms
31: learn: 0.2536914 total: 11.7ms remaining: 6.56ms
32: learn: 0.2520893 total: 12.1ms remaining: 6.22ms
33: learn: 0.2475817 total: 12.5ms remaining: 5.9ms
34: learn: 0.2444968 total: 13ms remaining: 5.59ms
35: learn: 0.2418765 total: 13.4ms remaining: 5.23ms
36: learn: 0.2382324 total: 13.9ms remaining: 4.9ms
37: learn: 0.2348878 total: 14.3ms remaining: 4.53ms
38: learn: 0.2327583 total: 14.8ms remaining: 4.16ms
39: learn: 0.2315955 total: 15.2ms remaining: 3.81ms
40: learn: 0.2295426 total: 15.6ms remaining: 3.42ms
41: learn: 0.2272825 total: 16.1ms remaining: 3.07ms
42: learn: 0.2246312 total: 16.7ms remaining: 2.72ms
43: learn: 0.2213682 total: 17.1ms remaining: 2.33ms
44: learn: 0.2189293 total: 17.5ms remaining: 1.95ms
45: learn: 0.2179273 total: 17.9ms remaining: 1.56ms
46: learn: 0.2159202 total: 18.3ms remaining: 1.17ms
47: learn: 0.2133740 total: 18.8ms remaining: 781us
48: learn: 0.2098472 total: 19.2ms remaining: 391us
49: learn: 0.2068154 total: 19.7ms remaining: 0us
0: learn: 0.6522701 total: 493us remaining: 24.2ms
1: learn: 0.6036416 total: 824us remaining: 19.8ms
2: learn: 0.5640355 total: 1.18ms remaining: 18.5ms
3: learn: 0.5322241 total: 1.62ms remaining: 18.7ms
4: learn: 0.5010241 total: 2.34ms remaining: 21ms
5: learn: 0.4775931 total: 3.19ms remaining: 23.4ms
6: learn: 0.4563549 total: 3.97ms remaining: 24.4ms
7: learn: 0.4387096 total: 4.79ms remaining: 25.1ms
8: learn: 0.4244739 total: 5.55ms remaining: 25.3ms
9: learn: 0.4063645 total: 6.12ms remaining: 24.5ms
10: learn: 0.3924550 total: 6.83ms remaining: 24.2ms
11: learn: 0.3783817 total: 7.58ms remaining: 24ms
12: learn: 0.3647220 total: 8.22ms remaining: 23.4ms
13: learn: 0.3577826 total: 8.91ms remaining: 22.9ms
14: learn: 0.3494593 total: 9.57ms remaining: 22.3ms
15: learn: 0.3400580 total: 10.1ms remaining: 21.5ms
16: learn: 0.3285507 total: 10.7ms remaining: 20.8ms
17: learn: 0.3217872 total: 11.3ms remaining: 20.1ms
18: learn: 0.3149769 total: 11.9ms remaining: 19.4ms
19: learn: 0.3077400 total: 12.6ms remaining: 18.8ms
20: learn: 0.3042261 total: 13.1ms remaining: 18.1ms
21: learn: 0.2983474 total: 13.7ms remaining: 17.4ms
22: learn: 0.2942262 total: 14.3ms remaining: 16.8ms
23: learn: 0.2882372 total: 14.8ms remaining: 16.1ms
24: learn: 0.2851206 total: 15.3ms remaining: 15.3ms
25: learn: 0.2815644 total: 15.9ms remaining: 14.6ms
26: learn: 0.2776790 total: 16.4ms remaining: 14ms
27: learn: 0.2734958 total: 17.1ms remaining: 13.5ms
28: learn: 0.2701786 total: 17.7ms remaining: 12.8ms
29: learn: 0.2667674 total: 18.3ms remaining: 12.2ms
30: learn: 0.2630019 total: 18.9ms remaining: 11.6ms
31: learn: 0.2581845 total: 19.6ms remaining: 11ms
32: learn: 0.2556435 total: 20.2ms remaining: 10.4ms
33: learn: 0.2538021 total: 20.8ms remaining: 9.78ms
34: learn: 0.2517359 total: 21.3ms remaining: 9.13ms
35: learn: 0.2485945 total: 22ms remaining: 8.55ms
36: learn: 0.2465127 total: 22.3ms remaining: 7.82ms
37: learn: 0.2438727 total: 22.9ms remaining: 7.24ms
38: learn: 0.2420634 total: 23.4ms remaining: 6.61ms
39: learn: 0.2404796 total: 24ms remaining: 6.01ms
40: learn: 0.2388046 total: 24.7ms remaining: 5.42ms
41: learn: 0.2371937 total: 25.3ms remaining: 4.81ms
42: learn: 0.2338179 total: 25.9ms remaining: 4.21ms
43: learn: 0.2308034 total: 26.3ms remaining: 3.58ms
44: learn: 0.2280634 total: 26.7ms remaining: 2.97ms
45: learn: 0.2253802 total: 27.1ms remaining: 2.36ms
46: learn: 0.2243953 total: 27.6ms remaining: 1.76ms
47: learn: 0.2231214 total: 28ms remaining: 1.17ms
48: learn: 0.2208584 total: 28.6ms remaining: 582us
49: learn: 0.2188559 total: 29.2ms remaining: 0us
0: learn: 0.6451451 total: 753us remaining: 36.9ms
1: learn: 0.6042992 total: 1.37ms remaining: 32.9ms
2: learn: 0.5723961 total: 1.92ms remaining: 30.1ms
3: learn: 0.5505362 total: 2.52ms remaining: 28.9ms
4: learn: 0.5203715 total: 3.07ms remaining: 27.6ms
5: learn: 0.4949788 total: 3.54ms remaining: 25.9ms
6: learn: 0.4705068 total: 4.11ms remaining: 25.2ms
7: learn: 0.4529208 total: 4.69ms remaining: 24.6ms
8: learn: 0.4367037 total: 5.36ms remaining: 24.4ms
9: learn: 0.4220916 total: 5.87ms remaining: 23.5ms
10: learn: 0.4072312 total: 6.5ms remaining: 23.1ms
11: learn: 0.3918750 total: 6.81ms remaining: 21.6ms
12: learn: 0.3797389 total: 7.46ms remaining: 21.2ms
13: learn: 0.3713153 total: 8.1ms remaining: 20.8ms
14: learn: 0.3621565 total: 8.71ms remaining: 20.3ms
15: learn: 0.3513609 total: 9.23ms remaining: 19.6ms
16: learn: 0.3437247 total: 9.8ms remaining: 19ms
17: learn: 0.3380817 total: 10.3ms remaining: 18.3ms
18: learn: 0.3312335 total: 11ms remaining: 17.9ms
19: learn: 0.3250979 total: 11.5ms remaining: 17.2ms
20: learn: 0.3185907 total: 12.2ms remaining: 16.8ms
21: learn: 0.3126966 total: 12.7ms remaining: 16.2ms
22: learn: 0.3073101 total: 13.2ms remaining: 15.5ms
23: learn: 0.3028373 total: 14.2ms remaining: 15.3ms
24: learn: 0.2980124 total: 14.6ms remaining: 14.6ms
25: learn: 0.2929595 total: 15.3ms remaining: 14.1ms
26: learn: 0.2896203 total: 15.8ms remaining: 13.5ms
27: learn: 0.2835319 total: 16.3ms remaining: 12.8ms
28: learn: 0.2797981 total: 16.8ms remaining: 12.2ms
29: learn: 0.2771595 total: 17.3ms remaining: 11.5ms
30: learn: 0.2729367 total: 17.7ms remaining: 10.9ms
31: learn: 0.2688679 total: 18.2ms remaining: 10.2ms
32: learn: 0.2653477 total: 18.6ms remaining: 9.6ms
33: learn: 0.2611816 total: 19.1ms remaining: 8.97ms
34: learn: 0.2576321 total: 19.4ms remaining: 8.32ms
35: learn: 0.2548372 total: 19.8ms remaining: 7.71ms
36: learn: 0.2529773 total: 20ms remaining: 7.02ms
37: learn: 0.2487806 total: 20.6ms remaining: 6.5ms
38: learn: 0.2461394 total: 21.2ms remaining: 5.96ms
39: learn: 0.2428244 total: 21.8ms remaining: 5.45ms
40: learn: 0.2400398 total: 22.3ms remaining: 4.9ms
41: learn: 0.2380082 total: 22.8ms remaining: 4.34ms
42: learn: 0.2364277 total: 23.3ms remaining: 3.79ms
43: learn: 0.2332589 total: 23.8ms remaining: 3.25ms
44: learn: 0.2325072 total: 24.4ms remaining: 2.71ms
45: learn: 0.2297279 total: 25ms remaining: 2.18ms
46: learn: 0.2262849 total: 25.7ms remaining: 1.64ms
47: learn: 0.2240035 total: 26.3ms remaining: 1.1ms
48: learn: 0.2228309 total: 26.9ms remaining: 548us
49: learn: 0.2212052 total: 27.6ms remaining: 0us
0: learn: 0.6447701 total: 470us remaining: 23.1ms
1: learn: 0.6056301 total: 765us remaining: 18.4ms
2: learn: 0.5733978 total: 1.27ms remaining: 19.9ms
3: learn: 0.5441743 total: 1.62ms remaining: 18.6ms
4: learn: 0.5177609 total: 2.23ms remaining: 20.1ms
5: learn: 0.4945935 total: 2.77ms remaining: 20.3ms
6: learn: 0.4688536 total: 3.38ms remaining: 20.8ms
7: learn: 0.4504243 total: 3.89ms remaining: 20.4ms
8: learn: 0.4310441 total: 4.45ms remaining: 20.3ms
9: learn: 0.4211420 total: 5.06ms remaining: 20.2ms
10: learn: 0.4040153 total: 5.82ms remaining: 20.6ms
11: learn: 0.3899990 total: 6.42ms remaining: 20.3ms
12: learn: 0.3811531 total: 6.89ms remaining: 19.6ms
13: learn: 0.3741547 total: 7.43ms remaining: 19.1ms
14: learn: 0.3608915 total: 7.99ms remaining: 18.6ms
15: learn: 0.3514996 total: 8.65ms remaining: 18.4ms
16: learn: 0.3453344 total: 8.88ms remaining: 17.2ms
17: learn: 0.3362904 total: 9.43ms remaining: 16.8ms
18: learn: 0.3315063 total: 9.93ms remaining: 16.2ms
19: learn: 0.3234206 total: 10.6ms remaining: 15.9ms
20: learn: 0.3207093 total: 11.2ms remaining: 15.4ms
21: learn: 0.3149737 total: 11.4ms remaining: 14.6ms
22: learn: 0.3079959 total: 12ms remaining: 14.1ms
23: learn: 0.3030842 total: 12.5ms remaining: 13.5ms
24: learn: 0.2982382 total: 13ms remaining: 13ms
25: learn: 0.2940517 total: 13.5ms remaining: 12.5ms
26: learn: 0.2880557 total: 14.1ms remaining: 12ms
27: learn: 0.2842384 total: 14.6ms remaining: 11.4ms
28: learn: 0.2790874 total: 15.1ms remaining: 10.9ms
29: learn: 0.2742982 total: 15.7ms remaining: 10.5ms
30: learn: 0.2681622 total: 16.4ms remaining: 10ms
31: learn: 0.2664259 total: 16.9ms remaining: 9.52ms
32: learn: 0.2647500 total: 17.4ms remaining: 8.98ms
33: learn: 0.2628096 total: 17.9ms remaining: 8.45ms
34: learn: 0.2588150 total: 18.5ms remaining: 7.92ms
35: learn: 0.2561726 total: 18.9ms remaining: 7.35ms
36: learn: 0.2541944 total: 19.2ms remaining: 6.75ms
37: learn: 0.2513833 total: 19.6ms remaining: 6.19ms
38: learn: 0.2476279 total: 20ms remaining: 5.63ms
39: learn: 0.2456028 total: 20.3ms remaining: 5.08ms
40: learn: 0.2430387 total: 20.8ms remaining: 4.58ms
41: learn: 0.2411154 total: 21.4ms remaining: 4.07ms
42: learn: 0.2384670 total: 22ms remaining: 3.59ms
43: learn: 0.2342631 total: 22.5ms remaining: 3.06ms
44: learn: 0.2306784 total: 22.9ms remaining: 2.55ms
45: learn: 0.2253848 total: 23.4ms remaining: 2.04ms
46: learn: 0.2227968 total: 23.8ms remaining: 1.52ms
47: learn: 0.2208583 total: 24.2ms remaining: 1.01ms
48: learn: 0.2187812 total: 24.7ms remaining: 505us
49: learn: 0.2158569 total: 25.3ms remaining: 0us
0: learn: 0.6458849 total: 425us remaining: 20.9ms
1: learn: 0.6133520 total: 737us remaining: 17.7ms
2: learn: 0.5788935 total: 1.14ms remaining: 17.8ms
3: learn: 0.5506121 total: 1.87ms remaining: 21.5ms
4: learn: 0.5188294 total: 2.32ms remaining: 20.9ms
5: learn: 0.4954000 total: 2.85ms remaining: 20.9ms
6: learn: 0.4734426 total: 3.29ms remaining: 20.2ms
7: learn: 0.4556870 total: 3.81ms remaining: 20ms
8: learn: 0.4339884 total: 4.37ms remaining: 19.9ms
9: learn: 0.4206988 total: 4.84ms remaining: 19.4ms
10: learn: 0.4079334 total: 5.24ms remaining: 18.6ms
11: learn: 0.3911875 total: 5.68ms remaining: 18ms
12: learn: 0.3788150 total: 6.34ms remaining: 18ms
13: learn: 0.3691222 total: 6.8ms remaining: 17.5ms
14: learn: 0.3599406 total: 7.26ms remaining: 16.9ms
15: learn: 0.3492508 total: 7.43ms remaining: 15.8ms
16: learn: 0.3372736 total: 7.76ms remaining: 15.1ms
17: learn: 0.3290084 total: 7.95ms remaining: 14.1ms
18: learn: 0.3184555 total: 8.33ms remaining: 13.6ms
19: learn: 0.3135967 total: 8.72ms remaining: 13.1ms
20: learn: 0.3097415 total: 9.04ms remaining: 12.5ms
21: learn: 0.3035698 total: 9.46ms remaining: 12ms
22: learn: 0.3004913 total: 9.82ms remaining: 11.5ms
23: learn: 0.2958960 total: 10.2ms remaining: 11.1ms
24: learn: 0.2910836 total: 10.7ms remaining: 10.7ms
25: learn: 0.2872216 total: 11ms remaining: 10.1ms
26: learn: 0.2824063 total: 11.3ms remaining: 9.67ms
27: learn: 0.2793386 total: 11.8ms remaining: 9.25ms
28: learn: 0.2753716 total: 12.1ms remaining: 8.79ms
29: learn: 0.2718755 total: 12.5ms remaining: 8.33ms
30: learn: 0.2690465 total: 12.8ms remaining: 7.87ms
31: learn: 0.2671851 total: 13.1ms remaining: 7.39ms
32: learn: 0.2623224 total: 13.5ms remaining: 6.97ms
33: learn: 0.2594564 total: 13.9ms remaining: 6.56ms
34: learn: 0.2556683 total: 14.4ms remaining: 6.16ms
35: learn: 0.2514976 total: 14.8ms remaining: 5.75ms
36: learn: 0.2476577 total: 15.1ms remaining: 5.31ms
37: learn: 0.2463112 total: 15.5ms remaining: 4.89ms
38: learn: 0.2431250 total: 15.8ms remaining: 4.46ms
39: learn: 0.2414802 total: 16.2ms remaining: 4.05ms
40: learn: 0.2386433 total: 16.6ms remaining: 3.65ms
41: learn: 0.2367208 total: 17ms remaining: 3.24ms
42: learn: 0.2339689 total: 17.4ms remaining: 2.83ms
43: learn: 0.2313215 total: 17.8ms remaining: 2.43ms
44: learn: 0.2287363 total: 18.2ms remaining: 2.02ms
45: learn: 0.2278756 total: 18.6ms remaining: 1.61ms
46: learn: 0.2254814 total: 18.9ms remaining: 1.21ms
47: learn: 0.2239846 total: 19.3ms remaining: 804us
48: learn: 0.2226685 total: 19.7ms remaining: 401us
49: learn: 0.2202107 total: 20.1ms remaining: 0us
0: learn: 0.6561469 total: 683us remaining: 33.5ms
1: learn: 0.6141428 total: 2.1ms remaining: 50.5ms
2: learn: 0.5806953 total: 2.74ms remaining: 42.9ms
3: learn: 0.5532450 total: 3.42ms remaining: 39.4ms
4: learn: 0.5284830 total: 5.28ms remaining: 47.6ms
5: learn: 0.5031883 total: 5.81ms remaining: 42.6ms
6: learn: 0.4813054 total: 6.38ms remaining: 39.2ms
7: learn: 0.4639089 total: 7.68ms remaining: 40.3ms
8: learn: 0.4440443 total: 10.1ms remaining: 46ms
9: learn: 0.4251806 total: 11.6ms remaining: 46.3ms
10: learn: 0.4131648 total: 13.1ms remaining: 46.4ms
11: learn: 0.3995829 total: 14.8ms remaining: 46.8ms
12: learn: 0.3895909 total: 16.3ms remaining: 46.4ms
13: learn: 0.3790789 total: 17.6ms remaining: 45.3ms
14: learn: 0.3702250 total: 18.3ms remaining: 42.7ms
15: learn: 0.3628179 total: 19.7ms remaining: 41.8ms
16: learn: 0.3521130 total: 21.7ms remaining: 42.1ms
17: learn: 0.3455921 total: 22.7ms remaining: 40.4ms
18: learn: 0.3399362 total: 23.6ms remaining: 38.6ms
19: learn: 0.3315420 total: 25.2ms remaining: 37.8ms
20: learn: 0.3265923 total: 26ms remaining: 36ms
21: learn: 0.3224172 total: 27.1ms remaining: 34.5ms
22: learn: 0.3211601 total: 28.1ms remaining: 33ms
23: learn: 0.3154900 total: 29.2ms remaining: 31.7ms
24: learn: 0.3131349 total: 30ms remaining: 30ms
25: learn: 0.3068798 total: 30.8ms remaining: 28.4ms
26: learn: 0.3016954 total: 31.5ms remaining: 26.8ms
27: learn: 0.2983040 total: 31.9ms remaining: 25ms
28: learn: 0.2934145 total: 33ms remaining: 23.9ms
29: learn: 0.2892725 total: 34.1ms remaining: 22.8ms
30: learn: 0.2831620 total: 35.2ms remaining: 21.6ms
31: learn: 0.2787553 total: 36.3ms remaining: 20.4ms
32: learn: 0.2756261 total: 37.7ms remaining: 19.4ms
33: learn: 0.2716675 total: 39.2ms remaining: 18.5ms
34: learn: 0.2672679 total: 40.7ms remaining: 17.4ms
35: learn: 0.2655949 total: 41.6ms remaining: 16.2ms
36: learn: 0.2626896 total: 42.9ms remaining: 15.1ms
37: learn: 0.2603952 total: 44.3ms remaining: 14ms
38: learn: 0.2570534 total: 45.8ms remaining: 12.9ms
39: learn: 0.2542723 total: 47.1ms remaining: 11.8ms
40: learn: 0.2506732 total: 48.4ms remaining: 10.6ms
41: learn: 0.2464743 total: 49.4ms remaining: 9.4ms
42: learn: 0.2426782 total: 50.4ms remaining: 8.21ms
43: learn: 0.2401948 total: 51.5ms remaining: 7.02ms
44: learn: 0.2385713 total: 52.6ms remaining: 5.85ms
45: learn: 0.2362299 total: 53.3ms remaining: 4.63ms
46: learn: 0.2326157 total: 53.7ms remaining: 3.43ms
47: learn: 0.2316490 total: 54.4ms remaining: 2.27ms
48: learn: 0.2282742 total: 55.4ms remaining: 1.13ms
49: learn: 0.2270968 total: 56.6ms remaining: 0us
0: learn: 0.6616346 total: 547us remaining: 26.8ms
1: learn: 0.6109973 total: 972us remaining: 23.3ms
2: learn: 0.5722396 total: 1.33ms remaining: 20.9ms
3: learn: 0.5408827 total: 1.7ms remaining: 19.6ms
4: learn: 0.5094934 total: 2.07ms remaining: 18.7ms
5: learn: 0.4906131 total: 2.5ms remaining: 18.3ms
6: learn: 0.4645653 total: 2.94ms remaining: 18ms
7: learn: 0.4444164 total: 3.1ms remaining: 16.3ms
8: learn: 0.4272058 total: 3.48ms remaining: 15.9ms
9: learn: 0.4115717 total: 3.91ms remaining: 15.6ms
10: learn: 0.3950856 total: 4.34ms remaining: 15.4ms
11: learn: 0.3835814 total: 4.72ms remaining: 15ms
12: learn: 0.3704893 total: 4.93ms remaining: 14ms
13: learn: 0.3607335 total: 5.39ms remaining: 13.9ms
14: learn: 0.3545259 total: 5.82ms remaining: 13.6ms
15: learn: 0.3461680 total: 6.26ms remaining: 13.3ms
16: learn: 0.3363363 total: 6.65ms remaining: 12.9ms
17: learn: 0.3290670 total: 7.06ms remaining: 12.6ms
18: learn: 0.3213655 total: 7.66ms remaining: 12.5ms
19: learn: 0.3164629 total: 8.15ms remaining: 12.2ms
20: learn: 0.3102984 total: 8.65ms remaining: 11.9ms
21: learn: 0.3031531 total: 9.17ms remaining: 11.7ms
22: learn: 0.2953113 total: 9.7ms remaining: 11.4ms
23: learn: 0.2912851 total: 10.1ms remaining: 11ms
24: learn: 0.2842226 total: 10.7ms remaining: 10.7ms
25: learn: 0.2815780 total: 11.1ms remaining: 10.3ms
26: learn: 0.2782092 total: 11.5ms remaining: 9.76ms
27: learn: 0.2749687 total: 11.7ms remaining: 9.22ms
28: learn: 0.2712762 total: 12.2ms remaining: 8.8ms
29: learn: 0.2651409 total: 12.6ms remaining: 8.4ms
30: learn: 0.2625774 total: 12.9ms remaining: 7.94ms
31: learn: 0.2607545 total: 13.3ms remaining: 7.49ms
32: learn: 0.2581615 total: 13.6ms remaining: 6.98ms
33: learn: 0.2530686 total: 14.1ms remaining: 6.65ms
34: learn: 0.2482771 total: 14.7ms remaining: 6.31ms
35: learn: 0.2443547 total: 15.3ms remaining: 5.96ms
36: learn: 0.2390791 total: 15.9ms remaining: 5.6ms
37: learn: 0.2359834 total: 16.4ms remaining: 5.17ms
38: learn: 0.2340394 total: 16.9ms remaining: 4.76ms
39: learn: 0.2312033 total: 17.4ms remaining: 4.34ms
40: learn: 0.2302284 total: 17.9ms remaining: 3.92ms
41: learn: 0.2263697 total: 18.4ms remaining: 3.5ms
42: learn: 0.2228828 total: 19ms remaining: 3.09ms
43: learn: 0.2206554 total: 19.5ms remaining: 2.67ms
44: learn: 0.2177805 total: 20.1ms remaining: 2.23ms
45: learn: 0.2166365 total: 20.4ms remaining: 1.77ms
46: learn: 0.2147782 total: 21.1ms remaining: 1.34ms
47: learn: 0.2117934 total: 21.6ms remaining: 899us
48: learn: 0.2099487 total: 22.1ms remaining: 452us
49: learn: 0.2071163 total: 22.6ms remaining: 0us
0: learn: 0.6589850 total: 615us remaining: 30.2ms
1: learn: 0.6173268 total: 1.03ms remaining: 24.7ms
2: learn: 0.5819228 total: 1.43ms remaining: 22.3ms
3: learn: 0.5574915 total: 1.88ms remaining: 21.6ms
4: learn: 0.5332271 total: 2.28ms remaining: 20.6ms
5: learn: 0.5109981 total: 2.69ms remaining: 19.8ms
6: learn: 0.4817409 total: 3.14ms remaining: 19.3ms
7: learn: 0.4578100 total: 3.44ms remaining: 18.1ms
8: learn: 0.4400720 total: 3.88ms remaining: 17.7ms
9: learn: 0.4247016 total: 4.32ms remaining: 17.3ms
10: learn: 0.4074585 total: 4.7ms remaining: 16.6ms
11: learn: 0.3976352 total: 5.06ms remaining: 16ms
12: learn: 0.3866986 total: 5.46ms remaining: 15.5ms
13: learn: 0.3755725 total: 5.93ms remaining: 15.2ms
14: learn: 0.3623562 total: 6.3ms remaining: 14.7ms
15: learn: 0.3535372 total: 6.77ms remaining: 14.4ms
16: learn: 0.3441112 total: 7.27ms remaining: 14.1ms
17: learn: 0.3373060 total: 7.73ms remaining: 13.7ms
18: learn: 0.3273168 total: 8.26ms remaining: 13.5ms
19: learn: 0.3180001 total: 8.85ms remaining: 13.3ms
20: learn: 0.3132445 total: 9.23ms remaining: 12.7ms
21: learn: 0.3055298 total: 9.66ms remaining: 12.3ms
22: learn: 0.3002391 total: 10.1ms remaining: 11.8ms
23: learn: 0.2953327 total: 10.4ms remaining: 11.3ms
24: learn: 0.2890278 total: 10.8ms remaining: 10.8ms
25: learn: 0.2838117 total: 11.1ms remaining: 10.3ms
26: learn: 0.2787583 total: 11.6ms remaining: 9.84ms
27: learn: 0.2758002 total: 11.8ms remaining: 9.26ms
28: learn: 0.2709398 total: 12.2ms remaining: 8.84ms
29: learn: 0.2657668 total: 12.6ms remaining: 8.39ms
30: learn: 0.2618131 total: 13ms remaining: 7.96ms
31: learn: 0.2588815 total: 13.4ms remaining: 7.51ms
32: learn: 0.2559394 total: 13.8ms remaining: 7.09ms
33: learn: 0.2534781 total: 14.2ms remaining: 6.66ms
34: learn: 0.2504927 total: 14.5ms remaining: 6.24ms
35: learn: 0.2487456 total: 14.9ms remaining: 5.78ms
36: learn: 0.2457432 total: 15.2ms remaining: 5.34ms
37: learn: 0.2436861 total: 15.6ms remaining: 4.93ms
38: learn: 0.2404680 total: 15.9ms remaining: 4.5ms
39: learn: 0.2375523 total: 16.4ms remaining: 4.1ms
40: learn: 0.2342454 total: 17ms remaining: 3.73ms
41: learn: 0.2318136 total: 17.5ms remaining: 3.33ms
42: learn: 0.2288749 total: 17.9ms remaining: 2.92ms
43: learn: 0.2265562 total: 18.5ms remaining: 2.53ms
44: learn: 0.2232778 total: 18.9ms remaining: 2.1ms
45: learn: 0.2213184 total: 19.3ms remaining: 1.68ms
46: learn: 0.2187823 total: 19.8ms remaining: 1.26ms
47: learn: 0.2166353 total: 20.2ms remaining: 841us
48: learn: 0.2147026 total: 20.7ms remaining: 421us
49: learn: 0.2128187 total: 21.1ms remaining: 0us
0: learn: 0.6452274 total: 640us remaining: 31.4ms
1: learn: 0.6007395 total: 1.09ms remaining: 26.2ms
2: learn: 0.5606860 total: 1.36ms remaining: 21.3ms
3: learn: 0.5356117 total: 1.86ms remaining: 21.4ms
4: learn: 0.5040476 total: 2.4ms remaining: 21.6ms
5: learn: 0.4782473 total: 2.74ms remaining: 20.1ms
6: learn: 0.4579484 total: 3.42ms remaining: 21ms
7: learn: 0.4339063 total: 4.13ms remaining: 21.7ms
8: learn: 0.4136929 total: 4.61ms remaining: 21ms
9: learn: 0.3964760 total: 5ms remaining: 20ms
10: learn: 0.3776982 total: 5.54ms remaining: 19.7ms
11: learn: 0.3631631 total: 5.87ms remaining: 18.6ms
12: learn: 0.3538404 total: 6.28ms remaining: 17.9ms
13: learn: 0.3402280 total: 6.71ms remaining: 17.3ms
14: learn: 0.3279063 total: 7.03ms remaining: 16.4ms
15: learn: 0.3190464 total: 7.39ms remaining: 15.7ms
16: learn: 0.3131871 total: 7.8ms remaining: 15.1ms
17: learn: 0.3048667 total: 8.12ms remaining: 14.4ms
18: learn: 0.2987186 total: 8.67ms remaining: 14.2ms
19: learn: 0.2904161 total: 9.22ms remaining: 13.8ms
20: learn: 0.2830408 total: 9.67ms remaining: 13.3ms
21: learn: 0.2757885 total: 10.1ms remaining: 12.8ms
22: learn: 0.2687860 total: 10.5ms remaining: 12.3ms
23: learn: 0.2623586 total: 11ms remaining: 11.9ms
24: learn: 0.2581471 total: 11.4ms remaining: 11.4ms
25: learn: 0.2539348 total: 11.9ms remaining: 11ms
26: learn: 0.2503573 total: 12.3ms remaining: 10.5ms
27: learn: 0.2460689 total: 12.8ms remaining: 10ms
28: learn: 0.2428545 total: 13ms remaining: 9.43ms
29: learn: 0.2395749 total: 13.7ms remaining: 9.16ms
30: learn: 0.2357687 total: 14.4ms remaining: 8.82ms
31: learn: 0.2310186 total: 14.7ms remaining: 8.26ms
32: learn: 0.2269526 total: 15.1ms remaining: 7.8ms
33: learn: 0.2230483 total: 15.6ms remaining: 7.34ms
34: learn: 0.2196256 total: 16.1ms remaining: 6.88ms
35: learn: 0.2164008 total: 16.4ms remaining: 6.37ms
36: learn: 0.2136322 total: 16.9ms remaining: 5.95ms
37: learn: 0.2115325 total: 17.4ms remaining: 5.49ms
38: learn: 0.2080654 total: 17.8ms remaining: 5.01ms
39: learn: 0.2058333 total: 18.3ms remaining: 4.58ms
40: learn: 0.2037221 total: 18.8ms remaining: 4.13ms
41: learn: 0.2015290 total: 19.3ms remaining: 3.68ms
42: learn: 0.1996680 total: 19.6ms remaining: 3.19ms
43: learn: 0.1985694 total: 20.2ms remaining: 2.75ms
44: learn: 0.1962434 total: 20.5ms remaining: 2.27ms
45: learn: 0.1949811 total: 21ms remaining: 1.83ms
46: learn: 0.1933688 total: 21.7ms remaining: 1.38ms
47: learn: 0.1904367 total: 22ms remaining: 916us
48: learn: 0.1886177 total: 22.4ms remaining: 457us
49: learn: 0.1877138 total: 22.7ms remaining: 0us
0: learn: 0.6413457 total: 455us remaining: 22.3ms
1: learn: 0.5900578 total: 800us remaining: 19.2ms
2: learn: 0.5519438 total: 1.08ms remaining: 17ms
3: learn: 0.5243187 total: 1.42ms remaining: 16.3ms
4: learn: 0.4950511 total: 1.72ms remaining: 15.5ms
5: learn: 0.4709211 total: 1.99ms remaining: 14.6ms
6: learn: 0.4495564 total: 2.38ms remaining: 14.6ms
7: learn: 0.4249429 total: 2.82ms remaining: 14.8ms
8: learn: 0.4057863 total: 3.32ms remaining: 15.1ms
9: learn: 0.3912050 total: 3.7ms remaining: 14.8ms
10: learn: 0.3781296 total: 4.19ms remaining: 14.9ms
11: learn: 0.3678321 total: 4.59ms remaining: 14.5ms
12: learn: 0.3538277 total: 4.97ms remaining: 14.2ms
13: learn: 0.3401897 total: 5.3ms remaining: 13.6ms
14: learn: 0.3271568 total: 5.74ms remaining: 13.4ms
15: learn: 0.3166289 total: 6.15ms remaining: 13.1ms
16: learn: 0.3094882 total: 6.34ms remaining: 12.3ms
17: learn: 0.3004746 total: 6.76ms remaining: 12ms
18: learn: 0.2952407 total: 7.24ms remaining: 11.8ms
19: learn: 0.2889081 total: 7.62ms remaining: 11.4ms
20: learn: 0.2864670 total: 7.93ms remaining: 11ms
21: learn: 0.2819508 total: 8.23ms remaining: 10.5ms
22: learn: 0.2767701 total: 8.57ms remaining: 10.1ms
23: learn: 0.2728949 total: 9.03ms remaining: 9.79ms
24: learn: 0.2690209 total: 9.45ms remaining: 9.45ms
25: learn: 0.2661912 total: 9.8ms remaining: 9.04ms
26: learn: 0.2623100 total: 10.1ms remaining: 8.63ms
27: learn: 0.2602555 total: 10.5ms remaining: 8.23ms
28: learn: 0.2581525 total: 10.8ms remaining: 7.83ms
29: learn: 0.2538615 total: 11.1ms remaining: 7.42ms
30: learn: 0.2516232 total: 11.5ms remaining: 7.08ms
31: learn: 0.2491004 total: 12ms remaining: 6.73ms
32: learn: 0.2458602 total: 12.5ms remaining: 6.45ms
33: learn: 0.2437416 total: 12.9ms remaining: 6.07ms
34: learn: 0.2418580 total: 13.4ms remaining: 5.73ms
35: learn: 0.2394861 total: 13.8ms remaining: 5.38ms
36: learn: 0.2365999 total: 14.4ms remaining: 5.04ms
37: learn: 0.2328293 total: 14.8ms remaining: 4.68ms
38: learn: 0.2304114 total: 15.2ms remaining: 4.29ms
39: learn: 0.2277712 total: 15.7ms remaining: 3.93ms
40: learn: 0.2248847 total: 16ms remaining: 3.52ms
41: learn: 0.2237335 total: 16.5ms remaining: 3.14ms
42: learn: 0.2203033 total: 16.9ms remaining: 2.75ms
43: learn: 0.2170414 total: 17.4ms remaining: 2.38ms
44: learn: 0.2140231 total: 17.9ms remaining: 1.99ms
45: learn: 0.2131041 total: 18.3ms remaining: 1.59ms
46: learn: 0.2109699 total: 18.7ms remaining: 1.19ms
47: learn: 0.2098659 total: 19ms remaining: 792us
48: learn: 0.2092036 total: 19.5ms remaining: 398us
49: learn: 0.2079761 total: 19.9ms remaining: 0us
0: learn: 0.6452260 total: 837us remaining: 41ms
1: learn: 0.5951641 total: 1.54ms remaining: 36.9ms
2: learn: 0.5576028 total: 2.11ms remaining: 33ms
3: learn: 0.5242696 total: 3.2ms remaining: 36.8ms
4: learn: 0.4943754 total: 5.06ms remaining: 45.5ms
5: learn: 0.4725212 total: 5.71ms remaining: 41.9ms
6: learn: 0.4484628 total: 7.11ms remaining: 43.7ms
7: learn: 0.4263461 total: 8.82ms remaining: 46.3ms
8: learn: 0.4072501 total: 10.6ms remaining: 48.3ms
9: learn: 0.3935810 total: 11.8ms remaining: 47.1ms
10: learn: 0.3762641 total: 13.1ms remaining: 46.6ms
11: learn: 0.3641626 total: 14.9ms remaining: 47.2ms
12: learn: 0.3522660 total: 15.9ms remaining: 45.4ms
13: learn: 0.3433186 total: 17.2ms remaining: 44.2ms
14: learn: 0.3338644 total: 18.8ms remaining: 43.8ms
15: learn: 0.3241537 total: 19.7ms remaining: 41.8ms
16: learn: 0.3143320 total: 20.4ms remaining: 39.5ms
17: learn: 0.3102218 total: 20.8ms remaining: 37ms
18: learn: 0.3024684 total: 21.3ms remaining: 34.7ms
19: learn: 0.2942581 total: 22ms remaining: 33ms
20: learn: 0.2864357 total: 22.5ms remaining: 31ms
21: learn: 0.2801061 total: 24ms remaining: 30.5ms
22: learn: 0.2754716 total: 25.3ms remaining: 29.7ms
23: learn: 0.2698246 total: 26.4ms remaining: 28.6ms
24: learn: 0.2647569 total: 27.4ms remaining: 27.4ms
25: learn: 0.2603520 total: 28.6ms remaining: 26.4ms
26: learn: 0.2562941 total: 29.9ms remaining: 25.5ms
27: learn: 0.2523354 total: 31.3ms remaining: 24.6ms
28: learn: 0.2471671 total: 31.9ms remaining: 23.1ms
29: learn: 0.2439060 total: 32.5ms remaining: 21.7ms
30: learn: 0.2400371 total: 33.8ms remaining: 20.7ms
31: learn: 0.2367053 total: 35.1ms remaining: 19.7ms
32: learn: 0.2346123 total: 36.2ms remaining: 18.7ms
33: learn: 0.2307711 total: 37.3ms remaining: 17.6ms
34: learn: 0.2275036 total: 38.7ms remaining: 16.6ms
35: learn: 0.2242418 total: 39.8ms remaining: 15.5ms
36: learn: 0.2214850 total: 41ms remaining: 14.4ms
37: learn: 0.2196511 total: 41.9ms remaining: 13.2ms
38: learn: 0.2176325 total: 42.7ms remaining: 12ms
39: learn: 0.2149849 total: 43.9ms remaining: 11ms
40: learn: 0.2134714 total: 44.8ms remaining: 9.84ms
41: learn: 0.2107307 total: 45.5ms remaining: 8.67ms
42: learn: 0.2093829 total: 46.3ms remaining: 7.54ms
43: learn: 0.2062526 total: 47.2ms remaining: 6.44ms
44: learn: 0.2041734 total: 48ms remaining: 5.33ms
45: learn: 0.2034350 total: 48.8ms remaining: 4.25ms
46: learn: 0.2008729 total: 49.9ms remaining: 3.19ms
47: learn: 0.1982945 total: 50.8ms remaining: 2.12ms
48: learn: 0.1964192 total: 51.6ms remaining: 1.05ms
49: learn: 0.1941753 total: 52.3ms remaining: 0us
0: learn: 0.6489590 total: 682us remaining: 33.4ms
1: learn: 0.6054729 total: 1.36ms remaining: 32.7ms
2: learn: 0.5713667 total: 1.84ms remaining: 28.9ms
3: learn: 0.5478161 total: 2.78ms remaining: 31.9ms
4: learn: 0.5216653 total: 3.71ms remaining: 33.4ms
5: learn: 0.4966537 total: 4.51ms remaining: 33.1ms
6: learn: 0.4782736 total: 5.24ms remaining: 32.2ms
7: learn: 0.4638061 total: 5.9ms remaining: 31ms
8: learn: 0.4486561 total: 6.92ms remaining: 31.5ms
9: learn: 0.4323845 total: 7.86ms remaining: 31.4ms
10: learn: 0.4182749 total: 8.66ms remaining: 30.7ms
11: learn: 0.4006948 total: 9.06ms remaining: 28.7ms
12: learn: 0.3868310 total: 9.88ms remaining: 28.1ms
13: learn: 0.3766702 total: 10.6ms remaining: 27.3ms
14: learn: 0.3630724 total: 11.3ms remaining: 26.4ms
15: learn: 0.3532909 total: 12ms remaining: 25.6ms
16: learn: 0.3404533 total: 12.9ms remaining: 25.1ms
17: learn: 0.3326679 total: 13.6ms remaining: 24.2ms
18: learn: 0.3261320 total: 14.3ms remaining: 23.3ms
19: learn: 0.3193806 total: 15.1ms remaining: 22.6ms
20: learn: 0.3129853 total: 15.6ms remaining: 21.6ms
21: learn: 0.3052955 total: 16.5ms remaining: 20.9ms
22: learn: 0.2995204 total: 17.1ms remaining: 20.1ms
23: learn: 0.2938979 total: 17.9ms remaining: 19.4ms
24: learn: 0.2905536 total: 18.5ms remaining: 18.5ms
25: learn: 0.2864712 total: 19.1ms remaining: 17.7ms
26: learn: 0.2807762 total: 19.7ms remaining: 16.8ms
27: learn: 0.2749710 total: 20.4ms remaining: 16ms
28: learn: 0.2681340 total: 21ms remaining: 15.2ms
29: learn: 0.2656587 total: 21.6ms remaining: 14.4ms
30: learn: 0.2628564 total: 22.4ms remaining: 13.7ms
31: learn: 0.2568914 total: 22.9ms remaining: 12.9ms
32: learn: 0.2537556 total: 23.5ms remaining: 12.1ms
33: learn: 0.2519176 total: 24ms remaining: 11.3ms
34: learn: 0.2473640 total: 24.6ms remaining: 10.6ms
35: learn: 0.2450667 total: 25.2ms remaining: 9.81ms
36: learn: 0.2436387 total: 25.7ms remaining: 9.02ms
37: learn: 0.2417667 total: 26.2ms remaining: 8.27ms
38: learn: 0.2383129 total: 26.9ms remaining: 7.58ms
39: learn: 0.2367566 total: 27.6ms remaining: 6.91ms
40: learn: 0.2354297 total: 28ms remaining: 6.14ms
41: learn: 0.2339993 total: 28.4ms remaining: 5.41ms
42: learn: 0.2309912 total: 28.9ms remaining: 4.71ms
43: learn: 0.2284946 total: 29.4ms remaining: 4.01ms
44: learn: 0.2269643 total: 30ms remaining: 3.33ms
45: learn: 0.2244732 total: 30.5ms remaining: 2.65ms
46: learn: 0.2206652 total: 30.9ms remaining: 1.98ms
47: learn: 0.2180727 total: 31.4ms remaining: 1.31ms
48: learn: 0.2169677 total: 32ms remaining: 653us
49: learn: 0.2159717 total: 32.5ms remaining: 0us
Evaluating param combo 8/16: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.1, 'boosted_cb_model__max_depth': 6, 'boosted_cb_model__num_leaves': 16}
Duplicate found for this combo (hash=9b979e7549e9eb8a3b749597ec04d2e7). Removing old run: d0c7999e98534e4b917beb6720051b67
0: learn: 0.6460564 total: 551us remaining: 27ms
1: learn: 0.6115491 total: 1.72ms remaining: 41.4ms
2: learn: 0.5773897 total: 2.85ms remaining: 44.6ms
3: learn: 0.5478549 total: 3.5ms remaining: 40.2ms
4: learn: 0.5182940 total: 4.29ms remaining: 38.6ms
5: learn: 0.4881346 total: 5.08ms remaining: 37.3ms
6: learn: 0.4636411 total: 5.83ms remaining: 35.8ms
7: learn: 0.4414647 total: 6.51ms remaining: 34.2ms
8: learn: 0.4212306 total: 7.59ms remaining: 34.6ms
9: learn: 0.4061705 total: 8.72ms remaining: 34.9ms
10: learn: 0.3929099 total: 9.54ms remaining: 33.8ms
11: learn: 0.3795535 total: 10.2ms remaining: 32.3ms
12: learn: 0.3676262 total: 10.7ms remaining: 30.4ms
13: learn: 0.3571708 total: 11.3ms remaining: 29ms
14: learn: 0.3438041 total: 12ms remaining: 28ms
15: learn: 0.3339876 total: 12.8ms remaining: 27.3ms
16: learn: 0.3256147 total: 13.6ms remaining: 26.3ms
17: learn: 0.3237170 total: 13.7ms remaining: 24.4ms
18: learn: 0.3154076 total: 14.4ms remaining: 23.6ms
19: learn: 0.3071222 total: 14.9ms remaining: 22.3ms
20: learn: 0.2995403 total: 15.5ms remaining: 21.4ms
21: learn: 0.2945462 total: 15.7ms remaining: 20ms
22: learn: 0.2896742 total: 16.3ms remaining: 19.1ms
23: learn: 0.2845711 total: 16.9ms remaining: 18.3ms
24: learn: 0.2800764 total: 17.7ms remaining: 17.7ms
25: learn: 0.2747643 total: 18.2ms remaining: 16.8ms
26: learn: 0.2706259 total: 18.7ms remaining: 15.9ms
27: learn: 0.2647070 total: 19.3ms remaining: 15.2ms
28: learn: 0.2612732 total: 19.9ms remaining: 14.4ms
29: learn: 0.2569599 total: 20.5ms remaining: 13.7ms
30: learn: 0.2539537 total: 21.2ms remaining: 13ms
31: learn: 0.2499569 total: 21.8ms remaining: 12.2ms
32: learn: 0.2464926 total: 22.2ms remaining: 11.5ms
33: learn: 0.2425367 total: 23.1ms remaining: 10.9ms
34: learn: 0.2380646 total: 23.7ms remaining: 10.2ms
35: learn: 0.2359067 total: 24.2ms remaining: 9.39ms
36: learn: 0.2324772 total: 24.8ms remaining: 8.7ms
37: learn: 0.2296507 total: 25.4ms remaining: 8.02ms
38: learn: 0.2279374 total: 25.7ms remaining: 7.25ms
39: learn: 0.2260783 total: 26.5ms remaining: 6.63ms
40: learn: 0.2234158 total: 27ms remaining: 5.92ms
41: learn: 0.2204111 total: 27.7ms remaining: 5.27ms
42: learn: 0.2189710 total: 28.2ms remaining: 4.59ms
43: learn: 0.2167972 total: 28.7ms remaining: 3.92ms
44: learn: 0.2140913 total: 29.5ms remaining: 3.28ms
45: learn: 0.2114060 total: 30.2ms remaining: 2.62ms
46: learn: 0.2096511 total: 30.9ms remaining: 1.97ms
47: learn: 0.2066832 total: 31.5ms remaining: 1.31ms
48: learn: 0.2035646 total: 32.3ms remaining: 659us
49: learn: 0.2009341 total: 32.8ms remaining: 0us
0: learn: 0.6424726 total: 656us remaining: 32.2ms
1: learn: 0.5994503 total: 999us remaining: 24ms
2: learn: 0.5650641 total: 1.59ms remaining: 25ms
3: learn: 0.5330205 total: 1.72ms remaining: 19.8ms
4: learn: 0.5040646 total: 2.26ms remaining: 20.4ms
5: learn: 0.4756208 total: 2.64ms remaining: 19.4ms
6: learn: 0.4556726 total: 3.29ms remaining: 20.2ms
7: learn: 0.4312891 total: 3.83ms remaining: 20.1ms
8: learn: 0.4153181 total: 4.37ms remaining: 19.9ms
9: learn: 0.4013725 total: 5ms remaining: 20ms
10: learn: 0.3841636 total: 5.58ms remaining: 19.8ms
11: learn: 0.3713251 total: 5.77ms remaining: 18.3ms
12: learn: 0.3553684 total: 6.24ms remaining: 17.8ms
13: learn: 0.3468555 total: 6.83ms remaining: 17.6ms
14: learn: 0.3368721 total: 7.38ms remaining: 17.2ms
15: learn: 0.3250555 total: 7.96ms remaining: 16.9ms
16: learn: 0.3135326 total: 8.58ms remaining: 16.6ms
17: learn: 0.3068917 total: 8.85ms remaining: 15.7ms
18: learn: 0.2983682 total: 9.4ms remaining: 15.3ms
19: learn: 0.2903195 total: 10.1ms remaining: 15.2ms
20: learn: 0.2825822 total: 10.7ms remaining: 14.8ms
21: learn: 0.2786885 total: 11.1ms remaining: 14.2ms
22: learn: 0.2713611 total: 11.7ms remaining: 13.7ms
23: learn: 0.2655422 total: 12.3ms remaining: 13.3ms
24: learn: 0.2595446 total: 12.9ms remaining: 12.9ms
25: learn: 0.2553193 total: 13.5ms remaining: 12.5ms
26: learn: 0.2485124 total: 14.2ms remaining: 12.1ms
27: learn: 0.2449750 total: 14.7ms remaining: 11.6ms
28: learn: 0.2407880 total: 15.3ms remaining: 11.1ms
29: learn: 0.2379387 total: 15.8ms remaining: 10.6ms
30: learn: 0.2331728 total: 16.4ms remaining: 10ms
31: learn: 0.2301211 total: 16.6ms remaining: 9.31ms
32: learn: 0.2274073 total: 17.2ms remaining: 8.85ms
33: learn: 0.2239642 total: 17.8ms remaining: 8.4ms
34: learn: 0.2218696 total: 18.4ms remaining: 7.88ms
35: learn: 0.2190630 total: 19ms remaining: 7.39ms
36: learn: 0.2162074 total: 19.5ms remaining: 6.86ms
37: learn: 0.2142116 total: 20.2ms remaining: 6.38ms
38: learn: 0.2122932 total: 20.7ms remaining: 5.84ms
39: learn: 0.2092948 total: 21.4ms remaining: 5.34ms
40: learn: 0.2081077 total: 21.7ms remaining: 4.76ms
41: learn: 0.2036558 total: 22.2ms remaining: 4.24ms
42: learn: 0.2020587 total: 22.9ms remaining: 3.73ms
43: learn: 0.1991714 total: 23.6ms remaining: 3.22ms
44: learn: 0.1963465 total: 24.3ms remaining: 2.7ms
45: learn: 0.1949122 total: 24.9ms remaining: 2.16ms
46: learn: 0.1922773 total: 25.6ms remaining: 1.64ms
47: learn: 0.1906400 total: 26.2ms remaining: 1.09ms
48: learn: 0.1875899 total: 26.8ms remaining: 546us
49: learn: 0.1859991 total: 27.5ms remaining: 0us
0: learn: 0.6425807 total: 659us remaining: 32.3ms
1: learn: 0.6095934 total: 1.88ms remaining: 45.2ms
2: learn: 0.5724604 total: 3.1ms remaining: 48.6ms
3: learn: 0.5389820 total: 3.9ms remaining: 44.9ms
4: learn: 0.5086857 total: 4.95ms remaining: 44.5ms
5: learn: 0.4798514 total: 6.01ms remaining: 44.1ms
6: learn: 0.4553136 total: 7.16ms remaining: 44ms
7: learn: 0.4379000 total: 7.82ms remaining: 41.1ms
8: learn: 0.4197578 total: 8.91ms remaining: 40.6ms
9: learn: 0.4020864 total: 9.29ms remaining: 37.1ms
10: learn: 0.3874267 total: 10ms remaining: 35.6ms
11: learn: 0.3753395 total: 10.9ms remaining: 34.6ms
12: learn: 0.3618715 total: 12.1ms remaining: 34.4ms
13: learn: 0.3506784 total: 13.2ms remaining: 34ms
14: learn: 0.3434627 total: 14.1ms remaining: 32.9ms
15: learn: 0.3329389 total: 15.2ms remaining: 32.3ms
16: learn: 0.3242477 total: 16.1ms remaining: 31.3ms
17: learn: 0.3125269 total: 17ms remaining: 30.2ms
18: learn: 0.3053817 total: 17.3ms remaining: 28.2ms
19: learn: 0.2966711 total: 18.1ms remaining: 27.1ms
20: learn: 0.2902346 total: 18.8ms remaining: 26ms
21: learn: 0.2821835 total: 19.7ms remaining: 25.1ms
22: learn: 0.2768605 total: 20.5ms remaining: 24ms
23: learn: 0.2723296 total: 20.7ms remaining: 22.4ms
24: learn: 0.2672881 total: 21.5ms remaining: 21.5ms
25: learn: 0.2625101 total: 22.2ms remaining: 20.5ms
26: learn: 0.2578836 total: 22.6ms remaining: 19.3ms
27: learn: 0.2509535 total: 23.4ms remaining: 18.3ms
28: learn: 0.2465789 total: 24ms remaining: 17.4ms
29: learn: 0.2428655 total: 24.7ms remaining: 16.5ms
30: learn: 0.2391947 total: 25.5ms remaining: 15.6ms
31: learn: 0.2370659 total: 25.8ms remaining: 14.5ms
32: learn: 0.2336989 total: 26.5ms remaining: 13.7ms
33: learn: 0.2316409 total: 27.2ms remaining: 12.8ms
34: learn: 0.2276796 total: 28.1ms remaining: 12ms
35: learn: 0.2242121 total: 28.6ms remaining: 11.1ms
36: learn: 0.2216698 total: 29.3ms remaining: 10.3ms
37: learn: 0.2202014 total: 29.7ms remaining: 9.37ms
38: learn: 0.2165851 total: 30.3ms remaining: 8.56ms
39: learn: 0.2155305 total: 30.9ms remaining: 7.72ms
40: learn: 0.2132991 total: 31.5ms remaining: 6.92ms
41: learn: 0.2118226 total: 32.3ms remaining: 6.16ms
42: learn: 0.2098110 total: 33ms remaining: 5.37ms
43: learn: 0.2078951 total: 33.5ms remaining: 4.57ms
44: learn: 0.2053172 total: 34ms remaining: 3.78ms
45: learn: 0.2028354 total: 34.7ms remaining: 3.02ms
46: learn: 0.2013843 total: 35.3ms remaining: 2.25ms
47: learn: 0.1991441 total: 35.9ms remaining: 1.5ms
48: learn: 0.1973173 total: 36.4ms remaining: 743us
49: learn: 0.1958110 total: 37.1ms remaining: 0us
0: learn: 0.6414042 total: 587us remaining: 28.8ms
1: learn: 0.6036809 total: 1.25ms remaining: 29.9ms
2: learn: 0.5672952 total: 1.9ms remaining: 29.7ms
3: learn: 0.5305474 total: 2.47ms remaining: 28.4ms
4: learn: 0.5054898 total: 3.09ms remaining: 27.8ms
5: learn: 0.4765417 total: 3.61ms remaining: 26.5ms
6: learn: 0.4545655 total: 4.14ms remaining: 25.5ms
7: learn: 0.4373139 total: 4.59ms remaining: 24.1ms
8: learn: 0.4180403 total: 5.16ms remaining: 23.5ms
9: learn: 0.4024810 total: 5.77ms remaining: 23.1ms
10: learn: 0.3849705 total: 6.42ms remaining: 22.8ms
11: learn: 0.3727187 total: 7.17ms remaining: 22.7ms
12: learn: 0.3586289 total: 7.88ms remaining: 22.4ms
13: learn: 0.3477836 total: 8.49ms remaining: 21.8ms
14: learn: 0.3371383 total: 9.12ms remaining: 21.3ms
15: learn: 0.3260366 total: 9.36ms remaining: 19.9ms
16: learn: 0.3151335 total: 9.91ms remaining: 19.2ms
17: learn: 0.3072938 total: 10.6ms remaining: 18.8ms
18: learn: 0.3010589 total: 10.8ms remaining: 17.7ms
19: learn: 0.2941139 total: 11.4ms remaining: 17ms
20: learn: 0.2866631 total: 12ms remaining: 16.5ms
21: learn: 0.2785329 total: 12.7ms remaining: 16.2ms
22: learn: 0.2711571 total: 13.4ms remaining: 15.7ms
23: learn: 0.2662387 total: 14.1ms remaining: 15.3ms
24: learn: 0.2614794 total: 14.6ms remaining: 14.6ms
25: learn: 0.2561155 total: 15.2ms remaining: 14.1ms
26: learn: 0.2525781 total: 15.6ms remaining: 13.3ms
27: learn: 0.2485784 total: 16.2ms remaining: 12.7ms
28: learn: 0.2443970 total: 16.7ms remaining: 12.1ms
29: learn: 0.2414316 total: 17.3ms remaining: 11.5ms
30: learn: 0.2370679 total: 18.1ms remaining: 11.1ms
31: learn: 0.2333710 total: 18.7ms remaining: 10.5ms
32: learn: 0.2297558 total: 19.3ms remaining: 9.92ms
33: learn: 0.2275985 total: 19.5ms remaining: 9.2ms
34: learn: 0.2261549 total: 20ms remaining: 8.58ms
35: learn: 0.2232823 total: 20.5ms remaining: 7.99ms
36: learn: 0.2214664 total: 21ms remaining: 7.38ms
37: learn: 0.2187996 total: 21.3ms remaining: 6.74ms
38: learn: 0.2167631 total: 21.9ms remaining: 6.17ms
39: learn: 0.2150543 total: 22.4ms remaining: 5.59ms
40: learn: 0.2123694 total: 23.1ms remaining: 5.07ms
41: learn: 0.2092893 total: 23.8ms remaining: 4.54ms
42: learn: 0.2077938 total: 24.4ms remaining: 3.96ms
43: learn: 0.2051896 total: 24.9ms remaining: 3.39ms
44: learn: 0.2020838 total: 25.5ms remaining: 2.83ms
45: learn: 0.1996024 total: 26.1ms remaining: 2.27ms
46: learn: 0.1977749 total: 26.7ms remaining: 1.71ms
47: learn: 0.1958991 total: 27.4ms remaining: 1.14ms
48: learn: 0.1944225 total: 27.5ms remaining: 561us
49: learn: 0.1931922 total: 28.4ms remaining: 0us
0: learn: 0.6483277 total: 656us remaining: 32.2ms
1: learn: 0.6155300 total: 1.58ms remaining: 37.9ms
2: learn: 0.5801133 total: 2.57ms remaining: 40.3ms
3: learn: 0.5438671 total: 3.67ms remaining: 42.2ms
4: learn: 0.5153476 total: 4.33ms remaining: 39ms
5: learn: 0.4930093 total: 5.18ms remaining: 38ms
6: learn: 0.4716985 total: 6.01ms remaining: 36.9ms
7: learn: 0.4519960 total: 7.21ms remaining: 37.9ms
8: learn: 0.4380235 total: 8.2ms remaining: 37.3ms
9: learn: 0.4193870 total: 9.07ms remaining: 36.3ms
10: learn: 0.4050681 total: 9.42ms remaining: 33.4ms
11: learn: 0.3896630 total: 10.3ms remaining: 32.8ms
12: learn: 0.3783375 total: 11.3ms remaining: 32.3ms
13: learn: 0.3636949 total: 12.7ms remaining: 32.7ms
14: learn: 0.3514784 total: 13.6ms remaining: 31.7ms
15: learn: 0.3406462 total: 14.4ms remaining: 30.5ms
16: learn: 0.3328499 total: 15.1ms remaining: 29.3ms
17: learn: 0.3269265 total: 15.8ms remaining: 28.2ms
18: learn: 0.3204205 total: 16.9ms remaining: 27.5ms
19: learn: 0.3124206 total: 17.9ms remaining: 26.8ms
20: learn: 0.3037066 total: 18.7ms remaining: 25.9ms
21: learn: 0.2972371 total: 19.7ms remaining: 25ms
22: learn: 0.2900100 total: 20.4ms remaining: 24ms
23: learn: 0.2832710 total: 21ms remaining: 22.7ms
24: learn: 0.2786096 total: 21.9ms remaining: 21.9ms
25: learn: 0.2745835 total: 23ms remaining: 21.2ms
26: learn: 0.2714739 total: 23.8ms remaining: 20.3ms
27: learn: 0.2660829 total: 24.8ms remaining: 19.5ms
28: learn: 0.2649405 total: 25.3ms remaining: 18.3ms
29: learn: 0.2617376 total: 26ms remaining: 17.3ms
30: learn: 0.2580553 total: 27ms remaining: 16.6ms
31: learn: 0.2532095 total: 27.9ms remaining: 15.7ms
32: learn: 0.2507447 total: 28.2ms remaining: 14.5ms
33: learn: 0.2467854 total: 28.9ms remaining: 13.6ms
34: learn: 0.2443274 total: 29.2ms remaining: 12.5ms
35: learn: 0.2410986 total: 30.1ms remaining: 11.7ms
36: learn: 0.2381974 total: 30.8ms remaining: 10.8ms
37: learn: 0.2342700 total: 31.5ms remaining: 9.96ms
38: learn: 0.2309641 total: 32.3ms remaining: 9.1ms
39: learn: 0.2275861 total: 33.1ms remaining: 8.27ms
40: learn: 0.2227911 total: 33.8ms remaining: 7.43ms
41: learn: 0.2206702 total: 34.3ms remaining: 6.54ms
42: learn: 0.2178427 total: 35ms remaining: 5.7ms
43: learn: 0.2155070 total: 35.7ms remaining: 4.87ms
44: learn: 0.2124377 total: 36.5ms remaining: 4.05ms
45: learn: 0.2095209 total: 37.1ms remaining: 3.23ms
46: learn: 0.2068874 total: 38ms remaining: 2.43ms
47: learn: 0.2049574 total: 38.8ms remaining: 1.61ms
48: learn: 0.2033372 total: 39.6ms remaining: 807us
49: learn: 0.2018192 total: 40.3ms remaining: 0us
0: learn: 0.6478540 total: 872us remaining: 42.7ms
1: learn: 0.6188368 total: 1.57ms remaining: 37.7ms
2: learn: 0.5812869 total: 2.3ms remaining: 36ms
3: learn: 0.5494925 total: 2.8ms remaining: 32.2ms
4: learn: 0.5199929 total: 3.33ms remaining: 29.9ms
5: learn: 0.4941833 total: 3.99ms remaining: 29.3ms
6: learn: 0.4742569 total: 4.18ms remaining: 25.7ms
7: learn: 0.4530433 total: 4.83ms remaining: 25.4ms
8: learn: 0.4320795 total: 5.23ms remaining: 23.8ms
9: learn: 0.4138967 total: 5.89ms remaining: 23.6ms
10: learn: 0.3986699 total: 6.52ms remaining: 23.1ms
11: learn: 0.3873607 total: 7.12ms remaining: 22.6ms
12: learn: 0.3746075 total: 7.76ms remaining: 22.1ms
13: learn: 0.3635151 total: 8.4ms remaining: 21.6ms
14: learn: 0.3541650 total: 9.12ms remaining: 21.3ms
15: learn: 0.3423315 total: 9.77ms remaining: 20.8ms
16: learn: 0.3322791 total: 10.7ms remaining: 20.7ms
17: learn: 0.3245152 total: 11.5ms remaining: 20.4ms
18: learn: 0.3141369 total: 12.3ms remaining: 20ms
19: learn: 0.3077105 total: 13ms remaining: 19.5ms
20: learn: 0.2985603 total: 14ms remaining: 19.3ms
21: learn: 0.2914161 total: 14.8ms remaining: 18.8ms
22: learn: 0.2847772 total: 15.6ms remaining: 18.3ms
23: learn: 0.2771363 total: 16.3ms remaining: 17.7ms
24: learn: 0.2718045 total: 17ms remaining: 17ms
25: learn: 0.2658817 total: 17.9ms remaining: 16.5ms
26: learn: 0.2597141 total: 18.6ms remaining: 15.8ms
27: learn: 0.2534277 total: 19.3ms remaining: 15.1ms
28: learn: 0.2484247 total: 19.9ms remaining: 14.4ms
29: learn: 0.2437953 total: 20.6ms remaining: 13.8ms
30: learn: 0.2402339 total: 21.4ms remaining: 13.1ms
31: learn: 0.2374675 total: 22.2ms remaining: 12.5ms
32: learn: 0.2352614 total: 22.4ms remaining: 11.5ms
33: learn: 0.2301768 total: 23.1ms remaining: 10.9ms
34: learn: 0.2275918 total: 23.3ms remaining: 9.98ms
35: learn: 0.2240243 total: 23.9ms remaining: 9.31ms
36: learn: 0.2212741 total: 24.5ms remaining: 8.61ms
37: learn: 0.2188742 total: 25.1ms remaining: 7.93ms
38: learn: 0.2153598 total: 25.8ms remaining: 7.27ms
39: learn: 0.2115964 total: 26.2ms remaining: 6.56ms
40: learn: 0.2089791 total: 26.8ms remaining: 5.88ms
41: learn: 0.2068259 total: 27.3ms remaining: 5.2ms
42: learn: 0.2045457 total: 28ms remaining: 4.55ms
43: learn: 0.2025532 total: 28.5ms remaining: 3.89ms
44: learn: 0.1992884 total: 29.2ms remaining: 3.24ms
45: learn: 0.1963081 total: 29.7ms remaining: 2.58ms
46: learn: 0.1936464 total: 30.3ms remaining: 1.94ms
47: learn: 0.1918183 total: 31ms remaining: 1.29ms
48: learn: 0.1896215 total: 31.7ms remaining: 647us
49: learn: 0.1881047 total: 32.3ms remaining: 0us
0: learn: 0.6452232 total: 728us remaining: 35.7ms
1: learn: 0.6130854 total: 1.41ms remaining: 33.9ms
2: learn: 0.5785534 total: 2.13ms remaining: 33.4ms
3: learn: 0.5487896 total: 2.74ms remaining: 31.6ms
4: learn: 0.5235638 total: 3.46ms remaining: 31.1ms
5: learn: 0.4958000 total: 3.95ms remaining: 29ms
6: learn: 0.4707181 total: 4.57ms remaining: 28.1ms
7: learn: 0.4489693 total: 4.92ms remaining: 25.8ms
8: learn: 0.4264777 total: 5.29ms remaining: 24.1ms
9: learn: 0.4124217 total: 6.16ms remaining: 24.6ms
10: learn: 0.3945337 total: 6.8ms remaining: 24.1ms
11: learn: 0.3795706 total: 7.48ms remaining: 23.7ms
12: learn: 0.3660203 total: 8.04ms remaining: 22.9ms
13: learn: 0.3555878 total: 8.68ms remaining: 22.3ms
14: learn: 0.3450628 total: 9.24ms remaining: 21.6ms
15: learn: 0.3352606 total: 9.78ms remaining: 20.8ms
16: learn: 0.3276190 total: 10.5ms remaining: 20.3ms
17: learn: 0.3212609 total: 11.1ms remaining: 19.8ms
18: learn: 0.3123439 total: 11.8ms remaining: 19.2ms
19: learn: 0.3031837 total: 12.4ms remaining: 18.6ms
20: learn: 0.2938529 total: 13ms remaining: 18ms
21: learn: 0.2866061 total: 13.7ms remaining: 17.4ms
22: learn: 0.2828694 total: 14.3ms remaining: 16.8ms
23: learn: 0.2753056 total: 15ms remaining: 16.2ms
24: learn: 0.2711134 total: 15.6ms remaining: 15.6ms
25: learn: 0.2653193 total: 16.3ms remaining: 15ms
26: learn: 0.2603788 total: 17.1ms remaining: 14.6ms
27: learn: 0.2568278 total: 17.4ms remaining: 13.7ms
28: learn: 0.2506470 total: 18.1ms remaining: 13.1ms
29: learn: 0.2446159 total: 19.1ms remaining: 12.7ms
30: learn: 0.2402115 total: 19.7ms remaining: 12.1ms
31: learn: 0.2359320 total: 20.3ms remaining: 11.4ms
32: learn: 0.2322850 total: 20.8ms remaining: 10.7ms
33: learn: 0.2288307 total: 21.4ms remaining: 10.1ms
34: learn: 0.2263137 total: 22ms remaining: 9.42ms
35: learn: 0.2215138 total: 22.6ms remaining: 8.78ms
36: learn: 0.2196910 total: 23.3ms remaining: 8.19ms
37: learn: 0.2177473 total: 23.7ms remaining: 7.48ms
38: learn: 0.2123156 total: 24.2ms remaining: 6.83ms
39: learn: 0.2082721 total: 24.8ms remaining: 6.19ms
40: learn: 0.2048000 total: 25.4ms remaining: 5.57ms
41: learn: 0.2015699 total: 26ms remaining: 4.94ms
42: learn: 0.1971835 total: 26.5ms remaining: 4.32ms
43: learn: 0.1961963 total: 27ms remaining: 3.68ms
44: learn: 0.1947997 total: 27.6ms remaining: 3.06ms
45: learn: 0.1912582 total: 28.2ms remaining: 2.45ms
46: learn: 0.1887218 total: 28.9ms remaining: 1.84ms
47: learn: 0.1865246 total: 29.5ms remaining: 1.23ms
48: learn: 0.1839188 total: 30.1ms remaining: 614us
49: learn: 0.1812008 total: 30.7ms remaining: 0us
0: learn: 0.6482935 total: 520us remaining: 25.5ms
1: learn: 0.6097817 total: 1.05ms remaining: 25.3ms
2: learn: 0.5723488 total: 1.72ms remaining: 26.9ms
3: learn: 0.5391228 total: 2.29ms remaining: 26.4ms
4: learn: 0.5138453 total: 3.04ms remaining: 27.4ms
5: learn: 0.4910075 total: 3.76ms remaining: 27.6ms
6: learn: 0.4707257 total: 4.53ms remaining: 27.8ms
7: learn: 0.4519516 total: 5.03ms remaining: 26.4ms
8: learn: 0.4328789 total: 5.57ms remaining: 25.4ms
9: learn: 0.4164797 total: 6.14ms remaining: 24.5ms
10: learn: 0.3997077 total: 6.86ms remaining: 24.3ms
11: learn: 0.3883643 total: 7.16ms remaining: 22.7ms
12: learn: 0.3787946 total: 7.82ms remaining: 22.3ms
13: learn: 0.3660023 total: 8.33ms remaining: 21.4ms
14: learn: 0.3557177 total: 9.21ms remaining: 21.5ms
15: learn: 0.3464166 total: 9.98ms remaining: 21.2ms
16: learn: 0.3349490 total: 10.5ms remaining: 20.4ms
17: learn: 0.3258922 total: 11.3ms remaining: 20ms
18: learn: 0.3195433 total: 11.8ms remaining: 19.2ms
19: learn: 0.3100072 total: 12.2ms remaining: 18.4ms
20: learn: 0.3025717 total: 13.1ms remaining: 18ms
21: learn: 0.2956294 total: 13.5ms remaining: 17.2ms
22: learn: 0.2888096 total: 14ms remaining: 16.4ms
23: learn: 0.2839482 total: 14.5ms remaining: 15.7ms
24: learn: 0.2786366 total: 14.9ms remaining: 14.9ms
25: learn: 0.2740122 total: 15.3ms remaining: 14.1ms
26: learn: 0.2699392 total: 15.8ms remaining: 13.5ms
27: learn: 0.2675302 total: 16.5ms remaining: 12.9ms
28: learn: 0.2624445 total: 18.1ms remaining: 13.1ms
29: learn: 0.2568874 total: 19.9ms remaining: 13.3ms
30: learn: 0.2517272 total: 21.4ms remaining: 13.1ms
31: learn: 0.2486361 total: 22.7ms remaining: 12.8ms
32: learn: 0.2462152 total: 23.9ms remaining: 12.3ms
33: learn: 0.2436858 total: 25.5ms remaining: 12ms
34: learn: 0.2408060 total: 27ms remaining: 11.6ms
35: learn: 0.2372277 total: 28.7ms remaining: 11.2ms
36: learn: 0.2355722 total: 30.8ms remaining: 10.8ms
37: learn: 0.2322521 total: 33.2ms remaining: 10.5ms
38: learn: 0.2292490 total: 35.7ms remaining: 10.1ms
39: learn: 0.2252908 total: 38.7ms remaining: 9.67ms
40: learn: 0.2220657 total: 41.5ms remaining: 9.11ms
41: learn: 0.2207384 total: 43.7ms remaining: 8.33ms
42: learn: 0.2181120 total: 47.3ms remaining: 7.7ms
43: learn: 0.2155424 total: 51ms remaining: 6.96ms
44: learn: 0.2122211 total: 53.4ms remaining: 5.93ms
45: learn: 0.2101936 total: 56.3ms remaining: 4.89ms
46: learn: 0.2074187 total: 59ms remaining: 3.77ms
47: learn: 0.2059315 total: 61.8ms remaining: 2.58ms
48: learn: 0.2035496 total: 64.9ms remaining: 1.32ms
49: learn: 0.2002119 total: 67.7ms remaining: 0us
0: learn: 0.6424092 total: 2.4ms remaining: 117ms
1: learn: 0.6044739 total: 5.26ms remaining: 126ms
2: learn: 0.5658378 total: 7.09ms remaining: 111ms
3: learn: 0.5331348 total: 9.7ms remaining: 111ms
4: learn: 0.5020048 total: 11.9ms remaining: 107ms
5: learn: 0.4785496 total: 14.6ms remaining: 107ms
6: learn: 0.4548111 total: 17.2ms remaining: 106ms
7: learn: 0.4343862 total: 19.4ms remaining: 102ms
8: learn: 0.4164076 total: 21.8ms remaining: 99.3ms
9: learn: 0.3980669 total: 24.3ms remaining: 97.3ms
10: learn: 0.3842137 total: 27.4ms remaining: 97.1ms
11: learn: 0.3709775 total: 30ms remaining: 95.1ms
12: learn: 0.3556386 total: 32.8ms remaining: 93.4ms
13: learn: 0.3451018 total: 35.5ms remaining: 91.2ms
14: learn: 0.3348098 total: 38.5ms remaining: 89.8ms
15: learn: 0.3271334 total: 41.2ms remaining: 87.5ms
16: learn: 0.3190411 total: 44.1ms remaining: 85.6ms
17: learn: 0.3107571 total: 44.8ms remaining: 79.6ms
18: learn: 0.3020451 total: 46.9ms remaining: 76.5ms
19: learn: 0.2965826 total: 50.1ms remaining: 75.2ms
20: learn: 0.2903581 total: 53.7ms remaining: 74.1ms
21: learn: 0.2825763 total: 55.9ms remaining: 71.2ms
22: learn: 0.2779718 total: 57.9ms remaining: 68ms
23: learn: 0.2721132 total: 59.7ms remaining: 64.6ms
24: learn: 0.2690395 total: 61.5ms remaining: 61.5ms
25: learn: 0.2656442 total: 63.7ms remaining: 58.8ms
26: learn: 0.2595123 total: 66.1ms remaining: 56.3ms
27: learn: 0.2550912 total: 69.3ms remaining: 54.5ms
28: learn: 0.2509144 total: 72.1ms remaining: 52.2ms
29: learn: 0.2469739 total: 73.1ms remaining: 48.8ms
30: learn: 0.2433197 total: 75.2ms remaining: 46.1ms
31: learn: 0.2407384 total: 77.7ms remaining: 43.7ms
32: learn: 0.2381498 total: 78.8ms remaining: 40.6ms
33: learn: 0.2341570 total: 80.4ms remaining: 37.8ms
34: learn: 0.2301784 total: 82.2ms remaining: 35.2ms
35: learn: 0.2271857 total: 84.1ms remaining: 32.7ms
36: learn: 0.2245278 total: 86ms remaining: 30.2ms
37: learn: 0.2220755 total: 87.4ms remaining: 27.6ms
38: learn: 0.2187753 total: 89.4ms remaining: 25.2ms
39: learn: 0.2156415 total: 91.2ms remaining: 22.8ms
40: learn: 0.2132895 total: 92.8ms remaining: 20.4ms
41: learn: 0.2109095 total: 94.5ms remaining: 18ms
42: learn: 0.2088499 total: 95.8ms remaining: 15.6ms
43: learn: 0.2062905 total: 97.2ms remaining: 13.3ms
44: learn: 0.2043945 total: 98.7ms remaining: 11ms
45: learn: 0.2021090 total: 100ms remaining: 8.69ms
46: learn: 0.2012857 total: 101ms remaining: 6.45ms
47: learn: 0.1985814 total: 102ms remaining: 4.26ms
48: learn: 0.1967447 total: 103ms remaining: 2.11ms
49: learn: 0.1953459 total: 104ms remaining: 0us
0: learn: 0.6495789 total: 2.73ms remaining: 134ms
1: learn: 0.6178464 total: 5.06ms remaining: 121ms
2: learn: 0.5852578 total: 7.5ms remaining: 118ms
3: learn: 0.5533435 total: 9.95ms remaining: 114ms
4: learn: 0.5300644 total: 12.7ms remaining: 114ms
5: learn: 0.5104585 total: 14.4ms remaining: 106ms
6: learn: 0.4862911 total: 16.9ms remaining: 104ms
7: learn: 0.4666907 total: 18ms remaining: 94.5ms
8: learn: 0.4496751 total: 18.6ms remaining: 84.7ms
9: learn: 0.4323048 total: 20.8ms remaining: 83.3ms
10: learn: 0.4180174 total: 23.4ms remaining: 83.1ms
11: learn: 0.4056119 total: 25.1ms remaining: 79.3ms
12: learn: 0.3933982 total: 26.4ms remaining: 75.2ms
13: learn: 0.3860414 total: 29ms remaining: 74.7ms
14: learn: 0.3734601 total: 30.3ms remaining: 70.6ms
15: learn: 0.3647702 total: 33.3ms remaining: 70.8ms
16: learn: 0.3555684 total: 35.5ms remaining: 69ms
17: learn: 0.3451648 total: 38.3ms remaining: 68.1ms
18: learn: 0.3365500 total: 39.6ms remaining: 64.7ms
19: learn: 0.3307781 total: 42.2ms remaining: 63.3ms
20: learn: 0.3238906 total: 44.1ms remaining: 60.9ms
21: learn: 0.3172724 total: 46ms remaining: 58.5ms
22: learn: 0.3102313 total: 48.3ms remaining: 56.7ms
23: learn: 0.3047438 total: 50.1ms remaining: 54.3ms
24: learn: 0.2978030 total: 52.1ms remaining: 52.1ms
25: learn: 0.2927210 total: 53.8ms remaining: 49.6ms
26: learn: 0.2895587 total: 55.6ms remaining: 47.4ms
27: learn: 0.2840902 total: 57.9ms remaining: 45.5ms
28: learn: 0.2795832 total: 60.3ms remaining: 43.7ms
29: learn: 0.2755125 total: 61.2ms remaining: 40.8ms
30: learn: 0.2705809 total: 62.5ms remaining: 38.3ms
31: learn: 0.2674857 total: 63.9ms remaining: 35.9ms
32: learn: 0.2630010 total: 65.3ms remaining: 33.6ms
33: learn: 0.2567622 total: 66.7ms remaining: 31.4ms
34: learn: 0.2541048 total: 67.9ms remaining: 29.1ms
35: learn: 0.2515091 total: 69.1ms remaining: 26.9ms
36: learn: 0.2496420 total: 69.7ms remaining: 24.5ms
37: learn: 0.2462492 total: 70.7ms remaining: 22.3ms
38: learn: 0.2432209 total: 71.7ms remaining: 20.2ms
39: learn: 0.2400406 total: 72.6ms remaining: 18.2ms
40: learn: 0.2370158 total: 73.6ms remaining: 16.2ms
41: learn: 0.2339843 total: 74.6ms remaining: 14.2ms
42: learn: 0.2316020 total: 75.6ms remaining: 12.3ms
43: learn: 0.2285619 total: 76.7ms remaining: 10.5ms
44: learn: 0.2263588 total: 77.7ms remaining: 8.63ms
45: learn: 0.2230401 total: 78.6ms remaining: 6.83ms
46: learn: 0.2209444 total: 79.6ms remaining: 5.08ms
47: learn: 0.2184339 total: 80.6ms remaining: 3.36ms
48: learn: 0.2155951 total: 81.6ms remaining: 1.67ms
49: learn: 0.2125427 total: 82.8ms remaining: 0us
0: learn: 0.6534941 total: 3.14ms remaining: 154ms
1: learn: 0.6122449 total: 5.63ms remaining: 135ms
2: learn: 0.5769827 total: 6.71ms remaining: 105ms
3: learn: 0.5450784 total: 7.72ms remaining: 88.8ms
4: learn: 0.5199399 total: 8.61ms remaining: 77.5ms
5: learn: 0.4947532 total: 9.66ms remaining: 70.9ms
6: learn: 0.4703112 total: 10.5ms remaining: 64.7ms
7: learn: 0.4494416 total: 14.1ms remaining: 74ms
8: learn: 0.4298333 total: 18.6ms remaining: 84.5ms
9: learn: 0.4160616 total: 22.4ms remaining: 89.6ms
10: learn: 0.4025642 total: 26.9ms remaining: 95.3ms
11: learn: 0.3853415 total: 31.3ms remaining: 99.2ms
12: learn: 0.3727685 total: 35.2ms remaining: 100ms
13: learn: 0.3608699 total: 39.1ms remaining: 101ms
14: learn: 0.3496626 total: 42.1ms remaining: 98.3ms
15: learn: 0.3392487 total: 44.9ms remaining: 95.3ms
16: learn: 0.3287677 total: 48.4ms remaining: 94ms
17: learn: 0.3236872 total: 51.6ms remaining: 91.8ms
18: learn: 0.3140516 total: 55.2ms remaining: 90.1ms
19: learn: 0.3051473 total: 58.3ms remaining: 87.5ms
20: learn: 0.2994371 total: 61.1ms remaining: 84.3ms
21: learn: 0.2929577 total: 65ms remaining: 82.7ms
22: learn: 0.2855741 total: 67.7ms remaining: 79.5ms
23: learn: 0.2793941 total: 70.5ms remaining: 76.4ms
24: learn: 0.2750060 total: 73.3ms remaining: 73.3ms
25: learn: 0.2691104 total: 76.3ms remaining: 70.4ms
26: learn: 0.2622555 total: 78.7ms remaining: 67ms
27: learn: 0.2564873 total: 82ms remaining: 64.5ms
28: learn: 0.2530226 total: 85.2ms remaining: 61.7ms
29: learn: 0.2480208 total: 87.3ms remaining: 58.2ms
30: learn: 0.2438530 total: 89.6ms remaining: 54.9ms
31: learn: 0.2406571 total: 92.2ms remaining: 51.8ms
32: learn: 0.2361800 total: 94.1ms remaining: 48.5ms
33: learn: 0.2335486 total: 96.9ms remaining: 45.6ms
34: learn: 0.2296192 total: 99.6ms remaining: 42.7ms
35: learn: 0.2280074 total: 101ms remaining: 39.4ms
36: learn: 0.2268101 total: 103ms remaining: 36.2ms
37: learn: 0.2246386 total: 106ms remaining: 33.5ms
38: learn: 0.2218528 total: 108ms remaining: 30.5ms
39: learn: 0.2191333 total: 110ms remaining: 27.6ms
40: learn: 0.2161464 total: 112ms remaining: 24.5ms
41: learn: 0.2136653 total: 114ms remaining: 21.7ms
42: learn: 0.2110949 total: 115ms remaining: 18.7ms
43: learn: 0.2093502 total: 117ms remaining: 15.9ms
44: learn: 0.2073207 total: 118ms remaining: 13.1ms
45: learn: 0.2050560 total: 119ms remaining: 10.4ms
46: learn: 0.2020952 total: 121ms remaining: 7.71ms
47: learn: 0.2004267 total: 122ms remaining: 5.09ms
48: learn: 0.1976483 total: 123ms remaining: 2.52ms
49: learn: 0.1969053 total: 125ms remaining: 0us
0: learn: 0.6476609 total: 2.59ms remaining: 127ms
1: learn: 0.6078913 total: 4.67ms remaining: 112ms
2: learn: 0.5838627 total: 6.91ms remaining: 108ms
3: learn: 0.5587682 total: 10.5ms remaining: 121ms
4: learn: 0.5268380 total: 13ms remaining: 117ms
5: learn: 0.5053589 total: 15.9ms remaining: 116ms
6: learn: 0.4802092 total: 18.4ms remaining: 113ms
7: learn: 0.4607218 total: 21.6ms remaining: 113ms
8: learn: 0.4412163 total: 23ms remaining: 105ms
9: learn: 0.4208723 total: 25.3ms remaining: 101ms
10: learn: 0.4055317 total: 26ms remaining: 92.3ms
11: learn: 0.3942299 total: 28.8ms remaining: 91.1ms
12: learn: 0.3861527 total: 31.9ms remaining: 90.7ms
13: learn: 0.3773984 total: 34.6ms remaining: 88.8ms
14: learn: 0.3650244 total: 38.1ms remaining: 88.8ms
15: learn: 0.3543091 total: 40.9ms remaining: 87ms
16: learn: 0.3475590 total: 42.2ms remaining: 81.9ms
17: learn: 0.3398008 total: 43.5ms remaining: 77.4ms
18: learn: 0.3337312 total: 45.9ms remaining: 75ms
19: learn: 0.3257317 total: 48.3ms remaining: 72.5ms
20: learn: 0.3176647 total: 50.4ms remaining: 69.6ms
21: learn: 0.3113245 total: 52.4ms remaining: 66.7ms
22: learn: 0.3060822 total: 54ms remaining: 63.4ms
23: learn: 0.3013323 total: 54.6ms remaining: 59.1ms
24: learn: 0.2961287 total: 56.8ms remaining: 56.8ms
25: learn: 0.2900945 total: 58.1ms remaining: 53.7ms
26: learn: 0.2839752 total: 59.5ms remaining: 50.7ms
27: learn: 0.2776371 total: 61.3ms remaining: 48.1ms
28: learn: 0.2747837 total: 61.8ms remaining: 44.7ms
29: learn: 0.2714390 total: 63.4ms remaining: 42.3ms
30: learn: 0.2677248 total: 64ms remaining: 39.2ms
31: learn: 0.2649790 total: 65.5ms remaining: 36.8ms
32: learn: 0.2620417 total: 66.8ms remaining: 34.4ms
33: learn: 0.2575582 total: 68.3ms remaining: 32.2ms
34: learn: 0.2541548 total: 69.7ms remaining: 29.9ms
35: learn: 0.2520526 total: 70.7ms remaining: 27.5ms
36: learn: 0.2479510 total: 71.8ms remaining: 25.2ms
37: learn: 0.2449521 total: 72.9ms remaining: 23ms
38: learn: 0.2421768 total: 74ms remaining: 20.9ms
39: learn: 0.2390914 total: 75ms remaining: 18.8ms
40: learn: 0.2378562 total: 75.9ms remaining: 16.7ms
41: learn: 0.2356405 total: 76.7ms remaining: 14.6ms
42: learn: 0.2335215 total: 78ms remaining: 12.7ms
43: learn: 0.2311720 total: 79.2ms remaining: 10.8ms
44: learn: 0.2283697 total: 80.3ms remaining: 8.92ms
45: learn: 0.2243870 total: 81.3ms remaining: 7.07ms
46: learn: 0.2211732 total: 82.1ms remaining: 5.24ms
47: learn: 0.2177466 total: 83ms remaining: 3.46ms
48: learn: 0.2156051 total: 83.5ms remaining: 1.7ms
49: learn: 0.2122104 total: 84.5ms remaining: 0us
0: learn: 0.6476069 total: 2.29ms remaining: 112ms
1: learn: 0.6099705 total: 5.08ms remaining: 122ms
2: learn: 0.5765838 total: 7.91ms remaining: 124ms
3: learn: 0.5427603 total: 10.4ms remaining: 120ms
4: learn: 0.5151544 total: 13.8ms remaining: 124ms
5: learn: 0.4924592 total: 16.6ms remaining: 122ms
6: learn: 0.4694414 total: 19.1ms remaining: 118ms
7: learn: 0.4535522 total: 22.4ms remaining: 118ms
8: learn: 0.4367589 total: 25.1ms remaining: 114ms
9: learn: 0.4192366 total: 29.2ms remaining: 117ms
10: learn: 0.4047310 total: 30.4ms remaining: 108ms
11: learn: 0.3939310 total: 32.6ms remaining: 103ms
12: learn: 0.3829631 total: 34.9ms remaining: 99.4ms
13: learn: 0.3716386 total: 38ms remaining: 97.8ms
14: learn: 0.3640746 total: 40.4ms remaining: 94.2ms
15: learn: 0.3538183 total: 43ms remaining: 91.3ms
16: learn: 0.3456950 total: 45.5ms remaining: 88.3ms
17: learn: 0.3368499 total: 48.1ms remaining: 85.6ms
18: learn: 0.3281077 total: 51.5ms remaining: 84.1ms
19: learn: 0.3206841 total: 53.5ms remaining: 80.3ms
20: learn: 0.3135813 total: 55.8ms remaining: 77.1ms
21: learn: 0.3073593 total: 58.2ms remaining: 74.1ms
22: learn: 0.3027511 total: 60.4ms remaining: 70.9ms
23: learn: 0.2978065 total: 62.8ms remaining: 68ms
24: learn: 0.2951878 total: 64.1ms remaining: 64.1ms
25: learn: 0.2918457 total: 65.7ms remaining: 60.6ms
26: learn: 0.2893576 total: 66.3ms remaining: 56.5ms
27: learn: 0.2841110 total: 67.7ms remaining: 53.2ms
28: learn: 0.2800618 total: 68.9ms remaining: 49.9ms
29: learn: 0.2749841 total: 70.7ms remaining: 47.2ms
30: learn: 0.2729425 total: 71.9ms remaining: 44ms
31: learn: 0.2681197 total: 73ms remaining: 41ms
32: learn: 0.2627012 total: 74.4ms remaining: 38.3ms
33: learn: 0.2591936 total: 75.5ms remaining: 35.5ms
34: learn: 0.2564770 total: 76.7ms remaining: 32.9ms
35: learn: 0.2527661 total: 77.6ms remaining: 30.2ms
36: learn: 0.2477277 total: 78.6ms remaining: 27.6ms
37: learn: 0.2461602 total: 79.8ms remaining: 25.2ms
38: learn: 0.2418578 total: 80.7ms remaining: 22.8ms
39: learn: 0.2390617 total: 81.9ms remaining: 20.5ms
40: learn: 0.2338773 total: 82.9ms remaining: 18.2ms
41: learn: 0.2302547 total: 83.9ms remaining: 16ms
42: learn: 0.2288855 total: 84.9ms remaining: 13.8ms
43: learn: 0.2262853 total: 85.8ms remaining: 11.7ms
44: learn: 0.2236002 total: 86.9ms remaining: 9.65ms
45: learn: 0.2200589 total: 87.8ms remaining: 7.64ms
46: learn: 0.2176929 total: 88.8ms remaining: 5.67ms
47: learn: 0.2143357 total: 89.7ms remaining: 3.74ms
48: learn: 0.2130031 total: 90.7ms remaining: 1.85ms
49: learn: 0.2103242 total: 91.1ms remaining: 0us
0: learn: 0.6419363 total: 2.41ms remaining: 118ms
1: learn: 0.6011176 total: 4.1ms remaining: 98.5ms
2: learn: 0.5650957 total: 6.19ms remaining: 96.9ms
3: learn: 0.5351090 total: 7.93ms remaining: 91.2ms
4: learn: 0.5040384 total: 9.58ms remaining: 86.2ms
5: learn: 0.4787169 total: 11.5ms remaining: 84.2ms
6: learn: 0.4553862 total: 12.9ms remaining: 79.3ms
7: learn: 0.4341118 total: 14.7ms remaining: 77.3ms
8: learn: 0.4170355 total: 15.9ms remaining: 72.4ms
9: learn: 0.4032727 total: 17.3ms remaining: 69.3ms
10: learn: 0.3912721 total: 19.1ms remaining: 67.9ms
11: learn: 0.3798536 total: 21ms remaining: 66.4ms
12: learn: 0.3686481 total: 22.7ms remaining: 64.7ms
13: learn: 0.3550926 total: 23.9ms remaining: 61.4ms
14: learn: 0.3444844 total: 25.2ms remaining: 58.7ms
15: learn: 0.3355957 total: 26.3ms remaining: 55.9ms
16: learn: 0.3291119 total: 27.4ms remaining: 53.1ms
17: learn: 0.3197235 total: 28.7ms remaining: 51ms
18: learn: 0.3112880 total: 29.7ms remaining: 48.5ms
19: learn: 0.3032755 total: 31ms remaining: 46.5ms
20: learn: 0.2962665 total: 32ms remaining: 44.2ms
21: learn: 0.2890773 total: 33.2ms remaining: 42.3ms
22: learn: 0.2838434 total: 34ms remaining: 40ms
23: learn: 0.2781875 total: 35.1ms remaining: 38ms
24: learn: 0.2731688 total: 36ms remaining: 36ms
25: learn: 0.2682044 total: 37.2ms remaining: 34.3ms
26: learn: 0.2636309 total: 37.9ms remaining: 32.3ms
27: learn: 0.2581415 total: 38.8ms remaining: 30.5ms
28: learn: 0.2540585 total: 39.9ms remaining: 28.9ms
29: learn: 0.2496847 total: 40.6ms remaining: 27.1ms
30: learn: 0.2484047 total: 41ms remaining: 25.1ms
31: learn: 0.2441142 total: 42ms remaining: 23.6ms
32: learn: 0.2380824 total: 43.1ms remaining: 22.2ms
33: learn: 0.2340262 total: 44ms remaining: 20.7ms
34: learn: 0.2297177 total: 44.7ms remaining: 19.2ms
35: learn: 0.2263603 total: 45.5ms remaining: 17.7ms
36: learn: 0.2233339 total: 46.7ms remaining: 16.4ms
37: learn: 0.2204648 total: 48ms remaining: 15.2ms
38: learn: 0.2164013 total: 48.8ms remaining: 13.8ms
39: learn: 0.2139482 total: 49.7ms remaining: 12.4ms
40: learn: 0.2108738 total: 50.8ms remaining: 11.1ms
41: learn: 0.2083536 total: 52.2ms remaining: 9.95ms
42: learn: 0.2045092 total: 53.3ms remaining: 8.68ms
43: learn: 0.2032248 total: 54.3ms remaining: 7.41ms
44: learn: 0.2018938 total: 55ms remaining: 6.11ms
45: learn: 0.1991045 total: 56.3ms remaining: 4.89ms
46: learn: 0.1973864 total: 57ms remaining: 3.64ms
47: learn: 0.1955079 total: 58ms remaining: 2.41ms
48: learn: 0.1931020 total: 58.9ms remaining: 1.2ms
49: learn: 0.1912296 total: 59.9ms remaining: 0us
0: learn: 0.6458911 total: 2.97ms remaining: 146ms
1: learn: 0.6098312 total: 4.17ms remaining: 100ms
2: learn: 0.5719195 total: 6.39ms remaining: 100ms
3: learn: 0.5372780 total: 6.93ms remaining: 79.7ms
4: learn: 0.5094653 total: 8.42ms remaining: 75.8ms
5: learn: 0.4869324 total: 10.6ms remaining: 77.9ms
6: learn: 0.4639285 total: 12.9ms remaining: 79ms
7: learn: 0.4424897 total: 14.9ms remaining: 78.2ms
8: learn: 0.4271859 total: 16.3ms remaining: 74.2ms
9: learn: 0.4113984 total: 18.1ms remaining: 72.4ms
10: learn: 0.3951133 total: 20.1ms remaining: 71.3ms
11: learn: 0.3832256 total: 22.3ms remaining: 70.6ms
12: learn: 0.3720227 total: 23.7ms remaining: 67.4ms
13: learn: 0.3609613 total: 25.2ms remaining: 64.9ms
14: learn: 0.3515607 total: 27.1ms remaining: 63.3ms
15: learn: 0.3432732 total: 28.6ms remaining: 60.7ms
16: learn: 0.3364668 total: 29.4ms remaining: 57.1ms
17: learn: 0.3283772 total: 30.5ms remaining: 54.3ms
18: learn: 0.3221569 total: 32ms remaining: 52.2ms
19: learn: 0.3176788 total: 33.2ms remaining: 49.9ms
20: learn: 0.3096443 total: 34.6ms remaining: 47.7ms
21: learn: 0.3010229 total: 35.8ms remaining: 45.6ms
22: learn: 0.2947529 total: 37.1ms remaining: 43.6ms
23: learn: 0.2893152 total: 38.6ms remaining: 41.8ms
24: learn: 0.2865792 total: 39.6ms remaining: 39.6ms
25: learn: 0.2797334 total: 40.8ms remaining: 37.7ms
26: learn: 0.2745505 total: 42ms remaining: 35.8ms
27: learn: 0.2689394 total: 43.1ms remaining: 33.9ms
28: learn: 0.2629653 total: 44.2ms remaining: 32ms
29: learn: 0.2583618 total: 45.3ms remaining: 30.2ms
30: learn: 0.2532520 total: 46.2ms remaining: 28.3ms
31: learn: 0.2483569 total: 47.1ms remaining: 26.5ms
32: learn: 0.2449190 total: 48ms remaining: 24.7ms
33: learn: 0.2427465 total: 48.6ms remaining: 22.9ms
34: learn: 0.2399617 total: 49.5ms remaining: 21.2ms
35: learn: 0.2367692 total: 50.4ms remaining: 19.6ms
36: learn: 0.2343335 total: 51.4ms remaining: 18.1ms
37: learn: 0.2318076 total: 52.6ms remaining: 16.6ms
38: learn: 0.2295396 total: 53.4ms remaining: 15.1ms
39: learn: 0.2278173 total: 54.4ms remaining: 13.6ms
40: learn: 0.2238035 total: 55ms remaining: 12.1ms
41: learn: 0.2214597 total: 55.9ms remaining: 10.7ms
42: learn: 0.2183419 total: 56.8ms remaining: 9.25ms
43: learn: 0.2163152 total: 57.8ms remaining: 7.88ms
44: learn: 0.2142766 total: 58.7ms remaining: 6.52ms
45: learn: 0.2114595 total: 59.6ms remaining: 5.19ms
46: learn: 0.2094141 total: 60.6ms remaining: 3.87ms
47: learn: 0.2076379 total: 61.1ms remaining: 2.54ms
48: learn: 0.2062048 total: 62.1ms remaining: 1.27ms
49: learn: 0.2037692 total: 63ms remaining: 0us
0: learn: 0.6449164 total: 2.1ms remaining: 103ms
1: learn: 0.6085246 total: 4.48ms remaining: 108ms
2: learn: 0.5715641 total: 6.06ms remaining: 95ms
3: learn: 0.5404080 total: 7.41ms remaining: 85.3ms
4: learn: 0.5092282 total: 8.51ms remaining: 76.6ms
5: learn: 0.4858956 total: 10.2ms remaining: 74.5ms
6: learn: 0.4624937 total: 11.5ms remaining: 70.9ms
7: learn: 0.4465534 total: 13.5ms remaining: 71ms
8: learn: 0.4296513 total: 15.5ms remaining: 70.6ms
9: learn: 0.4162316 total: 16.5ms remaining: 65.9ms
10: learn: 0.4039378 total: 18.2ms remaining: 64.6ms
11: learn: 0.3929279 total: 19.5ms remaining: 61.9ms
12: learn: 0.3783309 total: 21.1ms remaining: 59.9ms
13: learn: 0.3689193 total: 21.8ms remaining: 56.2ms
14: learn: 0.3599247 total: 22.6ms remaining: 52.8ms
15: learn: 0.3482234 total: 23.6ms remaining: 50.1ms
16: learn: 0.3409746 total: 24.6ms remaining: 47.8ms
17: learn: 0.3310210 total: 26ms remaining: 46.2ms
18: learn: 0.3223923 total: 27ms remaining: 44.1ms
19: learn: 0.3150966 total: 28.2ms remaining: 42.3ms
20: learn: 0.3096095 total: 29.6ms remaining: 40.9ms
21: learn: 0.3043243 total: 30.8ms remaining: 39.2ms
22: learn: 0.2982197 total: 31.6ms remaining: 37.1ms
23: learn: 0.2937462 total: 32.6ms remaining: 35.3ms
24: learn: 0.2894376 total: 33.8ms remaining: 33.8ms
25: learn: 0.2841506 total: 34.8ms remaining: 32.2ms
26: learn: 0.2805311 total: 35.8ms remaining: 30.5ms
27: learn: 0.2743572 total: 37ms remaining: 29.1ms
28: learn: 0.2709245 total: 37.8ms remaining: 27.4ms
29: learn: 0.2672337 total: 39ms remaining: 26ms
30: learn: 0.2644080 total: 40ms remaining: 24.5ms
31: learn: 0.2585723 total: 40.9ms remaining: 23ms
32: learn: 0.2560024 total: 41.9ms remaining: 21.6ms
33: learn: 0.2526009 total: 42.7ms remaining: 20.1ms
34: learn: 0.2492032 total: 43.7ms remaining: 18.7ms
35: learn: 0.2470315 total: 44.3ms remaining: 17.2ms
36: learn: 0.2431673 total: 45.3ms remaining: 15.9ms
37: learn: 0.2408352 total: 46.2ms remaining: 14.6ms
38: learn: 0.2378579 total: 47.1ms remaining: 13.3ms
39: learn: 0.2358564 total: 47.9ms remaining: 12ms
40: learn: 0.2325985 total: 49ms remaining: 10.7ms
41: learn: 0.2308968 total: 49.7ms remaining: 9.47ms
42: learn: 0.2281575 total: 50.6ms remaining: 8.24ms
43: learn: 0.2270003 total: 51.4ms remaining: 7.01ms
44: learn: 0.2243739 total: 52.4ms remaining: 5.82ms
45: learn: 0.2216083 total: 53.3ms remaining: 4.63ms
46: learn: 0.2184681 total: 54.1ms remaining: 3.45ms
47: learn: 0.2168333 total: 54.8ms remaining: 2.28ms
48: learn: 0.2134635 total: 55.9ms remaining: 1.14ms
49: learn: 0.2105444 total: 57.6ms remaining: 0us
0: learn: 0.6440574 total: 2.59ms remaining: 127ms
1: learn: 0.6071082 total: 5.39ms remaining: 129ms
2: learn: 0.5809191 total: 8.92ms remaining: 140ms
3: learn: 0.5513366 total: 11.1ms remaining: 128ms
4: learn: 0.5225763 total: 13.6ms remaining: 122ms
5: learn: 0.5004805 total: 15.6ms remaining: 115ms
6: learn: 0.4775513 total: 18.3ms remaining: 113ms
7: learn: 0.4622070 total: 19.6ms remaining: 103ms
8: learn: 0.4410241 total: 21.4ms remaining: 97.3ms
9: learn: 0.4249438 total: 23.2ms remaining: 92.8ms
10: learn: 0.4078417 total: 25.8ms remaining: 91.4ms
11: learn: 0.3959296 total: 28.3ms remaining: 89.6ms
12: learn: 0.3861366 total: 29.7ms remaining: 84.4ms
13: learn: 0.3747309 total: 31ms remaining: 79.8ms
14: learn: 0.3643013 total: 33.1ms remaining: 77.3ms
15: learn: 0.3549483 total: 34.8ms remaining: 74ms
16: learn: 0.3464547 total: 36.4ms remaining: 70.7ms
17: learn: 0.3386374 total: 37.4ms remaining: 66.6ms
18: learn: 0.3312841 total: 38.6ms remaining: 62.9ms
19: learn: 0.3235724 total: 39.9ms remaining: 59.8ms
20: learn: 0.3169947 total: 41.1ms remaining: 56.8ms
21: learn: 0.3096418 total: 42.2ms remaining: 53.8ms
22: learn: 0.3042768 total: 43.1ms remaining: 50.6ms
23: learn: 0.2978684 total: 43.9ms remaining: 47.6ms
24: learn: 0.2933065 total: 45.5ms remaining: 45.5ms
25: learn: 0.2842634 total: 46.3ms remaining: 42.8ms
26: learn: 0.2772658 total: 47.4ms remaining: 40.4ms
27: learn: 0.2736495 total: 48.4ms remaining: 38ms
28: learn: 0.2698878 total: 49.4ms remaining: 35.8ms
29: learn: 0.2629201 total: 50.3ms remaining: 33.6ms
30: learn: 0.2585210 total: 51.4ms remaining: 31.5ms
31: learn: 0.2557255 total: 52.2ms remaining: 29.3ms
32: learn: 0.2529692 total: 53.4ms remaining: 27.5ms
33: learn: 0.2489882 total: 54.3ms remaining: 25.6ms
34: learn: 0.2452763 total: 55.2ms remaining: 23.7ms
35: learn: 0.2415789 total: 55.8ms remaining: 21.7ms
36: learn: 0.2387921 total: 56.8ms remaining: 19.9ms
37: learn: 0.2351807 total: 57.7ms remaining: 18.2ms
38: learn: 0.2320092 total: 59.2ms remaining: 16.7ms
39: learn: 0.2298565 total: 60.1ms remaining: 15ms
40: learn: 0.2263351 total: 61ms remaining: 13.4ms
41: learn: 0.2238749 total: 61.8ms remaining: 11.8ms
42: learn: 0.2210930 total: 62.8ms remaining: 10.2ms
43: learn: 0.2183586 total: 63.7ms remaining: 8.68ms
44: learn: 0.2160132 total: 64.7ms remaining: 7.19ms
45: learn: 0.2131334 total: 65.4ms remaining: 5.69ms
46: learn: 0.2119702 total: 65.8ms remaining: 4.2ms
47: learn: 0.2102321 total: 66.7ms remaining: 2.78ms
48: learn: 0.2069067 total: 67.7ms remaining: 1.38ms
49: learn: 0.2032386 total: 68.6ms remaining: 0us
0: learn: 0.6461176 total: 2.21ms remaining: 108ms
1: learn: 0.6074389 total: 4.01ms remaining: 96.3ms
2: learn: 0.5663744 total: 6.73ms remaining: 105ms
3: learn: 0.5338644 total: 8.69ms remaining: 99.9ms
4: learn: 0.5036071 total: 11.6ms remaining: 104ms
5: learn: 0.4810570 total: 14.5ms remaining: 106ms
6: learn: 0.4610225 total: 16.9ms remaining: 104ms
7: learn: 0.4382829 total: 19ms remaining: 99.8ms
8: learn: 0.4184979 total: 20.6ms remaining: 93.9ms
9: learn: 0.4040368 total: 23.7ms remaining: 94.7ms
10: learn: 0.3879856 total: 26.5ms remaining: 93.9ms
11: learn: 0.3755899 total: 27.4ms remaining: 86.8ms
12: learn: 0.3652345 total: 29.2ms remaining: 83ms
13: learn: 0.3522070 total: 31.3ms remaining: 80.5ms
14: learn: 0.3402084 total: 34.6ms remaining: 80.6ms
15: learn: 0.3334455 total: 37.9ms remaining: 80.6ms
16: learn: 0.3269060 total: 40.4ms remaining: 78.4ms
17: learn: 0.3184363 total: 41.5ms remaining: 73.8ms
18: learn: 0.3122238 total: 43.4ms remaining: 70.9ms
19: learn: 0.3061585 total: 44.6ms remaining: 66.9ms
20: learn: 0.2989587 total: 47.3ms remaining: 65.3ms
21: learn: 0.2926460 total: 49.6ms remaining: 63.2ms
22: learn: 0.2872523 total: 51.4ms remaining: 60.4ms
23: learn: 0.2818606 total: 53.1ms remaining: 57.5ms
24: learn: 0.2739065 total: 54.6ms remaining: 54.6ms
25: learn: 0.2690171 total: 56.8ms remaining: 52.4ms
26: learn: 0.2649473 total: 58ms remaining: 49.4ms
27: learn: 0.2615369 total: 59.6ms remaining: 46.9ms
28: learn: 0.2582622 total: 60.8ms remaining: 44.1ms
29: learn: 0.2524443 total: 62.2ms remaining: 41.5ms
30: learn: 0.2490535 total: 63.8ms remaining: 39.1ms
31: learn: 0.2465288 total: 65.2ms remaining: 36.7ms
32: learn: 0.2423547 total: 66ms remaining: 34ms
33: learn: 0.2383376 total: 67.4ms remaining: 31.7ms
34: learn: 0.2348481 total: 68.3ms remaining: 29.3ms
35: learn: 0.2314557 total: 69.3ms remaining: 27ms
36: learn: 0.2293154 total: 70ms remaining: 24.6ms
37: learn: 0.2269631 total: 70.8ms remaining: 22.3ms
38: learn: 0.2233239 total: 72ms remaining: 20.3ms
39: learn: 0.2219447 total: 73.4ms remaining: 18.4ms
40: learn: 0.2197608 total: 74.3ms remaining: 16.3ms
41: learn: 0.2179991 total: 75.2ms remaining: 14.3ms
42: learn: 0.2153602 total: 76.2ms remaining: 12.4ms
43: learn: 0.2128584 total: 77.3ms remaining: 10.5ms
44: learn: 0.2110075 total: 78.2ms remaining: 8.69ms
45: learn: 0.2076444 total: 79.1ms remaining: 6.88ms
46: learn: 0.2059115 total: 79.8ms remaining: 5.09ms
47: learn: 0.2039299 total: 80.8ms remaining: 3.37ms
48: learn: 0.2021505 total: 81.8ms remaining: 1.67ms
49: learn: 0.1995858 total: 82.9ms remaining: 0us
0: learn: 0.6584029 total: 2.93ms remaining: 143ms
1: learn: 0.6158136 total: 5.32ms remaining: 128ms
2: learn: 0.5789053 total: 7.42ms remaining: 116ms
3: learn: 0.5476814 total: 10.4ms remaining: 120ms
4: learn: 0.5177018 total: 13ms remaining: 117ms
5: learn: 0.4899133 total: 15.7ms remaining: 115ms
6: learn: 0.4664993 total: 18.6ms remaining: 114ms
7: learn: 0.4507562 total: 21.2ms remaining: 112ms
8: learn: 0.4301009 total: 24.2ms remaining: 110ms
9: learn: 0.4091667 total: 26.7ms remaining: 107ms
10: learn: 0.3923362 total: 29.7ms remaining: 105ms
11: learn: 0.3813501 total: 32.7ms remaining: 103ms
12: learn: 0.3700872 total: 34.1ms remaining: 97.1ms
13: learn: 0.3595115 total: 35.3ms remaining: 90.7ms
14: learn: 0.3496026 total: 37.8ms remaining: 88.2ms
15: learn: 0.3430776 total: 39.7ms remaining: 84.3ms
16: learn: 0.3324861 total: 42.1ms remaining: 81.7ms
17: learn: 0.3249253 total: 44ms remaining: 78.2ms
18: learn: 0.3175635 total: 45.8ms remaining: 74.7ms
19: learn: 0.3094936 total: 47.7ms remaining: 71.6ms
20: learn: 0.3045673 total: 49.1ms remaining: 67.9ms
21: learn: 0.2987827 total: 51.3ms remaining: 65.3ms
22: learn: 0.2937094 total: 53.5ms remaining: 62.8ms
23: learn: 0.2869886 total: 55.6ms remaining: 60.3ms
24: learn: 0.2814709 total: 57.4ms remaining: 57.4ms
25: learn: 0.2753705 total: 59.2ms remaining: 54.6ms
26: learn: 0.2707435 total: 60.1ms remaining: 51.2ms
27: learn: 0.2654776 total: 62.1ms remaining: 48.8ms
28: learn: 0.2604934 total: 63.2ms remaining: 45.8ms
29: learn: 0.2571748 total: 63.7ms remaining: 42.5ms
30: learn: 0.2529820 total: 65.3ms remaining: 40ms
31: learn: 0.2471451 total: 66.5ms remaining: 37.4ms
32: learn: 0.2430683 total: 68.1ms remaining: 35.1ms
33: learn: 0.2399410 total: 69.3ms remaining: 32.6ms
34: learn: 0.2359258 total: 70.4ms remaining: 30.2ms
35: learn: 0.2337736 total: 71.8ms remaining: 27.9ms
36: learn: 0.2311579 total: 73.3ms remaining: 25.8ms
37: learn: 0.2289602 total: 74.5ms remaining: 23.5ms
38: learn: 0.2259417 total: 75.6ms remaining: 21.3ms
39: learn: 0.2229631 total: 76.3ms remaining: 19.1ms
40: learn: 0.2201452 total: 77.2ms remaining: 16.9ms
41: learn: 0.2183124 total: 78.5ms remaining: 15ms
42: learn: 0.2167107 total: 79.5ms remaining: 12.9ms
43: learn: 0.2145715 total: 80.6ms remaining: 11ms
44: learn: 0.2138454 total: 81.4ms remaining: 9.05ms
45: learn: 0.2116082 total: 82.5ms remaining: 7.18ms
46: learn: 0.2103168 total: 83.2ms remaining: 5.31ms
47: learn: 0.2066677 total: 84.2ms remaining: 3.51ms
48: learn: 0.2035156 total: 85.2ms remaining: 1.74ms
49: learn: 0.2011849 total: 86.1ms remaining: 0us
0: learn: 0.6619530 total: 2.86ms remaining: 140ms
1: learn: 0.6283297 total: 5.97ms remaining: 143ms
2: learn: 0.5960954 total: 9.68ms remaining: 152ms
3: learn: 0.5627508 total: 12.3ms remaining: 141ms
4: learn: 0.5359547 total: 15.1ms remaining: 136ms
5: learn: 0.5055974 total: 18.4ms remaining: 135ms
6: learn: 0.4784684 total: 21ms remaining: 129ms
7: learn: 0.4624911 total: 23.3ms remaining: 122ms
8: learn: 0.4376804 total: 24.8ms remaining: 113ms
9: learn: 0.4158659 total: 27.5ms remaining: 110ms
10: learn: 0.3980852 total: 29.8ms remaining: 106ms
11: learn: 0.3829023 total: 32.2ms remaining: 102ms
12: learn: 0.3654256 total: 34.2ms remaining: 97.4ms
13: learn: 0.3569122 total: 36.1ms remaining: 92.9ms
14: learn: 0.3479229 total: 37.8ms remaining: 88.2ms
15: learn: 0.3372314 total: 39.5ms remaining: 83.9ms
16: learn: 0.3236670 total: 40.9ms remaining: 79.4ms
17: learn: 0.3132976 total: 42.5ms remaining: 75.5ms
18: learn: 0.3066007 total: 43.9ms remaining: 71.6ms
19: learn: 0.2992773 total: 45.6ms remaining: 68.4ms
20: learn: 0.2903921 total: 47.1ms remaining: 65ms
21: learn: 0.2843295 total: 48.5ms remaining: 61.8ms
22: learn: 0.2770463 total: 49.9ms remaining: 58.6ms
23: learn: 0.2721898 total: 51ms remaining: 55.2ms
24: learn: 0.2671702 total: 52ms remaining: 52ms
25: learn: 0.2615728 total: 53.2ms remaining: 49.1ms
26: learn: 0.2559444 total: 54.2ms remaining: 46.1ms
27: learn: 0.2522468 total: 55.3ms remaining: 43.4ms
28: learn: 0.2466611 total: 56.4ms remaining: 40.8ms
29: learn: 0.2428671 total: 57.2ms remaining: 38.2ms
30: learn: 0.2399610 total: 57.6ms remaining: 35.3ms
31: learn: 0.2362310 total: 58.4ms remaining: 32.9ms
32: learn: 0.2325890 total: 59.4ms remaining: 30.6ms
33: learn: 0.2291176 total: 60.4ms remaining: 28.4ms
34: learn: 0.2243576 total: 61.5ms remaining: 26.3ms
35: learn: 0.2212875 total: 62.4ms remaining: 24.3ms
36: learn: 0.2175959 total: 63.3ms remaining: 22.2ms
37: learn: 0.2138491 total: 64.3ms remaining: 20.3ms
38: learn: 0.2101711 total: 65ms remaining: 18.3ms
39: learn: 0.2061384 total: 65.8ms remaining: 16.5ms
40: learn: 0.2041421 total: 66.7ms remaining: 14.6ms
41: learn: 0.2009166 total: 67.6ms remaining: 12.9ms
42: learn: 0.1967390 total: 68.4ms remaining: 11.1ms
43: learn: 0.1937388 total: 69.6ms remaining: 9.49ms
44: learn: 0.1912718 total: 70.8ms remaining: 7.87ms
45: learn: 0.1891110 total: 71.7ms remaining: 6.24ms
46: learn: 0.1865737 total: 72.7ms remaining: 4.64ms
47: learn: 0.1845496 total: 73.6ms remaining: 3.07ms
48: learn: 0.1824034 total: 74.7ms remaining: 1.52ms
49: learn: 0.1796541 total: 75.9ms remaining: 0us
0: learn: 0.6581386 total: 2.98ms remaining: 146ms
1: learn: 0.6167948 total: 4.97ms remaining: 119ms
2: learn: 0.5790476 total: 7.97ms remaining: 125ms
3: learn: 0.5488848 total: 10.3ms remaining: 119ms
4: learn: 0.5230009 total: 13ms remaining: 117ms
5: learn: 0.4964807 total: 15.5ms remaining: 114ms
6: learn: 0.4712568 total: 17.8ms remaining: 109ms
7: learn: 0.4526747 total: 20.6ms remaining: 108ms
8: learn: 0.4333489 total: 23ms remaining: 105ms
9: learn: 0.4163775 total: 25.5ms remaining: 102ms
10: learn: 0.4031902 total: 28.3ms remaining: 100ms
11: learn: 0.3862230 total: 31.7ms remaining: 100ms
12: learn: 0.3749004 total: 32.7ms remaining: 93ms
13: learn: 0.3627256 total: 35.2ms remaining: 90.5ms
14: learn: 0.3513545 total: 38.1ms remaining: 88.8ms
15: learn: 0.3412036 total: 40.8ms remaining: 86.6ms
16: learn: 0.3338192 total: 42.9ms remaining: 83.2ms
17: learn: 0.3247744 total: 45.2ms remaining: 80.3ms
18: learn: 0.3152335 total: 46.3ms remaining: 75.5ms
19: learn: 0.3101617 total: 47.8ms remaining: 71.7ms
20: learn: 0.3029026 total: 49.6ms remaining: 68.6ms
21: learn: 0.2956389 total: 51.5ms remaining: 65.5ms
22: learn: 0.2879109 total: 52.9ms remaining: 62.1ms
23: learn: 0.2820702 total: 54.8ms remaining: 59.3ms
24: learn: 0.2771588 total: 56ms remaining: 56ms
25: learn: 0.2726471 total: 57.5ms remaining: 53.1ms
26: learn: 0.2659988 total: 58.9ms remaining: 50.2ms
27: learn: 0.2600717 total: 61.3ms remaining: 48.1ms
28: learn: 0.2585335 total: 62.2ms remaining: 45ms
29: learn: 0.2543809 total: 63.8ms remaining: 42.5ms
30: learn: 0.2510589 total: 65.1ms remaining: 39.9ms
31: learn: 0.2476309 total: 66.2ms remaining: 37.2ms
32: learn: 0.2446868 total: 67.3ms remaining: 34.7ms
33: learn: 0.2413541 total: 68.4ms remaining: 32.2ms
34: learn: 0.2375438 total: 69.4ms remaining: 29.8ms
35: learn: 0.2350143 total: 70.2ms remaining: 27.3ms
36: learn: 0.2316015 total: 71.4ms remaining: 25.1ms
37: learn: 0.2283235 total: 72.3ms remaining: 22.8ms
38: learn: 0.2246981 total: 73.3ms remaining: 20.7ms
39: learn: 0.2227796 total: 74.4ms remaining: 18.6ms
40: learn: 0.2213225 total: 75ms remaining: 16.5ms
41: learn: 0.2192394 total: 75.8ms remaining: 14.4ms
42: learn: 0.2178576 total: 76ms remaining: 12.4ms
43: learn: 0.2155307 total: 76.8ms remaining: 10.5ms
44: learn: 0.2119431 total: 77.8ms remaining: 8.64ms
45: learn: 0.2100281 total: 78.6ms remaining: 6.84ms
46: learn: 0.2069576 total: 79.5ms remaining: 5.07ms
47: learn: 0.2036339 total: 80.4ms remaining: 3.35ms
48: learn: 0.2008020 total: 81.4ms remaining: 1.66ms
49: learn: 0.1984541 total: 82.5ms remaining: 0us
0: learn: 0.6450008 total: 2.78ms remaining: 136ms
1: learn: 0.5991432 total: 4.52ms remaining: 108ms
2: learn: 0.5618368 total: 7.18ms remaining: 113ms
3: learn: 0.5318991 total: 10.1ms remaining: 116ms
4: learn: 0.5072157 total: 12.5ms remaining: 112ms
5: learn: 0.4774205 total: 15.2ms remaining: 111ms
6: learn: 0.4542422 total: 16.8ms remaining: 103ms
7: learn: 0.4335540 total: 19.1ms remaining: 100ms
8: learn: 0.4192534 total: 21.4ms remaining: 97.7ms
9: learn: 0.4003498 total: 24.9ms remaining: 99.6ms
10: learn: 0.3843162 total: 28ms remaining: 99.1ms
11: learn: 0.3682167 total: 31.2ms remaining: 98.7ms
12: learn: 0.3542211 total: 31.8ms remaining: 90.6ms
13: learn: 0.3399096 total: 34.6ms remaining: 89ms
14: learn: 0.3288582 total: 37.6ms remaining: 87.6ms
15: learn: 0.3173883 total: 40.7ms remaining: 86.4ms
16: learn: 0.3096656 total: 43.5ms remaining: 84.4ms
17: learn: 0.3012150 total: 44.7ms remaining: 79.4ms
18: learn: 0.2922121 total: 47ms remaining: 76.7ms
19: learn: 0.2846976 total: 49.4ms remaining: 74.1ms
20: learn: 0.2777799 total: 52.9ms remaining: 73ms
21: learn: 0.2716195 total: 55.3ms remaining: 70.3ms
22: learn: 0.2661617 total: 57ms remaining: 66.9ms
23: learn: 0.2633045 total: 58.5ms remaining: 63.4ms
24: learn: 0.2569821 total: 60.2ms remaining: 60.2ms
25: learn: 0.2508427 total: 62ms remaining: 57.3ms
26: learn: 0.2446129 total: 64.3ms remaining: 54.7ms
27: learn: 0.2398813 total: 65.7ms remaining: 51.6ms
28: learn: 0.2347845 total: 67.6ms remaining: 48.9ms
29: learn: 0.2305331 total: 69.4ms remaining: 46.3ms
30: learn: 0.2248650 total: 70.4ms remaining: 43.1ms
31: learn: 0.2210912 total: 71.8ms remaining: 40.4ms
32: learn: 0.2184215 total: 72.9ms remaining: 37.6ms
33: learn: 0.2132052 total: 74.1ms remaining: 34.9ms
34: learn: 0.2106714 total: 75.2ms remaining: 32.2ms
35: learn: 0.2080676 total: 76.5ms remaining: 29.7ms
36: learn: 0.2050681 total: 77.4ms remaining: 27.2ms
37: learn: 0.2022172 total: 78.8ms remaining: 24.9ms
38: learn: 0.1983147 total: 79.6ms remaining: 22.5ms
39: learn: 0.1956122 total: 80.5ms remaining: 20.1ms
40: learn: 0.1929652 total: 81.5ms remaining: 17.9ms
41: learn: 0.1906725 total: 82.5ms remaining: 15.7ms
42: learn: 0.1876575 total: 83.5ms remaining: 13.6ms
43: learn: 0.1857988 total: 84.5ms remaining: 11.5ms
44: learn: 0.1830635 total: 85.2ms remaining: 9.46ms
45: learn: 0.1815324 total: 86.1ms remaining: 7.49ms
46: learn: 0.1788223 total: 87ms remaining: 5.55ms
47: learn: 0.1760001 total: 88ms remaining: 3.67ms
48: learn: 0.1747437 total: 88.9ms remaining: 1.81ms
49: learn: 0.1737366 total: 89.2ms remaining: 0us
0: learn: 0.6424707 total: 2.89ms remaining: 142ms
1: learn: 0.6093615 total: 5.28ms remaining: 127ms
2: learn: 0.5728187 total: 8.45ms remaining: 132ms
3: learn: 0.5402520 total: 11.1ms remaining: 128ms
4: learn: 0.5081451 total: 14.3ms remaining: 129ms
5: learn: 0.4814130 total: 16.6ms remaining: 122ms
6: learn: 0.4585083 total: 19.5ms remaining: 120ms
7: learn: 0.4367014 total: 21.1ms remaining: 111ms
8: learn: 0.4201252 total: 23.5ms remaining: 107ms
9: learn: 0.4062633 total: 25.6ms remaining: 102ms
10: learn: 0.3902766 total: 26.6ms remaining: 94.2ms
11: learn: 0.3733889 total: 29.3ms remaining: 92.6ms
12: learn: 0.3591882 total: 31.8ms remaining: 90.6ms
13: learn: 0.3489420 total: 34.1ms remaining: 87.6ms
14: learn: 0.3384339 total: 36.7ms remaining: 85.7ms
15: learn: 0.3286939 total: 39.1ms remaining: 83.1ms
16: learn: 0.3207029 total: 41.2ms remaining: 79.9ms
17: learn: 0.3121512 total: 43.6ms remaining: 77.5ms
18: learn: 0.3071762 total: 46.2ms remaining: 75.3ms
19: learn: 0.3015041 total: 47.8ms remaining: 71.8ms
20: learn: 0.2945076 total: 50ms remaining: 69.1ms
21: learn: 0.2897907 total: 51.6ms remaining: 65.6ms
22: learn: 0.2811226 total: 53.5ms remaining: 62.8ms
23: learn: 0.2738252 total: 55.1ms remaining: 59.7ms
24: learn: 0.2675024 total: 56.4ms remaining: 56.4ms
25: learn: 0.2637681 total: 58.1ms remaining: 53.7ms
26: learn: 0.2604760 total: 59.5ms remaining: 50.7ms
27: learn: 0.2545391 total: 60.7ms remaining: 47.7ms
28: learn: 0.2506947 total: 62.1ms remaining: 45ms
29: learn: 0.2453228 total: 63.3ms remaining: 42.2ms
30: learn: 0.2412747 total: 64.7ms remaining: 39.6ms
31: learn: 0.2382940 total: 65.7ms remaining: 36.9ms
32: learn: 0.2333589 total: 67ms remaining: 34.5ms
33: learn: 0.2309363 total: 68ms remaining: 32ms
34: learn: 0.2267844 total: 68.9ms remaining: 29.5ms
35: learn: 0.2240942 total: 69.6ms remaining: 27.1ms
36: learn: 0.2214865 total: 70.5ms remaining: 24.8ms
37: learn: 0.2187099 total: 71.1ms remaining: 22.5ms
38: learn: 0.2172117 total: 72ms remaining: 20.3ms
39: learn: 0.2149843 total: 73.1ms remaining: 18.3ms
40: learn: 0.2123648 total: 73.9ms remaining: 16.2ms
41: learn: 0.2094861 total: 74.8ms remaining: 14.2ms
42: learn: 0.2062167 total: 75.5ms remaining: 12.3ms
43: learn: 0.2044708 total: 76.4ms remaining: 10.4ms
44: learn: 0.2019504 total: 77.1ms remaining: 8.57ms
45: learn: 0.1990791 total: 78.1ms remaining: 6.79ms
46: learn: 0.1979083 total: 79.1ms remaining: 5.05ms
47: learn: 0.1945802 total: 80ms remaining: 3.33ms
48: learn: 0.1920564 total: 80.9ms remaining: 1.65ms
49: learn: 0.1900021 total: 81.7ms remaining: 0us
0: learn: 0.6442230 total: 3.04ms remaining: 149ms
1: learn: 0.6048965 total: 5.55ms remaining: 133ms
2: learn: 0.5655809 total: 8.55ms remaining: 134ms
3: learn: 0.5350456 total: 11.2ms remaining: 128ms
4: learn: 0.5072746 total: 13.7ms remaining: 123ms
5: learn: 0.4781308 total: 16.2ms remaining: 119ms
6: learn: 0.4601451 total: 17.9ms remaining: 110ms
7: learn: 0.4348007 total: 19.8ms remaining: 104ms
8: learn: 0.4156452 total: 22.7ms remaining: 104ms
9: learn: 0.3961522 total: 25.9ms remaining: 104ms
10: learn: 0.3820850 total: 26.8ms remaining: 95.1ms
11: learn: 0.3679453 total: 28.1ms remaining: 89.1ms
12: learn: 0.3552289 total: 31.1ms remaining: 88.6ms
13: learn: 0.3430151 total: 33.7ms remaining: 86.7ms
14: learn: 0.3345714 total: 36.6ms remaining: 85.3ms
15: learn: 0.3238381 total: 38.4ms remaining: 81.7ms
16: learn: 0.3166062 total: 40.7ms remaining: 79.1ms
17: learn: 0.3115804 total: 41.7ms remaining: 74.1ms
18: learn: 0.3034827 total: 44.9ms remaining: 73.2ms
19: learn: 0.2976907 total: 47.4ms remaining: 71.1ms
20: learn: 0.2906328 total: 48.8ms remaining: 67.3ms
21: learn: 0.2840429 total: 51.1ms remaining: 65ms
22: learn: 0.2768625 total: 53.8ms remaining: 63.2ms
23: learn: 0.2716351 total: 55.9ms remaining: 60.6ms
24: learn: 0.2664229 total: 59.2ms remaining: 59.2ms
25: learn: 0.2614586 total: 61.2ms remaining: 56.5ms
26: learn: 0.2561204 total: 63.6ms remaining: 54.2ms
27: learn: 0.2513686 total: 65.9ms remaining: 51.8ms
28: learn: 0.2468270 total: 68.5ms remaining: 49.6ms
29: learn: 0.2422397 total: 70.3ms remaining: 46.9ms
30: learn: 0.2389905 total: 71.6ms remaining: 43.9ms
31: learn: 0.2367266 total: 73.5ms remaining: 41.3ms
32: learn: 0.2344441 total: 74.8ms remaining: 38.5ms
33: learn: 0.2309710 total: 76.2ms remaining: 35.8ms
34: learn: 0.2294099 total: 76.6ms remaining: 32.8ms
35: learn: 0.2260055 total: 78.2ms remaining: 30.4ms
36: learn: 0.2219051 total: 79.8ms remaining: 28.1ms
37: learn: 0.2194785 total: 80.8ms remaining: 25.5ms
38: learn: 0.2168878 total: 81.7ms remaining: 23.1ms
39: learn: 0.2138163 total: 82.7ms remaining: 20.7ms
40: learn: 0.2104126 total: 83.6ms remaining: 18.3ms
41: learn: 0.2084811 total: 84.4ms remaining: 16.1ms
42: learn: 0.2062840 total: 85.4ms remaining: 13.9ms
43: learn: 0.2034978 total: 86.6ms remaining: 11.8ms
44: learn: 0.2009875 total: 88ms remaining: 9.78ms
45: learn: 0.1979982 total: 89ms remaining: 7.74ms
46: learn: 0.1964768 total: 89.8ms remaining: 5.73ms
47: learn: 0.1949001 total: 90.6ms remaining: 3.77ms
48: learn: 0.1921654 total: 91.4ms remaining: 1.86ms
49: learn: 0.1907001 total: 92.3ms remaining: 0us
0: learn: 0.6447617 total: 1.96ms remaining: 96.1ms
1: learn: 0.6051152 total: 3.79ms remaining: 90.9ms
2: learn: 0.5714000 total: 5.93ms remaining: 92.9ms
3: learn: 0.5414568 total: 7.75ms remaining: 89.1ms
4: learn: 0.5109503 total: 9.41ms remaining: 84.7ms
5: learn: 0.4857484 total: 11.1ms remaining: 81.2ms
6: learn: 0.4643829 total: 12.5ms remaining: 76.6ms
7: learn: 0.4466605 total: 14.2ms remaining: 74.4ms
8: learn: 0.4343357 total: 15.3ms remaining: 69.6ms
9: learn: 0.4134473 total: 16.8ms remaining: 67.1ms
10: learn: 0.4020706 total: 18.6ms remaining: 65.8ms
11: learn: 0.3879785 total: 20.4ms remaining: 64.5ms
12: learn: 0.3722573 total: 21.5ms remaining: 61.2ms
13: learn: 0.3585459 total: 23.1ms remaining: 59.5ms
14: learn: 0.3485039 total: 24.9ms remaining: 58.1ms
15: learn: 0.3390197 total: 27.1ms remaining: 57.6ms
16: learn: 0.3285231 total: 28.9ms remaining: 56ms
17: learn: 0.3189429 total: 31.7ms remaining: 56.4ms
18: learn: 0.3085253 total: 33.8ms remaining: 55.1ms
19: learn: 0.3030259 total: 36.3ms remaining: 54.4ms
20: learn: 0.2977019 total: 38.5ms remaining: 53.1ms
21: learn: 0.2911362 total: 41.1ms remaining: 52.2ms
22: learn: 0.2851606 total: 43.8ms remaining: 51.5ms
23: learn: 0.2788896 total: 47.3ms remaining: 51.3ms
24: learn: 0.2728411 total: 50ms remaining: 50ms
25: learn: 0.2678508 total: 52.8ms remaining: 48.7ms
26: learn: 0.2639602 total: 55.2ms remaining: 47ms
27: learn: 0.2581845 total: 58.5ms remaining: 46ms
28: learn: 0.2549563 total: 60.6ms remaining: 43.9ms
29: learn: 0.2512371 total: 62.8ms remaining: 41.8ms
30: learn: 0.2482787 total: 63.7ms remaining: 39.1ms
31: learn: 0.2454422 total: 65.7ms remaining: 36.9ms
32: learn: 0.2425382 total: 67.5ms remaining: 34.8ms
33: learn: 0.2382371 total: 69.1ms remaining: 32.5ms
34: learn: 0.2352638 total: 71.5ms remaining: 30.6ms
35: learn: 0.2323701 total: 73.3ms remaining: 28.5ms
36: learn: 0.2306640 total: 75.7ms remaining: 26.6ms
37: learn: 0.2270014 total: 77.4ms remaining: 24.4ms
38: learn: 0.2227674 total: 78.4ms remaining: 22.1ms
39: learn: 0.2198379 total: 79.9ms remaining: 20ms
40: learn: 0.2175182 total: 81.9ms remaining: 18ms
41: learn: 0.2151720 total: 83.5ms remaining: 15.9ms
42: learn: 0.2130668 total: 85.2ms remaining: 13.9ms
43: learn: 0.2090422 total: 86.5ms remaining: 11.8ms
44: learn: 0.2073105 total: 88.3ms remaining: 9.81ms
45: learn: 0.2044258 total: 89.3ms remaining: 7.77ms
46: learn: 0.2008996 total: 90.8ms remaining: 5.79ms
47: learn: 0.1985224 total: 92ms remaining: 3.83ms
48: learn: 0.1963995 total: 93ms remaining: 1.9ms
49: learn: 0.1938656 total: 94ms remaining: 0us
Evaluating param combo 9/16: {'boosted_cb_model__iterations': 100, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 8}
Duplicate found for this combo (hash=001b0a8dcc441eff9e6b8fad1fb158e6). Removing old run: b6a90667611a4b1baa788329ec47cdcb
0: learn: 0.6882794 total: 1.08ms remaining: 107ms
1: learn: 0.6829408 total: 2.58ms remaining: 126ms
2: learn: 0.6779769 total: 3.59ms remaining: 116ms
3: learn: 0.6731590 total: 4.88ms remaining: 117ms
4: learn: 0.6685559 total: 5.95ms remaining: 113ms
5: learn: 0.6636779 total: 7.35ms remaining: 115ms
6: learn: 0.6591763 total: 8.54ms remaining: 113ms
7: learn: 0.6551357 total: 9.52ms remaining: 109ms
8: learn: 0.6504101 total: 10.5ms remaining: 106ms
9: learn: 0.6464580 total: 11.8ms remaining: 106ms
10: learn: 0.6425359 total: 13ms remaining: 105ms
11: learn: 0.6387708 total: 14.3ms remaining: 105ms
12: learn: 0.6346059 total: 15.8ms remaining: 106ms
13: learn: 0.6304314 total: 17.1ms remaining: 105ms
14: learn: 0.6268009 total: 18.1ms remaining: 103ms
15: learn: 0.6229881 total: 19.9ms remaining: 104ms
16: learn: 0.6190015 total: 21.2ms remaining: 104ms
17: learn: 0.6151072 total: 22.9ms remaining: 104ms
18: learn: 0.6113699 total: 24.1ms remaining: 103ms
19: learn: 0.6075444 total: 25.4ms remaining: 101ms
20: learn: 0.6037564 total: 26.4ms remaining: 99.4ms
21: learn: 0.5999778 total: 27.8ms remaining: 98.4ms
22: learn: 0.5958785 total: 29ms remaining: 97ms
23: learn: 0.5922311 total: 29.7ms remaining: 94.1ms
24: learn: 0.5892157 total: 30.8ms remaining: 92.4ms
25: learn: 0.5861289 total: 32.2ms remaining: 91.7ms
26: learn: 0.5829753 total: 33.4ms remaining: 90.2ms
27: learn: 0.5798042 total: 34.6ms remaining: 88.9ms
28: learn: 0.5767399 total: 35.7ms remaining: 87.4ms
29: learn: 0.5739181 total: 36.6ms remaining: 85.4ms
30: learn: 0.5712722 total: 37.6ms remaining: 83.8ms
31: learn: 0.5679889 total: 38.3ms remaining: 81.3ms
32: learn: 0.5647182 total: 39.7ms remaining: 80.7ms
33: learn: 0.5613468 total: 40.9ms remaining: 79.4ms
34: learn: 0.5577149 total: 41.8ms remaining: 77.6ms
35: learn: 0.5547437 total: 42.7ms remaining: 75.9ms
36: learn: 0.5517524 total: 43.9ms remaining: 74.8ms
37: learn: 0.5484946 total: 45ms remaining: 73.4ms
38: learn: 0.5451312 total: 46.1ms remaining: 72.1ms
39: learn: 0.5421510 total: 47ms remaining: 70.4ms
40: learn: 0.5387318 total: 48.2ms remaining: 69.3ms
41: learn: 0.5355684 total: 48.8ms remaining: 67.4ms
42: learn: 0.5324215 total: 49.1ms remaining: 65.1ms
43: learn: 0.5298921 total: 50.1ms remaining: 63.7ms
44: learn: 0.5271650 total: 51.4ms remaining: 62.9ms
45: learn: 0.5248260 total: 53.3ms remaining: 62.5ms
46: learn: 0.5222534 total: 54.5ms remaining: 61.4ms
47: learn: 0.5193675 total: 55.5ms remaining: 60.1ms
48: learn: 0.5168600 total: 56.5ms remaining: 58.8ms
49: learn: 0.5145437 total: 57.2ms remaining: 57.2ms
50: learn: 0.5114851 total: 58.1ms remaining: 55.8ms
51: learn: 0.5093087 total: 59.3ms remaining: 54.7ms
52: learn: 0.5067977 total: 60.2ms remaining: 53.3ms
53: learn: 0.5046903 total: 61.6ms remaining: 52.5ms
54: learn: 0.5020617 total: 62.5ms remaining: 51.2ms
55: learn: 0.4998635 total: 63.4ms remaining: 49.8ms
56: learn: 0.4974511 total: 64.2ms remaining: 48.5ms
57: learn: 0.4953198 total: 65.1ms remaining: 47.2ms
58: learn: 0.4931948 total: 65.8ms remaining: 45.7ms
59: learn: 0.4911060 total: 66.7ms remaining: 44.5ms
60: learn: 0.4889797 total: 67.6ms remaining: 43.2ms
61: learn: 0.4866935 total: 68.5ms remaining: 42ms
62: learn: 0.4843003 total: 69.4ms remaining: 40.8ms
63: learn: 0.4820957 total: 70.1ms remaining: 39.4ms
64: learn: 0.4798245 total: 70.8ms remaining: 38.1ms
65: learn: 0.4777677 total: 71.4ms remaining: 36.8ms
66: learn: 0.4756431 total: 72ms remaining: 35.5ms
67: learn: 0.4735064 total: 72.2ms remaining: 34ms
68: learn: 0.4711289 total: 72.8ms remaining: 32.7ms
69: learn: 0.4686863 total: 73.5ms remaining: 31.5ms
70: learn: 0.4665532 total: 74.1ms remaining: 30.3ms
71: learn: 0.4646554 total: 74.9ms remaining: 29.1ms
72: learn: 0.4626431 total: 75.5ms remaining: 27.9ms
73: learn: 0.4606925 total: 76.1ms remaining: 26.8ms
74: learn: 0.4587245 total: 76.8ms remaining: 25.6ms
75: learn: 0.4565481 total: 77.4ms remaining: 24.4ms
76: learn: 0.4547888 total: 77.8ms remaining: 23.2ms
77: learn: 0.4528959 total: 78.5ms remaining: 22.1ms
78: learn: 0.4508121 total: 79.1ms remaining: 21ms
79: learn: 0.4490872 total: 79.9ms remaining: 20ms
80: learn: 0.4471421 total: 80.7ms remaining: 18.9ms
81: learn: 0.4453254 total: 81.1ms remaining: 17.8ms
82: learn: 0.4432644 total: 81.7ms remaining: 16.7ms
83: learn: 0.4413600 total: 82ms remaining: 15.6ms
84: learn: 0.4394892 total: 82.6ms remaining: 14.6ms
85: learn: 0.4377549 total: 83ms remaining: 13.5ms
86: learn: 0.4359258 total: 83.4ms remaining: 12.5ms
87: learn: 0.4339840 total: 83.8ms remaining: 11.4ms
88: learn: 0.4318123 total: 84.1ms remaining: 10.4ms
89: learn: 0.4300119 total: 84.4ms remaining: 9.38ms
90: learn: 0.4281025 total: 84.8ms remaining: 8.39ms
91: learn: 0.4265387 total: 85.3ms remaining: 7.42ms
92: learn: 0.4249134 total: 85.6ms remaining: 6.44ms
93: learn: 0.4233160 total: 86ms remaining: 5.49ms
94: learn: 0.4217480 total: 86.4ms remaining: 4.54ms
95: learn: 0.4200962 total: 86.8ms remaining: 3.62ms
96: learn: 0.4187601 total: 87.2ms remaining: 2.69ms
97: learn: 0.4172840 total: 87.5ms remaining: 1.78ms
98: learn: 0.4160567 total: 87.9ms remaining: 888us
99: learn: 0.4143777 total: 88.3ms remaining: 0us
0: learn: 0.6880102 total: 1.27ms remaining: 126ms
1: learn: 0.6822828 total: 2.62ms remaining: 128ms
2: learn: 0.6772106 total: 3.76ms remaining: 122ms
3: learn: 0.6726730 total: 5.24ms remaining: 126ms
4: learn: 0.6682095 total: 6.63ms remaining: 126ms
5: learn: 0.6635268 total: 7.95ms remaining: 125ms
6: learn: 0.6588811 total: 8.81ms remaining: 117ms
7: learn: 0.6545703 total: 10.1ms remaining: 117ms
8: learn: 0.6497856 total: 11ms remaining: 111ms
9: learn: 0.6448858 total: 12.4ms remaining: 112ms
10: learn: 0.6405230 total: 13.3ms remaining: 108ms
11: learn: 0.6361860 total: 14.5ms remaining: 107ms
12: learn: 0.6314977 total: 16ms remaining: 107ms
13: learn: 0.6272630 total: 17.5ms remaining: 107ms
14: learn: 0.6237927 total: 18.7ms remaining: 106ms
15: learn: 0.6202946 total: 19.7ms remaining: 104ms
16: learn: 0.6160996 total: 21.2ms remaining: 103ms
17: learn: 0.6121987 total: 22.3ms remaining: 102ms
18: learn: 0.6085887 total: 23.2ms remaining: 99ms
19: learn: 0.6053875 total: 24.7ms remaining: 98.7ms
20: learn: 0.6012749 total: 26.2ms remaining: 98.5ms
21: learn: 0.5970959 total: 27.2ms remaining: 96.4ms
22: learn: 0.5929697 total: 27.9ms remaining: 93.4ms
23: learn: 0.5894176 total: 28.7ms remaining: 90.9ms
24: learn: 0.5848522 total: 29.5ms remaining: 88.4ms
25: learn: 0.5821273 total: 31.1ms remaining: 88.6ms
26: learn: 0.5789455 total: 32.2ms remaining: 87.2ms
27: learn: 0.5754969 total: 33.4ms remaining: 85.9ms
28: learn: 0.5721862 total: 34.7ms remaining: 84.9ms
29: learn: 0.5691136 total: 35.8ms remaining: 83.5ms
30: learn: 0.5660278 total: 36.9ms remaining: 82.2ms
31: learn: 0.5628754 total: 37.7ms remaining: 80.1ms
32: learn: 0.5589497 total: 38.8ms remaining: 78.7ms
33: learn: 0.5553905 total: 40ms remaining: 77.7ms
34: learn: 0.5515082 total: 41.2ms remaining: 76.5ms
35: learn: 0.5482866 total: 42.1ms remaining: 74.8ms
36: learn: 0.5449903 total: 43.1ms remaining: 73.4ms
37: learn: 0.5423647 total: 44.5ms remaining: 72.6ms
38: learn: 0.5386898 total: 45.4ms remaining: 71ms
39: learn: 0.5350966 total: 46.3ms remaining: 69.5ms
40: learn: 0.5319492 total: 46.9ms remaining: 67.5ms
41: learn: 0.5288603 total: 47.8ms remaining: 65.9ms
42: learn: 0.5260202 total: 48.7ms remaining: 64.6ms
43: learn: 0.5230047 total: 49.5ms remaining: 63ms
44: learn: 0.5198876 total: 50.2ms remaining: 61.4ms
45: learn: 0.5168284 total: 51.2ms remaining: 60.1ms
46: learn: 0.5136389 total: 52ms remaining: 58.7ms
47: learn: 0.5112892 total: 52.7ms remaining: 57.1ms
48: learn: 0.5085121 total: 53.2ms remaining: 55.4ms
49: learn: 0.5054328 total: 54.2ms remaining: 54.2ms
50: learn: 0.5026563 total: 54.9ms remaining: 52.7ms
51: learn: 0.4999174 total: 55.6ms remaining: 51.3ms
52: learn: 0.4972964 total: 56.4ms remaining: 50ms
53: learn: 0.4949977 total: 57ms remaining: 48.5ms
54: learn: 0.4919354 total: 57.5ms remaining: 47.1ms
55: learn: 0.4898954 total: 58.1ms remaining: 45.7ms
56: learn: 0.4878160 total: 58.6ms remaining: 44.2ms
57: learn: 0.4855045 total: 59.1ms remaining: 42.8ms
58: learn: 0.4829743 total: 59.6ms remaining: 41.4ms
59: learn: 0.4804930 total: 60.1ms remaining: 40.1ms
60: learn: 0.4780999 total: 61ms remaining: 39ms
61: learn: 0.4758416 total: 61.5ms remaining: 37.7ms
62: learn: 0.4728837 total: 62.2ms remaining: 36.5ms
63: learn: 0.4706892 total: 62.7ms remaining: 35.3ms
64: learn: 0.4683178 total: 63.3ms remaining: 34.1ms
65: learn: 0.4656676 total: 63.7ms remaining: 32.8ms
66: learn: 0.4632440 total: 64.1ms remaining: 31.6ms
67: learn: 0.4611426 total: 64.6ms remaining: 30.4ms
68: learn: 0.4588472 total: 65.1ms remaining: 29.2ms
69: learn: 0.4565314 total: 65.5ms remaining: 28.1ms
70: learn: 0.4548869 total: 66ms remaining: 27ms
71: learn: 0.4526554 total: 66.5ms remaining: 25.9ms
72: learn: 0.4499787 total: 66.9ms remaining: 24.8ms
73: learn: 0.4475357 total: 67.3ms remaining: 23.7ms
74: learn: 0.4455755 total: 67.8ms remaining: 22.6ms
75: learn: 0.4440104 total: 68.2ms remaining: 21.6ms
76: learn: 0.4417982 total: 68.7ms remaining: 20.5ms
77: learn: 0.4398600 total: 69.1ms remaining: 19.5ms
78: learn: 0.4379151 total: 70.2ms remaining: 18.7ms
79: learn: 0.4361857 total: 70.6ms remaining: 17.7ms
80: learn: 0.4338838 total: 71ms remaining: 16.7ms
81: learn: 0.4319658 total: 71.7ms remaining: 15.7ms
82: learn: 0.4302179 total: 72ms remaining: 14.7ms
83: learn: 0.4280088 total: 72.3ms remaining: 13.8ms
84: learn: 0.4261117 total: 72.7ms remaining: 12.8ms
85: learn: 0.4244622 total: 73.2ms remaining: 11.9ms
86: learn: 0.4222954 total: 73.6ms remaining: 11ms
87: learn: 0.4202130 total: 74ms remaining: 10.1ms
88: learn: 0.4181864 total: 74.5ms remaining: 9.21ms
89: learn: 0.4164203 total: 74.9ms remaining: 8.33ms
90: learn: 0.4149126 total: 75.3ms remaining: 7.45ms
91: learn: 0.4133189 total: 75.7ms remaining: 6.58ms
92: learn: 0.4112811 total: 76.1ms remaining: 5.73ms
93: learn: 0.4094290 total: 76.5ms remaining: 4.88ms
94: learn: 0.4081478 total: 76.9ms remaining: 4.05ms
95: learn: 0.4062874 total: 77.3ms remaining: 3.22ms
96: learn: 0.4044458 total: 77.7ms remaining: 2.4ms
97: learn: 0.4030461 total: 78ms remaining: 1.59ms
98: learn: 0.4012621 total: 78.6ms remaining: 793us
99: learn: 0.3996016 total: 79ms remaining: 0us
0: learn: 0.6881915 total: 1.15ms remaining: 114ms
1: learn: 0.6830903 total: 2.51ms remaining: 123ms
2: learn: 0.6779156 total: 3.56ms remaining: 115ms
3: learn: 0.6727870 total: 5.13ms remaining: 123ms
4: learn: 0.6680191 total: 6.3ms remaining: 120ms
5: learn: 0.6632566 total: 7.32ms remaining: 115ms
6: learn: 0.6589348 total: 8.36ms remaining: 111ms
7: learn: 0.6548497 total: 9.61ms remaining: 110ms
8: learn: 0.6499422 total: 10.7ms remaining: 108ms
9: learn: 0.6456086 total: 11.7ms remaining: 106ms
10: learn: 0.6416798 total: 12.6ms remaining: 102ms
11: learn: 0.6376258 total: 13.7ms remaining: 100ms
12: learn: 0.6328452 total: 14.8ms remaining: 99.1ms
13: learn: 0.6291112 total: 16.3ms remaining: 100ms
14: learn: 0.6252126 total: 17.4ms remaining: 98.7ms
15: learn: 0.6213698 total: 18.7ms remaining: 98.1ms
16: learn: 0.6172627 total: 19.9ms remaining: 97.2ms
17: learn: 0.6136354 total: 20.7ms remaining: 94.1ms
18: learn: 0.6099157 total: 21.7ms remaining: 92.5ms
19: learn: 0.6065275 total: 22.8ms remaining: 91ms
20: learn: 0.6026046 total: 24.1ms remaining: 90.6ms
21: learn: 0.5986205 total: 24.7ms remaining: 87.7ms
22: learn: 0.5943580 total: 25.4ms remaining: 84.9ms
23: learn: 0.5906964 total: 26.2ms remaining: 83.1ms
24: learn: 0.5869638 total: 27.5ms remaining: 82.4ms
25: learn: 0.5838294 total: 28.8ms remaining: 81.9ms
26: learn: 0.5809812 total: 29.8ms remaining: 80.6ms
27: learn: 0.5776571 total: 31.1ms remaining: 79.9ms
28: learn: 0.5743650 total: 32.3ms remaining: 79.1ms
29: learn: 0.5714944 total: 33.3ms remaining: 77.7ms
30: learn: 0.5687434 total: 34.4ms remaining: 76.5ms
31: learn: 0.5653821 total: 35.2ms remaining: 74.8ms
32: learn: 0.5620526 total: 36.5ms remaining: 74.1ms
33: learn: 0.5584545 total: 37.2ms remaining: 72.2ms
34: learn: 0.5550520 total: 38.1ms remaining: 70.8ms
35: learn: 0.5514777 total: 39.2ms remaining: 69.7ms
36: learn: 0.5483220 total: 40.4ms remaining: 68.8ms
37: learn: 0.5453927 total: 41.3ms remaining: 67.4ms
38: learn: 0.5418823 total: 42.1ms remaining: 65.8ms
39: learn: 0.5394460 total: 43.1ms remaining: 64.7ms
40: learn: 0.5362960 total: 44.2ms remaining: 63.6ms
41: learn: 0.5328204 total: 44.9ms remaining: 62ms
42: learn: 0.5296236 total: 45.7ms remaining: 60.6ms
43: learn: 0.5272600 total: 46.6ms remaining: 59.3ms
44: learn: 0.5244234 total: 47.4ms remaining: 57.9ms
45: learn: 0.5224995 total: 48.1ms remaining: 56.5ms
46: learn: 0.5199309 total: 48.9ms remaining: 55.1ms
47: learn: 0.5169806 total: 50ms remaining: 54.2ms
48: learn: 0.5142186 total: 50.9ms remaining: 53ms
49: learn: 0.5113786 total: 51.6ms remaining: 51.6ms
50: learn: 0.5086399 total: 52.4ms remaining: 50.3ms
51: learn: 0.5061395 total: 53ms remaining: 49ms
52: learn: 0.5038713 total: 53.4ms remaining: 47.4ms
53: learn: 0.5014624 total: 53.8ms remaining: 45.9ms
54: learn: 0.4989674 total: 54.2ms remaining: 44.4ms
55: learn: 0.4964659 total: 54.6ms remaining: 42.9ms
56: learn: 0.4937484 total: 55.1ms remaining: 41.6ms
57: learn: 0.4914684 total: 55.7ms remaining: 40.4ms
58: learn: 0.4891281 total: 56.3ms remaining: 39.1ms
59: learn: 0.4867757 total: 56.9ms remaining: 37.9ms
60: learn: 0.4845035 total: 57.4ms remaining: 36.7ms
61: learn: 0.4819540 total: 57.9ms remaining: 35.5ms
62: learn: 0.4796578 total: 58.4ms remaining: 34.3ms
63: learn: 0.4774683 total: 58.9ms remaining: 33.1ms
64: learn: 0.4752972 total: 59.5ms remaining: 32ms
65: learn: 0.4730619 total: 60ms remaining: 30.9ms
66: learn: 0.4708604 total: 60.6ms remaining: 29.8ms
67: learn: 0.4685726 total: 61.1ms remaining: 28.7ms
68: learn: 0.4664067 total: 61.4ms remaining: 27.6ms
69: learn: 0.4643398 total: 61.9ms remaining: 26.5ms
70: learn: 0.4624445 total: 62.6ms remaining: 25.5ms
71: learn: 0.4605617 total: 63ms remaining: 24.5ms
72: learn: 0.4584894 total: 63.5ms remaining: 23.5ms
73: learn: 0.4563449 total: 63.9ms remaining: 22.5ms
74: learn: 0.4543194 total: 64.5ms remaining: 21.5ms
75: learn: 0.4523466 total: 64.9ms remaining: 20.5ms
76: learn: 0.4500421 total: 65.3ms remaining: 19.5ms
77: learn: 0.4482336 total: 65.8ms remaining: 18.6ms
78: learn: 0.4461296 total: 66.2ms remaining: 17.6ms
79: learn: 0.4442134 total: 66.5ms remaining: 16.6ms
80: learn: 0.4420610 total: 66.8ms remaining: 15.7ms
81: learn: 0.4400215 total: 67.1ms remaining: 14.7ms
82: learn: 0.4381004 total: 67.5ms remaining: 13.8ms
83: learn: 0.4365113 total: 68.1ms remaining: 13ms
84: learn: 0.4348688 total: 68.4ms remaining: 12.1ms
85: learn: 0.4327667 total: 68.8ms remaining: 11.2ms
86: learn: 0.4305854 total: 69.1ms remaining: 10.3ms
87: learn: 0.4286623 total: 69.5ms remaining: 9.48ms
88: learn: 0.4267661 total: 69.8ms remaining: 8.63ms
89: learn: 0.4250506 total: 70.2ms remaining: 7.8ms
90: learn: 0.4231133 total: 70.5ms remaining: 6.98ms
91: learn: 0.4214037 total: 70.9ms remaining: 6.17ms
92: learn: 0.4197256 total: 71.3ms remaining: 5.36ms
93: learn: 0.4179794 total: 71.7ms remaining: 4.58ms
94: learn: 0.4162980 total: 72.1ms remaining: 3.79ms
95: learn: 0.4145354 total: 72.5ms remaining: 3.02ms
96: learn: 0.4127874 total: 72.8ms remaining: 2.25ms
97: learn: 0.4112734 total: 73.3ms remaining: 1.5ms
98: learn: 0.4096648 total: 73.7ms remaining: 743us
99: learn: 0.4080795 total: 74ms remaining: 0us
0: learn: 0.6883581 total: 1.37ms remaining: 136ms
1: learn: 0.6829490 total: 1.8ms remaining: 88.1ms
2: learn: 0.6777105 total: 2.75ms remaining: 88.8ms
3: learn: 0.6724965 total: 3.84ms remaining: 92.3ms
4: learn: 0.6678869 total: 4.94ms remaining: 94ms
5: learn: 0.6630227 total: 6.32ms remaining: 98.9ms
6: learn: 0.6580845 total: 7.58ms remaining: 101ms
7: learn: 0.6539384 total: 8.64ms remaining: 99.4ms
8: learn: 0.6488837 total: 9.51ms remaining: 96.2ms
9: learn: 0.6442866 total: 10.5ms remaining: 94.1ms
10: learn: 0.6404207 total: 11.3ms remaining: 91.2ms
11: learn: 0.6359348 total: 12.4ms remaining: 90.7ms
12: learn: 0.6315682 total: 13.7ms remaining: 92ms
13: learn: 0.6273393 total: 14.3ms remaining: 87.9ms
14: learn: 0.6234264 total: 14.8ms remaining: 83.8ms
15: learn: 0.6197946 total: 16.1ms remaining: 84.4ms
16: learn: 0.6155398 total: 17.4ms remaining: 84.7ms
17: learn: 0.6113889 total: 18.6ms remaining: 84.7ms
18: learn: 0.6079857 total: 19.8ms remaining: 84.5ms
19: learn: 0.6040984 total: 20.6ms remaining: 82.6ms
20: learn: 0.6001675 total: 21.4ms remaining: 80.5ms
21: learn: 0.5959995 total: 22.3ms remaining: 78.9ms
22: learn: 0.5919796 total: 23.3ms remaining: 77.9ms
23: learn: 0.5884148 total: 24.3ms remaining: 76.9ms
24: learn: 0.5841205 total: 25.1ms remaining: 75.4ms
25: learn: 0.5809535 total: 26ms remaining: 74ms
26: learn: 0.5776310 total: 26.9ms remaining: 72.7ms
27: learn: 0.5743591 total: 27.8ms remaining: 71.6ms
28: learn: 0.5712718 total: 28.6ms remaining: 70.1ms
29: learn: 0.5684340 total: 29.4ms remaining: 68.7ms
30: learn: 0.5657478 total: 30.4ms remaining: 67.7ms
31: learn: 0.5623463 total: 31.1ms remaining: 66ms
32: learn: 0.5591812 total: 32ms remaining: 64.9ms
33: learn: 0.5555487 total: 32.9ms remaining: 63.9ms
34: learn: 0.5525218 total: 33.7ms remaining: 62.6ms
35: learn: 0.5495863 total: 34.5ms remaining: 61.4ms
36: learn: 0.5463044 total: 35.4ms remaining: 60.3ms
37: learn: 0.5429413 total: 36.4ms remaining: 59.3ms
38: learn: 0.5394540 total: 37.1ms remaining: 58ms
39: learn: 0.5363956 total: 37.9ms remaining: 56.8ms
40: learn: 0.5330491 total: 38.7ms remaining: 55.7ms
41: learn: 0.5306820 total: 39.4ms remaining: 54.4ms
42: learn: 0.5275766 total: 40.2ms remaining: 53.3ms
43: learn: 0.5246236 total: 41ms remaining: 52.2ms
44: learn: 0.5218134 total: 42ms remaining: 51.3ms
45: learn: 0.5189606 total: 42.7ms remaining: 50.2ms
46: learn: 0.5159627 total: 43.5ms remaining: 49ms
47: learn: 0.5128221 total: 44.3ms remaining: 48ms
48: learn: 0.5096912 total: 44.9ms remaining: 46.8ms
49: learn: 0.5072024 total: 45.7ms remaining: 45.7ms
50: learn: 0.5043210 total: 46.3ms remaining: 44.5ms
51: learn: 0.5022366 total: 46.7ms remaining: 43.1ms
52: learn: 0.4997103 total: 47.3ms remaining: 42ms
53: learn: 0.4970926 total: 48.1ms remaining: 40.9ms
54: learn: 0.4948878 total: 48.8ms remaining: 39.9ms
55: learn: 0.4921710 total: 49.3ms remaining: 38.8ms
56: learn: 0.4894281 total: 49.8ms remaining: 37.5ms
57: learn: 0.4868875 total: 50.3ms remaining: 36.4ms
58: learn: 0.4842999 total: 50.8ms remaining: 35.3ms
59: learn: 0.4821379 total: 51.3ms remaining: 34.2ms
60: learn: 0.4793659 total: 51.7ms remaining: 33.1ms
61: learn: 0.4765558 total: 52.3ms remaining: 32ms
62: learn: 0.4739383 total: 52.9ms remaining: 31.1ms
63: learn: 0.4714391 total: 53.4ms remaining: 30.1ms
64: learn: 0.4689153 total: 54ms remaining: 29.1ms
65: learn: 0.4669516 total: 54.5ms remaining: 28.1ms
66: learn: 0.4649550 total: 54.9ms remaining: 27ms
67: learn: 0.4626150 total: 55.3ms remaining: 26ms
68: learn: 0.4608829 total: 55.5ms remaining: 25ms
69: learn: 0.4585578 total: 56.2ms remaining: 24.1ms
70: learn: 0.4567417 total: 56.6ms remaining: 23.1ms
71: learn: 0.4546172 total: 57ms remaining: 22.2ms
72: learn: 0.4525670 total: 57.5ms remaining: 21.3ms
73: learn: 0.4505951 total: 57.8ms remaining: 20.3ms
74: learn: 0.4487806 total: 58.1ms remaining: 19.4ms
75: learn: 0.4468307 total: 58.5ms remaining: 18.5ms
76: learn: 0.4445511 total: 59.2ms remaining: 17.7ms
77: learn: 0.4425293 total: 59.6ms remaining: 16.8ms
78: learn: 0.4403919 total: 60ms remaining: 15.9ms
79: learn: 0.4385428 total: 60.2ms remaining: 15.1ms
80: learn: 0.4362771 total: 60.8ms remaining: 14.3ms
81: learn: 0.4341435 total: 61.1ms remaining: 13.4ms
82: learn: 0.4320750 total: 61.4ms remaining: 12.6ms
83: learn: 0.4304187 total: 61.8ms remaining: 11.8ms
84: learn: 0.4287434 total: 62.2ms remaining: 11ms
85: learn: 0.4267036 total: 62.5ms remaining: 10.2ms
86: learn: 0.4245506 total: 62.8ms remaining: 9.39ms
87: learn: 0.4226060 total: 63.1ms remaining: 8.61ms
88: learn: 0.4209438 total: 63.7ms remaining: 7.88ms
89: learn: 0.4191627 total: 64.1ms remaining: 7.12ms
90: learn: 0.4175165 total: 64.4ms remaining: 6.37ms
91: learn: 0.4157530 total: 64.8ms remaining: 5.63ms
92: learn: 0.4139980 total: 65.2ms remaining: 4.91ms
93: learn: 0.4120571 total: 65.6ms remaining: 4.19ms
94: learn: 0.4106134 total: 66ms remaining: 3.47ms
95: learn: 0.4088877 total: 66.3ms remaining: 2.76ms
96: learn: 0.4074953 total: 67ms remaining: 2.07ms
97: learn: 0.4057214 total: 67.4ms remaining: 1.38ms
98: learn: 0.4042434 total: 67.7ms remaining: 683us
99: learn: 0.4024514 total: 68.1ms remaining: 0us
0: learn: 0.6883175 total: 952us remaining: 94.3ms
1: learn: 0.6833177 total: 1.68ms remaining: 82.2ms
2: learn: 0.6787594 total: 2.35ms remaining: 76.1ms
3: learn: 0.6742785 total: 3.11ms remaining: 74.7ms
4: learn: 0.6698376 total: 3.7ms remaining: 70.3ms
5: learn: 0.6655790 total: 4.61ms remaining: 72.3ms
6: learn: 0.6606753 total: 5.24ms remaining: 69.6ms
7: learn: 0.6567153 total: 6ms remaining: 69ms
8: learn: 0.6521844 total: 6.55ms remaining: 66.2ms
9: learn: 0.6476670 total: 7.59ms remaining: 68.3ms
10: learn: 0.6436401 total: 8.15ms remaining: 65.9ms
11: learn: 0.6392506 total: 8.73ms remaining: 64ms
12: learn: 0.6352223 total: 9.4ms remaining: 62.9ms
13: learn: 0.6314012 total: 10.5ms remaining: 64.5ms
14: learn: 0.6271130 total: 11.3ms remaining: 63.9ms
15: learn: 0.6228527 total: 11.8ms remaining: 62.2ms
16: learn: 0.6193636 total: 12.5ms remaining: 61.1ms
17: learn: 0.6149698 total: 13.7ms remaining: 62.4ms
18: learn: 0.6115084 total: 14.2ms remaining: 60.6ms
19: learn: 0.6080555 total: 15.2ms remaining: 60.9ms
20: learn: 0.6044442 total: 16.4ms remaining: 61.6ms
21: learn: 0.6011192 total: 17.5ms remaining: 62.1ms
22: learn: 0.5974142 total: 18.6ms remaining: 62.4ms
23: learn: 0.5941231 total: 19.6ms remaining: 62.1ms
24: learn: 0.5900927 total: 20.7ms remaining: 62ms
25: learn: 0.5870085 total: 21.8ms remaining: 62ms
26: learn: 0.5840114 total: 22.9ms remaining: 61.9ms
27: learn: 0.5808997 total: 24.4ms remaining: 62.6ms
28: learn: 0.5778788 total: 25.3ms remaining: 62ms
29: learn: 0.5750842 total: 26.1ms remaining: 60.8ms
30: learn: 0.5722879 total: 26.8ms remaining: 59.7ms
31: learn: 0.5693116 total: 27.3ms remaining: 58ms
32: learn: 0.5661397 total: 28.5ms remaining: 57.9ms
33: learn: 0.5627994 total: 29.6ms remaining: 57.5ms
34: learn: 0.5597978 total: 31.2ms remaining: 57.9ms
35: learn: 0.5564847 total: 32.4ms remaining: 57.5ms
36: learn: 0.5535813 total: 33.4ms remaining: 56.9ms
37: learn: 0.5511907 total: 34.3ms remaining: 56ms
38: learn: 0.5477524 total: 35.4ms remaining: 55.3ms
39: learn: 0.5446551 total: 36.8ms remaining: 55.1ms
40: learn: 0.5418243 total: 37.8ms remaining: 54.4ms
41: learn: 0.5384306 total: 38.9ms remaining: 53.7ms
42: learn: 0.5355158 total: 40.1ms remaining: 53.2ms
43: learn: 0.5330304 total: 41.2ms remaining: 52.4ms
44: learn: 0.5303464 total: 42.5ms remaining: 51.9ms
45: learn: 0.5282582 total: 43.3ms remaining: 50.8ms
46: learn: 0.5258402 total: 44.7ms remaining: 50.4ms
47: learn: 0.5235785 total: 45.6ms remaining: 49.4ms
48: learn: 0.5207689 total: 46.5ms remaining: 48.4ms
49: learn: 0.5183663 total: 48.1ms remaining: 48.1ms
50: learn: 0.5158919 total: 49.3ms remaining: 47.3ms
51: learn: 0.5139315 total: 50.3ms remaining: 46.4ms
52: learn: 0.5116847 total: 51.5ms remaining: 45.7ms
53: learn: 0.5090818 total: 53ms remaining: 45.1ms
54: learn: 0.5074963 total: 54ms remaining: 44.2ms
55: learn: 0.5046849 total: 55.3ms remaining: 43.4ms
56: learn: 0.5021928 total: 56.3ms remaining: 42.5ms
57: learn: 0.4999986 total: 58.1ms remaining: 42.1ms
58: learn: 0.4977483 total: 59.1ms remaining: 41.1ms
59: learn: 0.4952915 total: 59.9ms remaining: 39.9ms
60: learn: 0.4936844 total: 61.9ms remaining: 39.6ms
61: learn: 0.4916698 total: 63ms remaining: 38.6ms
62: learn: 0.4893568 total: 64.6ms remaining: 38ms
63: learn: 0.4871933 total: 65.7ms remaining: 36.9ms
64: learn: 0.4850710 total: 67.4ms remaining: 36.3ms
65: learn: 0.4829277 total: 68.3ms remaining: 35.2ms
66: learn: 0.4809395 total: 69.6ms remaining: 34.3ms
67: learn: 0.4784928 total: 70.6ms remaining: 33.2ms
68: learn: 0.4763350 total: 72ms remaining: 32.4ms
69: learn: 0.4741475 total: 72.9ms remaining: 31.2ms
70: learn: 0.4726073 total: 73.6ms remaining: 30.1ms
71: learn: 0.4705962 total: 74.8ms remaining: 29.1ms
72: learn: 0.4689538 total: 75.8ms remaining: 28ms
73: learn: 0.4668614 total: 76.6ms remaining: 26.9ms
74: learn: 0.4645647 total: 77.6ms remaining: 25.9ms
75: learn: 0.4623719 total: 78.6ms remaining: 24.8ms
76: learn: 0.4604691 total: 79.7ms remaining: 23.8ms
77: learn: 0.4585415 total: 80.4ms remaining: 22.7ms
78: learn: 0.4563981 total: 81.5ms remaining: 21.7ms
79: learn: 0.4544774 total: 82.3ms remaining: 20.6ms
80: learn: 0.4523978 total: 83ms remaining: 19.5ms
81: learn: 0.4507069 total: 83.9ms remaining: 18.4ms
82: learn: 0.4484920 total: 84.7ms remaining: 17.3ms
83: learn: 0.4466815 total: 85.6ms remaining: 16.3ms
84: learn: 0.4449599 total: 86.3ms remaining: 15.2ms
85: learn: 0.4430525 total: 86.9ms remaining: 14.1ms
86: learn: 0.4413869 total: 87.6ms remaining: 13.1ms
87: learn: 0.4399236 total: 88.2ms remaining: 12ms
88: learn: 0.4380750 total: 89ms remaining: 11ms
89: learn: 0.4362257 total: 89.9ms remaining: 9.99ms
90: learn: 0.4345031 total: 90.4ms remaining: 8.94ms
91: learn: 0.4328117 total: 91.2ms remaining: 7.93ms
92: learn: 0.4313235 total: 91.9ms remaining: 6.91ms
93: learn: 0.4295387 total: 92.5ms remaining: 5.9ms
94: learn: 0.4281810 total: 93ms remaining: 4.9ms
95: learn: 0.4265977 total: 93.7ms remaining: 3.9ms
96: learn: 0.4253384 total: 94.1ms remaining: 2.91ms
97: learn: 0.4236111 total: 94.7ms remaining: 1.93ms
98: learn: 0.4216073 total: 95.4ms remaining: 963us
99: learn: 0.4200482 total: 95.8ms remaining: 0us
0: learn: 0.6886824 total: 1.13ms remaining: 112ms
1: learn: 0.6836439 total: 2.33ms remaining: 114ms
2: learn: 0.6783803 total: 3.22ms remaining: 104ms
3: learn: 0.6732528 total: 4.82ms remaining: 116ms
4: learn: 0.6684937 total: 5.73ms remaining: 109ms
5: learn: 0.6634511 total: 6.83ms remaining: 107ms
6: learn: 0.6589192 total: 7.72ms remaining: 103ms
7: learn: 0.6551883 total: 9.11ms remaining: 105ms
8: learn: 0.6502473 total: 10.4ms remaining: 105ms
9: learn: 0.6460178 total: 11.4ms remaining: 102ms
10: learn: 0.6419694 total: 12.4ms remaining: 100ms
11: learn: 0.6383164 total: 13.7ms remaining: 101ms
12: learn: 0.6337982 total: 14.8ms remaining: 99.1ms
13: learn: 0.6295016 total: 16.2ms remaining: 99.8ms
14: learn: 0.6253754 total: 17.1ms remaining: 97.2ms
15: learn: 0.6213047 total: 18.4ms remaining: 96.3ms
16: learn: 0.6172331 total: 19.7ms remaining: 96.3ms
17: learn: 0.6140067 total: 20.9ms remaining: 95ms
18: learn: 0.6101040 total: 22ms remaining: 93.7ms
19: learn: 0.6066469 total: 23.7ms remaining: 94.8ms
20: learn: 0.6029030 total: 25.2ms remaining: 94.9ms
21: learn: 0.5987959 total: 26.4ms remaining: 93.7ms
22: learn: 0.5948275 total: 27.4ms remaining: 91.9ms
23: learn: 0.5915322 total: 28.2ms remaining: 89.4ms
24: learn: 0.5872507 total: 29.7ms remaining: 89.1ms
25: learn: 0.5839759 total: 31.6ms remaining: 90ms
26: learn: 0.5805313 total: 33.3ms remaining: 89.9ms
27: learn: 0.5778302 total: 34.5ms remaining: 88.6ms
28: learn: 0.5752130 total: 35.8ms remaining: 87.6ms
29: learn: 0.5727888 total: 36.7ms remaining: 85.7ms
30: learn: 0.5702654 total: 38.3ms remaining: 85.3ms
31: learn: 0.5666849 total: 39.5ms remaining: 84ms
32: learn: 0.5634739 total: 41ms remaining: 83.2ms
33: learn: 0.5599432 total: 42.3ms remaining: 82.2ms
34: learn: 0.5569637 total: 43.9ms remaining: 81.5ms
35: learn: 0.5542632 total: 45.2ms remaining: 80.3ms
36: learn: 0.5511816 total: 46.1ms remaining: 78.4ms
37: learn: 0.5479280 total: 47.3ms remaining: 77.2ms
38: learn: 0.5446114 total: 48.3ms remaining: 75.5ms
39: learn: 0.5416442 total: 49.9ms remaining: 74.9ms
40: learn: 0.5388472 total: 51.1ms remaining: 73.6ms
41: learn: 0.5353803 total: 52ms remaining: 71.8ms
42: learn: 0.5323092 total: 53.5ms remaining: 70.9ms
43: learn: 0.5297938 total: 55.3ms remaining: 70.4ms
44: learn: 0.5270777 total: 56ms remaining: 68.5ms
45: learn: 0.5242021 total: 57.4ms remaining: 67.4ms
46: learn: 0.5210269 total: 59.2ms remaining: 66.8ms
47: learn: 0.5183354 total: 60.3ms remaining: 65.4ms
48: learn: 0.5155134 total: 61.7ms remaining: 64.3ms
49: learn: 0.5125593 total: 63ms remaining: 63ms
50: learn: 0.5100516 total: 63.6ms remaining: 61.1ms
51: learn: 0.5082201 total: 64.1ms remaining: 59.2ms
52: learn: 0.5057142 total: 65.3ms remaining: 57.9ms
53: learn: 0.5033139 total: 66.6ms remaining: 56.8ms
54: learn: 0.5008812 total: 68ms remaining: 55.6ms
55: learn: 0.4980920 total: 68.8ms remaining: 54.1ms
56: learn: 0.4952230 total: 70.1ms remaining: 52.9ms
57: learn: 0.4929389 total: 71ms remaining: 51.4ms
58: learn: 0.4904993 total: 72.5ms remaining: 50.4ms
59: learn: 0.4881358 total: 73.6ms remaining: 49.1ms
60: learn: 0.4858069 total: 74.8ms remaining: 47.8ms
61: learn: 0.4832165 total: 75.6ms remaining: 46.3ms
62: learn: 0.4808001 total: 76.8ms remaining: 45.1ms
63: learn: 0.4789522 total: 77.8ms remaining: 43.8ms
64: learn: 0.4769107 total: 79.2ms remaining: 42.7ms
65: learn: 0.4744882 total: 80.5ms remaining: 41.5ms
66: learn: 0.4720492 total: 81.2ms remaining: 40ms
67: learn: 0.4698849 total: 82.1ms remaining: 38.6ms
68: learn: 0.4680298 total: 83ms remaining: 37.3ms
69: learn: 0.4657194 total: 84ms remaining: 36ms
70: learn: 0.4637653 total: 84.7ms remaining: 34.6ms
71: learn: 0.4614873 total: 85.7ms remaining: 33.3ms
72: learn: 0.4593273 total: 86.5ms remaining: 32ms
73: learn: 0.4572340 total: 87.2ms remaining: 30.7ms
74: learn: 0.4552783 total: 88.4ms remaining: 29.5ms
75: learn: 0.4531864 total: 89.2ms remaining: 28.2ms
76: learn: 0.4514435 total: 90.2ms remaining: 26.9ms
77: learn: 0.4495506 total: 90.8ms remaining: 25.6ms
78: learn: 0.4474527 total: 91.7ms remaining: 24.4ms
79: learn: 0.4457739 total: 92.1ms remaining: 23ms
80: learn: 0.4442183 total: 92.8ms remaining: 21.8ms
81: learn: 0.4421743 total: 93.2ms remaining: 20.5ms
82: learn: 0.4403339 total: 93.6ms remaining: 19.2ms
83: learn: 0.4385316 total: 94ms remaining: 17.9ms
84: learn: 0.4369359 total: 94.4ms remaining: 16.7ms
85: learn: 0.4355767 total: 94.8ms remaining: 15.4ms
86: learn: 0.4339461 total: 95.3ms remaining: 14.2ms
87: learn: 0.4322172 total: 95.8ms remaining: 13.1ms
88: learn: 0.4304228 total: 96.6ms remaining: 11.9ms
89: learn: 0.4289271 total: 97.2ms remaining: 10.8ms
90: learn: 0.4272516 total: 97.7ms remaining: 9.66ms
91: learn: 0.4256348 total: 98.3ms remaining: 8.54ms
92: learn: 0.4237505 total: 98.9ms remaining: 7.44ms
93: learn: 0.4220555 total: 99.4ms remaining: 6.34ms
94: learn: 0.4203653 total: 99.9ms remaining: 5.26ms
95: learn: 0.4185166 total: 100ms remaining: 4.19ms
96: learn: 0.4168763 total: 101ms remaining: 3.12ms
97: learn: 0.4157957 total: 102ms remaining: 2.07ms
98: learn: 0.4140381 total: 102ms remaining: 1.03ms
99: learn: 0.4123918 total: 103ms remaining: 0us
0: learn: 0.6884710 total: 1.28ms remaining: 127ms
1: learn: 0.6831483 total: 2.12ms remaining: 104ms
2: learn: 0.6781742 total: 2.48ms remaining: 80.1ms
3: learn: 0.6735979 total: 3.94ms remaining: 94.5ms
4: learn: 0.6690825 total: 4.97ms remaining: 94.4ms
5: learn: 0.6643177 total: 6.41ms remaining: 100ms
6: learn: 0.6602321 total: 7.44ms remaining: 98.9ms
7: learn: 0.6561625 total: 8.4ms remaining: 96.7ms
8: learn: 0.6513684 total: 9.4ms remaining: 95.1ms
9: learn: 0.6464446 total: 10.7ms remaining: 95.9ms
10: learn: 0.6425160 total: 11.4ms remaining: 92.1ms
11: learn: 0.6379029 total: 12.2ms remaining: 89.6ms
12: learn: 0.6338682 total: 13.2ms remaining: 88.7ms
13: learn: 0.6299456 total: 14.6ms remaining: 89.5ms
14: learn: 0.6261431 total: 15.3ms remaining: 86.7ms
15: learn: 0.6214318 total: 16.1ms remaining: 84.6ms
16: learn: 0.6176888 total: 17.3ms remaining: 84.5ms
17: learn: 0.6136496 total: 18.7ms remaining: 85.1ms
18: learn: 0.6098567 total: 19.2ms remaining: 81.7ms
19: learn: 0.6061197 total: 19.9ms remaining: 79.6ms
20: learn: 0.6021095 total: 21.1ms remaining: 79.4ms
21: learn: 0.5977883 total: 22.3ms remaining: 79.1ms
22: learn: 0.5936190 total: 23.1ms remaining: 77.5ms
23: learn: 0.5899167 total: 24.1ms remaining: 76.2ms
24: learn: 0.5858649 total: 25.1ms remaining: 75.2ms
25: learn: 0.5829470 total: 26.3ms remaining: 74.8ms
26: learn: 0.5798447 total: 27.8ms remaining: 75.2ms
27: learn: 0.5765984 total: 29.1ms remaining: 74.8ms
28: learn: 0.5737242 total: 30.2ms remaining: 74ms
29: learn: 0.5710620 total: 31.1ms remaining: 72.5ms
30: learn: 0.5684374 total: 32.4ms remaining: 72.2ms
31: learn: 0.5652138 total: 33.1ms remaining: 70.3ms
32: learn: 0.5622089 total: 34.1ms remaining: 69.1ms
33: learn: 0.5588240 total: 35.3ms remaining: 68.5ms
34: learn: 0.5553290 total: 36ms remaining: 66.9ms
35: learn: 0.5523463 total: 36.5ms remaining: 64.9ms
36: learn: 0.5491377 total: 37.7ms remaining: 64.1ms
37: learn: 0.5464623 total: 38.7ms remaining: 63.1ms
38: learn: 0.5430188 total: 39.4ms remaining: 61.7ms
39: learn: 0.5400183 total: 40.7ms remaining: 61ms
40: learn: 0.5375189 total: 41.5ms remaining: 59.7ms
41: learn: 0.5339443 total: 42.4ms remaining: 58.5ms
42: learn: 0.5309167 total: 43.2ms remaining: 57.3ms
43: learn: 0.5285461 total: 44.3ms remaining: 56.4ms
44: learn: 0.5257915 total: 45.4ms remaining: 55.4ms
45: learn: 0.5235061 total: 46.1ms remaining: 54.2ms
46: learn: 0.5208134 total: 47.2ms remaining: 53.2ms
47: learn: 0.5181357 total: 47.8ms remaining: 51.8ms
48: learn: 0.5152125 total: 48.5ms remaining: 50.4ms
49: learn: 0.5127882 total: 49.4ms remaining: 49.4ms
50: learn: 0.5103441 total: 50.3ms remaining: 48.3ms
51: learn: 0.5085697 total: 50.9ms remaining: 47ms
52: learn: 0.5060762 total: 51.7ms remaining: 45.9ms
53: learn: 0.5036028 total: 52.5ms remaining: 44.8ms
54: learn: 0.5011696 total: 53.3ms remaining: 43.6ms
55: learn: 0.4985615 total: 54.3ms remaining: 42.6ms
56: learn: 0.4958495 total: 54.8ms remaining: 41.3ms
57: learn: 0.4934627 total: 55.7ms remaining: 40.4ms
58: learn: 0.4911664 total: 56.3ms remaining: 39.1ms
59: learn: 0.4888431 total: 57ms remaining: 38ms
60: learn: 0.4867067 total: 57.6ms remaining: 36.9ms
61: learn: 0.4843027 total: 58.1ms remaining: 35.6ms
62: learn: 0.4819804 total: 58.8ms remaining: 34.5ms
63: learn: 0.4799181 total: 59.2ms remaining: 33.3ms
64: learn: 0.4777196 total: 59.9ms remaining: 32.3ms
65: learn: 0.4752769 total: 60.4ms remaining: 31.1ms
66: learn: 0.4731355 total: 60.9ms remaining: 30ms
67: learn: 0.4709208 total: 61.4ms remaining: 28.9ms
68: learn: 0.4688675 total: 61.7ms remaining: 27.7ms
69: learn: 0.4669209 total: 62.3ms remaining: 26.7ms
70: learn: 0.4646306 total: 63.1ms remaining: 25.8ms
71: learn: 0.4624272 total: 63.6ms remaining: 24.7ms
72: learn: 0.4603330 total: 64.1ms remaining: 23.7ms
73: learn: 0.4585022 total: 64.7ms remaining: 22.7ms
74: learn: 0.4566034 total: 65.2ms remaining: 21.7ms
75: learn: 0.4548515 total: 65.8ms remaining: 20.8ms
76: learn: 0.4527723 total: 66.3ms remaining: 19.8ms
77: learn: 0.4508659 total: 67ms remaining: 18.9ms
78: learn: 0.4486937 total: 67.5ms remaining: 17.9ms
79: learn: 0.4468623 total: 67.8ms remaining: 17ms
80: learn: 0.4453247 total: 68.4ms remaining: 16ms
81: learn: 0.4434634 total: 68.7ms remaining: 15.1ms
82: learn: 0.4412807 total: 69.1ms remaining: 14.2ms
83: learn: 0.4393264 total: 69.4ms remaining: 13.2ms
84: learn: 0.4374024 total: 69.9ms remaining: 12.3ms
85: learn: 0.4357541 total: 70.2ms remaining: 11.4ms
86: learn: 0.4340766 total: 70.5ms remaining: 10.5ms
87: learn: 0.4326258 total: 70.9ms remaining: 9.67ms
88: learn: 0.4303947 total: 71.3ms remaining: 8.81ms
89: learn: 0.4288562 total: 71.6ms remaining: 7.95ms
90: learn: 0.4273194 total: 72.1ms remaining: 7.13ms
91: learn: 0.4257218 total: 72.5ms remaining: 6.3ms
92: learn: 0.4239834 total: 72.8ms remaining: 5.48ms
93: learn: 0.4226384 total: 73.2ms remaining: 4.67ms
94: learn: 0.4206514 total: 73.7ms remaining: 3.88ms
95: learn: 0.4190590 total: 74.1ms remaining: 3.09ms
96: learn: 0.4174809 total: 74.5ms remaining: 2.3ms
97: learn: 0.4161426 total: 75ms remaining: 1.53ms
98: learn: 0.4143139 total: 75.4ms remaining: 761us
99: learn: 0.4126026 total: 75.7ms remaining: 0us
0: learn: 0.6888251 total: 1.54ms remaining: 152ms
1: learn: 0.6838743 total: 2.82ms remaining: 138ms
2: learn: 0.6790688 total: 3.92ms remaining: 127ms
3: learn: 0.6744890 total: 4.98ms remaining: 119ms
4: learn: 0.6697935 total: 5.82ms remaining: 111ms
5: learn: 0.6652243 total: 6.82ms remaining: 107ms
6: learn: 0.6616198 total: 8.47ms remaining: 112ms
7: learn: 0.6576694 total: 9.41ms remaining: 108ms
8: learn: 0.6532890 total: 10.9ms remaining: 110ms
9: learn: 0.6492571 total: 12.4ms remaining: 111ms
10: learn: 0.6463276 total: 13.7ms remaining: 111ms
11: learn: 0.6419683 total: 14.7ms remaining: 108ms
12: learn: 0.6383123 total: 15.7ms remaining: 105ms
13: learn: 0.6346641 total: 17.5ms remaining: 107ms
14: learn: 0.6310619 total: 19ms remaining: 108ms
15: learn: 0.6268264 total: 20.5ms remaining: 108ms
16: learn: 0.6232008 total: 21.6ms remaining: 105ms
17: learn: 0.6188903 total: 22.9ms remaining: 104ms
18: learn: 0.6154295 total: 23.9ms remaining: 102ms
19: learn: 0.6110983 total: 24.9ms remaining: 99.8ms
20: learn: 0.6074348 total: 27ms remaining: 101ms
21: learn: 0.6035654 total: 29ms remaining: 103ms
22: learn: 0.6001629 total: 30.2ms remaining: 101ms
23: learn: 0.5972153 total: 31.9ms remaining: 101ms
24: learn: 0.5938984 total: 33ms remaining: 98.9ms
25: learn: 0.5914189 total: 33.9ms remaining: 96.5ms
26: learn: 0.5879412 total: 34.6ms remaining: 93.7ms
27: learn: 0.5844532 total: 35.7ms remaining: 91.7ms
28: learn: 0.5812474 total: 36.2ms remaining: 88.7ms
29: learn: 0.5780829 total: 37.2ms remaining: 86.7ms
30: learn: 0.5747267 total: 37.9ms remaining: 84.5ms
31: learn: 0.5714414 total: 38.7ms remaining: 82.2ms
32: learn: 0.5686548 total: 39.6ms remaining: 80.3ms
33: learn: 0.5656884 total: 40ms remaining: 77.7ms
34: learn: 0.5628498 total: 40.8ms remaining: 75.7ms
35: learn: 0.5601182 total: 41.8ms remaining: 74.2ms
36: learn: 0.5571363 total: 42.5ms remaining: 72.4ms
37: learn: 0.5543120 total: 43.2ms remaining: 70.5ms
38: learn: 0.5508372 total: 43.9ms remaining: 68.6ms
39: learn: 0.5482084 total: 44.8ms remaining: 67.1ms
40: learn: 0.5451572 total: 45.4ms remaining: 65.4ms
41: learn: 0.5418393 total: 45.9ms remaining: 63.4ms
42: learn: 0.5393336 total: 46.5ms remaining: 61.7ms
43: learn: 0.5368743 total: 47.3ms remaining: 60.2ms
44: learn: 0.5341000 total: 47.9ms remaining: 58.5ms
45: learn: 0.5315797 total: 48.5ms remaining: 56.9ms
46: learn: 0.5288963 total: 49.1ms remaining: 55.4ms
47: learn: 0.5262389 total: 49.7ms remaining: 53.8ms
48: learn: 0.5234348 total: 50ms remaining: 52ms
49: learn: 0.5209916 total: 50.6ms remaining: 50.6ms
50: learn: 0.5185438 total: 51.1ms remaining: 49.1ms
51: learn: 0.5166256 total: 51.6ms remaining: 47.7ms
52: learn: 0.5142948 total: 52ms remaining: 46.1ms
53: learn: 0.5119237 total: 52.6ms remaining: 44.8ms
54: learn: 0.5099750 total: 53.1ms remaining: 43.5ms
55: learn: 0.5073470 total: 53.8ms remaining: 42.2ms
56: learn: 0.5049048 total: 54.2ms remaining: 40.9ms
57: learn: 0.5025382 total: 54.8ms remaining: 39.7ms
58: learn: 0.5001287 total: 55.2ms remaining: 38.4ms
59: learn: 0.4976328 total: 55.9ms remaining: 37.3ms
60: learn: 0.4954885 total: 56.5ms remaining: 36.1ms
61: learn: 0.4929791 total: 57.1ms remaining: 35ms
62: learn: 0.4906066 total: 57.7ms remaining: 33.9ms
63: learn: 0.4883619 total: 58.2ms remaining: 32.7ms
64: learn: 0.4861991 total: 58.7ms remaining: 31.6ms
65: learn: 0.4838713 total: 59.2ms remaining: 30.5ms
66: learn: 0.4817598 total: 59.8ms remaining: 29.5ms
67: learn: 0.4797222 total: 60.3ms remaining: 28.4ms
68: learn: 0.4776118 total: 60.9ms remaining: 27.3ms
69: learn: 0.4758566 total: 61.4ms remaining: 26.3ms
70: learn: 0.4737977 total: 61.9ms remaining: 25.3ms
71: learn: 0.4716287 total: 62.3ms remaining: 24.2ms
72: learn: 0.4696145 total: 62.8ms remaining: 23.2ms
73: learn: 0.4676496 total: 63.2ms remaining: 22.2ms
74: learn: 0.4655607 total: 63.7ms remaining: 21.2ms
75: learn: 0.4636208 total: 64.2ms remaining: 20.3ms
76: learn: 0.4618838 total: 64.7ms remaining: 19.3ms
77: learn: 0.4598499 total: 65.2ms remaining: 18.4ms
78: learn: 0.4579391 total: 65.6ms remaining: 17.4ms
79: learn: 0.4558926 total: 66ms remaining: 16.5ms
80: learn: 0.4538738 total: 66.6ms remaining: 15.6ms
81: learn: 0.4520528 total: 67ms remaining: 14.7ms
82: learn: 0.4502268 total: 67.3ms remaining: 13.8ms
83: learn: 0.4484919 total: 67.8ms remaining: 12.9ms
84: learn: 0.4468533 total: 68.2ms remaining: 12ms
85: learn: 0.4450997 total: 68.5ms remaining: 11.2ms
86: learn: 0.4432632 total: 68.9ms remaining: 10.3ms
87: learn: 0.4416660 total: 69.3ms remaining: 9.45ms
88: learn: 0.4401842 total: 69.7ms remaining: 8.61ms
89: learn: 0.4386951 total: 70ms remaining: 7.78ms
90: learn: 0.4370228 total: 70.4ms remaining: 6.96ms
91: learn: 0.4353236 total: 70.8ms remaining: 6.16ms
92: learn: 0.4339369 total: 71.3ms remaining: 5.36ms
93: learn: 0.4326707 total: 71.6ms remaining: 4.57ms
94: learn: 0.4309095 total: 72ms remaining: 3.79ms
95: learn: 0.4293567 total: 72.5ms remaining: 3.02ms
96: learn: 0.4277585 total: 72.9ms remaining: 2.25ms
97: learn: 0.4259049 total: 73.2ms remaining: 1.49ms
98: learn: 0.4241491 total: 73.6ms remaining: 743us
99: learn: 0.4228214 total: 74ms remaining: 0us
0: learn: 0.6881993 total: 1.31ms remaining: 130ms
1: learn: 0.6827008 total: 2.59ms remaining: 127ms
2: learn: 0.6774722 total: 3.88ms remaining: 125ms
3: learn: 0.6721781 total: 5.48ms remaining: 132ms
4: learn: 0.6675501 total: 6.58ms remaining: 125ms
5: learn: 0.6623863 total: 8.45ms remaining: 132ms
6: learn: 0.6575695 total: 9.9ms remaining: 132ms
7: learn: 0.6535074 total: 11.5ms remaining: 132ms
8: learn: 0.6485427 total: 12.9ms remaining: 130ms
9: learn: 0.6436104 total: 14.7ms remaining: 132ms
10: learn: 0.6395245 total: 15.9ms remaining: 129ms
11: learn: 0.6347631 total: 17.1ms remaining: 126ms
12: learn: 0.6305070 total: 18.5ms remaining: 124ms
13: learn: 0.6265698 total: 20.2ms remaining: 124ms
14: learn: 0.6229825 total: 21.7ms remaining: 123ms
15: learn: 0.6182567 total: 22.8ms remaining: 120ms
16: learn: 0.6144702 total: 24.6ms remaining: 120ms
17: learn: 0.6102756 total: 26.6ms remaining: 121ms
18: learn: 0.6063688 total: 27.3ms remaining: 116ms
19: learn: 0.6025233 total: 29.2ms remaining: 117ms
20: learn: 0.5986269 total: 30.8ms remaining: 116ms
21: learn: 0.5942607 total: 32.9ms remaining: 117ms
22: learn: 0.5899548 total: 35.4ms remaining: 119ms
23: learn: 0.5862924 total: 36.6ms remaining: 116ms
24: learn: 0.5830002 total: 38.2ms remaining: 115ms
25: learn: 0.5798113 total: 39.7ms remaining: 113ms
26: learn: 0.5764879 total: 41.2ms remaining: 111ms
27: learn: 0.5732837 total: 42.6ms remaining: 110ms
28: learn: 0.5701657 total: 43.5ms remaining: 106ms
29: learn: 0.5673144 total: 44.6ms remaining: 104ms
30: learn: 0.5645907 total: 46.3ms remaining: 103ms
31: learn: 0.5612450 total: 47.3ms remaining: 100ms
32: learn: 0.5579183 total: 49.2ms remaining: 99.8ms
33: learn: 0.5543718 total: 50.7ms remaining: 98.4ms
34: learn: 0.5513997 total: 51.5ms remaining: 95.7ms
35: learn: 0.5484174 total: 52.7ms remaining: 93.7ms
36: learn: 0.5451814 total: 54.4ms remaining: 92.7ms
37: learn: 0.5421966 total: 55.9ms remaining: 91.2ms
38: learn: 0.5386374 total: 57.2ms remaining: 89.4ms
39: learn: 0.5359418 total: 58.4ms remaining: 87.5ms
40: learn: 0.5328272 total: 59.7ms remaining: 85.9ms
41: learn: 0.5292787 total: 60.6ms remaining: 83.7ms
42: learn: 0.5261061 total: 61.7ms remaining: 81.8ms
43: learn: 0.5237163 total: 62.9ms remaining: 80ms
44: learn: 0.5208345 total: 64.2ms remaining: 78.4ms
45: learn: 0.5184355 total: 65.1ms remaining: 76.4ms
46: learn: 0.5158247 total: 66.2ms remaining: 74.7ms
47: learn: 0.5135288 total: 67ms remaining: 72.6ms
48: learn: 0.5104705 total: 67.7ms remaining: 70.4ms
49: learn: 0.5081160 total: 68.2ms remaining: 68.2ms
50: learn: 0.5056550 total: 68.8ms remaining: 66.1ms
51: learn: 0.5038595 total: 69.6ms remaining: 64.2ms
52: learn: 0.5013747 total: 70.6ms remaining: 62.6ms
53: learn: 0.4986649 total: 71.2ms remaining: 60.7ms
54: learn: 0.4965716 total: 72ms remaining: 58.9ms
55: learn: 0.4939157 total: 72.7ms remaining: 57.1ms
56: learn: 0.4910497 total: 73.4ms remaining: 55.4ms
57: learn: 0.4884881 total: 74.3ms remaining: 53.8ms
58: learn: 0.4862442 total: 75ms remaining: 52.1ms
59: learn: 0.4839807 total: 75.6ms remaining: 50.4ms
60: learn: 0.4813368 total: 76.2ms remaining: 48.7ms
61: learn: 0.4787983 total: 76.8ms remaining: 47.1ms
62: learn: 0.4763822 total: 77.4ms remaining: 45.5ms
63: learn: 0.4742053 total: 77.8ms remaining: 43.8ms
64: learn: 0.4720609 total: 78.5ms remaining: 42.3ms
65: learn: 0.4695373 total: 79.3ms remaining: 40.8ms
66: learn: 0.4674749 total: 79.8ms remaining: 39.3ms
67: learn: 0.4652863 total: 80.4ms remaining: 37.8ms
68: learn: 0.4631123 total: 80.7ms remaining: 36.3ms
69: learn: 0.4611278 total: 81.3ms remaining: 34.8ms
70: learn: 0.4589138 total: 81.8ms remaining: 33.4ms
71: learn: 0.4566025 total: 82.4ms remaining: 32.1ms
72: learn: 0.4542314 total: 83ms remaining: 30.7ms
73: learn: 0.4520691 total: 83.5ms remaining: 29.3ms
74: learn: 0.4500480 total: 83.9ms remaining: 28ms
75: learn: 0.4478340 total: 84.3ms remaining: 26.6ms
76: learn: 0.4463365 total: 84.8ms remaining: 25.3ms
77: learn: 0.4445490 total: 85.3ms remaining: 24.1ms
78: learn: 0.4427074 total: 85.8ms remaining: 22.8ms
79: learn: 0.4405743 total: 86.3ms remaining: 21.6ms
80: learn: 0.4388167 total: 86.9ms remaining: 20.4ms
81: learn: 0.4366750 total: 87.2ms remaining: 19.1ms
82: learn: 0.4348196 total: 87.6ms remaining: 17.9ms
83: learn: 0.4329126 total: 88.1ms remaining: 16.8ms
84: learn: 0.4311000 total: 88.7ms remaining: 15.7ms
85: learn: 0.4293995 total: 89.2ms remaining: 14.5ms
86: learn: 0.4272472 total: 89.5ms remaining: 13.4ms
87: learn: 0.4252671 total: 90ms remaining: 12.3ms
88: learn: 0.4234933 total: 90.5ms remaining: 11.2ms
89: learn: 0.4218135 total: 90.8ms remaining: 10.1ms
90: learn: 0.4200800 total: 91.3ms remaining: 9.03ms
91: learn: 0.4184939 total: 92ms remaining: 8ms
92: learn: 0.4165901 total: 92.4ms remaining: 6.96ms
93: learn: 0.4152496 total: 92.8ms remaining: 5.92ms
94: learn: 0.4135666 total: 93.3ms remaining: 4.91ms
95: learn: 0.4122281 total: 93.8ms remaining: 3.91ms
96: learn: 0.4103071 total: 94.1ms remaining: 2.91ms
97: learn: 0.4088729 total: 94.4ms remaining: 1.93ms
98: learn: 0.4071209 total: 94.8ms remaining: 957us
99: learn: 0.4055178 total: 95.2ms remaining: 0us
0: learn: 0.6886959 total: 1.36ms remaining: 135ms
1: learn: 0.6838265 total: 2.73ms remaining: 134ms
2: learn: 0.6793184 total: 3.69ms remaining: 119ms
3: learn: 0.6746587 total: 5ms remaining: 120ms
4: learn: 0.6698654 total: 6.23ms remaining: 118ms
5: learn: 0.6655969 total: 7.84ms remaining: 123ms
6: learn: 0.6613624 total: 9.4ms remaining: 125ms
7: learn: 0.6574805 total: 10.8ms remaining: 124ms
8: learn: 0.6531921 total: 11.8ms remaining: 120ms
9: learn: 0.6488752 total: 13.3ms remaining: 120ms
10: learn: 0.6451668 total: 14.6ms remaining: 118ms
11: learn: 0.6411035 total: 16.1ms remaining: 118ms
12: learn: 0.6370053 total: 17.9ms remaining: 120ms
13: learn: 0.6329368 total: 19.7ms remaining: 121ms
14: learn: 0.6292349 total: 21.2ms remaining: 120ms
15: learn: 0.6251452 total: 22.2ms remaining: 116ms
16: learn: 0.6212621 total: 23.5ms remaining: 115ms
17: learn: 0.6170321 total: 25.3ms remaining: 115ms
18: learn: 0.6132722 total: 26.2ms remaining: 112ms
19: learn: 0.6099714 total: 27.7ms remaining: 111ms
20: learn: 0.6066178 total: 29.2ms remaining: 110ms
21: learn: 0.6027816 total: 31.4ms remaining: 111ms
22: learn: 0.5990405 total: 32.9ms remaining: 110ms
23: learn: 0.5957426 total: 33.8ms remaining: 107ms
24: learn: 0.5922791 total: 35.1ms remaining: 105ms
25: learn: 0.5893376 total: 36.2ms remaining: 103ms
26: learn: 0.5861754 total: 37.7ms remaining: 102ms
27: learn: 0.5829950 total: 38.9ms remaining: 100ms
28: learn: 0.5794016 total: 40.2ms remaining: 98.5ms
29: learn: 0.5765496 total: 41.9ms remaining: 97.8ms
30: learn: 0.5736307 total: 43.2ms remaining: 96.2ms
31: learn: 0.5704244 total: 44.3ms remaining: 94.2ms
32: learn: 0.5676356 total: 46ms remaining: 93.3ms
33: learn: 0.5646941 total: 46.7ms remaining: 90.6ms
34: learn: 0.5620194 total: 48.2ms remaining: 89.6ms
35: learn: 0.5600655 total: 49.2ms remaining: 87.5ms
36: learn: 0.5572839 total: 50.5ms remaining: 85.9ms
37: learn: 0.5547866 total: 51.2ms remaining: 83.5ms
38: learn: 0.5521352 total: 52ms remaining: 81.4ms
39: learn: 0.5493307 total: 53.5ms remaining: 80.3ms
40: learn: 0.5467472 total: 54.6ms remaining: 78.5ms
41: learn: 0.5446479 total: 55.1ms remaining: 76.1ms
42: learn: 0.5426825 total: 56ms remaining: 74.2ms
43: learn: 0.5399677 total: 57.6ms remaining: 73.3ms
44: learn: 0.5379476 total: 59.1ms remaining: 72.3ms
45: learn: 0.5354392 total: 60.3ms remaining: 70.8ms
46: learn: 0.5325750 total: 61.2ms remaining: 69ms
47: learn: 0.5300571 total: 61.9ms remaining: 67.1ms
48: learn: 0.5276968 total: 63ms remaining: 65.6ms
49: learn: 0.5250418 total: 63.7ms remaining: 63.7ms
50: learn: 0.5225769 total: 65ms remaining: 62.4ms
51: learn: 0.5200919 total: 66.1ms remaining: 61ms
52: learn: 0.5178163 total: 67ms remaining: 59.4ms
53: learn: 0.5153453 total: 68.1ms remaining: 58ms
54: learn: 0.5127813 total: 68.9ms remaining: 56.4ms
55: learn: 0.5104769 total: 69.9ms remaining: 54.9ms
56: learn: 0.5081644 total: 70.8ms remaining: 53.4ms
57: learn: 0.5056439 total: 71.7ms remaining: 51.9ms
58: learn: 0.5031357 total: 72.5ms remaining: 50.4ms
59: learn: 0.5007311 total: 73.2ms remaining: 48.8ms
60: learn: 0.4983959 total: 74ms remaining: 47.3ms
61: learn: 0.4962703 total: 74.7ms remaining: 45.8ms
62: learn: 0.4946152 total: 75.4ms remaining: 44.3ms
63: learn: 0.4924402 total: 76.1ms remaining: 42.8ms
64: learn: 0.4904118 total: 76.7ms remaining: 41.3ms
65: learn: 0.4883486 total: 77.2ms remaining: 39.8ms
66: learn: 0.4863043 total: 77.8ms remaining: 38.3ms
67: learn: 0.4838890 total: 78.6ms remaining: 37ms
68: learn: 0.4820407 total: 79.2ms remaining: 35.6ms
69: learn: 0.4799651 total: 79.8ms remaining: 34.2ms
70: learn: 0.4783326 total: 80.2ms remaining: 32.7ms
71: learn: 0.4762731 total: 80.9ms remaining: 31.5ms
72: learn: 0.4745866 total: 81.3ms remaining: 30.1ms
73: learn: 0.4723220 total: 81.9ms remaining: 28.8ms
74: learn: 0.4704764 total: 82.3ms remaining: 27.4ms
75: learn: 0.4689428 total: 82.7ms remaining: 26.1ms
76: learn: 0.4667857 total: 83.3ms remaining: 24.9ms
77: learn: 0.4649443 total: 83.9ms remaining: 23.7ms
78: learn: 0.4629698 total: 84.4ms remaining: 22.4ms
79: learn: 0.4614328 total: 85ms remaining: 21.2ms
80: learn: 0.4599062 total: 85.4ms remaining: 20ms
81: learn: 0.4583270 total: 85.8ms remaining: 18.8ms
82: learn: 0.4562689 total: 86.1ms remaining: 17.6ms
83: learn: 0.4546344 total: 86.6ms remaining: 16.5ms
84: learn: 0.4530122 total: 87.1ms remaining: 15.4ms
85: learn: 0.4512857 total: 87.5ms remaining: 14.2ms
86: learn: 0.4495425 total: 87.9ms remaining: 13.1ms
87: learn: 0.4477200 total: 88.2ms remaining: 12ms
88: learn: 0.4460820 total: 88.6ms remaining: 11ms
89: learn: 0.4445896 total: 89.1ms remaining: 9.89ms
90: learn: 0.4427587 total: 89.4ms remaining: 8.84ms
91: learn: 0.4412886 total: 89.7ms remaining: 7.8ms
92: learn: 0.4396576 total: 90.1ms remaining: 6.78ms
93: learn: 0.4380537 total: 90.6ms remaining: 5.79ms
94: learn: 0.4365266 total: 91.1ms remaining: 4.79ms
95: learn: 0.4351730 total: 91.5ms remaining: 3.81ms
96: learn: 0.4337504 total: 92ms remaining: 2.84ms
97: learn: 0.4323788 total: 92.4ms remaining: 1.89ms
98: learn: 0.4307403 total: 92.8ms remaining: 936us
99: learn: 0.4292391 total: 93.1ms remaining: 0us
0: learn: 0.6888430 total: 951us remaining: 94.2ms
1: learn: 0.6844696 total: 1.62ms remaining: 79.3ms
2: learn: 0.6801295 total: 2ms remaining: 64.7ms
3: learn: 0.6752420 total: 2.78ms remaining: 66.8ms
4: learn: 0.6701626 total: 3.8ms remaining: 72.3ms
5: learn: 0.6655182 total: 4.95ms remaining: 77.5ms
6: learn: 0.6620344 total: 6.23ms remaining: 82.8ms
7: learn: 0.6584874 total: 7.03ms remaining: 80.8ms
8: learn: 0.6539240 total: 7.91ms remaining: 80ms
9: learn: 0.6499828 total: 8.89ms remaining: 80ms
10: learn: 0.6455269 total: 9.49ms remaining: 76.8ms
11: learn: 0.6414986 total: 10.4ms remaining: 76.3ms
12: learn: 0.6374259 total: 11.7ms remaining: 78.2ms
13: learn: 0.6333841 total: 13ms remaining: 79.9ms
14: learn: 0.6287552 total: 13.6ms remaining: 76.9ms
15: learn: 0.6255913 total: 14.2ms remaining: 74.6ms
16: learn: 0.6228745 total: 14.7ms remaining: 72ms
17: learn: 0.6192158 total: 15.2ms remaining: 69.1ms
18: learn: 0.6151053 total: 16.4ms remaining: 69.9ms
19: learn: 0.6117255 total: 17.8ms remaining: 71.2ms
20: learn: 0.6077871 total: 18.7ms remaining: 70.2ms
21: learn: 0.6036369 total: 19.8ms remaining: 70.2ms
22: learn: 0.5997223 total: 20.8ms remaining: 69.6ms
23: learn: 0.5957430 total: 21.8ms remaining: 69ms
24: learn: 0.5924717 total: 22.6ms remaining: 67.7ms
25: learn: 0.5891835 total: 23.6ms remaining: 67.1ms
26: learn: 0.5859911 total: 24.5ms remaining: 66.3ms
27: learn: 0.5830599 total: 25.6ms remaining: 65.8ms
28: learn: 0.5804551 total: 26.8ms remaining: 65.5ms
29: learn: 0.5777595 total: 27.3ms remaining: 63.7ms
30: learn: 0.5746008 total: 27.6ms remaining: 61.4ms
31: learn: 0.5713796 total: 28.1ms remaining: 59.6ms
32: learn: 0.5679259 total: 28.5ms remaining: 57.9ms
33: learn: 0.5650754 total: 29.5ms remaining: 57.3ms
34: learn: 0.5622872 total: 30.5ms remaining: 56.7ms
35: learn: 0.5591263 total: 31.4ms remaining: 55.9ms
36: learn: 0.5563465 total: 32.7ms remaining: 55.7ms
37: learn: 0.5529493 total: 33.4ms remaining: 54.6ms
38: learn: 0.5501690 total: 34.3ms remaining: 53.6ms
39: learn: 0.5469190 total: 35.2ms remaining: 52.8ms
40: learn: 0.5432410 total: 36ms remaining: 51.8ms
41: learn: 0.5401389 total: 36.8ms remaining: 50.9ms
42: learn: 0.5378797 total: 37.6ms remaining: 49.9ms
43: learn: 0.5350410 total: 38.4ms remaining: 48.9ms
44: learn: 0.5326489 total: 39ms remaining: 47.7ms
45: learn: 0.5300776 total: 39.4ms remaining: 46.3ms
46: learn: 0.5270868 total: 40.1ms remaining: 45.3ms
47: learn: 0.5249808 total: 40.8ms remaining: 44.2ms
48: learn: 0.5227876 total: 41.4ms remaining: 43.1ms
49: learn: 0.5200878 total: 41.8ms remaining: 41.8ms
50: learn: 0.5172000 total: 43.3ms remaining: 41.6ms
51: learn: 0.5149013 total: 43.9ms remaining: 40.5ms
52: learn: 0.5120194 total: 44.3ms remaining: 39.3ms
53: learn: 0.5093406 total: 44.9ms remaining: 38.2ms
54: learn: 0.5066302 total: 45.6ms remaining: 37.3ms
55: learn: 0.5037123 total: 46ms remaining: 36.1ms
56: learn: 0.5013412 total: 46.4ms remaining: 35ms
57: learn: 0.4989405 total: 46.9ms remaining: 34ms
58: learn: 0.4965496 total: 47.5ms remaining: 33ms
59: learn: 0.4937551 total: 48.1ms remaining: 32.1ms
60: learn: 0.4912467 total: 48.6ms remaining: 31.1ms
61: learn: 0.4885967 total: 49.3ms remaining: 30.2ms
62: learn: 0.4859458 total: 49.6ms remaining: 29.1ms
63: learn: 0.4835383 total: 50ms remaining: 28.1ms
64: learn: 0.4811866 total: 50.4ms remaining: 27.1ms
65: learn: 0.4790421 total: 50.8ms remaining: 26.2ms
66: learn: 0.4767719 total: 51.4ms remaining: 25.3ms
67: learn: 0.4747126 total: 52ms remaining: 24.4ms
68: learn: 0.4723738 total: 52.6ms remaining: 23.6ms
69: learn: 0.4706107 total: 53ms remaining: 22.7ms
70: learn: 0.4684780 total: 53.6ms remaining: 21.9ms
71: learn: 0.4663688 total: 54.2ms remaining: 21.1ms
72: learn: 0.4647644 total: 54.7ms remaining: 20.2ms
73: learn: 0.4625221 total: 55.3ms remaining: 19.4ms
74: learn: 0.4604456 total: 55.8ms remaining: 18.6ms
75: learn: 0.4586607 total: 56.2ms remaining: 17.8ms
76: learn: 0.4567422 total: 56.8ms remaining: 17ms
77: learn: 0.4550168 total: 57.3ms remaining: 16.2ms
78: learn: 0.4529971 total: 57.7ms remaining: 15.3ms
79: learn: 0.4510502 total: 58.1ms remaining: 14.5ms
80: learn: 0.4495286 total: 58.7ms remaining: 13.8ms
81: learn: 0.4478815 total: 59.5ms remaining: 13.1ms
82: learn: 0.4457721 total: 59.8ms remaining: 12.2ms
83: learn: 0.4434536 total: 60.3ms remaining: 11.5ms
84: learn: 0.4415553 total: 61ms remaining: 10.8ms
85: learn: 0.4396112 total: 61.6ms remaining: 10ms
86: learn: 0.4376530 total: 61.9ms remaining: 9.26ms
87: learn: 0.4359785 total: 62.2ms remaining: 8.49ms
88: learn: 0.4342797 total: 62.6ms remaining: 7.74ms
89: learn: 0.4324861 total: 63.2ms remaining: 7.03ms
90: learn: 0.4306033 total: 63.6ms remaining: 6.29ms
91: learn: 0.4291357 total: 64ms remaining: 5.57ms
92: learn: 0.4275348 total: 64.6ms remaining: 4.86ms
93: learn: 0.4261666 total: 65ms remaining: 4.15ms
94: learn: 0.4243422 total: 65.4ms remaining: 3.44ms
95: learn: 0.4225094 total: 65.8ms remaining: 2.74ms
96: learn: 0.4206795 total: 66.2ms remaining: 2.05ms
97: learn: 0.4193637 total: 66.6ms remaining: 1.36ms
98: learn: 0.4176426 total: 66.9ms remaining: 675us
99: learn: 0.4156633 total: 67.2ms remaining: 0us
0: learn: 0.6886167 total: 1.31ms remaining: 129ms
1: learn: 0.6839296 total: 2.54ms remaining: 124ms
2: learn: 0.6790130 total: 3.37ms remaining: 109ms
3: learn: 0.6745981 total: 4.48ms remaining: 107ms
4: learn: 0.6700984 total: 5.6ms remaining: 106ms
5: learn: 0.6652168 total: 6.65ms remaining: 104ms
6: learn: 0.6606527 total: 7.67ms remaining: 102ms
7: learn: 0.6566447 total: 8.76ms remaining: 101ms
8: learn: 0.6519140 total: 9.74ms remaining: 98.5ms
9: learn: 0.6475038 total: 10.8ms remaining: 97.2ms
10: learn: 0.6444133 total: 11.8ms remaining: 95.7ms
11: learn: 0.6400321 total: 12.6ms remaining: 92.2ms
12: learn: 0.6361821 total: 13ms remaining: 87ms
13: learn: 0.6324678 total: 13.6ms remaining: 83.8ms
14: learn: 0.6284970 total: 14.2ms remaining: 80.4ms
15: learn: 0.6241548 total: 14.8ms remaining: 77.7ms
16: learn: 0.6212588 total: 15.2ms remaining: 74.4ms
17: learn: 0.6181208 total: 16.1ms remaining: 73.4ms
18: learn: 0.6149727 total: 17.1ms remaining: 72.8ms
19: learn: 0.6112675 total: 17.8ms remaining: 71.3ms
20: learn: 0.6076084 total: 18.4ms remaining: 69.2ms
21: learn: 0.6038614 total: 19.3ms remaining: 68.4ms
22: learn: 0.5996882 total: 20ms remaining: 67.1ms
23: learn: 0.5962705 total: 20.8ms remaining: 65.8ms
24: learn: 0.5923546 total: 21.3ms remaining: 63.8ms
25: learn: 0.5894756 total: 22.2ms remaining: 63.3ms
26: learn: 0.5864478 total: 23.1ms remaining: 62.3ms
27: learn: 0.5833874 total: 23.7ms remaining: 60.9ms
28: learn: 0.5804593 total: 24.3ms remaining: 59.5ms
29: learn: 0.5777455 total: 24.9ms remaining: 58.2ms
30: learn: 0.5751136 total: 25.9ms remaining: 57.6ms
31: learn: 0.5718058 total: 26.4ms remaining: 56ms
32: learn: 0.5689155 total: 27ms remaining: 54.8ms
33: learn: 0.5658437 total: 27.5ms remaining: 53.3ms
34: learn: 0.5625370 total: 28.1ms remaining: 52.1ms
35: learn: 0.5600906 total: 28.6ms remaining: 50.9ms
36: learn: 0.5574840 total: 29.3ms remaining: 49.9ms
37: learn: 0.5548446 total: 30ms remaining: 48.9ms
38: learn: 0.5521241 total: 30.7ms remaining: 48.1ms
39: learn: 0.5493534 total: 31.3ms remaining: 47ms
40: learn: 0.5464643 total: 31.7ms remaining: 45.7ms
41: learn: 0.5439402 total: 32.3ms remaining: 44.6ms
42: learn: 0.5407503 total: 32.8ms remaining: 43.5ms
43: learn: 0.5383021 total: 33.2ms remaining: 42.2ms
44: learn: 0.5357105 total: 33.7ms remaining: 41.2ms
45: learn: 0.5332736 total: 34.1ms remaining: 40ms
46: learn: 0.5308820 total: 35ms remaining: 39.5ms
47: learn: 0.5280463 total: 35.8ms remaining: 38.8ms
48: learn: 0.5251939 total: 36.2ms remaining: 37.7ms
49: learn: 0.5226601 total: 36.7ms remaining: 36.7ms
50: learn: 0.5200151 total: 36.9ms remaining: 35.5ms
51: learn: 0.5174533 total: 37.3ms remaining: 34.4ms
52: learn: 0.5148804 total: 37.7ms remaining: 33.4ms
53: learn: 0.5125037 total: 38.1ms remaining: 32.5ms
54: learn: 0.5103436 total: 38.5ms remaining: 31.5ms
55: learn: 0.5079663 total: 38.8ms remaining: 30.5ms
56: learn: 0.5058975 total: 39.2ms remaining: 29.5ms
57: learn: 0.5036559 total: 39.6ms remaining: 28.7ms
58: learn: 0.5016062 total: 39.9ms remaining: 27.7ms
59: learn: 0.4994963 total: 40.3ms remaining: 26.9ms
60: learn: 0.4971523 total: 40.7ms remaining: 26ms
61: learn: 0.4950265 total: 41.1ms remaining: 25.2ms
62: learn: 0.4927465 total: 41.6ms remaining: 24.5ms
63: learn: 0.4903560 total: 41.9ms remaining: 23.6ms
64: learn: 0.4881810 total: 42.3ms remaining: 22.8ms
65: learn: 0.4861520 total: 42.7ms remaining: 22ms
66: learn: 0.4841875 total: 43.2ms remaining: 21.3ms
67: learn: 0.4821350 total: 43.5ms remaining: 20.4ms
68: learn: 0.4798477 total: 43.8ms remaining: 19.7ms
69: learn: 0.4774413 total: 44.2ms remaining: 18.9ms
70: learn: 0.4754794 total: 44.5ms remaining: 18.2ms
71: learn: 0.4733676 total: 44.7ms remaining: 17.4ms
72: learn: 0.4710711 total: 45.1ms remaining: 16.7ms
73: learn: 0.4693374 total: 45.4ms remaining: 16ms
74: learn: 0.4675932 total: 45.9ms remaining: 15.3ms
75: learn: 0.4657995 total: 46.3ms remaining: 14.6ms
76: learn: 0.4640184 total: 46.6ms remaining: 13.9ms
77: learn: 0.4623541 total: 47ms remaining: 13.3ms
78: learn: 0.4605428 total: 47.4ms remaining: 12.6ms
79: learn: 0.4584769 total: 47.9ms remaining: 12ms
80: learn: 0.4566638 total: 48.2ms remaining: 11.3ms
81: learn: 0.4548536 total: 48.6ms remaining: 10.7ms
82: learn: 0.4530710 total: 49ms remaining: 10ms
83: learn: 0.4510168 total: 49.3ms remaining: 9.39ms
84: learn: 0.4493297 total: 49.7ms remaining: 8.76ms
85: learn: 0.4474483 total: 50ms remaining: 8.14ms
86: learn: 0.4457039 total: 50.5ms remaining: 7.54ms
87: learn: 0.4440623 total: 50.8ms remaining: 6.93ms
88: learn: 0.4424507 total: 51.2ms remaining: 6.33ms
89: learn: 0.4406137 total: 51.6ms remaining: 5.73ms
90: learn: 0.4388783 total: 51.9ms remaining: 5.13ms
91: learn: 0.4372664 total: 52.3ms remaining: 4.55ms
92: learn: 0.4356385 total: 52.8ms remaining: 3.97ms
93: learn: 0.4339450 total: 53.2ms remaining: 3.39ms
94: learn: 0.4321958 total: 53.5ms remaining: 2.82ms
95: learn: 0.4305977 total: 53.9ms remaining: 2.25ms
96: learn: 0.4292602 total: 54.4ms remaining: 1.68ms
97: learn: 0.4277594 total: 54.8ms remaining: 1.12ms
98: learn: 0.4264082 total: 55.2ms remaining: 558us
99: learn: 0.4250075 total: 55.7ms remaining: 0us
0: learn: 0.6885339 total: 1.31ms remaining: 130ms
1: learn: 0.6834227 total: 2.38ms remaining: 117ms
2: learn: 0.6786232 total: 3.55ms remaining: 115ms
3: learn: 0.6739003 total: 5.04ms remaining: 121ms
4: learn: 0.6693001 total: 5.92ms remaining: 112ms
5: learn: 0.6644451 total: 6.94ms remaining: 109ms
6: learn: 0.6598747 total: 7.97ms remaining: 106ms
7: learn: 0.6558602 total: 9.18ms remaining: 106ms
8: learn: 0.6512912 total: 10.4ms remaining: 105ms
9: learn: 0.6467205 total: 11.5ms remaining: 103ms
10: learn: 0.6428079 total: 12.2ms remaining: 98.7ms
11: learn: 0.6384207 total: 13ms remaining: 95.4ms
12: learn: 0.6344698 total: 14.3ms remaining: 95.9ms
13: learn: 0.6304828 total: 15.3ms remaining: 94.2ms
14: learn: 0.6266173 total: 16.4ms remaining: 93.2ms
15: learn: 0.6223299 total: 17.6ms remaining: 92.6ms
16: learn: 0.6186546 total: 18.5ms remaining: 90.3ms
17: learn: 0.6147792 total: 19.4ms remaining: 88.3ms
18: learn: 0.6110091 total: 20ms remaining: 85.1ms
19: learn: 0.6073387 total: 21.1ms remaining: 84.6ms
20: learn: 0.6038272 total: 22.1ms remaining: 83.3ms
21: learn: 0.6001991 total: 23ms remaining: 81.5ms
22: learn: 0.5964357 total: 23.7ms remaining: 79.2ms
23: learn: 0.5929511 total: 24.3ms remaining: 76.9ms
24: learn: 0.5893719 total: 25ms remaining: 75ms
25: learn: 0.5864672 total: 25.8ms remaining: 73.5ms
26: learn: 0.5837256 total: 26.6ms remaining: 71.9ms
27: learn: 0.5805511 total: 27.4ms remaining: 70.5ms
28: learn: 0.5772513 total: 28.2ms remaining: 69ms
29: learn: 0.5745393 total: 28.8ms remaining: 67.2ms
30: learn: 0.5717751 total: 29.5ms remaining: 65.7ms
31: learn: 0.5685652 total: 30.1ms remaining: 64ms
32: learn: 0.5654609 total: 30.8ms remaining: 62.5ms
33: learn: 0.5622746 total: 31.4ms remaining: 60.9ms
34: learn: 0.5588724 total: 32ms remaining: 59.5ms
35: learn: 0.5555821 total: 32.8ms remaining: 58.3ms
36: learn: 0.5527355 total: 33.4ms remaining: 56.9ms
37: learn: 0.5500608 total: 34ms remaining: 55.5ms
38: learn: 0.5467145 total: 34.6ms remaining: 54.1ms
39: learn: 0.5441104 total: 35.2ms remaining: 52.8ms
40: learn: 0.5417590 total: 35.8ms remaining: 51.5ms
41: learn: 0.5389472 total: 36.2ms remaining: 50ms
42: learn: 0.5365060 total: 36.8ms remaining: 48.8ms
43: learn: 0.5333535 total: 37.3ms remaining: 47.5ms
44: learn: 0.5309882 total: 37.9ms remaining: 46.4ms
45: learn: 0.5286618 total: 38.5ms remaining: 45.2ms
46: learn: 0.5259226 total: 39ms remaining: 43.9ms
47: learn: 0.5235263 total: 39.6ms remaining: 42.9ms
48: learn: 0.5210188 total: 40ms remaining: 41.6ms
49: learn: 0.5185356 total: 40.4ms remaining: 40.4ms
50: learn: 0.5160011 total: 40.7ms remaining: 39.1ms
51: learn: 0.5139627 total: 41.2ms remaining: 38.1ms
52: learn: 0.5115609 total: 41.7ms remaining: 37ms
53: learn: 0.5092764 total: 42.3ms remaining: 36ms
54: learn: 0.5076697 total: 42.9ms remaining: 35.1ms
55: learn: 0.5051063 total: 43.4ms remaining: 34.1ms
56: learn: 0.5025289 total: 43.8ms remaining: 33ms
57: learn: 0.5004309 total: 44.2ms remaining: 32ms
58: learn: 0.4982484 total: 44.7ms remaining: 31.1ms
59: learn: 0.4958368 total: 45.2ms remaining: 30.1ms
60: learn: 0.4935093 total: 45.6ms remaining: 29.1ms
61: learn: 0.4909244 total: 46ms remaining: 28.2ms
62: learn: 0.4885892 total: 46.5ms remaining: 27.3ms
63: learn: 0.4863330 total: 46.8ms remaining: 26.3ms
64: learn: 0.4840421 total: 47.2ms remaining: 25.4ms
65: learn: 0.4819510 total: 47.7ms remaining: 24.5ms
66: learn: 0.4799421 total: 48.1ms remaining: 23.7ms
67: learn: 0.4779539 total: 48.5ms remaining: 22.8ms
68: learn: 0.4757741 total: 48.8ms remaining: 21.9ms
69: learn: 0.4734062 total: 49.2ms remaining: 21.1ms
70: learn: 0.4718209 total: 49.6ms remaining: 20.3ms
71: learn: 0.4698129 total: 49.9ms remaining: 19.4ms
72: learn: 0.4678973 total: 50.3ms remaining: 18.6ms
73: learn: 0.4660518 total: 50.7ms remaining: 17.8ms
74: learn: 0.4641532 total: 51.1ms remaining: 17ms
75: learn: 0.4623182 total: 51.5ms remaining: 16.3ms
76: learn: 0.4606424 total: 52ms remaining: 15.5ms
77: learn: 0.4589035 total: 52.5ms remaining: 14.8ms
78: learn: 0.4572527 total: 52.8ms remaining: 14ms
79: learn: 0.4554347 total: 53ms remaining: 13.3ms
80: learn: 0.4534556 total: 53.3ms remaining: 12.5ms
81: learn: 0.4516094 total: 53.7ms remaining: 11.8ms
82: learn: 0.4501233 total: 54ms remaining: 11.1ms
83: learn: 0.4489080 total: 54.4ms remaining: 10.4ms
84: learn: 0.4471864 total: 54.9ms remaining: 9.68ms
85: learn: 0.4452958 total: 55.2ms remaining: 8.98ms
86: learn: 0.4433208 total: 55.5ms remaining: 8.29ms
87: learn: 0.4415841 total: 55.8ms remaining: 7.61ms
88: learn: 0.4399392 total: 56.4ms remaining: 6.97ms
89: learn: 0.4381792 total: 56.7ms remaining: 6.3ms
90: learn: 0.4363048 total: 57ms remaining: 5.64ms
91: learn: 0.4346445 total: 57.6ms remaining: 5.01ms
92: learn: 0.4330773 total: 58ms remaining: 4.36ms
93: learn: 0.4313780 total: 58.4ms remaining: 3.73ms
94: learn: 0.4299629 total: 58.7ms remaining: 3.09ms
95: learn: 0.4284760 total: 59.2ms remaining: 2.47ms
96: learn: 0.4269777 total: 59.6ms remaining: 1.84ms
97: learn: 0.4254477 total: 60ms remaining: 1.22ms
98: learn: 0.4240495 total: 60.3ms remaining: 609us
99: learn: 0.4224510 total: 60.8ms remaining: 0us
0: learn: 0.6883550 total: 748us remaining: 74.1ms
1: learn: 0.6836109 total: 1.26ms remaining: 61.9ms
2: learn: 0.6787323 total: 1.67ms remaining: 53.9ms
3: learn: 0.6738335 total: 2.52ms remaining: 60.6ms
4: learn: 0.6693032 total: 3.2ms remaining: 60.8ms
5: learn: 0.6644562 total: 4.02ms remaining: 63ms
6: learn: 0.6602992 total: 4.64ms remaining: 61.7ms
7: learn: 0.6565796 total: 5.35ms remaining: 61.6ms
8: learn: 0.6517652 total: 5.87ms remaining: 59.4ms
9: learn: 0.6474219 total: 6.81ms remaining: 61.3ms
10: learn: 0.6444322 total: 7.35ms remaining: 59.4ms
11: learn: 0.6397700 total: 7.92ms remaining: 58.1ms
12: learn: 0.6359370 total: 8.55ms remaining: 57.2ms
13: learn: 0.6324723 total: 9.3ms remaining: 57.1ms
14: learn: 0.6285007 total: 9.84ms remaining: 55.8ms
15: learn: 0.6241371 total: 10.4ms remaining: 54.5ms
16: learn: 0.6205886 total: 11.1ms remaining: 54ms
17: learn: 0.6165610 total: 11.7ms remaining: 53.5ms
18: learn: 0.6127567 total: 12.1ms remaining: 51.5ms
19: learn: 0.6088093 total: 12.8ms remaining: 51ms
20: learn: 0.6051968 total: 13.3ms remaining: 50.2ms
21: learn: 0.6014306 total: 13.9ms remaining: 49.3ms
22: learn: 0.5974358 total: 14.4ms remaining: 48.1ms
23: learn: 0.5939927 total: 14.9ms remaining: 47ms
24: learn: 0.5909375 total: 15.5ms remaining: 46.6ms
25: learn: 0.5873963 total: 16.1ms remaining: 45.9ms
26: learn: 0.5844064 total: 16.8ms remaining: 45.4ms
27: learn: 0.5813962 total: 17.5ms remaining: 44.9ms
28: learn: 0.5785490 total: 18ms remaining: 44ms
29: learn: 0.5758908 total: 18.4ms remaining: 42.9ms
30: learn: 0.5732910 total: 18.9ms remaining: 42.1ms
31: learn: 0.5700190 total: 19.4ms remaining: 41.1ms
32: learn: 0.5669492 total: 19.8ms remaining: 40.1ms
33: learn: 0.5635924 total: 20.2ms remaining: 39.3ms
34: learn: 0.5600908 total: 20.8ms remaining: 38.6ms
35: learn: 0.5572768 total: 21.5ms remaining: 38.1ms
36: learn: 0.5542501 total: 21.9ms remaining: 37.4ms
37: learn: 0.5515929 total: 22.5ms remaining: 36.7ms
38: learn: 0.5480918 total: 23ms remaining: 35.9ms
39: learn: 0.5449589 total: 23.5ms remaining: 35.3ms
40: learn: 0.5420133 total: 24ms remaining: 34.6ms
41: learn: 0.5389790 total: 24.4ms remaining: 33.7ms
42: learn: 0.5360152 total: 24.9ms remaining: 33ms
43: learn: 0.5336202 total: 25.5ms remaining: 32.4ms
44: learn: 0.5308643 total: 25.9ms remaining: 31.7ms
45: learn: 0.5285430 total: 26.3ms remaining: 30.9ms
46: learn: 0.5260419 total: 26.9ms remaining: 30.3ms
47: learn: 0.5236528 total: 27.2ms remaining: 29.5ms
48: learn: 0.5206597 total: 27.6ms remaining: 28.7ms
49: learn: 0.5182717 total: 28ms remaining: 28ms
50: learn: 0.5159021 total: 28.4ms remaining: 27.3ms
51: learn: 0.5142012 total: 28.8ms remaining: 26.5ms
52: learn: 0.5118869 total: 29.2ms remaining: 25.9ms
53: learn: 0.5092385 total: 29.7ms remaining: 25.3ms
54: learn: 0.5074476 total: 30.1ms remaining: 24.6ms
55: learn: 0.5049388 total: 30.6ms remaining: 24.1ms
56: learn: 0.5023647 total: 30.9ms remaining: 23.3ms
57: learn: 0.5003344 total: 31.3ms remaining: 22.7ms
58: learn: 0.4976284 total: 31.7ms remaining: 22ms
59: learn: 0.4952153 total: 32.2ms remaining: 21.5ms
60: learn: 0.4925942 total: 32.6ms remaining: 20.8ms
61: learn: 0.4899387 total: 32.9ms remaining: 20.2ms
62: learn: 0.4879824 total: 33.3ms remaining: 19.6ms
63: learn: 0.4857219 total: 33.8ms remaining: 19ms
64: learn: 0.4833536 total: 34.1ms remaining: 18.4ms
65: learn: 0.4812883 total: 34.5ms remaining: 17.8ms
66: learn: 0.4790204 total: 34.9ms remaining: 17.2ms
67: learn: 0.4769817 total: 35.4ms remaining: 16.6ms
68: learn: 0.4746428 total: 35.7ms remaining: 16ms
69: learn: 0.4722181 total: 36ms remaining: 15.4ms
70: learn: 0.4704845 total: 36.4ms remaining: 14.9ms
71: learn: 0.4681145 total: 36.9ms remaining: 14.4ms
72: learn: 0.4659689 total: 37.3ms remaining: 13.8ms
73: learn: 0.4638687 total: 37.7ms remaining: 13.2ms
74: learn: 0.4614369 total: 38.2ms remaining: 12.7ms
75: learn: 0.4592970 total: 38.5ms remaining: 12.2ms
76: learn: 0.4577747 total: 38.9ms remaining: 11.6ms
77: learn: 0.4553532 total: 39.3ms remaining: 11.1ms
78: learn: 0.4536303 total: 39.8ms remaining: 10.6ms
79: learn: 0.4514065 total: 40.1ms remaining: 10ms
80: learn: 0.4494436 total: 40.4ms remaining: 9.48ms
81: learn: 0.4477547 total: 40.8ms remaining: 8.95ms
82: learn: 0.4461889 total: 41.2ms remaining: 8.43ms
83: learn: 0.4445026 total: 41.5ms remaining: 7.91ms
84: learn: 0.4426023 total: 41.9ms remaining: 7.39ms
85: learn: 0.4405358 total: 42.4ms remaining: 6.9ms
86: learn: 0.4386933 total: 42.8ms remaining: 6.39ms
87: learn: 0.4368974 total: 43.7ms remaining: 5.96ms
88: learn: 0.4351774 total: 44.4ms remaining: 5.49ms
89: learn: 0.4331364 total: 44.8ms remaining: 4.98ms
90: learn: 0.4315189 total: 45.2ms remaining: 4.47ms
91: learn: 0.4297491 total: 45.6ms remaining: 3.97ms
92: learn: 0.4282855 total: 46.1ms remaining: 3.47ms
93: learn: 0.4269431 total: 46.5ms remaining: 2.97ms
94: learn: 0.4256240 total: 46.9ms remaining: 2.47ms
95: learn: 0.4240214 total: 47.2ms remaining: 1.97ms
96: learn: 0.4226613 total: 47.6ms remaining: 1.47ms
97: learn: 0.4208426 total: 48ms remaining: 980us
98: learn: 0.4195020 total: 48.3ms remaining: 488us
99: learn: 0.4177747 total: 48.6ms remaining: 0us
0: learn: 0.6889933 total: 953us remaining: 94.4ms
1: learn: 0.6836164 total: 1.73ms remaining: 84.5ms
2: learn: 0.6784902 total: 2.09ms remaining: 67.7ms
3: learn: 0.6738344 total: 3.15ms remaining: 75.6ms
4: learn: 0.6690759 total: 4.03ms remaining: 76.5ms
5: learn: 0.6641179 total: 5ms remaining: 78.3ms
6: learn: 0.6609183 total: 6.04ms remaining: 80.2ms
7: learn: 0.6564302 total: 6.73ms remaining: 77.4ms
8: learn: 0.6518274 total: 7.72ms remaining: 78ms
9: learn: 0.6472279 total: 8.26ms remaining: 74.4ms
10: learn: 0.6439145 total: 8.84ms remaining: 71.5ms
11: learn: 0.6400426 total: 9.81ms remaining: 71.9ms
12: learn: 0.6356838 total: 10.6ms remaining: 70.6ms
13: learn: 0.6314445 total: 11.4ms remaining: 70.1ms
14: learn: 0.6280712 total: 12ms remaining: 68.2ms
15: learn: 0.6240877 total: 12.8ms remaining: 67.3ms
16: learn: 0.6200849 total: 13.6ms remaining: 66.2ms
17: learn: 0.6168229 total: 14.2ms remaining: 64.6ms
18: learn: 0.6129056 total: 14.5ms remaining: 61.9ms
19: learn: 0.6101052 total: 15.2ms remaining: 60.6ms
20: learn: 0.6064010 total: 15.8ms remaining: 59.3ms
21: learn: 0.6022683 total: 16.4ms remaining: 58.3ms
22: learn: 0.5980306 total: 16.8ms remaining: 56.3ms
23: learn: 0.5949120 total: 17.2ms remaining: 54.6ms
24: learn: 0.5912501 total: 17.9ms remaining: 53.6ms
25: learn: 0.5879653 total: 18.8ms remaining: 53.4ms
26: learn: 0.5845873 total: 19.4ms remaining: 52.4ms
27: learn: 0.5814684 total: 20.1ms remaining: 51.6ms
28: learn: 0.5788440 total: 20.7ms remaining: 50.6ms
29: learn: 0.5759054 total: 21.4ms remaining: 49.9ms
30: learn: 0.5723048 total: 22.1ms remaining: 49.2ms
31: learn: 0.5690872 total: 22.8ms remaining: 48.5ms
32: learn: 0.5660702 total: 23.6ms remaining: 47.8ms
33: learn: 0.5628655 total: 24ms remaining: 46.6ms
34: learn: 0.5599171 total: 24.7ms remaining: 45.9ms
35: learn: 0.5573603 total: 25.3ms remaining: 45ms
36: learn: 0.5551515 total: 25.8ms remaining: 43.9ms
37: learn: 0.5521042 total: 26.5ms remaining: 43.2ms
38: learn: 0.5486314 total: 27ms remaining: 42.3ms
39: learn: 0.5453812 total: 27.7ms remaining: 41.6ms
40: learn: 0.5422646 total: 28.2ms remaining: 40.6ms
41: learn: 0.5396314 total: 28.8ms remaining: 39.8ms
42: learn: 0.5369313 total: 29.5ms remaining: 39.2ms
43: learn: 0.5341681 total: 29.9ms remaining: 38.1ms
44: learn: 0.5311134 total: 30.4ms remaining: 37.2ms
45: learn: 0.5279754 total: 31.2ms remaining: 36.7ms
46: learn: 0.5252587 total: 31.5ms remaining: 35.5ms
47: learn: 0.5224699 total: 31.9ms remaining: 34.5ms
48: learn: 0.5196144 total: 32.2ms remaining: 33.5ms
49: learn: 0.5168594 total: 32.6ms remaining: 32.6ms
50: learn: 0.5145349 total: 33.1ms remaining: 31.8ms
51: learn: 0.5120538 total: 33.5ms remaining: 30.9ms
52: learn: 0.5094141 total: 33.9ms remaining: 30.1ms
53: learn: 0.5067369 total: 34.4ms remaining: 29.3ms
54: learn: 0.5046851 total: 34.9ms remaining: 28.5ms
55: learn: 0.5021771 total: 35.4ms remaining: 27.8ms
56: learn: 0.5001128 total: 35.7ms remaining: 26.9ms
57: learn: 0.4976841 total: 36.1ms remaining: 26.1ms
58: learn: 0.4952075 total: 36.6ms remaining: 25.4ms
59: learn: 0.4934166 total: 37ms remaining: 24.7ms
60: learn: 0.4908600 total: 37.3ms remaining: 23.8ms
61: learn: 0.4884676 total: 37.7ms remaining: 23.1ms
62: learn: 0.4865485 total: 38ms remaining: 22.3ms
63: learn: 0.4844235 total: 38.4ms remaining: 21.6ms
64: learn: 0.4824806 total: 38.6ms remaining: 20.8ms
65: learn: 0.4798479 total: 38.9ms remaining: 20.1ms
66: learn: 0.4773723 total: 39.4ms remaining: 19.4ms
67: learn: 0.4753494 total: 39.8ms remaining: 18.7ms
68: learn: 0.4732775 total: 40.2ms remaining: 18.1ms
69: learn: 0.4708137 total: 40.6ms remaining: 17.4ms
70: learn: 0.4690268 total: 41ms remaining: 16.7ms
71: learn: 0.4671688 total: 41.4ms remaining: 16.1ms
72: learn: 0.4652406 total: 41.7ms remaining: 15.4ms
73: learn: 0.4632691 total: 42.1ms remaining: 14.8ms
74: learn: 0.4610572 total: 42.5ms remaining: 14.2ms
75: learn: 0.4593666 total: 42.8ms remaining: 13.5ms
76: learn: 0.4575386 total: 43.2ms remaining: 12.9ms
77: learn: 0.4554613 total: 43.8ms remaining: 12.4ms
78: learn: 0.4531759 total: 44.2ms remaining: 11.7ms
79: learn: 0.4516175 total: 44.6ms remaining: 11.1ms
80: learn: 0.4497692 total: 44.9ms remaining: 10.5ms
81: learn: 0.4477158 total: 45.2ms remaining: 9.92ms
82: learn: 0.4458186 total: 45.6ms remaining: 9.33ms
83: learn: 0.4440212 total: 45.9ms remaining: 8.75ms
84: learn: 0.4421802 total: 46.5ms remaining: 8.2ms
85: learn: 0.4406457 total: 46.9ms remaining: 7.63ms
86: learn: 0.4386856 total: 47.3ms remaining: 7.07ms
87: learn: 0.4367161 total: 47.7ms remaining: 6.5ms
88: learn: 0.4348507 total: 48.2ms remaining: 5.96ms
89: learn: 0.4332635 total: 48.6ms remaining: 5.4ms
90: learn: 0.4317573 total: 49ms remaining: 4.84ms
91: learn: 0.4299747 total: 49.3ms remaining: 4.29ms
92: learn: 0.4284364 total: 49.9ms remaining: 3.75ms
93: learn: 0.4272723 total: 50.1ms remaining: 3.2ms
94: learn: 0.4254917 total: 50.4ms remaining: 2.65ms
95: learn: 0.4237938 total: 50.8ms remaining: 2.12ms
96: learn: 0.4223160 total: 51.2ms remaining: 1.58ms
97: learn: 0.4205233 total: 51.5ms remaining: 1.05ms
98: learn: 0.4187850 total: 51.8ms remaining: 523us
99: learn: 0.4171327 total: 52.2ms remaining: 0us
0: learn: 0.6890031 total: 1.22ms remaining: 121ms
1: learn: 0.6839941 total: 1.82ms remaining: 89.4ms
2: learn: 0.6790950 total: 2.48ms remaining: 80.4ms
3: learn: 0.6741892 total: 5.54ms remaining: 133ms
4: learn: 0.6699145 total: 5.96ms remaining: 113ms
5: learn: 0.6651736 total: 6.5ms remaining: 102ms
6: learn: 0.6605127 total: 7.82ms remaining: 104ms
7: learn: 0.6563352 total: 8.99ms remaining: 103ms
8: learn: 0.6525859 total: 9.58ms remaining: 96.9ms
9: learn: 0.6483100 total: 10.8ms remaining: 97.3ms
10: learn: 0.6443013 total: 11.6ms remaining: 93.5ms
11: learn: 0.6404553 total: 12.5ms remaining: 91.8ms
12: learn: 0.6363750 total: 13.8ms remaining: 92.4ms
13: learn: 0.6320174 total: 14.5ms remaining: 89ms
14: learn: 0.6288432 total: 15ms remaining: 84.9ms
15: learn: 0.6252655 total: 16.2ms remaining: 84.8ms
16: learn: 0.6214597 total: 17.2ms remaining: 84.2ms
17: learn: 0.6174034 total: 18ms remaining: 82.1ms
18: learn: 0.6143807 total: 19ms remaining: 81.1ms
19: learn: 0.6108119 total: 20ms remaining: 79.9ms
20: learn: 0.6071982 total: 21ms remaining: 79.1ms
21: learn: 0.6035162 total: 21.8ms remaining: 77.4ms
22: learn: 0.5995561 total: 22.5ms remaining: 75.2ms
23: learn: 0.5967885 total: 23.3ms remaining: 73.7ms
24: learn: 0.5928977 total: 24.1ms remaining: 72.4ms
25: learn: 0.5896768 total: 25ms remaining: 71.1ms
26: learn: 0.5864794 total: 25.7ms remaining: 69.6ms
27: learn: 0.5831156 total: 26.5ms remaining: 68.1ms
28: learn: 0.5804258 total: 27.1ms remaining: 66.4ms
29: learn: 0.5780639 total: 27.6ms remaining: 64.5ms
30: learn: 0.5757587 total: 28ms remaining: 62.3ms
31: learn: 0.5725405 total: 28.4ms remaining: 60.3ms
32: learn: 0.5693987 total: 29.2ms remaining: 59.2ms
33: learn: 0.5660367 total: 30ms remaining: 58.3ms
34: learn: 0.5626815 total: 30.7ms remaining: 57ms
35: learn: 0.5601347 total: 31.4ms remaining: 55.8ms
36: learn: 0.5570503 total: 31.9ms remaining: 54.3ms
37: learn: 0.5542878 total: 32.7ms remaining: 53.3ms
38: learn: 0.5511386 total: 33.1ms remaining: 51.8ms
39: learn: 0.5481846 total: 33.9ms remaining: 50.8ms
40: learn: 0.5450162 total: 34.4ms remaining: 49.5ms
41: learn: 0.5426760 total: 35ms remaining: 48.3ms
42: learn: 0.5395370 total: 35.4ms remaining: 46.9ms
43: learn: 0.5373680 total: 35.8ms remaining: 45.6ms
44: learn: 0.5345015 total: 36.7ms remaining: 44.9ms
45: learn: 0.5318743 total: 37.4ms remaining: 43.9ms
46: learn: 0.5292160 total: 37.8ms remaining: 42.6ms
47: learn: 0.5267250 total: 38.3ms remaining: 41.5ms
48: learn: 0.5243110 total: 38.9ms remaining: 40.5ms
49: learn: 0.5214591 total: 39.6ms remaining: 39.6ms
50: learn: 0.5186970 total: 40.1ms remaining: 38.6ms
51: learn: 0.5158365 total: 40.7ms remaining: 37.6ms
52: learn: 0.5136888 total: 41.2ms remaining: 36.5ms
53: learn: 0.5112052 total: 41.8ms remaining: 35.6ms
54: learn: 0.5089564 total: 42.2ms remaining: 34.5ms
55: learn: 0.5065765 total: 42.7ms remaining: 33.5ms
56: learn: 0.5040682 total: 43ms remaining: 32.5ms
57: learn: 0.5015641 total: 43.6ms remaining: 31.6ms
58: learn: 0.4991199 total: 44ms remaining: 30.6ms
59: learn: 0.4967366 total: 44.5ms remaining: 29.7ms
60: learn: 0.4942500 total: 44.9ms remaining: 28.7ms
61: learn: 0.4921693 total: 45.4ms remaining: 27.8ms
62: learn: 0.4901854 total: 45.9ms remaining: 26.9ms
63: learn: 0.4879617 total: 46.3ms remaining: 26ms
64: learn: 0.4857070 total: 46.7ms remaining: 25.1ms
65: learn: 0.4837889 total: 47.1ms remaining: 24.3ms
66: learn: 0.4814533 total: 47.7ms remaining: 23.5ms
67: learn: 0.4798315 total: 48.1ms remaining: 22.6ms
68: learn: 0.4775769 total: 48.4ms remaining: 21.7ms
69: learn: 0.4754393 total: 48.8ms remaining: 20.9ms
70: learn: 0.4739529 total: 49.3ms remaining: 20.2ms
71: learn: 0.4717598 total: 49.7ms remaining: 19.3ms
72: learn: 0.4696536 total: 50.1ms remaining: 18.5ms
73: learn: 0.4676443 total: 50.5ms remaining: 17.7ms
74: learn: 0.4655447 total: 51ms remaining: 17ms
75: learn: 0.4634157 total: 51.5ms remaining: 16.3ms
76: learn: 0.4614728 total: 51.8ms remaining: 15.5ms
77: learn: 0.4593235 total: 52.3ms remaining: 14.7ms
78: learn: 0.4575239 total: 52.7ms remaining: 14ms
79: learn: 0.4556075 total: 53ms remaining: 13.3ms
80: learn: 0.4535342 total: 53.4ms remaining: 12.5ms
81: learn: 0.4515864 total: 53.9ms remaining: 11.8ms
82: learn: 0.4496692 total: 54.3ms remaining: 11.1ms
83: learn: 0.4484274 total: 54.6ms remaining: 10.4ms
84: learn: 0.4469008 total: 54.8ms remaining: 9.68ms
85: learn: 0.4455647 total: 55.4ms remaining: 9.02ms
86: learn: 0.4436224 total: 55.7ms remaining: 8.32ms
87: learn: 0.4421467 total: 56ms remaining: 7.63ms
88: learn: 0.4405282 total: 56.4ms remaining: 6.97ms
89: learn: 0.4391787 total: 56.9ms remaining: 6.32ms
90: learn: 0.4374349 total: 57.2ms remaining: 5.66ms
91: learn: 0.4359123 total: 57.6ms remaining: 5.01ms
92: learn: 0.4344414 total: 57.9ms remaining: 4.36ms
93: learn: 0.4327461 total: 58.4ms remaining: 3.73ms
94: learn: 0.4311738 total: 58.8ms remaining: 3.1ms
95: learn: 0.4295184 total: 59.3ms remaining: 2.47ms
96: learn: 0.4276992 total: 59.7ms remaining: 1.84ms
97: learn: 0.4264722 total: 60ms remaining: 1.22ms
98: learn: 0.4249954 total: 60.3ms remaining: 609us
99: learn: 0.4234596 total: 60.7ms remaining: 0us
0: learn: 0.6884444 total: 1.3ms remaining: 129ms
1: learn: 0.6833777 total: 2.09ms remaining: 102ms
2: learn: 0.6789418 total: 2.68ms remaining: 86.6ms
3: learn: 0.6740883 total: 3.65ms remaining: 87.5ms
4: learn: 0.6695991 total: 4.26ms remaining: 81ms
5: learn: 0.6649560 total: 5.17ms remaining: 81.1ms
6: learn: 0.6607099 total: 6.41ms remaining: 85.2ms
7: learn: 0.6568861 total: 7.71ms remaining: 88.7ms
8: learn: 0.6526495 total: 8.41ms remaining: 85.1ms
9: learn: 0.6486747 total: 8.88ms remaining: 79.9ms
10: learn: 0.6446790 total: 10ms remaining: 80.9ms
11: learn: 0.6408419 total: 10.8ms remaining: 79.5ms
12: learn: 0.6366769 total: 11.8ms remaining: 79.2ms
13: learn: 0.6327758 total: 12.8ms remaining: 78.4ms
14: learn: 0.6289897 total: 13.5ms remaining: 76.3ms
15: learn: 0.6254446 total: 14.7ms remaining: 77ms
16: learn: 0.6214642 total: 16.2ms remaining: 79ms
17: learn: 0.6179222 total: 17.2ms remaining: 78.3ms
18: learn: 0.6145156 total: 18.3ms remaining: 77.9ms
19: learn: 0.6111632 total: 19ms remaining: 76ms
20: learn: 0.6075448 total: 20.4ms remaining: 76.9ms
21: learn: 0.6041593 total: 21.4ms remaining: 75.8ms
22: learn: 0.6005087 total: 22.4ms remaining: 75.1ms
23: learn: 0.5967117 total: 23.6ms remaining: 74.7ms
24: learn: 0.5937134 total: 24.7ms remaining: 74.1ms
25: learn: 0.5902819 total: 25.4ms remaining: 72.2ms
26: learn: 0.5876279 total: 26.4ms remaining: 71.3ms
27: learn: 0.5847773 total: 27.4ms remaining: 70.5ms
28: learn: 0.5817003 total: 28.1ms remaining: 68.8ms
29: learn: 0.5787036 total: 28.5ms remaining: 66.4ms
30: learn: 0.5760609 total: 29.3ms remaining: 65.2ms
31: learn: 0.5734735 total: 30.5ms remaining: 64.8ms
32: learn: 0.5707002 total: 31ms remaining: 62.9ms
33: learn: 0.5675329 total: 31.9ms remaining: 61.8ms
34: learn: 0.5643493 total: 32.8ms remaining: 60.9ms
35: learn: 0.5610712 total: 33.9ms remaining: 60.2ms
36: learn: 0.5582207 total: 34.6ms remaining: 59ms
37: learn: 0.5559611 total: 35.5ms remaining: 57.9ms
38: learn: 0.5537050 total: 36.3ms remaining: 56.7ms
39: learn: 0.5503936 total: 37ms remaining: 55.5ms
40: learn: 0.5478983 total: 38.5ms remaining: 55.4ms
41: learn: 0.5451844 total: 39.3ms remaining: 54.2ms
42: learn: 0.5418663 total: 39.8ms remaining: 52.8ms
43: learn: 0.5390675 total: 40.7ms remaining: 51.8ms
44: learn: 0.5366144 total: 41.7ms remaining: 51ms
45: learn: 0.5341339 total: 42.7ms remaining: 50.1ms
46: learn: 0.5320371 total: 43.3ms remaining: 48.8ms
47: learn: 0.5297100 total: 44.2ms remaining: 47.9ms
48: learn: 0.5275120 total: 44.7ms remaining: 46.5ms
49: learn: 0.5249431 total: 45.1ms remaining: 45.1ms
50: learn: 0.5222452 total: 45.8ms remaining: 44ms
51: learn: 0.5196150 total: 46.4ms remaining: 42.9ms
52: learn: 0.5174188 total: 47ms remaining: 41.7ms
53: learn: 0.5150613 total: 47.3ms remaining: 40.3ms
54: learn: 0.5126465 total: 48.1ms remaining: 39.3ms
55: learn: 0.5104492 total: 48.9ms remaining: 38.4ms
56: learn: 0.5080920 total: 49.7ms remaining: 37.5ms
57: learn: 0.5056335 total: 50.2ms remaining: 36.3ms
58: learn: 0.5035476 total: 50.6ms remaining: 35.2ms
59: learn: 0.5013622 total: 51.2ms remaining: 34.2ms
60: learn: 0.4988686 total: 51.7ms remaining: 33.1ms
61: learn: 0.4972610 total: 52.3ms remaining: 32ms
62: learn: 0.4953017 total: 52.8ms remaining: 31ms
63: learn: 0.4927863 total: 53.4ms remaining: 30ms
64: learn: 0.4907355 total: 53.7ms remaining: 28.9ms
65: learn: 0.4887132 total: 54.2ms remaining: 27.9ms
66: learn: 0.4864048 total: 54.6ms remaining: 26.9ms
67: learn: 0.4841681 total: 55.1ms remaining: 25.9ms
68: learn: 0.4822257 total: 55.5ms remaining: 24.9ms
69: learn: 0.4802451 total: 55.8ms remaining: 23.9ms
70: learn: 0.4782052 total: 56.3ms remaining: 23ms
71: learn: 0.4761668 total: 56.9ms remaining: 22.1ms
72: learn: 0.4740465 total: 57.3ms remaining: 21.2ms
73: learn: 0.4721290 total: 57.8ms remaining: 20.3ms
74: learn: 0.4701994 total: 58.3ms remaining: 19.4ms
75: learn: 0.4681550 total: 58.8ms remaining: 18.6ms
76: learn: 0.4665667 total: 59.4ms remaining: 17.7ms
77: learn: 0.4645251 total: 59.9ms remaining: 16.9ms
78: learn: 0.4625883 total: 60.3ms remaining: 16ms
79: learn: 0.4605779 total: 60.6ms remaining: 15.2ms
80: learn: 0.4587713 total: 60.8ms remaining: 14.3ms
81: learn: 0.4568585 total: 61.3ms remaining: 13.5ms
82: learn: 0.4551630 total: 61.6ms remaining: 12.6ms
83: learn: 0.4534023 total: 62ms remaining: 11.8ms
84: learn: 0.4519195 total: 62.5ms remaining: 11ms
85: learn: 0.4503519 total: 62.9ms remaining: 10.2ms
86: learn: 0.4485513 total: 63.2ms remaining: 9.45ms
87: learn: 0.4469570 total: 63.6ms remaining: 8.67ms
88: learn: 0.4453554 total: 64.1ms remaining: 7.93ms
89: learn: 0.4437199 total: 64.5ms remaining: 7.17ms
90: learn: 0.4418570 total: 64.9ms remaining: 6.41ms
91: learn: 0.4406584 total: 65.2ms remaining: 5.67ms
92: learn: 0.4389602 total: 65.6ms remaining: 4.94ms
93: learn: 0.4371791 total: 66ms remaining: 4.21ms
94: learn: 0.4354748 total: 66.4ms remaining: 3.49ms
95: learn: 0.4339965 total: 66.7ms remaining: 2.78ms
96: learn: 0.4323266 total: 67.2ms remaining: 2.08ms
97: learn: 0.4307838 total: 67.7ms remaining: 1.38ms
98: learn: 0.4293552 total: 68.1ms remaining: 687us
99: learn: 0.4278470 total: 68.3ms remaining: 0us
0: learn: 0.6885566 total: 1.21ms remaining: 120ms
1: learn: 0.6836071 total: 2.66ms remaining: 130ms
2: learn: 0.6782460 total: 3.67ms remaining: 119ms
3: learn: 0.6743132 total: 4.99ms remaining: 120ms
4: learn: 0.6702237 total: 6.19ms remaining: 118ms
5: learn: 0.6660752 total: 7.77ms remaining: 122ms
6: learn: 0.6616702 total: 8.92ms remaining: 119ms
7: learn: 0.6566263 total: 10.2ms remaining: 118ms
8: learn: 0.6518664 total: 11.6ms remaining: 117ms
9: learn: 0.6490527 total: 12.8ms remaining: 115ms
10: learn: 0.6444509 total: 13.9ms remaining: 113ms
11: learn: 0.6403671 total: 15.5ms remaining: 114ms
12: learn: 0.6363304 total: 16.6ms remaining: 111ms
13: learn: 0.6327678 total: 17.5ms remaining: 107ms
14: learn: 0.6282780 total: 18.5ms remaining: 105ms
15: learn: 0.6244872 total: 19.6ms remaining: 103ms
16: learn: 0.6203105 total: 21ms remaining: 103ms
17: learn: 0.6165432 total: 21.7ms remaining: 99ms
18: learn: 0.6128502 total: 22.9ms remaining: 97.8ms
19: learn: 0.6090656 total: 24.4ms remaining: 97.7ms
20: learn: 0.6050572 total: 25.7ms remaining: 96.6ms
21: learn: 0.6007740 total: 26.5ms remaining: 93.9ms
22: learn: 0.5970249 total: 27.1ms remaining: 90.7ms
23: learn: 0.5935481 total: 27.9ms remaining: 88.3ms
24: learn: 0.5906288 total: 29ms remaining: 87ms
25: learn: 0.5877041 total: 30ms remaining: 85.5ms
26: learn: 0.5839478 total: 31.1ms remaining: 84ms
27: learn: 0.5811081 total: 32.1ms remaining: 82.4ms
28: learn: 0.5773239 total: 33.1ms remaining: 81.1ms
29: learn: 0.5742893 total: 34.5ms remaining: 80.4ms
30: learn: 0.5706335 total: 35.5ms remaining: 79.1ms
31: learn: 0.5674696 total: 36.5ms remaining: 77.6ms
32: learn: 0.5645132 total: 37.4ms remaining: 75.9ms
33: learn: 0.5608553 total: 38.8ms remaining: 75.4ms
34: learn: 0.5579249 total: 39.6ms remaining: 73.5ms
35: learn: 0.5548740 total: 41ms remaining: 72.8ms
36: learn: 0.5521033 total: 42.1ms remaining: 71.6ms
37: learn: 0.5487798 total: 43.1ms remaining: 70.3ms
38: learn: 0.5458292 total: 44.1ms remaining: 69.1ms
39: learn: 0.5434128 total: 45.3ms remaining: 68ms
40: learn: 0.5398872 total: 46.2ms remaining: 66.5ms
41: learn: 0.5372327 total: 47.3ms remaining: 65.4ms
42: learn: 0.5348076 total: 48.6ms remaining: 64.4ms
43: learn: 0.5319729 total: 49.2ms remaining: 62.6ms
44: learn: 0.5296587 total: 49.7ms remaining: 60.8ms
45: learn: 0.5271298 total: 50.6ms remaining: 59.4ms
46: learn: 0.5243558 total: 51.6ms remaining: 58.2ms
47: learn: 0.5216964 total: 52.6ms remaining: 57ms
48: learn: 0.5191863 total: 53.5ms remaining: 55.7ms
49: learn: 0.5165567 total: 54.4ms remaining: 54.4ms
50: learn: 0.5141248 total: 55ms remaining: 52.8ms
51: learn: 0.5116138 total: 55.7ms remaining: 51.4ms
52: learn: 0.5092609 total: 56.2ms remaining: 49.8ms
53: learn: 0.5070869 total: 56.9ms remaining: 48.5ms
54: learn: 0.5046551 total: 57.9ms remaining: 47.4ms
55: learn: 0.5022862 total: 58.7ms remaining: 46.1ms
56: learn: 0.4996988 total: 59.6ms remaining: 45ms
57: learn: 0.4972926 total: 60.2ms remaining: 43.6ms
58: learn: 0.4957858 total: 61ms remaining: 42.4ms
59: learn: 0.4937636 total: 61.7ms remaining: 41.1ms
60: learn: 0.4914307 total: 62.5ms remaining: 39.9ms
61: learn: 0.4892601 total: 62.9ms remaining: 38.6ms
62: learn: 0.4870957 total: 63.6ms remaining: 37.4ms
63: learn: 0.4849294 total: 64.1ms remaining: 36ms
64: learn: 0.4828281 total: 64.8ms remaining: 34.9ms
65: learn: 0.4801656 total: 65.4ms remaining: 33.7ms
66: learn: 0.4779788 total: 66ms remaining: 32.5ms
67: learn: 0.4757880 total: 66.5ms remaining: 31.3ms
68: learn: 0.4741170 total: 67ms remaining: 30.1ms
69: learn: 0.4721005 total: 67.4ms remaining: 28.9ms
70: learn: 0.4703547 total: 67.8ms remaining: 27.7ms
71: learn: 0.4682511 total: 68.4ms remaining: 26.6ms
72: learn: 0.4666105 total: 68.9ms remaining: 25.5ms
73: learn: 0.4643244 total: 69.4ms remaining: 24.4ms
74: learn: 0.4620866 total: 70ms remaining: 23.3ms
75: learn: 0.4599344 total: 70.6ms remaining: 22.3ms
76: learn: 0.4578568 total: 71.4ms remaining: 21.3ms
77: learn: 0.4562268 total: 72.1ms remaining: 20.3ms
78: learn: 0.4540555 total: 72.6ms remaining: 19.3ms
79: learn: 0.4518526 total: 73.1ms remaining: 18.3ms
80: learn: 0.4498825 total: 73.5ms remaining: 17.2ms
81: learn: 0.4476596 total: 74.2ms remaining: 16.3ms
82: learn: 0.4463192 total: 74.6ms remaining: 15.3ms
83: learn: 0.4445282 total: 74.9ms remaining: 14.3ms
84: learn: 0.4429586 total: 75.5ms remaining: 13.3ms
85: learn: 0.4412175 total: 75.8ms remaining: 12.3ms
86: learn: 0.4394992 total: 76.1ms remaining: 11.4ms
87: learn: 0.4378353 total: 76.5ms remaining: 10.4ms
88: learn: 0.4362414 total: 76.9ms remaining: 9.5ms
89: learn: 0.4346269 total: 77.3ms remaining: 8.58ms
90: learn: 0.4330993 total: 77.6ms remaining: 7.67ms
91: learn: 0.4314341 total: 77.9ms remaining: 6.77ms
92: learn: 0.4296886 total: 78.6ms remaining: 5.91ms
93: learn: 0.4281258 total: 79.1ms remaining: 5.05ms
94: learn: 0.4266854 total: 79.6ms remaining: 4.19ms
95: learn: 0.4247964 total: 80.1ms remaining: 3.34ms
96: learn: 0.4232147 total: 80.5ms remaining: 2.49ms
97: learn: 0.4217981 total: 80.9ms remaining: 1.65ms
98: learn: 0.4198929 total: 81.2ms remaining: 820us
99: learn: 0.4181065 total: 81.8ms remaining: 0us
0: learn: 0.6896203 total: 1.55ms remaining: 153ms
1: learn: 0.6850714 total: 2.75ms remaining: 135ms
2: learn: 0.6803862 total: 3.73ms remaining: 121ms
3: learn: 0.6758516 total: 5.28ms remaining: 127ms
4: learn: 0.6716479 total: 6.48ms remaining: 123ms
5: learn: 0.6663871 total: 7.64ms remaining: 120ms
6: learn: 0.6619637 total: 9.07ms remaining: 121ms
7: learn: 0.6576289 total: 9.89ms remaining: 114ms
8: learn: 0.6535165 total: 10.8ms remaining: 110ms
9: learn: 0.6494451 total: 12.6ms remaining: 113ms
10: learn: 0.6454696 total: 13.6ms remaining: 110ms
11: learn: 0.6411167 total: 15ms remaining: 110ms
12: learn: 0.6375680 total: 16.2ms remaining: 108ms
13: learn: 0.6336478 total: 17.6ms remaining: 108ms
14: learn: 0.6296995 total: 19.2ms remaining: 109ms
15: learn: 0.6252287 total: 20.1ms remaining: 106ms
16: learn: 0.6216951 total: 21.3ms remaining: 104ms
17: learn: 0.6177813 total: 22.5ms remaining: 103ms
18: learn: 0.6140884 total: 23.3ms remaining: 99.3ms
19: learn: 0.6108468 total: 24.5ms remaining: 98.1ms
20: learn: 0.6072031 total: 25.9ms remaining: 97.5ms
21: learn: 0.6030750 total: 27.2ms remaining: 96.4ms
22: learn: 0.5992488 total: 28.3ms remaining: 94.7ms
23: learn: 0.5955617 total: 29.2ms remaining: 92.3ms
24: learn: 0.5922429 total: 29.9ms remaining: 89.6ms
25: learn: 0.5884927 total: 31.1ms remaining: 88.6ms
26: learn: 0.5856491 total: 32.1ms remaining: 86.9ms
27: learn: 0.5820865 total: 33.6ms remaining: 86.5ms
28: learn: 0.5795715 total: 34.6ms remaining: 84.7ms
29: learn: 0.5761366 total: 35.5ms remaining: 82.9ms
30: learn: 0.5728400 total: 36.5ms remaining: 81.2ms
31: learn: 0.5697248 total: 37.7ms remaining: 80.1ms
32: learn: 0.5668872 total: 38.6ms remaining: 78.4ms
33: learn: 0.5633521 total: 39.4ms remaining: 76.4ms
34: learn: 0.5601400 total: 40.5ms remaining: 75.2ms
35: learn: 0.5574012 total: 41.6ms remaining: 73.9ms
36: learn: 0.5543751 total: 42.4ms remaining: 72.2ms
37: learn: 0.5520800 total: 43.2ms remaining: 70.6ms
38: learn: 0.5495015 total: 43.8ms remaining: 68.5ms
39: learn: 0.5462074 total: 44.7ms remaining: 67ms
40: learn: 0.5434947 total: 45.4ms remaining: 65.4ms
41: learn: 0.5412224 total: 46.9ms remaining: 64.8ms
42: learn: 0.5385021 total: 47.5ms remaining: 63ms
43: learn: 0.5355544 total: 48.1ms remaining: 61.2ms
44: learn: 0.5328038 total: 49.5ms remaining: 60.5ms
45: learn: 0.5305187 total: 50.8ms remaining: 59.6ms
46: learn: 0.5281554 total: 51.5ms remaining: 58.1ms
47: learn: 0.5251024 total: 52.6ms remaining: 57ms
48: learn: 0.5225982 total: 53ms remaining: 55.2ms
49: learn: 0.5203373 total: 53.8ms remaining: 53.8ms
50: learn: 0.5181042 total: 54.7ms remaining: 52.6ms
51: learn: 0.5153619 total: 56ms remaining: 51.7ms
52: learn: 0.5130063 total: 56.9ms remaining: 50.5ms
53: learn: 0.5106676 total: 57.8ms remaining: 49.2ms
54: learn: 0.5085983 total: 58.5ms remaining: 47.9ms
55: learn: 0.5064403 total: 59.2ms remaining: 46.5ms
56: learn: 0.5042995 total: 59.9ms remaining: 45.2ms
57: learn: 0.5018453 total: 60.8ms remaining: 44ms
58: learn: 0.4999030 total: 61.5ms remaining: 42.7ms
59: learn: 0.4970939 total: 62.2ms remaining: 41.5ms
60: learn: 0.4956242 total: 63.1ms remaining: 40.4ms
61: learn: 0.4936966 total: 63.7ms remaining: 39ms
62: learn: 0.4915250 total: 64.5ms remaining: 37.9ms
63: learn: 0.4894404 total: 65.3ms remaining: 36.7ms
64: learn: 0.4869456 total: 66.1ms remaining: 35.6ms
65: learn: 0.4846882 total: 66.6ms remaining: 34.3ms
66: learn: 0.4827632 total: 67.5ms remaining: 33.2ms
67: learn: 0.4802655 total: 68ms remaining: 32ms
68: learn: 0.4782428 total: 68.8ms remaining: 30.9ms
69: learn: 0.4760303 total: 69.6ms remaining: 29.8ms
70: learn: 0.4745876 total: 70.1ms remaining: 28.6ms
71: learn: 0.4721742 total: 70.7ms remaining: 27.5ms
72: learn: 0.4706441 total: 71.3ms remaining: 26.4ms
73: learn: 0.4686661 total: 72ms remaining: 25.3ms
74: learn: 0.4671855 total: 72.6ms remaining: 24.2ms
75: learn: 0.4653315 total: 73.4ms remaining: 23.2ms
76: learn: 0.4632093 total: 73.9ms remaining: 22.1ms
77: learn: 0.4609629 total: 74.4ms remaining: 21ms
78: learn: 0.4590973 total: 74.8ms remaining: 19.9ms
79: learn: 0.4575248 total: 76.1ms remaining: 19ms
80: learn: 0.4557242 total: 76.6ms remaining: 18ms
81: learn: 0.4537280 total: 77.3ms remaining: 17ms
82: learn: 0.4518387 total: 77.7ms remaining: 15.9ms
83: learn: 0.4497253 total: 78.1ms remaining: 14.9ms
84: learn: 0.4482405 total: 78.6ms remaining: 13.9ms
85: learn: 0.4465275 total: 79ms remaining: 12.9ms
86: learn: 0.4450708 total: 79.5ms remaining: 11.9ms
87: learn: 0.4429419 total: 79.9ms remaining: 10.9ms
88: learn: 0.4413283 total: 80.4ms remaining: 9.93ms
89: learn: 0.4397207 total: 80.8ms remaining: 8.98ms
90: learn: 0.4384493 total: 81.2ms remaining: 8.03ms
91: learn: 0.4367745 total: 81.7ms remaining: 7.11ms
92: learn: 0.4353104 total: 82.2ms remaining: 6.18ms
93: learn: 0.4333906 total: 82.6ms remaining: 5.27ms
94: learn: 0.4317244 total: 83.1ms remaining: 4.38ms
95: learn: 0.4302170 total: 83.5ms remaining: 3.48ms
96: learn: 0.4286522 total: 84ms remaining: 2.6ms
97: learn: 0.4268209 total: 84.5ms remaining: 1.72ms
98: learn: 0.4253835 total: 84.8ms remaining: 856us
99: learn: 0.4237831 total: 85.2ms remaining: 0us
0: learn: 0.6894871 total: 1.71ms remaining: 169ms
1: learn: 0.6842252 total: 2.62ms remaining: 129ms
2: learn: 0.6798605 total: 4.09ms remaining: 132ms
3: learn: 0.6746510 total: 5.27ms remaining: 126ms
4: learn: 0.6695916 total: 6.7ms remaining: 127ms
5: learn: 0.6646934 total: 7.7ms remaining: 121ms
6: learn: 0.6608368 total: 9ms remaining: 120ms
7: learn: 0.6560198 total: 9.8ms remaining: 113ms
8: learn: 0.6514894 total: 10.9ms remaining: 111ms
9: learn: 0.6484876 total: 12.7ms remaining: 114ms
10: learn: 0.6438394 total: 13.8ms remaining: 111ms
11: learn: 0.6400430 total: 15.1ms remaining: 110ms
12: learn: 0.6364403 total: 16.4ms remaining: 110ms
13: learn: 0.6328070 total: 18.1ms remaining: 111ms
14: learn: 0.6285253 total: 19.4ms remaining: 110ms
15: learn: 0.6244703 total: 21.4ms remaining: 112ms
16: learn: 0.6203785 total: 23ms remaining: 112ms
17: learn: 0.6170779 total: 23.7ms remaining: 108ms
18: learn: 0.6134879 total: 25.3ms remaining: 108ms
19: learn: 0.6098244 total: 26.6ms remaining: 106ms
20: learn: 0.6058554 total: 27.8ms remaining: 105ms
21: learn: 0.6020761 total: 28.6ms remaining: 102ms
22: learn: 0.5989954 total: 28.9ms remaining: 96.9ms
23: learn: 0.5950739 total: 30.3ms remaining: 95.8ms
24: learn: 0.5921883 total: 31.7ms remaining: 95.1ms
25: learn: 0.5889009 total: 33.2ms remaining: 94.6ms
26: learn: 0.5860139 total: 34.3ms remaining: 92.8ms
27: learn: 0.5833418 total: 35.6ms remaining: 91.5ms
28: learn: 0.5810032 total: 36.5ms remaining: 89.3ms
29: learn: 0.5784543 total: 37.7ms remaining: 88ms
30: learn: 0.5750472 total: 38.7ms remaining: 86.2ms
31: learn: 0.5717814 total: 40.3ms remaining: 85.6ms
32: learn: 0.5686437 total: 41.5ms remaining: 84.3ms
33: learn: 0.5653167 total: 42.4ms remaining: 82.3ms
34: learn: 0.5630180 total: 43.6ms remaining: 81ms
35: learn: 0.5603828 total: 44.7ms remaining: 79.5ms
36: learn: 0.5575529 total: 45.9ms remaining: 78.2ms
37: learn: 0.5553556 total: 47ms remaining: 76.7ms
38: learn: 0.5525472 total: 48.3ms remaining: 75.5ms
39: learn: 0.5495731 total: 49ms remaining: 73.5ms
40: learn: 0.5473859 total: 49.8ms remaining: 71.7ms
41: learn: 0.5444052 total: 50.8ms remaining: 70.1ms
42: learn: 0.5422100 total: 51.7ms remaining: 68.6ms
43: learn: 0.5393793 total: 53.1ms remaining: 67.5ms
44: learn: 0.5367392 total: 54ms remaining: 66ms
45: learn: 0.5339108 total: 54.8ms remaining: 64.3ms
46: learn: 0.5312177 total: 55.8ms remaining: 62.9ms
47: learn: 0.5287355 total: 56.9ms remaining: 61.7ms
48: learn: 0.5265024 total: 58ms remaining: 60.4ms
49: learn: 0.5235980 total: 58.9ms remaining: 58.9ms
50: learn: 0.5207756 total: 59.9ms remaining: 57.6ms
51: learn: 0.5177716 total: 61.2ms remaining: 56.5ms
52: learn: 0.5154983 total: 62ms remaining: 55ms
53: learn: 0.5126700 total: 62.8ms remaining: 53.5ms
54: learn: 0.5101043 total: 63.2ms remaining: 51.7ms
55: learn: 0.5078914 total: 64.5ms remaining: 50.7ms
56: learn: 0.5052694 total: 65.2ms remaining: 49.2ms
57: learn: 0.5027824 total: 66.1ms remaining: 47.8ms
58: learn: 0.5004041 total: 66.8ms remaining: 46.4ms
59: learn: 0.4979322 total: 67.1ms remaining: 44.8ms
60: learn: 0.4954362 total: 67.4ms remaining: 43.1ms
61: learn: 0.4929859 total: 67.9ms remaining: 41.6ms
62: learn: 0.4911638 total: 68.2ms remaining: 40.1ms
63: learn: 0.4890122 total: 68.6ms remaining: 38.6ms
64: learn: 0.4866211 total: 69.1ms remaining: 37.2ms
65: learn: 0.4845082 total: 69.5ms remaining: 35.8ms
66: learn: 0.4821234 total: 70.1ms remaining: 34.5ms
67: learn: 0.4798660 total: 70.4ms remaining: 33.1ms
68: learn: 0.4779862 total: 70.9ms remaining: 31.9ms
69: learn: 0.4762877 total: 71.6ms remaining: 30.7ms
70: learn: 0.4741045 total: 72ms remaining: 29.4ms
71: learn: 0.4722938 total: 72.7ms remaining: 28.3ms
72: learn: 0.4700146 total: 73.1ms remaining: 27ms
73: learn: 0.4680951 total: 73.7ms remaining: 25.9ms
74: learn: 0.4659906 total: 74.2ms remaining: 24.7ms
75: learn: 0.4640971 total: 74.7ms remaining: 23.6ms
76: learn: 0.4620922 total: 75.2ms remaining: 22.5ms
77: learn: 0.4600803 total: 75.5ms remaining: 21.3ms
78: learn: 0.4586681 total: 75.9ms remaining: 20.2ms
79: learn: 0.4566174 total: 76.2ms remaining: 19.1ms
80: learn: 0.4545173 total: 76.8ms remaining: 18ms
81: learn: 0.4524986 total: 77.3ms remaining: 17ms
82: learn: 0.4505864 total: 77.8ms remaining: 15.9ms
83: learn: 0.4489424 total: 78.1ms remaining: 14.9ms
84: learn: 0.4474078 total: 78.4ms remaining: 13.8ms
85: learn: 0.4460705 total: 78.8ms remaining: 12.8ms
86: learn: 0.4445821 total: 79.3ms remaining: 11.8ms
87: learn: 0.4428882 total: 79.6ms remaining: 10.8ms
88: learn: 0.4411782 total: 79.9ms remaining: 9.88ms
89: learn: 0.4392787 total: 80.5ms remaining: 8.94ms
90: learn: 0.4375469 total: 80.9ms remaining: 8ms
91: learn: 0.4358815 total: 81.2ms remaining: 7.06ms
92: learn: 0.4342353 total: 81.6ms remaining: 6.14ms
93: learn: 0.4325697 total: 82.1ms remaining: 5.24ms
94: learn: 0.4309381 total: 82.5ms remaining: 4.34ms
95: learn: 0.4293767 total: 82.9ms remaining: 3.45ms
96: learn: 0.4278734 total: 83.2ms remaining: 2.57ms
97: learn: 0.4261342 total: 83.7ms remaining: 1.71ms
98: learn: 0.4245995 total: 84ms remaining: 848us
99: learn: 0.4228280 total: 84.4ms remaining: 0us
0: learn: 0.6895315 total: 1.38ms remaining: 137ms
1: learn: 0.6844180 total: 2.71ms remaining: 133ms
2: learn: 0.6797461 total: 3.73ms remaining: 121ms
3: learn: 0.6750861 total: 4.83ms remaining: 116ms
4: learn: 0.6707944 total: 5.56ms remaining: 106ms
5: learn: 0.6661817 total: 5.93ms remaining: 92.9ms
6: learn: 0.6616283 total: 6.88ms remaining: 91.3ms
7: learn: 0.6576498 total: 8.11ms remaining: 93.3ms
8: learn: 0.6531931 total: 8.85ms remaining: 89.5ms
9: learn: 0.6487036 total: 10.1ms remaining: 91ms
10: learn: 0.6449099 total: 11ms remaining: 89ms
11: learn: 0.6406385 total: 12ms remaining: 88.1ms
12: learn: 0.6364890 total: 12.8ms remaining: 85.7ms
13: learn: 0.6327426 total: 14.1ms remaining: 86.5ms
14: learn: 0.6293871 total: 15.4ms remaining: 87.5ms
15: learn: 0.6251434 total: 16.6ms remaining: 87ms
16: learn: 0.6214766 total: 17.6ms remaining: 86.1ms
17: learn: 0.6178709 total: 18.6ms remaining: 85ms
18: learn: 0.6143560 total: 19.7ms remaining: 83.9ms
19: learn: 0.6108690 total: 20.8ms remaining: 83.3ms
20: learn: 0.6073974 total: 22ms remaining: 82.7ms
21: learn: 0.6036627 total: 22.9ms remaining: 81.1ms
22: learn: 0.5997537 total: 23.6ms remaining: 79.1ms
23: learn: 0.5964076 total: 24.5ms remaining: 77.6ms
24: learn: 0.5934630 total: 25.6ms remaining: 76.7ms
25: learn: 0.5903772 total: 26.5ms remaining: 75.4ms
26: learn: 0.5873292 total: 27.4ms remaining: 74ms
27: learn: 0.5843486 total: 28.1ms remaining: 72.2ms
28: learn: 0.5815989 total: 28.7ms remaining: 70.2ms
29: learn: 0.5790148 total: 29.4ms remaining: 68.5ms
30: learn: 0.5764376 total: 30.2ms remaining: 67.2ms
31: learn: 0.5734280 total: 30.9ms remaining: 65.6ms
32: learn: 0.5703529 total: 31.6ms remaining: 64.2ms
33: learn: 0.5671999 total: 32.4ms remaining: 62.9ms
34: learn: 0.5637969 total: 33ms remaining: 61.4ms
35: learn: 0.5609537 total: 33.8ms remaining: 60.1ms
36: learn: 0.5583443 total: 34.5ms remaining: 58.7ms
37: learn: 0.5554906 total: 35.1ms remaining: 57.3ms
38: learn: 0.5523799 total: 35.7ms remaining: 55.8ms
39: learn: 0.5496638 total: 37ms remaining: 55.5ms
40: learn: 0.5467338 total: 37.7ms remaining: 54.2ms
41: learn: 0.5437414 total: 38.1ms remaining: 52.6ms
42: learn: 0.5410515 total: 38.7ms remaining: 51.3ms
43: learn: 0.5385706 total: 39.4ms remaining: 50.1ms
44: learn: 0.5359205 total: 40ms remaining: 48.9ms
45: learn: 0.5335821 total: 40.4ms remaining: 47.5ms
46: learn: 0.5311304 total: 41.1ms remaining: 46.3ms
47: learn: 0.5287105 total: 41.8ms remaining: 45.2ms
48: learn: 0.5262265 total: 42.2ms remaining: 43.9ms
49: learn: 0.5235670 total: 42.8ms remaining: 42.8ms
50: learn: 0.5209126 total: 43.3ms remaining: 41.6ms
51: learn: 0.5187759 total: 43.9ms remaining: 40.5ms
52: learn: 0.5164240 total: 44.4ms remaining: 39.4ms
53: learn: 0.5139137 total: 45ms remaining: 38.3ms
54: learn: 0.5115121 total: 45.5ms remaining: 37.2ms
55: learn: 0.5090840 total: 46.2ms remaining: 36.3ms
56: learn: 0.5065030 total: 46.6ms remaining: 35.2ms
57: learn: 0.5039316 total: 47.2ms remaining: 34.2ms
58: learn: 0.5014690 total: 47.6ms remaining: 33.1ms
59: learn: 0.4988399 total: 48.2ms remaining: 32.1ms
60: learn: 0.4965204 total: 48.7ms remaining: 31.1ms
61: learn: 0.4942442 total: 49ms remaining: 30ms
62: learn: 0.4920413 total: 49.4ms remaining: 29ms
63: learn: 0.4900332 total: 49.9ms remaining: 28.1ms
64: learn: 0.4879750 total: 50.3ms remaining: 27.1ms
65: learn: 0.4859368 total: 50.6ms remaining: 26ms
66: learn: 0.4841339 total: 50.9ms remaining: 25.1ms
67: learn: 0.4817868 total: 51.3ms remaining: 24.2ms
68: learn: 0.4798315 total: 51.7ms remaining: 23.2ms
69: learn: 0.4776966 total: 52.1ms remaining: 22.3ms
70: learn: 0.4754465 total: 52.6ms remaining: 21.5ms
71: learn: 0.4733520 total: 53ms remaining: 20.6ms
72: learn: 0.4714535 total: 53.4ms remaining: 19.8ms
73: learn: 0.4695847 total: 53.8ms remaining: 18.9ms
74: learn: 0.4679735 total: 54.3ms remaining: 18.1ms
75: learn: 0.4661721 total: 54.7ms remaining: 17.3ms
76: learn: 0.4641415 total: 55ms remaining: 16.4ms
77: learn: 0.4622109 total: 55.6ms remaining: 15.7ms
78: learn: 0.4602079 total: 56ms remaining: 14.9ms
79: learn: 0.4584789 total: 56.2ms remaining: 14ms
80: learn: 0.4564984 total: 56.5ms remaining: 13.2ms
81: learn: 0.4546838 total: 56.9ms remaining: 12.5ms
82: learn: 0.4528079 total: 57.3ms remaining: 11.7ms
83: learn: 0.4513652 total: 57.7ms remaining: 11ms
84: learn: 0.4497571 total: 58ms remaining: 10.2ms
85: learn: 0.4479051 total: 58.5ms remaining: 9.52ms
86: learn: 0.4458026 total: 58.9ms remaining: 8.8ms
87: learn: 0.4441070 total: 59.2ms remaining: 8.07ms
88: learn: 0.4425226 total: 59.6ms remaining: 7.37ms
89: learn: 0.4407601 total: 60.1ms remaining: 6.68ms
90: learn: 0.4391970 total: 60.4ms remaining: 5.97ms
91: learn: 0.4375876 total: 60.8ms remaining: 5.29ms
92: learn: 0.4359720 total: 61.3ms remaining: 4.61ms
93: learn: 0.4342904 total: 61.6ms remaining: 3.93ms
94: learn: 0.4329354 total: 62ms remaining: 3.26ms
95: learn: 0.4312499 total: 62.5ms remaining: 2.6ms
96: learn: 0.4294859 total: 63ms remaining: 1.95ms
97: learn: 0.4281853 total: 63.4ms remaining: 1.29ms
98: learn: 0.4265789 total: 63.7ms remaining: 643us
99: learn: 0.4249783 total: 64.1ms remaining: 0us
0: learn: 0.6884251 total: 1.62ms remaining: 160ms
1: learn: 0.6837882 total: 2.72ms remaining: 133ms
2: learn: 0.6788893 total: 3.75ms remaining: 121ms
3: learn: 0.6742531 total: 4.76ms remaining: 114ms
4: learn: 0.6694941 total: 6.24ms remaining: 119ms
5: learn: 0.6644974 total: 8.19ms remaining: 128ms
6: learn: 0.6602396 total: 9.74ms remaining: 129ms
7: learn: 0.6546575 total: 10.9ms remaining: 126ms
8: learn: 0.6495477 total: 12.3ms remaining: 124ms
9: learn: 0.6460980 total: 12.9ms remaining: 116ms
10: learn: 0.6407818 total: 13.5ms remaining: 109ms
11: learn: 0.6358886 total: 14.6ms remaining: 107ms
12: learn: 0.6319770 total: 16.2ms remaining: 109ms
13: learn: 0.6280995 total: 17.8ms remaining: 109ms
14: learn: 0.6233202 total: 18.8ms remaining: 106ms
15: learn: 0.6192099 total: 20.1ms remaining: 106ms
16: learn: 0.6147762 total: 21.3ms remaining: 104ms
17: learn: 0.6103238 total: 22ms remaining: 100ms
18: learn: 0.6060995 total: 23.5ms remaining: 100ms
19: learn: 0.6020556 total: 24.7ms remaining: 98.7ms
20: learn: 0.5977037 total: 25.4ms remaining: 95.7ms
21: learn: 0.5931581 total: 26ms remaining: 92.2ms
22: learn: 0.5890359 total: 26.7ms remaining: 89.4ms
23: learn: 0.5846377 total: 27.5ms remaining: 87.1ms
24: learn: 0.5814286 total: 28.3ms remaining: 84.9ms
25: learn: 0.5780560 total: 29.2ms remaining: 83.2ms
26: learn: 0.5739783 total: 30.1ms remaining: 81.3ms
27: learn: 0.5709059 total: 31ms remaining: 79.7ms
28: learn: 0.5681418 total: 31.5ms remaining: 77.2ms
29: learn: 0.5650585 total: 31.9ms remaining: 74.5ms
30: learn: 0.5612944 total: 32.4ms remaining: 72.2ms
31: learn: 0.5579837 total: 33.3ms remaining: 70.8ms
32: learn: 0.5545437 total: 33.9ms remaining: 68.8ms
33: learn: 0.5508624 total: 34.6ms remaining: 67.2ms
34: learn: 0.5476947 total: 35.3ms remaining: 65.5ms
35: learn: 0.5443277 total: 35.9ms remaining: 63.9ms
36: learn: 0.5410425 total: 36.7ms remaining: 62.5ms
37: learn: 0.5381051 total: 37.4ms remaining: 61ms
38: learn: 0.5349005 total: 38.1ms remaining: 59.5ms
39: learn: 0.5320597 total: 38.7ms remaining: 58.1ms
40: learn: 0.5288238 total: 39.2ms remaining: 56.4ms
41: learn: 0.5252659 total: 39.8ms remaining: 54.9ms
42: learn: 0.5227562 total: 40.1ms remaining: 53.2ms
43: learn: 0.5199700 total: 40.8ms remaining: 52ms
44: learn: 0.5171837 total: 41.6ms remaining: 50.9ms
45: learn: 0.5143990 total: 42.2ms remaining: 49.6ms
46: learn: 0.5116150 total: 42.8ms remaining: 48.3ms
47: learn: 0.5085206 total: 43.4ms remaining: 47ms
48: learn: 0.5052346 total: 44ms remaining: 45.8ms
49: learn: 0.5021518 total: 44.7ms remaining: 44.7ms
50: learn: 0.4995699 total: 45.2ms remaining: 43.4ms
51: learn: 0.4966323 total: 45.8ms remaining: 42.3ms
52: learn: 0.4942124 total: 46.2ms remaining: 41ms
53: learn: 0.4918257 total: 46.9ms remaining: 39.9ms
54: learn: 0.4890696 total: 47.2ms remaining: 38.6ms
55: learn: 0.4870031 total: 47.7ms remaining: 37.5ms
56: learn: 0.4843219 total: 48.2ms remaining: 36.3ms
57: learn: 0.4816078 total: 48.6ms remaining: 35.2ms
58: learn: 0.4785999 total: 49.1ms remaining: 34.1ms
59: learn: 0.4760945 total: 49.6ms remaining: 33ms
60: learn: 0.4733829 total: 49.9ms remaining: 31.9ms
61: learn: 0.4708449 total: 50.4ms remaining: 30.9ms
62: learn: 0.4687759 total: 50.7ms remaining: 29.8ms
63: learn: 0.4666086 total: 51.2ms remaining: 28.8ms
64: learn: 0.4640723 total: 51.6ms remaining: 27.8ms
65: learn: 0.4615086 total: 52.1ms remaining: 26.8ms
66: learn: 0.4593938 total: 52.5ms remaining: 25.8ms
67: learn: 0.4573500 total: 52.9ms remaining: 24.9ms
68: learn: 0.4549034 total: 53.2ms remaining: 23.9ms
69: learn: 0.4531617 total: 53.7ms remaining: 23ms
70: learn: 0.4508314 total: 54.1ms remaining: 22.1ms
71: learn: 0.4484736 total: 54.7ms remaining: 21.3ms
72: learn: 0.4462847 total: 55ms remaining: 20.3ms
73: learn: 0.4440720 total: 55.6ms remaining: 19.5ms
74: learn: 0.4418016 total: 56ms remaining: 18.7ms
75: learn: 0.4398148 total: 56.4ms remaining: 17.8ms
76: learn: 0.4376869 total: 56.8ms remaining: 17ms
77: learn: 0.4353386 total: 57.2ms remaining: 16.1ms
78: learn: 0.4331077 total: 57.5ms remaining: 15.3ms
79: learn: 0.4309541 total: 57.9ms remaining: 14.5ms
80: learn: 0.4292700 total: 58.4ms remaining: 13.7ms
81: learn: 0.4273715 total: 58.8ms remaining: 12.9ms
82: learn: 0.4253411 total: 59.2ms remaining: 12.1ms
83: learn: 0.4232837 total: 59.5ms remaining: 11.3ms
84: learn: 0.4217062 total: 60ms remaining: 10.6ms
85: learn: 0.4202087 total: 60.5ms remaining: 9.84ms
86: learn: 0.4181963 total: 60.9ms remaining: 9.1ms
87: learn: 0.4162303 total: 61.3ms remaining: 8.36ms
88: learn: 0.4145317 total: 61.6ms remaining: 7.62ms
89: learn: 0.4126805 total: 62.2ms remaining: 6.91ms
90: learn: 0.4109249 total: 62.6ms remaining: 6.19ms
91: learn: 0.4090370 total: 62.9ms remaining: 5.47ms
92: learn: 0.4075740 total: 63.4ms remaining: 4.77ms
93: learn: 0.4056473 total: 63.9ms remaining: 4.08ms
94: learn: 0.4043221 total: 64.2ms remaining: 3.38ms
95: learn: 0.4023332 total: 64.6ms remaining: 2.69ms
96: learn: 0.4003395 total: 65.1ms remaining: 2.01ms
97: learn: 0.3984095 total: 65.5ms remaining: 1.34ms
98: learn: 0.3966427 total: 65.8ms remaining: 664us
99: learn: 0.3947914 total: 66.1ms remaining: 0us
0: learn: 0.6878984 total: 1.42ms remaining: 141ms
1: learn: 0.6825637 total: 2.39ms remaining: 117ms
2: learn: 0.6778136 total: 3.44ms remaining: 111ms
3: learn: 0.6730922 total: 4.77ms remaining: 115ms
4: learn: 0.6683979 total: 5.65ms remaining: 107ms
5: learn: 0.6639298 total: 6.44ms remaining: 101ms
6: learn: 0.6588845 total: 7.02ms remaining: 93.3ms
7: learn: 0.6547090 total: 8.07ms remaining: 92.9ms
8: learn: 0.6501215 total: 9.01ms remaining: 91.1ms
9: learn: 0.6455656 total: 10.1ms remaining: 90.8ms
10: learn: 0.6411502 total: 11.2ms remaining: 90.4ms
11: learn: 0.6367748 total: 12.1ms remaining: 88.4ms
12: learn: 0.6324957 total: 13.1ms remaining: 87.5ms
13: learn: 0.6282916 total: 14ms remaining: 85.9ms
14: learn: 0.6244643 total: 15.2ms remaining: 85.9ms
15: learn: 0.6197827 total: 16ms remaining: 84ms
16: learn: 0.6162918 total: 17.3ms remaining: 84.6ms
17: learn: 0.6117805 total: 18.1ms remaining: 82.5ms
18: learn: 0.6076094 total: 18.8ms remaining: 80.3ms
19: learn: 0.6039850 total: 20ms remaining: 80.1ms
20: learn: 0.6001150 total: 21.2ms remaining: 79.6ms
21: learn: 0.5961289 total: 22.2ms remaining: 78.9ms
22: learn: 0.5919174 total: 23.4ms remaining: 78.4ms
23: learn: 0.5880261 total: 24.1ms remaining: 76.4ms
24: learn: 0.5845686 total: 25.8ms remaining: 77.3ms
25: learn: 0.5814502 total: 26.9ms remaining: 76.6ms
26: learn: 0.5783653 total: 28.1ms remaining: 76ms
27: learn: 0.5749444 total: 29ms remaining: 74.6ms
28: learn: 0.5717356 total: 30.1ms remaining: 73.6ms
29: learn: 0.5686947 total: 30.7ms remaining: 71.6ms
30: learn: 0.5658574 total: 31.3ms remaining: 69.7ms
31: learn: 0.5627563 total: 32.1ms remaining: 68.2ms
32: learn: 0.5594251 total: 33.4ms remaining: 67.9ms
33: learn: 0.5561764 total: 34.1ms remaining: 66.3ms
34: learn: 0.5529952 total: 34.6ms remaining: 64.2ms
35: learn: 0.5508336 total: 35.5ms remaining: 63ms
36: learn: 0.5478067 total: 37.9ms remaining: 64.5ms
37: learn: 0.5449646 total: 38.6ms remaining: 62.9ms
38: learn: 0.5420459 total: 39.8ms remaining: 62.2ms
39: learn: 0.5388165 total: 40.8ms remaining: 61.2ms
40: learn: 0.5358141 total: 41.9ms remaining: 60.3ms
41: learn: 0.5331245 total: 42.9ms remaining: 59.3ms
42: learn: 0.5300696 total: 43.7ms remaining: 58ms
43: learn: 0.5273135 total: 44.5ms remaining: 56.7ms
44: learn: 0.5247408 total: 45.5ms remaining: 55.7ms
45: learn: 0.5220816 total: 46.4ms remaining: 54.5ms
46: learn: 0.5198094 total: 47.1ms remaining: 53.2ms
47: learn: 0.5168347 total: 48.2ms remaining: 52.2ms
48: learn: 0.5144139 total: 49.2ms remaining: 51.2ms
49: learn: 0.5116065 total: 49.8ms remaining: 49.8ms
50: learn: 0.5090462 total: 50.5ms remaining: 48.5ms
51: learn: 0.5064657 total: 51.3ms remaining: 47.4ms
52: learn: 0.5045712 total: 51.9ms remaining: 46ms
53: learn: 0.5023182 total: 52.6ms remaining: 44.8ms
54: learn: 0.4998693 total: 53.3ms remaining: 43.6ms
55: learn: 0.4978502 total: 53.9ms remaining: 42.4ms
56: learn: 0.4956555 total: 54.4ms remaining: 41ms
57: learn: 0.4933164 total: 55.1ms remaining: 39.9ms
58: learn: 0.4907979 total: 55.7ms remaining: 38.7ms
59: learn: 0.4880570 total: 56.3ms remaining: 37.5ms
60: learn: 0.4857546 total: 56.7ms remaining: 36.3ms
61: learn: 0.4835209 total: 57.5ms remaining: 35.2ms
62: learn: 0.4807113 total: 58.1ms remaining: 34.1ms
63: learn: 0.4785444 total: 58.8ms remaining: 33.1ms
64: learn: 0.4762250 total: 59.2ms remaining: 31.9ms
65: learn: 0.4739229 total: 59.8ms remaining: 30.8ms
66: learn: 0.4715654 total: 60.2ms remaining: 29.6ms
67: learn: 0.4690922 total: 60.7ms remaining: 28.6ms
68: learn: 0.4664602 total: 61.3ms remaining: 27.5ms
69: learn: 0.4642567 total: 61.9ms remaining: 26.5ms
70: learn: 0.4621358 total: 62.4ms remaining: 25.5ms
71: learn: 0.4598338 total: 63ms remaining: 24.5ms
72: learn: 0.4581320 total: 63.7ms remaining: 23.6ms
73: learn: 0.4560829 total: 64.1ms remaining: 22.5ms
74: learn: 0.4539861 total: 64.6ms remaining: 21.5ms
75: learn: 0.4522336 total: 65.1ms remaining: 20.6ms
76: learn: 0.4502632 total: 65.7ms remaining: 19.6ms
77: learn: 0.4484178 total: 66.2ms remaining: 18.7ms
78: learn: 0.4465482 total: 66.7ms remaining: 17.7ms
79: learn: 0.4445991 total: 67.2ms remaining: 16.8ms
80: learn: 0.4427816 total: 67.6ms remaining: 15.9ms
81: learn: 0.4407815 total: 67.8ms remaining: 14.9ms
82: learn: 0.4389686 total: 68.3ms remaining: 14ms
83: learn: 0.4371507 total: 68.9ms remaining: 13.1ms
84: learn: 0.4352346 total: 69.3ms remaining: 12.2ms
85: learn: 0.4339660 total: 69.8ms remaining: 11.4ms
86: learn: 0.4323103 total: 70.3ms remaining: 10.5ms
87: learn: 0.4302934 total: 70.9ms remaining: 9.67ms
88: learn: 0.4281793 total: 71.3ms remaining: 8.81ms
89: learn: 0.4264841 total: 71.6ms remaining: 7.95ms
90: learn: 0.4245832 total: 72ms remaining: 7.12ms
91: learn: 0.4228376 total: 72.5ms remaining: 6.3ms
92: learn: 0.4210649 total: 72.8ms remaining: 5.48ms
93: learn: 0.4194267 total: 73.2ms remaining: 4.67ms
94: learn: 0.4178059 total: 73.6ms remaining: 3.87ms
95: learn: 0.4160512 total: 74.1ms remaining: 3.09ms
96: learn: 0.4147059 total: 74.5ms remaining: 2.3ms
97: learn: 0.4131006 total: 74.9ms remaining: 1.53ms
98: learn: 0.4116573 total: 75.3ms remaining: 760us
99: learn: 0.4098773 total: 75.6ms remaining: 0us
0: learn: 0.6882230 total: 1.13ms remaining: 112ms
1: learn: 0.6830130 total: 2.02ms remaining: 99.2ms
2: learn: 0.6777502 total: 3.07ms remaining: 99.4ms
3: learn: 0.6725869 total: 4.42ms remaining: 106ms
4: learn: 0.6678779 total: 5.36ms remaining: 102ms
5: learn: 0.6627342 total: 6.34ms remaining: 99.4ms
6: learn: 0.6578700 total: 7.36ms remaining: 97.8ms
7: learn: 0.6539456 total: 8.69ms remaining: 100ms
8: learn: 0.6489295 total: 9.6ms remaining: 97ms
9: learn: 0.6447199 total: 10.3ms remaining: 92.8ms
10: learn: 0.6410565 total: 11.1ms remaining: 89.9ms
11: learn: 0.6370334 total: 12.3ms remaining: 89.9ms
12: learn: 0.6323019 total: 13.5ms remaining: 90.2ms
13: learn: 0.6278074 total: 14.9ms remaining: 91.3ms
14: learn: 0.6244961 total: 15.9ms remaining: 90.1ms
15: learn: 0.6205365 total: 17.2ms remaining: 90.3ms
16: learn: 0.6164157 total: 18.4ms remaining: 89.9ms
17: learn: 0.6125643 total: 19.7ms remaining: 89.6ms
18: learn: 0.6087596 total: 20.6ms remaining: 87.8ms
19: learn: 0.6051407 total: 21.8ms remaining: 87.2ms
20: learn: 0.6012506 total: 22.9ms remaining: 86.1ms
21: learn: 0.5969953 total: 24ms remaining: 85.1ms
22: learn: 0.5926725 total: 24.9ms remaining: 83.2ms
23: learn: 0.5892457 total: 25.4ms remaining: 80.5ms
24: learn: 0.5861760 total: 26.5ms remaining: 79.6ms
25: learn: 0.5827887 total: 27.6ms remaining: 78.5ms
26: learn: 0.5792932 total: 28.9ms remaining: 78ms
27: learn: 0.5751741 total: 29.6ms remaining: 76.1ms
28: learn: 0.5722502 total: 30.3ms remaining: 74.1ms
29: learn: 0.5696262 total: 30.9ms remaining: 72ms
30: learn: 0.5670091 total: 32.1ms remaining: 71.4ms
31: learn: 0.5635108 total: 32.5ms remaining: 69.1ms
32: learn: 0.5600613 total: 33.4ms remaining: 67.8ms
33: learn: 0.5568582 total: 34ms remaining: 65.9ms
34: learn: 0.5533880 total: 34.6ms remaining: 64.4ms
35: learn: 0.5503392 total: 35.3ms remaining: 62.8ms
36: learn: 0.5472394 total: 36.1ms remaining: 61.4ms
37: learn: 0.5443595 total: 36.9ms remaining: 60.1ms
38: learn: 0.5412247 total: 37.8ms remaining: 59.2ms
39: learn: 0.5382340 total: 38.5ms remaining: 57.8ms
40: learn: 0.5349641 total: 39.4ms remaining: 56.8ms
41: learn: 0.5321799 total: 40.1ms remaining: 55.4ms
42: learn: 0.5289777 total: 40.9ms remaining: 54.2ms
43: learn: 0.5259768 total: 41.5ms remaining: 52.8ms
44: learn: 0.5230374 total: 42.1ms remaining: 51.5ms
45: learn: 0.5207025 total: 42.7ms remaining: 50.2ms
46: learn: 0.5181586 total: 43.4ms remaining: 48.9ms
47: learn: 0.5151432 total: 44.2ms remaining: 47.9ms
48: learn: 0.5125814 total: 44.8ms remaining: 46.7ms
49: learn: 0.5098965 total: 45.6ms remaining: 45.6ms
50: learn: 0.5069506 total: 46ms remaining: 44.2ms
51: learn: 0.5041681 total: 46.4ms remaining: 42.8ms
52: learn: 0.5016870 total: 47.1ms remaining: 41.8ms
53: learn: 0.4994037 total: 47.7ms remaining: 40.6ms
54: learn: 0.4966429 total: 48.6ms remaining: 39.8ms
55: learn: 0.4946069 total: 49ms remaining: 38.5ms
56: learn: 0.4919759 total: 49.7ms remaining: 37.5ms
57: learn: 0.4890635 total: 50.2ms remaining: 36.3ms
58: learn: 0.4867096 total: 50.9ms remaining: 35.4ms
59: learn: 0.4841801 total: 51.4ms remaining: 34.3ms
60: learn: 0.4816752 total: 52.1ms remaining: 33.3ms
61: learn: 0.4793108 total: 52.8ms remaining: 32.3ms
62: learn: 0.4767157 total: 53.5ms remaining: 31.4ms
63: learn: 0.4742852 total: 54.1ms remaining: 30.4ms
64: learn: 0.4723054 total: 54.6ms remaining: 29.4ms
65: learn: 0.4700578 total: 55.1ms remaining: 28.4ms
66: learn: 0.4679812 total: 55.5ms remaining: 27.4ms
67: learn: 0.4653752 total: 55.9ms remaining: 26.3ms
68: learn: 0.4629021 total: 56.5ms remaining: 25.4ms
69: learn: 0.4608863 total: 57.1ms remaining: 24.5ms
70: learn: 0.4589026 total: 57.7ms remaining: 23.6ms
71: learn: 0.4564408 total: 58.4ms remaining: 22.7ms
72: learn: 0.4540118 total: 58.9ms remaining: 21.8ms
73: learn: 0.4517610 total: 59.5ms remaining: 20.9ms
74: learn: 0.4497752 total: 59.8ms remaining: 19.9ms
75: learn: 0.4478640 total: 60.4ms remaining: 19.1ms
76: learn: 0.4457896 total: 60.9ms remaining: 18.2ms
77: learn: 0.4437100 total: 61.9ms remaining: 17.5ms
78: learn: 0.4415177 total: 62.4ms remaining: 16.6ms
79: learn: 0.4393996 total: 62.9ms remaining: 15.7ms
80: learn: 0.4373214 total: 63.4ms remaining: 14.9ms
81: learn: 0.4352059 total: 63.8ms remaining: 14ms
82: learn: 0.4336374 total: 64.2ms remaining: 13.1ms
83: learn: 0.4320289 total: 64.8ms remaining: 12.3ms
84: learn: 0.4300997 total: 65.3ms remaining: 11.5ms
85: learn: 0.4281833 total: 65.7ms remaining: 10.7ms
86: learn: 0.4263781 total: 66.2ms remaining: 9.89ms
87: learn: 0.4250742 total: 66.5ms remaining: 9.07ms
88: learn: 0.4231440 total: 67ms remaining: 8.28ms
89: learn: 0.4210817 total: 67.4ms remaining: 7.49ms
90: learn: 0.4192910 total: 67.8ms remaining: 6.71ms
91: learn: 0.4175057 total: 68.3ms remaining: 5.94ms
92: learn: 0.4157031 total: 68.7ms remaining: 5.17ms
93: learn: 0.4137212 total: 69.3ms remaining: 4.42ms
94: learn: 0.4120097 total: 69.7ms remaining: 3.67ms
95: learn: 0.4104197 total: 70.3ms remaining: 2.93ms
96: learn: 0.4086656 total: 70.7ms remaining: 2.19ms
97: learn: 0.4068281 total: 71.1ms remaining: 1.45ms
98: learn: 0.4053149 total: 71.4ms remaining: 720us
99: learn: 0.4035909 total: 71.7ms remaining: 0us
0: learn: 0.6886159 total: 1.46ms remaining: 145ms
1: learn: 0.6833054 total: 2.75ms remaining: 135ms
2: learn: 0.6784831 total: 3.62ms remaining: 117ms
3: learn: 0.6744239 total: 4.82ms remaining: 116ms
4: learn: 0.6703678 total: 5.86ms remaining: 111ms
5: learn: 0.6657698 total: 7.27ms remaining: 114ms
6: learn: 0.6613446 total: 8.4ms remaining: 112ms
7: learn: 0.6575104 total: 9.59ms remaining: 110ms
8: learn: 0.6529390 total: 10.7ms remaining: 108ms
9: learn: 0.6485341 total: 12.3ms remaining: 111ms
10: learn: 0.6445752 total: 13.2ms remaining: 106ms
11: learn: 0.6403021 total: 14.4ms remaining: 105ms
12: learn: 0.6365289 total: 15.8ms remaining: 106ms
13: learn: 0.6324526 total: 17ms remaining: 105ms
14: learn: 0.6290089 total: 18.3ms remaining: 103ms
15: learn: 0.6245414 total: 19.2ms remaining: 101ms
16: learn: 0.6210188 total: 20.2ms remaining: 98.7ms
17: learn: 0.6174320 total: 21.3ms remaining: 97.2ms
18: learn: 0.6140296 total: 22.1ms remaining: 94.1ms
19: learn: 0.6105599 total: 23.2ms remaining: 92.8ms
20: learn: 0.6069129 total: 24.4ms remaining: 91.9ms
21: learn: 0.6030373 total: 25.6ms remaining: 90.9ms
22: learn: 0.5995989 total: 26.8ms remaining: 89.8ms
23: learn: 0.5963684 total: 28.2ms remaining: 89.2ms
24: learn: 0.5922892 total: 29.2ms remaining: 87.6ms
25: learn: 0.5894788 total: 30.4ms remaining: 86.4ms
26: learn: 0.5863141 total: 31.6ms remaining: 85.6ms
27: learn: 0.5833325 total: 32.8ms remaining: 84.4ms
28: learn: 0.5804530 total: 33.8ms remaining: 82.7ms
29: learn: 0.5778179 total: 34.7ms remaining: 80.9ms
30: learn: 0.5753587 total: 36.2ms remaining: 80.5ms
31: learn: 0.5724407 total: 37ms remaining: 78.6ms
32: learn: 0.5690258 total: 38.1ms remaining: 77.4ms
33: learn: 0.5657888 total: 39.1ms remaining: 75.9ms
34: learn: 0.5621902 total: 40.8ms remaining: 75.7ms
35: learn: 0.5593933 total: 41.9ms remaining: 74.6ms
36: learn: 0.5564423 total: 43.1ms remaining: 73.3ms
37: learn: 0.5537762 total: 44.3ms remaining: 72.3ms
38: learn: 0.5504946 total: 45.7ms remaining: 71.5ms
39: learn: 0.5481516 total: 47.2ms remaining: 70.9ms
40: learn: 0.5452933 total: 48.4ms remaining: 69.6ms
41: learn: 0.5425361 total: 49.5ms remaining: 68.3ms
42: learn: 0.5398155 total: 50.9ms remaining: 67.5ms
43: learn: 0.5365296 total: 51.5ms remaining: 65.5ms
44: learn: 0.5338784 total: 51.9ms remaining: 63.5ms
45: learn: 0.5312372 total: 53ms remaining: 62.2ms
46: learn: 0.5285292 total: 54ms remaining: 60.9ms
47: learn: 0.5257266 total: 54.9ms remaining: 59.5ms
48: learn: 0.5232061 total: 55.2ms remaining: 57.4ms
49: learn: 0.5203838 total: 56.3ms remaining: 56.3ms
50: learn: 0.5178191 total: 57.3ms remaining: 55ms
51: learn: 0.5153748 total: 58.1ms remaining: 53.6ms
52: learn: 0.5132619 total: 58.6ms remaining: 52ms
53: learn: 0.5111186 total: 59.5ms remaining: 50.6ms
54: learn: 0.5089255 total: 60.3ms remaining: 49.4ms
55: learn: 0.5064440 total: 60.9ms remaining: 47.9ms
56: learn: 0.5038217 total: 61.3ms remaining: 46.2ms
57: learn: 0.5015302 total: 62.4ms remaining: 45.2ms
58: learn: 0.4997467 total: 63.2ms remaining: 43.9ms
59: learn: 0.4974990 total: 64.2ms remaining: 42.8ms
60: learn: 0.4953613 total: 65.1ms remaining: 41.6ms
61: learn: 0.4926778 total: 65.8ms remaining: 40.3ms
62: learn: 0.4903292 total: 66.9ms remaining: 39.3ms
63: learn: 0.4884905 total: 67.6ms remaining: 38ms
64: learn: 0.4859898 total: 68.5ms remaining: 36.9ms
65: learn: 0.4837754 total: 69.3ms remaining: 35.7ms
66: learn: 0.4818255 total: 70.2ms remaining: 34.6ms
67: learn: 0.4798123 total: 70.9ms remaining: 33.4ms
68: learn: 0.4778911 total: 72.1ms remaining: 32.4ms
69: learn: 0.4760780 total: 72.7ms remaining: 31.2ms
70: learn: 0.4738256 total: 73.4ms remaining: 30ms
71: learn: 0.4720699 total: 74ms remaining: 28.8ms
72: learn: 0.4699009 total: 74.7ms remaining: 27.6ms
73: learn: 0.4679986 total: 75.5ms remaining: 26.5ms
74: learn: 0.4660636 total: 76.1ms remaining: 25.4ms
75: learn: 0.4639535 total: 76.7ms remaining: 24.2ms
76: learn: 0.4622288 total: 77.3ms remaining: 23.1ms
77: learn: 0.4603284 total: 77.8ms remaining: 22ms
78: learn: 0.4585539 total: 78.6ms remaining: 20.9ms
79: learn: 0.4570284 total: 79.1ms remaining: 19.8ms
80: learn: 0.4549886 total: 79.7ms remaining: 18.7ms
81: learn: 0.4530752 total: 80.2ms remaining: 17.6ms
82: learn: 0.4514332 total: 80.6ms remaining: 16.5ms
83: learn: 0.4498867 total: 81.2ms remaining: 15.5ms
84: learn: 0.4478527 total: 81.6ms remaining: 14.4ms
85: learn: 0.4457519 total: 81.9ms remaining: 13.3ms
86: learn: 0.4439592 total: 82.3ms remaining: 12.3ms
87: learn: 0.4423116 total: 82.9ms remaining: 11.3ms
88: learn: 0.4404342 total: 83.2ms remaining: 10.3ms
89: learn: 0.4388356 total: 83.6ms remaining: 9.29ms
90: learn: 0.4372264 total: 84.3ms remaining: 8.34ms
91: learn: 0.4357569 total: 84.9ms remaining: 7.38ms
92: learn: 0.4341066 total: 85.3ms remaining: 6.42ms
93: learn: 0.4328156 total: 85.8ms remaining: 5.47ms
94: learn: 0.4311393 total: 86.2ms remaining: 4.54ms
95: learn: 0.4296042 total: 86.7ms remaining: 3.61ms
96: learn: 0.4279636 total: 87.4ms remaining: 2.7ms
97: learn: 0.4267022 total: 87.7ms remaining: 1.79ms
98: learn: 0.4249715 total: 88.1ms remaining: 890us
99: learn: 0.4236659 total: 88.6ms remaining: 0us
Evaluating param combo 10/16: {'boosted_cb_model__iterations': 100, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 16}
Duplicate found for this combo (hash=b668790ad9b40503a17675e2a080e82b). Removing old run: b3a1f2962c4543b1aa2cca90b09d2b3e
0: learn: 0.6882794 total: 1.29ms remaining: 127ms
1: learn: 0.6829408 total: 2.72ms remaining: 133ms
2: learn: 0.6779769 total: 3.59ms remaining: 116ms
3: learn: 0.6731590 total: 4.81ms remaining: 116ms
4: learn: 0.6685559 total: 5.73ms remaining: 109ms
5: learn: 0.6636779 total: 7.15ms remaining: 112ms
6: learn: 0.6591763 total: 8.38ms remaining: 111ms
7: learn: 0.6551357 total: 9.48ms remaining: 109ms
8: learn: 0.6504101 total: 10.5ms remaining: 106ms
9: learn: 0.6464580 total: 11.9ms remaining: 107ms
10: learn: 0.6425359 total: 12.9ms remaining: 104ms
11: learn: 0.6387708 total: 14.3ms remaining: 105ms
12: learn: 0.6346059 total: 15.5ms remaining: 104ms
13: learn: 0.6304314 total: 16.7ms remaining: 103ms
14: learn: 0.6268009 total: 17.8ms remaining: 101ms
15: learn: 0.6229881 total: 19.1ms remaining: 100ms
16: learn: 0.6190015 total: 20.5ms remaining: 99.9ms
17: learn: 0.6151072 total: 21.8ms remaining: 99.4ms
18: learn: 0.6113699 total: 23.1ms remaining: 98.5ms
19: learn: 0.6075444 total: 24.7ms remaining: 98.6ms
20: learn: 0.6037564 total: 25.7ms remaining: 96.8ms
21: learn: 0.5999778 total: 27.3ms remaining: 96.9ms
22: learn: 0.5958785 total: 28.4ms remaining: 94.9ms
23: learn: 0.5922311 total: 29.3ms remaining: 92.8ms
24: learn: 0.5892157 total: 30.6ms remaining: 91.7ms
25: learn: 0.5861289 total: 32ms remaining: 91.2ms
26: learn: 0.5829753 total: 33.4ms remaining: 90.4ms
27: learn: 0.5798042 total: 34.8ms remaining: 89.4ms
28: learn: 0.5767399 total: 36.2ms remaining: 88.6ms
29: learn: 0.5739181 total: 37.4ms remaining: 87.2ms
30: learn: 0.5712722 total: 38.8ms remaining: 86.3ms
31: learn: 0.5679889 total: 39.5ms remaining: 83.9ms
32: learn: 0.5647182 total: 40.9ms remaining: 83.1ms
33: learn: 0.5613468 total: 42.2ms remaining: 81.9ms
34: learn: 0.5577149 total: 43.2ms remaining: 80.2ms
35: learn: 0.5547437 total: 44ms remaining: 78.2ms
36: learn: 0.5517524 total: 45.3ms remaining: 77.1ms
37: learn: 0.5484946 total: 46.2ms remaining: 75.4ms
38: learn: 0.5451312 total: 47.3ms remaining: 74ms
39: learn: 0.5421510 total: 48.1ms remaining: 72.2ms
40: learn: 0.5387318 total: 49.4ms remaining: 71.1ms
41: learn: 0.5355684 total: 50.2ms remaining: 69.3ms
42: learn: 0.5324215 total: 51.1ms remaining: 67.7ms
43: learn: 0.5298921 total: 52.1ms remaining: 66.3ms
44: learn: 0.5271650 total: 53.5ms remaining: 65.4ms
45: learn: 0.5248260 total: 54.6ms remaining: 64.1ms
46: learn: 0.5222534 total: 55.4ms remaining: 62.5ms
47: learn: 0.5193675 total: 56.3ms remaining: 61ms
48: learn: 0.5168600 total: 57ms remaining: 59.3ms
49: learn: 0.5145437 total: 57.7ms remaining: 57.7ms
50: learn: 0.5114851 total: 58.4ms remaining: 56.1ms
51: learn: 0.5093087 total: 58.9ms remaining: 54.4ms
52: learn: 0.5067977 total: 59.4ms remaining: 52.7ms
53: learn: 0.5046903 total: 59.9ms remaining: 51ms
54: learn: 0.5020617 total: 61.1ms remaining: 50ms
55: learn: 0.4998635 total: 61.7ms remaining: 48.5ms
56: learn: 0.4974511 total: 62.3ms remaining: 47ms
57: learn: 0.4953198 total: 62.8ms remaining: 45.5ms
58: learn: 0.4931948 total: 63.5ms remaining: 44.1ms
59: learn: 0.4911060 total: 64.1ms remaining: 42.8ms
60: learn: 0.4889797 total: 64.6ms remaining: 41.3ms
61: learn: 0.4866935 total: 65.1ms remaining: 39.9ms
62: learn: 0.4843003 total: 65.5ms remaining: 38.5ms
63: learn: 0.4820957 total: 66ms remaining: 37.1ms
64: learn: 0.4798245 total: 66.4ms remaining: 35.8ms
65: learn: 0.4777677 total: 66.7ms remaining: 34.4ms
66: learn: 0.4756431 total: 67.2ms remaining: 33.1ms
67: learn: 0.4735064 total: 67.5ms remaining: 31.8ms
68: learn: 0.4711289 total: 68.1ms remaining: 30.6ms
69: learn: 0.4686863 total: 68.8ms remaining: 29.5ms
70: learn: 0.4665532 total: 69.6ms remaining: 28.4ms
71: learn: 0.4646554 total: 70.3ms remaining: 27.3ms
72: learn: 0.4626431 total: 71.1ms remaining: 26.3ms
73: learn: 0.4606925 total: 71.7ms remaining: 25.2ms
74: learn: 0.4587245 total: 72.3ms remaining: 24.1ms
75: learn: 0.4565481 total: 72.9ms remaining: 23ms
76: learn: 0.4547888 total: 73.7ms remaining: 22ms
77: learn: 0.4528959 total: 74.1ms remaining: 20.9ms
78: learn: 0.4508121 total: 74.4ms remaining: 19.8ms
79: learn: 0.4490872 total: 74.9ms remaining: 18.7ms
80: learn: 0.4471421 total: 75.8ms remaining: 17.8ms
81: learn: 0.4453254 total: 76.1ms remaining: 16.7ms
82: learn: 0.4432644 total: 76.4ms remaining: 15.6ms
83: learn: 0.4413600 total: 76.7ms remaining: 14.6ms
84: learn: 0.4394892 total: 77.2ms remaining: 13.6ms
85: learn: 0.4377549 total: 77.6ms remaining: 12.6ms
86: learn: 0.4359258 total: 78ms remaining: 11.7ms
87: learn: 0.4339840 total: 78.3ms remaining: 10.7ms
88: learn: 0.4318123 total: 78.8ms remaining: 9.74ms
89: learn: 0.4300119 total: 79.3ms remaining: 8.81ms
90: learn: 0.4281025 total: 79.7ms remaining: 7.89ms
91: learn: 0.4265387 total: 80.4ms remaining: 6.99ms
92: learn: 0.4249134 total: 80.7ms remaining: 6.08ms
93: learn: 0.4233160 total: 81.1ms remaining: 5.18ms
94: learn: 0.4217480 total: 81.6ms remaining: 4.29ms
95: learn: 0.4200962 total: 82.2ms remaining: 3.42ms
96: learn: 0.4187601 total: 82.7ms remaining: 2.56ms
97: learn: 0.4172840 total: 83ms remaining: 1.69ms
98: learn: 0.4160567 total: 83.6ms remaining: 844us
99: learn: 0.4143777 total: 83.9ms remaining: 0us
0: learn: 0.6880102 total: 1.92ms remaining: 190ms
1: learn: 0.6822828 total: 3.11ms remaining: 152ms
2: learn: 0.6772106 total: 3.93ms remaining: 127ms
3: learn: 0.6726730 total: 5.38ms remaining: 129ms
4: learn: 0.6682095 total: 6.38ms remaining: 121ms
5: learn: 0.6635268 total: 7.41ms remaining: 116ms
6: learn: 0.6588811 total: 8.39ms remaining: 111ms
7: learn: 0.6545703 total: 9.47ms remaining: 109ms
8: learn: 0.6497856 total: 10.4ms remaining: 105ms
9: learn: 0.6448858 total: 11.7ms remaining: 105ms
10: learn: 0.6405230 total: 12.7ms remaining: 103ms
11: learn: 0.6361860 total: 14ms remaining: 102ms
12: learn: 0.6314977 total: 15.2ms remaining: 102ms
13: learn: 0.6272630 total: 16.5ms remaining: 101ms
14: learn: 0.6237927 total: 17.7ms remaining: 101ms
15: learn: 0.6202946 total: 18.9ms remaining: 99.5ms
16: learn: 0.6160996 total: 20ms remaining: 97.9ms
17: learn: 0.6121987 total: 21.1ms remaining: 96.1ms
18: learn: 0.6085887 total: 22.4ms remaining: 95.5ms
19: learn: 0.6053875 total: 23.6ms remaining: 94.2ms
20: learn: 0.6012749 total: 24.8ms remaining: 93.2ms
21: learn: 0.5970959 total: 26ms remaining: 92.1ms
22: learn: 0.5929697 total: 27.1ms remaining: 90.6ms
23: learn: 0.5894176 total: 28.2ms remaining: 89.4ms
24: learn: 0.5848522 total: 29ms remaining: 87.1ms
25: learn: 0.5821273 total: 30.5ms remaining: 86.8ms
26: learn: 0.5789455 total: 31.6ms remaining: 85.3ms
27: learn: 0.5754969 total: 32.5ms remaining: 83.7ms
28: learn: 0.5721862 total: 33.5ms remaining: 82.1ms
29: learn: 0.5691136 total: 34.6ms remaining: 80.8ms
30: learn: 0.5660278 total: 36.2ms remaining: 80.5ms
31: learn: 0.5628754 total: 37ms remaining: 78.5ms
32: learn: 0.5589497 total: 38ms remaining: 77.2ms
33: learn: 0.5553905 total: 39.7ms remaining: 77ms
34: learn: 0.5515082 total: 40.8ms remaining: 75.8ms
35: learn: 0.5482866 total: 41.8ms remaining: 74.2ms
36: learn: 0.5449903 total: 42.9ms remaining: 73ms
37: learn: 0.5423647 total: 44.1ms remaining: 72ms
38: learn: 0.5386898 total: 44.9ms remaining: 70.2ms
39: learn: 0.5350966 total: 45.7ms remaining: 68.5ms
40: learn: 0.5319492 total: 46.4ms remaining: 66.8ms
41: learn: 0.5288603 total: 46.9ms remaining: 64.7ms
42: learn: 0.5260202 total: 47.6ms remaining: 63ms
43: learn: 0.5230047 total: 48.2ms remaining: 61.3ms
44: learn: 0.5198876 total: 48.9ms remaining: 59.8ms
45: learn: 0.5168284 total: 49.3ms remaining: 57.8ms
46: learn: 0.5136389 total: 50.1ms remaining: 56.5ms
47: learn: 0.5112892 total: 50.8ms remaining: 55.1ms
48: learn: 0.5085121 total: 51.5ms remaining: 53.6ms
49: learn: 0.5054328 total: 52.2ms remaining: 52.2ms
50: learn: 0.5026563 total: 52.9ms remaining: 50.8ms
51: learn: 0.4999174 total: 53.5ms remaining: 49.4ms
52: learn: 0.4972964 total: 53.9ms remaining: 47.8ms
53: learn: 0.4949977 total: 54.3ms remaining: 46.3ms
54: learn: 0.4919354 total: 54.9ms remaining: 44.9ms
55: learn: 0.4898954 total: 55.5ms remaining: 43.6ms
56: learn: 0.4878160 total: 56.1ms remaining: 42.3ms
57: learn: 0.4855045 total: 56.6ms remaining: 41ms
58: learn: 0.4829743 total: 57.2ms remaining: 39.8ms
59: learn: 0.4804930 total: 57.7ms remaining: 38.5ms
60: learn: 0.4780999 total: 58.5ms remaining: 37.4ms
61: learn: 0.4758416 total: 59ms remaining: 36.2ms
62: learn: 0.4728837 total: 59.6ms remaining: 35ms
63: learn: 0.4706892 total: 60.1ms remaining: 33.8ms
64: learn: 0.4683178 total: 60.6ms remaining: 32.6ms
65: learn: 0.4656676 total: 61ms remaining: 31.4ms
66: learn: 0.4632440 total: 61.4ms remaining: 30.2ms
67: learn: 0.4611426 total: 61.7ms remaining: 29.1ms
68: learn: 0.4588472 total: 62.1ms remaining: 27.9ms
69: learn: 0.4565314 total: 62.6ms remaining: 26.8ms
70: learn: 0.4548869 total: 63.2ms remaining: 25.8ms
71: learn: 0.4526554 total: 63.7ms remaining: 24.8ms
72: learn: 0.4499787 total: 64.1ms remaining: 23.7ms
73: learn: 0.4475357 total: 65ms remaining: 22.9ms
74: learn: 0.4455755 total: 65.6ms remaining: 21.9ms
75: learn: 0.4440104 total: 66.1ms remaining: 20.9ms
76: learn: 0.4417982 total: 66.5ms remaining: 19.9ms
77: learn: 0.4398600 total: 67ms remaining: 18.9ms
78: learn: 0.4379151 total: 67.4ms remaining: 17.9ms
79: learn: 0.4361857 total: 67.8ms remaining: 16.9ms
80: learn: 0.4338838 total: 68.3ms remaining: 16ms
81: learn: 0.4319658 total: 68.6ms remaining: 15.1ms
82: learn: 0.4302179 total: 69ms remaining: 14.1ms
83: learn: 0.4280088 total: 69.6ms remaining: 13.3ms
84: learn: 0.4261117 total: 70ms remaining: 12.4ms
85: learn: 0.4244622 total: 70.4ms remaining: 11.5ms
86: learn: 0.4222954 total: 70.8ms remaining: 10.6ms
87: learn: 0.4202130 total: 71.3ms remaining: 9.72ms
88: learn: 0.4181864 total: 71.7ms remaining: 8.86ms
89: learn: 0.4164203 total: 72ms remaining: 8.01ms
90: learn: 0.4149126 total: 72.5ms remaining: 7.17ms
91: learn: 0.4133189 total: 72.9ms remaining: 6.34ms
92: learn: 0.4112811 total: 73.2ms remaining: 5.51ms
93: learn: 0.4094290 total: 73.7ms remaining: 4.7ms
94: learn: 0.4081478 total: 74.1ms remaining: 3.9ms
95: learn: 0.4062874 total: 74.4ms remaining: 3.1ms
96: learn: 0.4044458 total: 74.8ms remaining: 2.31ms
97: learn: 0.4030461 total: 75.2ms remaining: 1.53ms
98: learn: 0.4012621 total: 75.6ms remaining: 763us
99: learn: 0.3996016 total: 76ms remaining: 0us
0: learn: 0.6881915 total: 1.22ms remaining: 121ms
1: learn: 0.6830903 total: 2.44ms remaining: 120ms
2: learn: 0.6779156 total: 3.42ms remaining: 111ms
3: learn: 0.6727870 total: 4.79ms remaining: 115ms
4: learn: 0.6680191 total: 6.04ms remaining: 115ms
5: learn: 0.6632566 total: 6.83ms remaining: 107ms
6: learn: 0.6589348 total: 7.61ms remaining: 101ms
7: learn: 0.6548497 total: 8.04ms remaining: 92.5ms
8: learn: 0.6499422 total: 8.89ms remaining: 89.9ms
9: learn: 0.6456086 total: 9.79ms remaining: 88.1ms
10: learn: 0.6416798 total: 10.8ms remaining: 87.5ms
11: learn: 0.6376258 total: 11.9ms remaining: 87.4ms
12: learn: 0.6328452 total: 12.9ms remaining: 86.2ms
13: learn: 0.6291112 total: 14.1ms remaining: 86.3ms
14: learn: 0.6252126 total: 14.9ms remaining: 84.2ms
15: learn: 0.6213698 total: 16.1ms remaining: 84.4ms
16: learn: 0.6172627 total: 17.3ms remaining: 84.6ms
17: learn: 0.6136354 total: 18.2ms remaining: 83.1ms
18: learn: 0.6099157 total: 19.4ms remaining: 82.8ms
19: learn: 0.6065275 total: 20.7ms remaining: 82.9ms
20: learn: 0.6026046 total: 21.8ms remaining: 82ms
21: learn: 0.5986205 total: 23ms remaining: 81.5ms
22: learn: 0.5943580 total: 24.1ms remaining: 80.8ms
23: learn: 0.5906964 total: 24.8ms remaining: 78.6ms
24: learn: 0.5869638 total: 26.1ms remaining: 78.4ms
25: learn: 0.5838294 total: 27.5ms remaining: 78.3ms
26: learn: 0.5809812 total: 28.7ms remaining: 77.6ms
27: learn: 0.5776571 total: 29.7ms remaining: 76.3ms
28: learn: 0.5743650 total: 31ms remaining: 75.9ms
29: learn: 0.5714944 total: 31.9ms remaining: 74.4ms
30: learn: 0.5687434 total: 33.2ms remaining: 73.8ms
31: learn: 0.5653821 total: 33.8ms remaining: 71.8ms
32: learn: 0.5620526 total: 34.8ms remaining: 70.7ms
33: learn: 0.5584545 total: 35.7ms remaining: 69.3ms
34: learn: 0.5550520 total: 37.1ms remaining: 68.9ms
35: learn: 0.5514777 total: 38.1ms remaining: 67.7ms
36: learn: 0.5483220 total: 39.4ms remaining: 67.1ms
37: learn: 0.5453927 total: 40.6ms remaining: 66.2ms
38: learn: 0.5418823 total: 41.5ms remaining: 64.9ms
39: learn: 0.5394460 total: 42.8ms remaining: 64.2ms
40: learn: 0.5362960 total: 44ms remaining: 63.3ms
41: learn: 0.5328204 total: 45ms remaining: 62.1ms
42: learn: 0.5296236 total: 45.9ms remaining: 60.9ms
43: learn: 0.5272600 total: 46.8ms remaining: 59.6ms
44: learn: 0.5244234 total: 48.2ms remaining: 58.9ms
45: learn: 0.5224995 total: 49.1ms remaining: 57.6ms
46: learn: 0.5199309 total: 50.3ms remaining: 56.7ms
47: learn: 0.5169806 total: 51.3ms remaining: 55.5ms
48: learn: 0.5142186 total: 52.7ms remaining: 54.8ms
49: learn: 0.5113786 total: 53.9ms remaining: 53.9ms
50: learn: 0.5086399 total: 54.9ms remaining: 52.7ms
51: learn: 0.5061395 total: 55.7ms remaining: 51.5ms
52: learn: 0.5038713 total: 56.7ms remaining: 50.3ms
53: learn: 0.5014624 total: 57.5ms remaining: 49ms
54: learn: 0.4989674 total: 58.2ms remaining: 47.6ms
55: learn: 0.4964659 total: 58.7ms remaining: 46.1ms
56: learn: 0.4937484 total: 59.5ms remaining: 44.9ms
57: learn: 0.4914684 total: 60.5ms remaining: 43.8ms
58: learn: 0.4891281 total: 61.4ms remaining: 42.6ms
59: learn: 0.4867757 total: 62.2ms remaining: 41.4ms
60: learn: 0.4845035 total: 63.1ms remaining: 40.3ms
61: learn: 0.4819540 total: 63.8ms remaining: 39.1ms
62: learn: 0.4796578 total: 64.5ms remaining: 37.9ms
63: learn: 0.4774683 total: 65ms remaining: 36.6ms
64: learn: 0.4752972 total: 65.7ms remaining: 35.4ms
65: learn: 0.4730619 total: 66.6ms remaining: 34.3ms
66: learn: 0.4708604 total: 67.2ms remaining: 33.1ms
67: learn: 0.4685726 total: 67.9ms remaining: 32ms
68: learn: 0.4664067 total: 68.4ms remaining: 30.7ms
69: learn: 0.4643398 total: 69.1ms remaining: 29.6ms
70: learn: 0.4624445 total: 69.7ms remaining: 28.5ms
71: learn: 0.4605617 total: 70.5ms remaining: 27.4ms
72: learn: 0.4584894 total: 71.3ms remaining: 26.4ms
73: learn: 0.4563449 total: 71.8ms remaining: 25.2ms
74: learn: 0.4543194 total: 72.3ms remaining: 24.1ms
75: learn: 0.4523466 total: 72.8ms remaining: 23ms
76: learn: 0.4500421 total: 73.2ms remaining: 21.9ms
77: learn: 0.4482336 total: 73.7ms remaining: 20.8ms
78: learn: 0.4461296 total: 74.2ms remaining: 19.7ms
79: learn: 0.4442134 total: 74.4ms remaining: 18.6ms
80: learn: 0.4420610 total: 74.7ms remaining: 17.5ms
81: learn: 0.4400215 total: 75.1ms remaining: 16.5ms
82: learn: 0.4381004 total: 75.6ms remaining: 15.5ms
83: learn: 0.4365113 total: 76ms remaining: 14.5ms
84: learn: 0.4348688 total: 76.4ms remaining: 13.5ms
85: learn: 0.4327667 total: 76.9ms remaining: 12.5ms
86: learn: 0.4305854 total: 77.3ms remaining: 11.6ms
87: learn: 0.4286623 total: 77.7ms remaining: 10.6ms
88: learn: 0.4267661 total: 78.2ms remaining: 9.67ms
89: learn: 0.4250506 total: 78.7ms remaining: 8.74ms
90: learn: 0.4231133 total: 79ms remaining: 7.82ms
91: learn: 0.4214037 total: 79.8ms remaining: 6.93ms
92: learn: 0.4197256 total: 80.2ms remaining: 6.04ms
93: learn: 0.4179794 total: 80.6ms remaining: 5.15ms
94: learn: 0.4162980 total: 81.2ms remaining: 4.27ms
95: learn: 0.4145354 total: 81.6ms remaining: 3.4ms
96: learn: 0.4127874 total: 82ms remaining: 2.54ms
97: learn: 0.4112734 total: 82.4ms remaining: 1.68ms
98: learn: 0.4096648 total: 82.9ms remaining: 837us
99: learn: 0.4080795 total: 83.4ms remaining: 0us
0: learn: 0.6883581 total: 612us remaining: 60.6ms
1: learn: 0.6829490 total: 1.06ms remaining: 52.2ms
2: learn: 0.6777105 total: 1.43ms remaining: 46.2ms
3: learn: 0.6724965 total: 2.34ms remaining: 56.2ms
4: learn: 0.6678869 total: 3.18ms remaining: 60.4ms
5: learn: 0.6630227 total: 4.26ms remaining: 66.8ms
6: learn: 0.6580845 total: 4.71ms remaining: 62.6ms
7: learn: 0.6539384 total: 5.37ms remaining: 61.8ms
8: learn: 0.6488837 total: 6.14ms remaining: 62.1ms
9: learn: 0.6442866 total: 7.16ms remaining: 64.4ms
10: learn: 0.6404207 total: 8.19ms remaining: 66.2ms
11: learn: 0.6359348 total: 9.01ms remaining: 66.1ms
12: learn: 0.6315682 total: 9.58ms remaining: 64.1ms
13: learn: 0.6273393 total: 10.1ms remaining: 61.9ms
14: learn: 0.6234264 total: 11ms remaining: 62.1ms
15: learn: 0.6197946 total: 12.1ms remaining: 63.3ms
16: learn: 0.6155398 total: 13.4ms remaining: 65.5ms
17: learn: 0.6113889 total: 14.4ms remaining: 65.8ms
18: learn: 0.6079857 total: 15.5ms remaining: 66ms
19: learn: 0.6040984 total: 16.5ms remaining: 66ms
20: learn: 0.6001675 total: 17.5ms remaining: 65.8ms
21: learn: 0.5959995 total: 18.5ms remaining: 65.4ms
22: learn: 0.5919796 total: 19.6ms remaining: 65.5ms
23: learn: 0.5884148 total: 21ms remaining: 66.6ms
24: learn: 0.5841205 total: 21.8ms remaining: 65.3ms
25: learn: 0.5809535 total: 23.1ms remaining: 65.8ms
26: learn: 0.5776310 total: 24.1ms remaining: 65.3ms
27: learn: 0.5743591 total: 25.5ms remaining: 65.5ms
28: learn: 0.5712718 total: 26.6ms remaining: 65.1ms
29: learn: 0.5684340 total: 27.3ms remaining: 63.8ms
30: learn: 0.5657478 total: 28.5ms remaining: 63.4ms
31: learn: 0.5623463 total: 29.1ms remaining: 61.8ms
32: learn: 0.5591812 total: 30.4ms remaining: 61.7ms
33: learn: 0.5555487 total: 31.6ms remaining: 61.4ms
34: learn: 0.5525218 total: 32.7ms remaining: 60.7ms
35: learn: 0.5495863 total: 33.9ms remaining: 60.2ms
36: learn: 0.5463044 total: 34.7ms remaining: 59.1ms
37: learn: 0.5429413 total: 36ms remaining: 58.7ms
38: learn: 0.5394540 total: 37ms remaining: 57.8ms
39: learn: 0.5363956 total: 38.2ms remaining: 57.4ms
40: learn: 0.5330491 total: 39.2ms remaining: 56.4ms
41: learn: 0.5306820 total: 40ms remaining: 55.2ms
42: learn: 0.5275766 total: 41.1ms remaining: 54.5ms
43: learn: 0.5246236 total: 42.1ms remaining: 53.6ms
44: learn: 0.5218134 total: 43.4ms remaining: 53.1ms
45: learn: 0.5189606 total: 44.6ms remaining: 52.3ms
46: learn: 0.5159627 total: 45.6ms remaining: 51.5ms
47: learn: 0.5128221 total: 47ms remaining: 50.9ms
48: learn: 0.5096912 total: 48ms remaining: 49.9ms
49: learn: 0.5072024 total: 49ms remaining: 49ms
50: learn: 0.5043210 total: 50ms remaining: 48ms
51: learn: 0.5022366 total: 51.1ms remaining: 47.2ms
52: learn: 0.4997103 total: 52.2ms remaining: 46.3ms
53: learn: 0.4970926 total: 53.2ms remaining: 45.3ms
54: learn: 0.4948878 total: 54.6ms remaining: 44.6ms
55: learn: 0.4921710 total: 55.9ms remaining: 44ms
56: learn: 0.4894281 total: 57ms remaining: 43ms
57: learn: 0.4868875 total: 58.2ms remaining: 42.1ms
58: learn: 0.4842999 total: 59ms remaining: 41ms
59: learn: 0.4821379 total: 60.1ms remaining: 40.1ms
60: learn: 0.4793659 total: 61.2ms remaining: 39.1ms
61: learn: 0.4765558 total: 62.3ms remaining: 38.2ms
62: learn: 0.4739383 total: 63.6ms remaining: 37.3ms
63: learn: 0.4714391 total: 64.6ms remaining: 36.3ms
64: learn: 0.4689153 total: 65.5ms remaining: 35.3ms
65: learn: 0.4669516 total: 66.4ms remaining: 34.2ms
66: learn: 0.4649550 total: 67.5ms remaining: 33.3ms
67: learn: 0.4626150 total: 68.6ms remaining: 32.3ms
68: learn: 0.4608829 total: 68.9ms remaining: 31ms
69: learn: 0.4585578 total: 69.5ms remaining: 29.8ms
70: learn: 0.4567417 total: 70.1ms remaining: 28.6ms
71: learn: 0.4546172 total: 70.6ms remaining: 27.5ms
72: learn: 0.4525670 total: 71.5ms remaining: 26.4ms
73: learn: 0.4505951 total: 72.2ms remaining: 25.4ms
74: learn: 0.4487806 total: 72.9ms remaining: 24.3ms
75: learn: 0.4468307 total: 73.4ms remaining: 23.2ms
76: learn: 0.4445511 total: 73.8ms remaining: 22ms
77: learn: 0.4425293 total: 74.4ms remaining: 21ms
78: learn: 0.4403919 total: 75ms remaining: 19.9ms
79: learn: 0.4385428 total: 75.5ms remaining: 18.9ms
80: learn: 0.4362771 total: 76.1ms remaining: 17.8ms
81: learn: 0.4341435 total: 76.6ms remaining: 16.8ms
82: learn: 0.4320750 total: 77.6ms remaining: 15.9ms
83: learn: 0.4304187 total: 78.2ms remaining: 14.9ms
84: learn: 0.4287434 total: 78.6ms remaining: 13.9ms
85: learn: 0.4267036 total: 79ms remaining: 12.9ms
86: learn: 0.4245506 total: 79.3ms remaining: 11.9ms
87: learn: 0.4226060 total: 79.6ms remaining: 10.9ms
88: learn: 0.4209438 total: 80.1ms remaining: 9.9ms
89: learn: 0.4191627 total: 80.6ms remaining: 8.96ms
90: learn: 0.4175165 total: 81ms remaining: 8.01ms
91: learn: 0.4157530 total: 81.6ms remaining: 7.09ms
92: learn: 0.4139980 total: 82.1ms remaining: 6.18ms
93: learn: 0.4120571 total: 82.7ms remaining: 5.28ms
94: learn: 0.4106134 total: 83.2ms remaining: 4.38ms
95: learn: 0.4088877 total: 83.6ms remaining: 3.48ms
96: learn: 0.4074953 total: 84.3ms remaining: 2.61ms
97: learn: 0.4057214 total: 84.8ms remaining: 1.73ms
98: learn: 0.4042434 total: 85.1ms remaining: 860us
99: learn: 0.4024514 total: 85.7ms remaining: 0us
0: learn: 0.6883175 total: 1.22ms remaining: 121ms
1: learn: 0.6833177 total: 2.41ms remaining: 118ms
2: learn: 0.6787594 total: 3.23ms remaining: 105ms
3: learn: 0.6742785 total: 4.48ms remaining: 108ms
4: learn: 0.6698376 total: 5.38ms remaining: 102ms
5: learn: 0.6655790 total: 6.83ms remaining: 107ms
6: learn: 0.6606753 total: 7.92ms remaining: 105ms
7: learn: 0.6567153 total: 10.4ms remaining: 120ms
8: learn: 0.6521844 total: 12ms remaining: 121ms
9: learn: 0.6476670 total: 13.3ms remaining: 120ms
10: learn: 0.6436401 total: 14.2ms remaining: 115ms
11: learn: 0.6392506 total: 15.4ms remaining: 113ms
12: learn: 0.6352223 total: 16.8ms remaining: 112ms
13: learn: 0.6314012 total: 18.3ms remaining: 112ms
14: learn: 0.6271130 total: 19.6ms remaining: 111ms
15: learn: 0.6228527 total: 20.6ms remaining: 108ms
16: learn: 0.6193636 total: 21.7ms remaining: 106ms
17: learn: 0.6149698 total: 22.7ms remaining: 104ms
18: learn: 0.6115084 total: 23.5ms remaining: 100ms
19: learn: 0.6080555 total: 24.8ms remaining: 99.1ms
20: learn: 0.6044442 total: 26ms remaining: 98ms
21: learn: 0.6011192 total: 27.3ms remaining: 96.8ms
22: learn: 0.5974142 total: 28.9ms remaining: 96.9ms
23: learn: 0.5941231 total: 30.5ms remaining: 96.5ms
24: learn: 0.5900927 total: 32ms remaining: 95.9ms
25: learn: 0.5870085 total: 33.4ms remaining: 95ms
26: learn: 0.5840114 total: 34.7ms remaining: 93.8ms
27: learn: 0.5808997 total: 36.4ms remaining: 93.5ms
28: learn: 0.5778788 total: 37.2ms remaining: 91ms
29: learn: 0.5750842 total: 37.8ms remaining: 88.2ms
30: learn: 0.5722879 total: 38.6ms remaining: 85.9ms
31: learn: 0.5693116 total: 39.2ms remaining: 83.3ms
32: learn: 0.5661397 total: 40.2ms remaining: 81.6ms
33: learn: 0.5627994 total: 41ms remaining: 79.6ms
34: learn: 0.5597978 total: 41.9ms remaining: 77.8ms
35: learn: 0.5564847 total: 43ms remaining: 76.4ms
36: learn: 0.5535813 total: 44ms remaining: 74.8ms
37: learn: 0.5511907 total: 44.9ms remaining: 73.2ms
38: learn: 0.5477524 total: 46ms remaining: 71.9ms
39: learn: 0.5446551 total: 46.6ms remaining: 69.8ms
40: learn: 0.5418243 total: 47.1ms remaining: 67.8ms
41: learn: 0.5384306 total: 47.7ms remaining: 65.8ms
42: learn: 0.5355158 total: 48.5ms remaining: 64.3ms
43: learn: 0.5330304 total: 49.4ms remaining: 62.8ms
44: learn: 0.5303464 total: 50.4ms remaining: 61.6ms
45: learn: 0.5282582 total: 51.1ms remaining: 60ms
46: learn: 0.5258402 total: 51.9ms remaining: 58.5ms
47: learn: 0.5235785 total: 52.5ms remaining: 56.8ms
48: learn: 0.5207689 total: 53.1ms remaining: 55.3ms
49: learn: 0.5183663 total: 54ms remaining: 54ms
50: learn: 0.5158919 total: 54.6ms remaining: 52.5ms
51: learn: 0.5139315 total: 55.2ms remaining: 51ms
52: learn: 0.5116847 total: 55.9ms remaining: 49.6ms
53: learn: 0.5090818 total: 56.7ms remaining: 48.3ms
54: learn: 0.5074963 total: 57ms remaining: 46.7ms
55: learn: 0.5046849 total: 57.5ms remaining: 45.1ms
56: learn: 0.5021928 total: 57.8ms remaining: 43.6ms
57: learn: 0.4999986 total: 58.2ms remaining: 42.2ms
58: learn: 0.4977483 total: 58.6ms remaining: 40.7ms
59: learn: 0.4952915 total: 58.9ms remaining: 39.3ms
60: learn: 0.4936844 total: 59.3ms remaining: 37.9ms
61: learn: 0.4916698 total: 59.9ms remaining: 36.7ms
62: learn: 0.4893568 total: 60.3ms remaining: 35.4ms
63: learn: 0.4871933 total: 60.8ms remaining: 34.2ms
64: learn: 0.4850710 total: 61.3ms remaining: 33ms
65: learn: 0.4829277 total: 61.8ms remaining: 31.8ms
66: learn: 0.4809395 total: 62.1ms remaining: 30.6ms
67: learn: 0.4784928 total: 62.5ms remaining: 29.4ms
68: learn: 0.4763350 total: 63.1ms remaining: 28.3ms
69: learn: 0.4741475 total: 63.6ms remaining: 27.3ms
70: learn: 0.4726073 total: 64ms remaining: 26.1ms
71: learn: 0.4705962 total: 64.4ms remaining: 25ms
72: learn: 0.4689538 total: 64.9ms remaining: 24ms
73: learn: 0.4668614 total: 65.6ms remaining: 23.1ms
74: learn: 0.4645647 total: 66.1ms remaining: 22ms
75: learn: 0.4623719 total: 66.5ms remaining: 21ms
76: learn: 0.4604691 total: 67.1ms remaining: 20ms
77: learn: 0.4585415 total: 67.4ms remaining: 19ms
78: learn: 0.4563981 total: 67.9ms remaining: 18.1ms
79: learn: 0.4544774 total: 68.3ms remaining: 17.1ms
80: learn: 0.4523978 total: 68.7ms remaining: 16.1ms
81: learn: 0.4507069 total: 69.1ms remaining: 15.2ms
82: learn: 0.4484920 total: 69.5ms remaining: 14.2ms
83: learn: 0.4466815 total: 70.1ms remaining: 13.4ms
84: learn: 0.4449599 total: 70.5ms remaining: 12.4ms
85: learn: 0.4430525 total: 70.8ms remaining: 11.5ms
86: learn: 0.4413869 total: 71.2ms remaining: 10.6ms
87: learn: 0.4399236 total: 71.6ms remaining: 9.76ms
88: learn: 0.4380750 total: 72.1ms remaining: 8.91ms
89: learn: 0.4362257 total: 72.4ms remaining: 8.05ms
90: learn: 0.4345031 total: 72.7ms remaining: 7.19ms
91: learn: 0.4328117 total: 73.2ms remaining: 6.37ms
92: learn: 0.4313235 total: 73.7ms remaining: 5.54ms
93: learn: 0.4295387 total: 74ms remaining: 4.72ms
94: learn: 0.4281810 total: 74.4ms remaining: 3.91ms
95: learn: 0.4265977 total: 74.7ms remaining: 3.11ms
96: learn: 0.4253384 total: 75.2ms remaining: 2.33ms
97: learn: 0.4236111 total: 75.6ms remaining: 1.54ms
98: learn: 0.4216073 total: 76ms remaining: 767us
99: learn: 0.4200482 total: 76.4ms remaining: 0us
0: learn: 0.6886824 total: 1.34ms remaining: 133ms
1: learn: 0.6836439 total: 2.65ms remaining: 130ms
2: learn: 0.6783803 total: 3.62ms remaining: 117ms
3: learn: 0.6732528 total: 5.29ms remaining: 127ms
4: learn: 0.6684937 total: 6.26ms remaining: 119ms
5: learn: 0.6634511 total: 8.3ms remaining: 130ms
6: learn: 0.6589192 total: 10.2ms remaining: 135ms
7: learn: 0.6551883 total: 11.4ms remaining: 131ms
8: learn: 0.6502473 total: 12.5ms remaining: 126ms
9: learn: 0.6460178 total: 13.5ms remaining: 121ms
10: learn: 0.6419694 total: 14.4ms remaining: 117ms
11: learn: 0.6383164 total: 15.7ms remaining: 115ms
12: learn: 0.6337982 total: 17.4ms remaining: 116ms
13: learn: 0.6295016 total: 18.8ms remaining: 115ms
14: learn: 0.6253754 total: 20.1ms remaining: 114ms
15: learn: 0.6213047 total: 21.5ms remaining: 113ms
16: learn: 0.6172331 total: 23.1ms remaining: 113ms
17: learn: 0.6140067 total: 24.4ms remaining: 111ms
18: learn: 0.6101040 total: 25.5ms remaining: 109ms
19: learn: 0.6066469 total: 27.4ms remaining: 109ms
20: learn: 0.6029030 total: 28.6ms remaining: 108ms
21: learn: 0.5987959 total: 30.1ms remaining: 107ms
22: learn: 0.5948275 total: 31ms remaining: 104ms
23: learn: 0.5915322 total: 32.3ms remaining: 102ms
24: learn: 0.5872507 total: 33.4ms remaining: 100ms
25: learn: 0.5839759 total: 34.6ms remaining: 98.4ms
26: learn: 0.5805313 total: 35.8ms remaining: 96.8ms
27: learn: 0.5778302 total: 37.3ms remaining: 95.8ms
28: learn: 0.5752130 total: 38.5ms remaining: 94.3ms
29: learn: 0.5727888 total: 39.4ms remaining: 92ms
30: learn: 0.5702654 total: 40.7ms remaining: 90.6ms
31: learn: 0.5666849 total: 41.6ms remaining: 88.4ms
32: learn: 0.5634739 total: 42.6ms remaining: 86.5ms
33: learn: 0.5599432 total: 43.3ms remaining: 84.1ms
34: learn: 0.5569637 total: 43.9ms remaining: 81.6ms
35: learn: 0.5542632 total: 44.5ms remaining: 79.1ms
36: learn: 0.5511816 total: 45.7ms remaining: 77.8ms
37: learn: 0.5479280 total: 46.4ms remaining: 75.7ms
38: learn: 0.5446114 total: 47.3ms remaining: 73.9ms
39: learn: 0.5416442 total: 47.9ms remaining: 71.8ms
40: learn: 0.5388472 total: 48.4ms remaining: 69.6ms
41: learn: 0.5353803 total: 48.8ms remaining: 67.4ms
42: learn: 0.5323092 total: 49.5ms remaining: 65.6ms
43: learn: 0.5297938 total: 50.2ms remaining: 63.9ms
44: learn: 0.5270777 total: 50.7ms remaining: 62ms
45: learn: 0.5242021 total: 51.4ms remaining: 60.3ms
46: learn: 0.5210269 total: 52.3ms remaining: 59ms
47: learn: 0.5183354 total: 53.1ms remaining: 57.5ms
48: learn: 0.5155134 total: 53.5ms remaining: 55.7ms
49: learn: 0.5125593 total: 53.9ms remaining: 53.9ms
50: learn: 0.5100516 total: 54.5ms remaining: 52.3ms
51: learn: 0.5082201 total: 55ms remaining: 50.8ms
52: learn: 0.5057142 total: 56ms remaining: 49.7ms
53: learn: 0.5033139 total: 56.7ms remaining: 48.3ms
54: learn: 0.5008812 total: 57.5ms remaining: 47.1ms
55: learn: 0.4980920 total: 58.1ms remaining: 45.7ms
56: learn: 0.4952230 total: 58.9ms remaining: 44.4ms
57: learn: 0.4929389 total: 59.4ms remaining: 43ms
58: learn: 0.4904993 total: 60.1ms remaining: 41.7ms
59: learn: 0.4881358 total: 60.8ms remaining: 40.5ms
60: learn: 0.4858069 total: 61.4ms remaining: 39.2ms
61: learn: 0.4832165 total: 62ms remaining: 38ms
62: learn: 0.4808001 total: 62.6ms remaining: 36.8ms
63: learn: 0.4789522 total: 63ms remaining: 35.4ms
64: learn: 0.4769107 total: 63.6ms remaining: 34.2ms
65: learn: 0.4744882 total: 64ms remaining: 33ms
66: learn: 0.4720492 total: 64.4ms remaining: 31.7ms
67: learn: 0.4698849 total: 65ms remaining: 30.6ms
68: learn: 0.4680298 total: 65.3ms remaining: 29.4ms
69: learn: 0.4657194 total: 65.7ms remaining: 28.2ms
70: learn: 0.4637653 total: 66.3ms remaining: 27.1ms
71: learn: 0.4614873 total: 66.8ms remaining: 26ms
72: learn: 0.4593273 total: 67.2ms remaining: 24.9ms
73: learn: 0.4572340 total: 67.7ms remaining: 23.8ms
74: learn: 0.4552783 total: 68.2ms remaining: 22.7ms
75: learn: 0.4531864 total: 68.6ms remaining: 21.7ms
76: learn: 0.4514435 total: 69.3ms remaining: 20.7ms
77: learn: 0.4495506 total: 69.6ms remaining: 19.6ms
78: learn: 0.4474527 total: 70ms remaining: 18.6ms
79: learn: 0.4457739 total: 70.3ms remaining: 17.6ms
80: learn: 0.4442183 total: 70.7ms remaining: 16.6ms
81: learn: 0.4421743 total: 71.1ms remaining: 15.6ms
82: learn: 0.4403339 total: 71.7ms remaining: 14.7ms
83: learn: 0.4385316 total: 72ms remaining: 13.7ms
84: learn: 0.4369359 total: 72.3ms remaining: 12.8ms
85: learn: 0.4355767 total: 72.7ms remaining: 11.8ms
86: learn: 0.4339461 total: 73ms remaining: 10.9ms
87: learn: 0.4322172 total: 73.3ms remaining: 10ms
88: learn: 0.4304228 total: 73.8ms remaining: 9.12ms
89: learn: 0.4289271 total: 74.3ms remaining: 8.25ms
90: learn: 0.4272516 total: 74.6ms remaining: 7.37ms
91: learn: 0.4256348 total: 74.9ms remaining: 6.51ms
92: learn: 0.4237505 total: 75.4ms remaining: 5.68ms
93: learn: 0.4220555 total: 75.8ms remaining: 4.84ms
94: learn: 0.4203653 total: 76.2ms remaining: 4.01ms
95: learn: 0.4185166 total: 76.7ms remaining: 3.19ms
96: learn: 0.4168763 total: 77.1ms remaining: 2.38ms
97: learn: 0.4157957 total: 77.4ms remaining: 1.58ms
98: learn: 0.4140381 total: 77.9ms remaining: 787us
99: learn: 0.4123918 total: 78.3ms remaining: 0us
0: learn: 0.6884710 total: 1.3ms remaining: 129ms
1: learn: 0.6831483 total: 2.3ms remaining: 113ms
2: learn: 0.6781742 total: 3.41ms remaining: 110ms
3: learn: 0.6735979 total: 4.75ms remaining: 114ms
4: learn: 0.6690825 total: 5.68ms remaining: 108ms
5: learn: 0.6643177 total: 6.7ms remaining: 105ms
6: learn: 0.6602321 total: 7.75ms remaining: 103ms
7: learn: 0.6561625 total: 8.8ms remaining: 101ms
8: learn: 0.6513684 total: 9.59ms remaining: 97ms
9: learn: 0.6464446 total: 10.7ms remaining: 96ms
10: learn: 0.6425160 total: 11.6ms remaining: 93.9ms
11: learn: 0.6379029 total: 12.6ms remaining: 92.6ms
12: learn: 0.6338682 total: 14ms remaining: 93.9ms
13: learn: 0.6299456 total: 17ms remaining: 104ms
14: learn: 0.6261431 total: 18.5ms remaining: 105ms
15: learn: 0.6214318 total: 19.4ms remaining: 102ms
16: learn: 0.6176888 total: 20.9ms remaining: 102ms
17: learn: 0.6136496 total: 22.2ms remaining: 101ms
18: learn: 0.6098567 total: 22.8ms remaining: 97.2ms
19: learn: 0.6061197 total: 24ms remaining: 96.1ms
20: learn: 0.6021095 total: 25.7ms remaining: 96.5ms
21: learn: 0.5977883 total: 27ms remaining: 95.7ms
22: learn: 0.5936190 total: 28ms remaining: 93.6ms
23: learn: 0.5899167 total: 28.9ms remaining: 91.7ms
24: learn: 0.5858649 total: 30.3ms remaining: 90.9ms
25: learn: 0.5829470 total: 31.8ms remaining: 90.6ms
26: learn: 0.5798447 total: 33.3ms remaining: 90ms
27: learn: 0.5765984 total: 34.8ms remaining: 89.5ms
28: learn: 0.5737242 total: 35.8ms remaining: 87.6ms
29: learn: 0.5710620 total: 36.6ms remaining: 85.4ms
30: learn: 0.5684374 total: 37.4ms remaining: 83.2ms
31: learn: 0.5652138 total: 38.1ms remaining: 80.9ms
32: learn: 0.5622089 total: 39.6ms remaining: 80.3ms
33: learn: 0.5588240 total: 40.8ms remaining: 79.2ms
34: learn: 0.5553290 total: 42.3ms remaining: 78.5ms
35: learn: 0.5523463 total: 43.3ms remaining: 77ms
36: learn: 0.5491377 total: 44.7ms remaining: 76.1ms
37: learn: 0.5464623 total: 45.8ms remaining: 74.8ms
38: learn: 0.5430188 total: 46.7ms remaining: 73.1ms
39: learn: 0.5400183 total: 48.3ms remaining: 72.5ms
40: learn: 0.5375189 total: 49.4ms remaining: 71ms
41: learn: 0.5339443 total: 50.1ms remaining: 69.2ms
42: learn: 0.5309167 total: 51.3ms remaining: 68ms
43: learn: 0.5285461 total: 52.5ms remaining: 66.8ms
44: learn: 0.5257915 total: 53.5ms remaining: 65.4ms
45: learn: 0.5235061 total: 54.2ms remaining: 63.6ms
46: learn: 0.5208134 total: 55ms remaining: 62ms
47: learn: 0.5181357 total: 55.6ms remaining: 60.3ms
48: learn: 0.5152125 total: 56.4ms remaining: 58.7ms
49: learn: 0.5127882 total: 57.1ms remaining: 57.1ms
50: learn: 0.5103441 total: 57.7ms remaining: 55.5ms
51: learn: 0.5085697 total: 58.3ms remaining: 53.8ms
52: learn: 0.5060762 total: 59.1ms remaining: 52.4ms
53: learn: 0.5036028 total: 59.7ms remaining: 50.9ms
54: learn: 0.5011696 total: 60.5ms remaining: 49.5ms
55: learn: 0.4985615 total: 61.1ms remaining: 48ms
56: learn: 0.4958495 total: 61.4ms remaining: 46.4ms
57: learn: 0.4934627 total: 61.9ms remaining: 44.8ms
58: learn: 0.4911664 total: 62.2ms remaining: 43.2ms
59: learn: 0.4888431 total: 62.8ms remaining: 41.8ms
60: learn: 0.4867067 total: 63.5ms remaining: 40.6ms
61: learn: 0.4843027 total: 64ms remaining: 39.2ms
62: learn: 0.4819804 total: 64.6ms remaining: 37.9ms
63: learn: 0.4799181 total: 64.9ms remaining: 36.5ms
64: learn: 0.4777196 total: 65.6ms remaining: 35.3ms
65: learn: 0.4752769 total: 66ms remaining: 34ms
66: learn: 0.4731355 total: 66.3ms remaining: 32.7ms
67: learn: 0.4709208 total: 66.8ms remaining: 31.4ms
68: learn: 0.4688675 total: 67.2ms remaining: 30.2ms
69: learn: 0.4669209 total: 67.6ms remaining: 29ms
70: learn: 0.4646306 total: 68ms remaining: 27.8ms
71: learn: 0.4624272 total: 68.6ms remaining: 26.7ms
72: learn: 0.4603330 total: 69.1ms remaining: 25.6ms
73: learn: 0.4585022 total: 69.6ms remaining: 24.4ms
74: learn: 0.4566034 total: 69.9ms remaining: 23.3ms
75: learn: 0.4548515 total: 70.3ms remaining: 22.2ms
76: learn: 0.4527723 total: 70.8ms remaining: 21.1ms
77: learn: 0.4508659 total: 71.2ms remaining: 20.1ms
78: learn: 0.4486937 total: 71.6ms remaining: 19ms
79: learn: 0.4468623 total: 71.9ms remaining: 18ms
80: learn: 0.4453247 total: 72.6ms remaining: 17ms
81: learn: 0.4434634 total: 73ms remaining: 16ms
82: learn: 0.4412807 total: 73.6ms remaining: 15.1ms
83: learn: 0.4393264 total: 74ms remaining: 14.1ms
84: learn: 0.4374024 total: 74.6ms remaining: 13.2ms
85: learn: 0.4357541 total: 74.9ms remaining: 12.2ms
86: learn: 0.4340766 total: 75.1ms remaining: 11.2ms
87: learn: 0.4326258 total: 75.5ms remaining: 10.3ms
88: learn: 0.4303947 total: 76ms remaining: 9.39ms
89: learn: 0.4288562 total: 76.3ms remaining: 8.47ms
90: learn: 0.4273194 total: 76.6ms remaining: 7.58ms
91: learn: 0.4257218 total: 77ms remaining: 6.7ms
92: learn: 0.4239834 total: 77.6ms remaining: 5.84ms
93: learn: 0.4226384 total: 78.1ms remaining: 4.99ms
94: learn: 0.4206514 total: 78.6ms remaining: 4.14ms
95: learn: 0.4190590 total: 79.2ms remaining: 3.3ms
96: learn: 0.4174809 total: 79.6ms remaining: 2.46ms
97: learn: 0.4161426 total: 79.9ms remaining: 1.63ms
98: learn: 0.4143139 total: 80.3ms remaining: 811us
99: learn: 0.4126026 total: 80.7ms remaining: 0us
0: learn: 0.6888251 total: 1.09ms remaining: 108ms
1: learn: 0.6838743 total: 2.45ms remaining: 120ms
2: learn: 0.6790688 total: 3.6ms remaining: 117ms
3: learn: 0.6744890 total: 5.3ms remaining: 127ms
4: learn: 0.6697935 total: 6.42ms remaining: 122ms
5: learn: 0.6652243 total: 7.6ms remaining: 119ms
6: learn: 0.6616198 total: 8.87ms remaining: 118ms
7: learn: 0.6576694 total: 9.9ms remaining: 114ms
8: learn: 0.6532890 total: 11.1ms remaining: 113ms
9: learn: 0.6492571 total: 12.5ms remaining: 113ms
10: learn: 0.6463276 total: 13.8ms remaining: 112ms
11: learn: 0.6419683 total: 14.9ms remaining: 109ms
12: learn: 0.6383123 total: 16.6ms remaining: 111ms
13: learn: 0.6346641 total: 18ms remaining: 110ms
14: learn: 0.6310619 total: 19.6ms remaining: 111ms
15: learn: 0.6268264 total: 20.6ms remaining: 108ms
16: learn: 0.6232008 total: 21.4ms remaining: 105ms
17: learn: 0.6188903 total: 22.7ms remaining: 104ms
18: learn: 0.6154295 total: 24.2ms remaining: 103ms
19: learn: 0.6110983 total: 25.2ms remaining: 101ms
20: learn: 0.6074348 total: 26ms remaining: 98ms
21: learn: 0.6035654 total: 26.7ms remaining: 94.8ms
22: learn: 0.6001629 total: 27.7ms remaining: 92.9ms
23: learn: 0.5972153 total: 29ms remaining: 91.8ms
24: learn: 0.5938984 total: 30.3ms remaining: 91ms
25: learn: 0.5914189 total: 32.9ms remaining: 93.7ms
26: learn: 0.5879412 total: 34.1ms remaining: 92.1ms
27: learn: 0.5844532 total: 35.4ms remaining: 91.1ms
28: learn: 0.5812474 total: 36.4ms remaining: 89.2ms
29: learn: 0.5780829 total: 37.7ms remaining: 87.9ms
30: learn: 0.5747267 total: 38.6ms remaining: 86ms
31: learn: 0.5714414 total: 39.4ms remaining: 83.8ms
32: learn: 0.5686548 total: 40.1ms remaining: 81.3ms
33: learn: 0.5656884 total: 40.7ms remaining: 79ms
34: learn: 0.5628498 total: 41.9ms remaining: 77.8ms
35: learn: 0.5601182 total: 42.7ms remaining: 75.9ms
36: learn: 0.5571363 total: 43.9ms remaining: 74.8ms
37: learn: 0.5543120 total: 45.2ms remaining: 73.7ms
38: learn: 0.5508372 total: 46.1ms remaining: 72.1ms
39: learn: 0.5482084 total: 47.1ms remaining: 70.6ms
40: learn: 0.5451572 total: 48.6ms remaining: 69.9ms
41: learn: 0.5418393 total: 49.4ms remaining: 68.2ms
42: learn: 0.5393336 total: 50.5ms remaining: 67ms
43: learn: 0.5368743 total: 51.7ms remaining: 65.9ms
44: learn: 0.5341000 total: 53ms remaining: 64.7ms
45: learn: 0.5315797 total: 53.6ms remaining: 62.9ms
46: learn: 0.5288963 total: 54.1ms remaining: 61ms
47: learn: 0.5262389 total: 55.2ms remaining: 59.8ms
48: learn: 0.5234348 total: 56.2ms remaining: 58.5ms
49: learn: 0.5209916 total: 56.8ms remaining: 56.8ms
50: learn: 0.5185438 total: 57.2ms remaining: 55ms
51: learn: 0.5166256 total: 58ms remaining: 53.6ms
52: learn: 0.5142948 total: 58.9ms remaining: 52.3ms
53: learn: 0.5119237 total: 59.8ms remaining: 50.9ms
54: learn: 0.5099750 total: 61ms remaining: 49.9ms
55: learn: 0.5073470 total: 62.2ms remaining: 48.8ms
56: learn: 0.5049048 total: 62.9ms remaining: 47.5ms
57: learn: 0.5025382 total: 63.7ms remaining: 46.1ms
58: learn: 0.5001287 total: 64.3ms remaining: 44.7ms
59: learn: 0.4976328 total: 65.2ms remaining: 43.5ms
60: learn: 0.4954885 total: 66.4ms remaining: 42.4ms
61: learn: 0.4929791 total: 67ms remaining: 41.1ms
62: learn: 0.4906066 total: 68.2ms remaining: 40ms
63: learn: 0.4883619 total: 68.9ms remaining: 38.8ms
64: learn: 0.4861991 total: 69.9ms remaining: 37.7ms
65: learn: 0.4838713 total: 70.7ms remaining: 36.4ms
66: learn: 0.4817598 total: 71.5ms remaining: 35.2ms
67: learn: 0.4797222 total: 72.4ms remaining: 34.1ms
68: learn: 0.4776118 total: 73.2ms remaining: 32.9ms
69: learn: 0.4758566 total: 74ms remaining: 31.7ms
70: learn: 0.4737977 total: 74.8ms remaining: 30.5ms
71: learn: 0.4716287 total: 75.2ms remaining: 29.3ms
72: learn: 0.4696145 total: 75.7ms remaining: 28ms
73: learn: 0.4676496 total: 76.3ms remaining: 26.8ms
74: learn: 0.4655607 total: 77.1ms remaining: 25.7ms
75: learn: 0.4636208 total: 77.8ms remaining: 24.6ms
76: learn: 0.4618838 total: 79ms remaining: 23.6ms
77: learn: 0.4598499 total: 79.7ms remaining: 22.5ms
78: learn: 0.4579391 total: 80.5ms remaining: 21.4ms
79: learn: 0.4558926 total: 81ms remaining: 20.3ms
80: learn: 0.4538738 total: 81.7ms remaining: 19.2ms
81: learn: 0.4520528 total: 82.5ms remaining: 18.1ms
82: learn: 0.4502268 total: 83.1ms remaining: 17ms
83: learn: 0.4484919 total: 84ms remaining: 16ms
84: learn: 0.4468533 total: 84.7ms remaining: 14.9ms
85: learn: 0.4450997 total: 85.3ms remaining: 13.9ms
86: learn: 0.4432632 total: 86ms remaining: 12.8ms
87: learn: 0.4416660 total: 86.5ms remaining: 11.8ms
88: learn: 0.4401842 total: 87.1ms remaining: 10.8ms
89: learn: 0.4386951 total: 87.6ms remaining: 9.73ms
90: learn: 0.4370228 total: 88.4ms remaining: 8.74ms
91: learn: 0.4353236 total: 88.8ms remaining: 7.72ms
92: learn: 0.4339369 total: 89.4ms remaining: 6.73ms
93: learn: 0.4326707 total: 90.1ms remaining: 5.75ms
94: learn: 0.4309095 total: 90.6ms remaining: 4.77ms
95: learn: 0.4293567 total: 91.1ms remaining: 3.79ms
96: learn: 0.4277585 total: 91.6ms remaining: 2.83ms
97: learn: 0.4259049 total: 92.2ms remaining: 1.88ms
98: learn: 0.4241491 total: 92.6ms remaining: 935us
99: learn: 0.4228214 total: 93.1ms remaining: 0us
0: learn: 0.6881993 total: 1.05ms remaining: 104ms
1: learn: 0.6827008 total: 1.57ms remaining: 76.8ms
2: learn: 0.6774722 total: 2.12ms remaining: 68.6ms
3: learn: 0.6721781 total: 3.52ms remaining: 84.4ms
4: learn: 0.6675501 total: 4.96ms remaining: 94.2ms
5: learn: 0.6623863 total: 6.09ms remaining: 95.4ms
6: learn: 0.6575695 total: 7.22ms remaining: 95.9ms
7: learn: 0.6535074 total: 8.17ms remaining: 94ms
8: learn: 0.6485427 total: 9.54ms remaining: 96.4ms
9: learn: 0.6436104 total: 10.9ms remaining: 98.2ms
10: learn: 0.6395245 total: 11.8ms remaining: 95.3ms
11: learn: 0.6347631 total: 12.6ms remaining: 92.7ms
12: learn: 0.6305070 total: 13.8ms remaining: 92.3ms
13: learn: 0.6265698 total: 14.9ms remaining: 91.3ms
14: learn: 0.6229825 total: 15.6ms remaining: 88.2ms
15: learn: 0.6182567 total: 16.6ms remaining: 86.9ms
16: learn: 0.6144702 total: 17.6ms remaining: 86.1ms
17: learn: 0.6102756 total: 18.7ms remaining: 85.1ms
18: learn: 0.6063688 total: 19.3ms remaining: 82.2ms
19: learn: 0.6025233 total: 20.3ms remaining: 81.2ms
20: learn: 0.5986269 total: 21.3ms remaining: 80ms
21: learn: 0.5942607 total: 22.6ms remaining: 80ms
22: learn: 0.5899548 total: 23.8ms remaining: 79.6ms
23: learn: 0.5862924 total: 24.4ms remaining: 77.3ms
24: learn: 0.5830002 total: 25.4ms remaining: 76.1ms
25: learn: 0.5798113 total: 26.4ms remaining: 75.2ms
26: learn: 0.5764879 total: 27.6ms remaining: 74.7ms
27: learn: 0.5732837 total: 28.5ms remaining: 73.2ms
28: learn: 0.5701657 total: 29.4ms remaining: 71.9ms
29: learn: 0.5673144 total: 30.2ms remaining: 70.4ms
30: learn: 0.5645907 total: 31ms remaining: 69ms
31: learn: 0.5612450 total: 31.7ms remaining: 67.3ms
32: learn: 0.5579183 total: 32.5ms remaining: 66ms
33: learn: 0.5543718 total: 33.4ms remaining: 64.9ms
34: learn: 0.5513997 total: 34.3ms remaining: 63.8ms
35: learn: 0.5484174 total: 35.1ms remaining: 62.4ms
36: learn: 0.5451814 total: 35.9ms remaining: 61.2ms
37: learn: 0.5421966 total: 36.7ms remaining: 59.8ms
38: learn: 0.5386374 total: 37.2ms remaining: 58.3ms
39: learn: 0.5359418 total: 38ms remaining: 57ms
40: learn: 0.5328272 total: 38.7ms remaining: 55.7ms
41: learn: 0.5292787 total: 39.2ms remaining: 54.2ms
42: learn: 0.5261061 total: 40.1ms remaining: 53.2ms
43: learn: 0.5237163 total: 40.9ms remaining: 52ms
44: learn: 0.5208345 total: 41.6ms remaining: 50.8ms
45: learn: 0.5184355 total: 42.1ms remaining: 49.4ms
46: learn: 0.5158247 total: 43.3ms remaining: 48.8ms
47: learn: 0.5135288 total: 44ms remaining: 47.7ms
48: learn: 0.5104705 total: 44.5ms remaining: 46.4ms
49: learn: 0.5081160 total: 45.4ms remaining: 45.4ms
50: learn: 0.5056550 total: 45.8ms remaining: 44ms
51: learn: 0.5038595 total: 46.5ms remaining: 42.9ms
52: learn: 0.5013747 total: 47.2ms remaining: 41.9ms
53: learn: 0.4986649 total: 47.9ms remaining: 40.8ms
54: learn: 0.4965716 total: 48.3ms remaining: 39.5ms
55: learn: 0.4939157 total: 48.8ms remaining: 38.4ms
56: learn: 0.4910497 total: 49.2ms remaining: 37.1ms
57: learn: 0.4884881 total: 49.9ms remaining: 36.1ms
58: learn: 0.4862442 total: 50.4ms remaining: 35.1ms
59: learn: 0.4839807 total: 50.9ms remaining: 33.9ms
60: learn: 0.4813368 total: 51.5ms remaining: 32.9ms
61: learn: 0.4787983 total: 52.1ms remaining: 31.9ms
62: learn: 0.4763822 total: 52.6ms remaining: 30.9ms
63: learn: 0.4742053 total: 53ms remaining: 29.8ms
64: learn: 0.4720609 total: 53.7ms remaining: 28.9ms
65: learn: 0.4695373 total: 54.2ms remaining: 27.9ms
66: learn: 0.4674749 total: 54.7ms remaining: 26.9ms
67: learn: 0.4652863 total: 55.2ms remaining: 26ms
68: learn: 0.4631123 total: 55.6ms remaining: 25ms
69: learn: 0.4611278 total: 56ms remaining: 24ms
70: learn: 0.4589138 total: 56.4ms remaining: 23.1ms
71: learn: 0.4566025 total: 56.9ms remaining: 22.1ms
72: learn: 0.4542314 total: 57.5ms remaining: 21.3ms
73: learn: 0.4520691 total: 57.8ms remaining: 20.3ms
74: learn: 0.4500480 total: 58.2ms remaining: 19.4ms
75: learn: 0.4478340 total: 58.8ms remaining: 18.6ms
76: learn: 0.4463365 total: 59.2ms remaining: 17.7ms
77: learn: 0.4445490 total: 59.6ms remaining: 16.8ms
78: learn: 0.4427074 total: 60ms remaining: 16ms
79: learn: 0.4405743 total: 60.5ms remaining: 15.1ms
80: learn: 0.4388167 total: 61.1ms remaining: 14.3ms
81: learn: 0.4366750 total: 61.5ms remaining: 13.5ms
82: learn: 0.4348196 total: 61.8ms remaining: 12.7ms
83: learn: 0.4329126 total: 62.2ms remaining: 11.9ms
84: learn: 0.4311000 total: 62.7ms remaining: 11.1ms
85: learn: 0.4293995 total: 63.1ms remaining: 10.3ms
86: learn: 0.4272472 total: 63.4ms remaining: 9.48ms
87: learn: 0.4252671 total: 63.9ms remaining: 8.71ms
88: learn: 0.4234933 total: 64.4ms remaining: 7.96ms
89: learn: 0.4218135 total: 64.7ms remaining: 7.19ms
90: learn: 0.4200800 total: 65.1ms remaining: 6.44ms
91: learn: 0.4184939 total: 65.7ms remaining: 5.71ms
92: learn: 0.4165901 total: 66.1ms remaining: 4.97ms
93: learn: 0.4152496 total: 66.5ms remaining: 4.24ms
94: learn: 0.4135666 total: 66.9ms remaining: 3.52ms
95: learn: 0.4122281 total: 67.4ms remaining: 2.81ms
96: learn: 0.4103071 total: 67.8ms remaining: 2.1ms
97: learn: 0.4088729 total: 68.1ms remaining: 1.39ms
98: learn: 0.4071209 total: 68.7ms remaining: 693us
99: learn: 0.4055178 total: 69.1ms remaining: 0us
0: learn: 0.6886959 total: 1.16ms remaining: 115ms
1: learn: 0.6838265 total: 2.41ms remaining: 118ms
2: learn: 0.6793184 total: 3.4ms remaining: 110ms
3: learn: 0.6746587 total: 4.52ms remaining: 109ms
4: learn: 0.6698654 total: 5.58ms remaining: 106ms
5: learn: 0.6655969 total: 6.76ms remaining: 106ms
6: learn: 0.6613624 total: 7.8ms remaining: 104ms
7: learn: 0.6574805 total: 8.97ms remaining: 103ms
8: learn: 0.6531921 total: 10.2ms remaining: 103ms
9: learn: 0.6488752 total: 11.5ms remaining: 104ms
10: learn: 0.6451668 total: 12.8ms remaining: 103ms
11: learn: 0.6411035 total: 13.8ms remaining: 101ms
12: learn: 0.6370053 total: 15ms remaining: 100ms
13: learn: 0.6329368 total: 16.1ms remaining: 98.7ms
14: learn: 0.6292349 total: 17.5ms remaining: 99.1ms
15: learn: 0.6251452 total: 18.6ms remaining: 97.5ms
16: learn: 0.6212621 total: 19.6ms remaining: 95.7ms
17: learn: 0.6170321 total: 20.6ms remaining: 94ms
18: learn: 0.6132722 total: 21.5ms remaining: 91.6ms
19: learn: 0.6099714 total: 22.5ms remaining: 89.8ms
20: learn: 0.6066178 total: 23.7ms remaining: 89.1ms
21: learn: 0.6027816 total: 24.9ms remaining: 88.3ms
22: learn: 0.5990405 total: 26.3ms remaining: 88.2ms
23: learn: 0.5957426 total: 27.1ms remaining: 85.8ms
24: learn: 0.5922791 total: 28.3ms remaining: 84.9ms
25: learn: 0.5893376 total: 29.5ms remaining: 84.1ms
26: learn: 0.5861754 total: 30.8ms remaining: 83.3ms
27: learn: 0.5829950 total: 32.2ms remaining: 82.8ms
28: learn: 0.5794016 total: 33.4ms remaining: 81.7ms
29: learn: 0.5765496 total: 34.8ms remaining: 81.2ms
30: learn: 0.5736307 total: 36.3ms remaining: 80.8ms
31: learn: 0.5704244 total: 37.2ms remaining: 79ms
32: learn: 0.5676356 total: 38.4ms remaining: 78ms
33: learn: 0.5646941 total: 39.5ms remaining: 76.7ms
34: learn: 0.5620194 total: 40.6ms remaining: 75.4ms
35: learn: 0.5600655 total: 41.4ms remaining: 73.7ms
36: learn: 0.5572839 total: 42.7ms remaining: 72.7ms
37: learn: 0.5547866 total: 43.6ms remaining: 71.1ms
38: learn: 0.5521352 total: 44.8ms remaining: 70.1ms
39: learn: 0.5493307 total: 46.1ms remaining: 69.1ms
40: learn: 0.5467472 total: 47.2ms remaining: 68ms
41: learn: 0.5446479 total: 47.9ms remaining: 66.2ms
42: learn: 0.5426825 total: 49.3ms remaining: 65.3ms
43: learn: 0.5399677 total: 50.4ms remaining: 64.2ms
44: learn: 0.5379476 total: 51.9ms remaining: 63.4ms
45: learn: 0.5354392 total: 53.2ms remaining: 62.4ms
46: learn: 0.5325750 total: 54ms remaining: 60.9ms
47: learn: 0.5300571 total: 54.9ms remaining: 59.5ms
48: learn: 0.5276968 total: 56.2ms remaining: 58.5ms
49: learn: 0.5250418 total: 56.8ms remaining: 56.8ms
50: learn: 0.5225769 total: 57.3ms remaining: 55.1ms
51: learn: 0.5200919 total: 58.4ms remaining: 53.9ms
52: learn: 0.5178163 total: 59.4ms remaining: 52.7ms
53: learn: 0.5153453 total: 60.2ms remaining: 51.3ms
54: learn: 0.5127813 total: 61ms remaining: 49.9ms
55: learn: 0.5104769 total: 62ms remaining: 48.7ms
56: learn: 0.5081644 total: 62.7ms remaining: 47.3ms
57: learn: 0.5056439 total: 64.6ms remaining: 46.8ms
58: learn: 0.5031357 total: 66ms remaining: 45.9ms
59: learn: 0.5007311 total: 67.1ms remaining: 44.7ms
60: learn: 0.4983959 total: 67.8ms remaining: 43.4ms
61: learn: 0.4962703 total: 68.6ms remaining: 42.1ms
62: learn: 0.4946152 total: 69.4ms remaining: 40.8ms
63: learn: 0.4924402 total: 70.4ms remaining: 39.6ms
64: learn: 0.4904118 total: 71.2ms remaining: 38.4ms
65: learn: 0.4883486 total: 71.9ms remaining: 37ms
66: learn: 0.4863043 total: 72.4ms remaining: 35.7ms
67: learn: 0.4838890 total: 73.2ms remaining: 34.4ms
68: learn: 0.4820407 total: 73.9ms remaining: 33.2ms
69: learn: 0.4799651 total: 74.5ms remaining: 31.9ms
70: learn: 0.4783326 total: 75.1ms remaining: 30.7ms
71: learn: 0.4762731 total: 75.9ms remaining: 29.5ms
72: learn: 0.4745866 total: 76.5ms remaining: 28.3ms
73: learn: 0.4723220 total: 77.2ms remaining: 27.1ms
74: learn: 0.4704764 total: 77.8ms remaining: 25.9ms
75: learn: 0.4689428 total: 78.4ms remaining: 24.7ms
76: learn: 0.4667857 total: 79ms remaining: 23.6ms
77: learn: 0.4649443 total: 79.5ms remaining: 22.4ms
78: learn: 0.4629698 total: 80.1ms remaining: 21.3ms
79: learn: 0.4614328 total: 80.9ms remaining: 20.2ms
80: learn: 0.4599062 total: 81.5ms remaining: 19.1ms
81: learn: 0.4583270 total: 81.9ms remaining: 18ms
82: learn: 0.4562689 total: 82.3ms remaining: 16.9ms
83: learn: 0.4546344 total: 83ms remaining: 15.8ms
84: learn: 0.4530122 total: 83.5ms remaining: 14.7ms
85: learn: 0.4512857 total: 84ms remaining: 13.7ms
86: learn: 0.4495425 total: 84.4ms remaining: 12.6ms
87: learn: 0.4477200 total: 84.8ms remaining: 11.6ms
88: learn: 0.4460820 total: 85.1ms remaining: 10.5ms
89: learn: 0.4445896 total: 85.6ms remaining: 9.52ms
90: learn: 0.4427587 total: 86.1ms remaining: 8.52ms
91: learn: 0.4412886 total: 86.5ms remaining: 7.52ms
92: learn: 0.4396576 total: 87ms remaining: 6.54ms
93: learn: 0.4380537 total: 87.5ms remaining: 5.58ms
94: learn: 0.4365266 total: 88ms remaining: 4.63ms
95: learn: 0.4351730 total: 88.4ms remaining: 3.68ms
96: learn: 0.4337504 total: 88.8ms remaining: 2.75ms
97: learn: 0.4323788 total: 89.4ms remaining: 1.82ms
98: learn: 0.4307403 total: 89.8ms remaining: 907us
99: learn: 0.4292391 total: 90.1ms remaining: 0us
0: learn: 0.6888430 total: 1.76ms remaining: 174ms
1: learn: 0.6844696 total: 2.74ms remaining: 134ms
2: learn: 0.6801295 total: 3.9ms remaining: 126ms
3: learn: 0.6752420 total: 5.07ms remaining: 122ms
4: learn: 0.6701626 total: 6.69ms remaining: 127ms
5: learn: 0.6655182 total: 7.89ms remaining: 124ms
6: learn: 0.6620344 total: 9.58ms remaining: 127ms
7: learn: 0.6584874 total: 10.4ms remaining: 120ms
8: learn: 0.6539240 total: 12.1ms remaining: 122ms
9: learn: 0.6499828 total: 13.2ms remaining: 119ms
10: learn: 0.6455269 total: 14.6ms remaining: 118ms
11: learn: 0.6414986 total: 16.1ms remaining: 118ms
12: learn: 0.6374259 total: 17.5ms remaining: 117ms
13: learn: 0.6333841 total: 18.7ms remaining: 115ms
14: learn: 0.6287552 total: 19.8ms remaining: 112ms
15: learn: 0.6255913 total: 20.9ms remaining: 110ms
16: learn: 0.6228745 total: 22ms remaining: 107ms
17: learn: 0.6192158 total: 23.3ms remaining: 106ms
18: learn: 0.6151053 total: 24.7ms remaining: 105ms
19: learn: 0.6117255 total: 26ms remaining: 104ms
20: learn: 0.6077871 total: 27.5ms remaining: 103ms
21: learn: 0.6036369 total: 28.5ms remaining: 101ms
22: learn: 0.5997223 total: 29.9ms remaining: 100ms
23: learn: 0.5957430 total: 30.8ms remaining: 97.7ms
24: learn: 0.5924717 total: 32.1ms remaining: 96.4ms
25: learn: 0.5891835 total: 33.4ms remaining: 95.1ms
26: learn: 0.5859911 total: 35.1ms remaining: 94.9ms
27: learn: 0.5830599 total: 36.3ms remaining: 93.2ms
28: learn: 0.5804551 total: 37.6ms remaining: 92ms
29: learn: 0.5777595 total: 39.2ms remaining: 91.4ms
30: learn: 0.5746008 total: 39.9ms remaining: 88.9ms
31: learn: 0.5713796 total: 41.4ms remaining: 87.9ms
32: learn: 0.5679259 total: 42.7ms remaining: 86.7ms
33: learn: 0.5650754 total: 44ms remaining: 85.3ms
34: learn: 0.5622872 total: 44.9ms remaining: 83.4ms
35: learn: 0.5591263 total: 46.2ms remaining: 82.2ms
36: learn: 0.5563465 total: 47.1ms remaining: 80.2ms
37: learn: 0.5529493 total: 48.4ms remaining: 78.9ms
38: learn: 0.5501690 total: 49.4ms remaining: 77.3ms
39: learn: 0.5469190 total: 50.4ms remaining: 75.6ms
40: learn: 0.5432410 total: 51.2ms remaining: 73.7ms
41: learn: 0.5401389 total: 52.3ms remaining: 72.2ms
42: learn: 0.5378797 total: 53.2ms remaining: 70.5ms
43: learn: 0.5350410 total: 54ms remaining: 68.8ms
44: learn: 0.5326489 total: 54.8ms remaining: 67ms
45: learn: 0.5300776 total: 55.4ms remaining: 65.1ms
46: learn: 0.5270868 total: 56.4ms remaining: 63.6ms
47: learn: 0.5249808 total: 56.9ms remaining: 61.7ms
48: learn: 0.5227876 total: 57.5ms remaining: 59.8ms
49: learn: 0.5200878 total: 58.2ms remaining: 58.2ms
50: learn: 0.5172000 total: 58.7ms remaining: 56.4ms
51: learn: 0.5149013 total: 59.4ms remaining: 54.8ms
52: learn: 0.5120194 total: 60.1ms remaining: 53.3ms
53: learn: 0.5093406 total: 61.5ms remaining: 52.4ms
54: learn: 0.5066302 total: 62.2ms remaining: 50.9ms
55: learn: 0.5037123 total: 62.8ms remaining: 49.3ms
56: learn: 0.5013412 total: 63.2ms remaining: 47.7ms
57: learn: 0.4989405 total: 63.8ms remaining: 46.2ms
58: learn: 0.4965496 total: 64.5ms remaining: 44.8ms
59: learn: 0.4937551 total: 64.9ms remaining: 43.3ms
60: learn: 0.4912467 total: 65.5ms remaining: 41.8ms
61: learn: 0.4885967 total: 65.9ms remaining: 40.4ms
62: learn: 0.4859458 total: 66.3ms remaining: 39ms
63: learn: 0.4835383 total: 66.8ms remaining: 37.6ms
64: learn: 0.4811866 total: 67.4ms remaining: 36.3ms
65: learn: 0.4790421 total: 67.8ms remaining: 34.9ms
66: learn: 0.4767719 total: 68.2ms remaining: 33.6ms
67: learn: 0.4747126 total: 68.8ms remaining: 32.4ms
68: learn: 0.4723738 total: 69.3ms remaining: 31.1ms
69: learn: 0.4706107 total: 69.6ms remaining: 29.8ms
70: learn: 0.4684780 total: 70.1ms remaining: 28.6ms
71: learn: 0.4663688 total: 70.5ms remaining: 27.4ms
72: learn: 0.4647644 total: 70.9ms remaining: 26.2ms
73: learn: 0.4625221 total: 71.4ms remaining: 25.1ms
74: learn: 0.4604456 total: 72.1ms remaining: 24ms
75: learn: 0.4586607 total: 72.4ms remaining: 22.9ms
76: learn: 0.4567422 total: 72.8ms remaining: 21.8ms
77: learn: 0.4550168 total: 73.2ms remaining: 20.6ms
78: learn: 0.4529971 total: 73.7ms remaining: 19.6ms
79: learn: 0.4510502 total: 74.1ms remaining: 18.5ms
80: learn: 0.4495286 total: 74.5ms remaining: 17.5ms
81: learn: 0.4478815 total: 74.9ms remaining: 16.4ms
82: learn: 0.4457721 total: 75.2ms remaining: 15.4ms
83: learn: 0.4434536 total: 75.5ms remaining: 14.4ms
84: learn: 0.4415553 total: 75.8ms remaining: 13.4ms
85: learn: 0.4396112 total: 76.3ms remaining: 12.4ms
86: learn: 0.4376530 total: 76.7ms remaining: 11.5ms
87: learn: 0.4359785 total: 77ms remaining: 10.5ms
88: learn: 0.4342797 total: 77.4ms remaining: 9.56ms
89: learn: 0.4324861 total: 77.9ms remaining: 8.66ms
90: learn: 0.4306033 total: 78.3ms remaining: 7.75ms
91: learn: 0.4291357 total: 78.7ms remaining: 6.84ms
92: learn: 0.4275348 total: 79.2ms remaining: 5.96ms
93: learn: 0.4261666 total: 79.7ms remaining: 5.08ms
94: learn: 0.4243422 total: 80ms remaining: 4.21ms
95: learn: 0.4225094 total: 80.5ms remaining: 3.35ms
96: learn: 0.4206795 total: 80.9ms remaining: 2.5ms
97: learn: 0.4193637 total: 81.3ms remaining: 1.66ms
98: learn: 0.4176426 total: 81.7ms remaining: 825us
99: learn: 0.4156633 total: 82ms remaining: 0us
0: learn: 0.6886167 total: 1.32ms remaining: 130ms
1: learn: 0.6839296 total: 2.34ms remaining: 115ms
2: learn: 0.6790130 total: 3.14ms remaining: 102ms
3: learn: 0.6745981 total: 4.53ms remaining: 109ms
4: learn: 0.6700984 total: 5.72ms remaining: 109ms
5: learn: 0.6652168 total: 7ms remaining: 110ms
6: learn: 0.6606527 total: 8.46ms remaining: 112ms
7: learn: 0.6566447 total: 9.54ms remaining: 110ms
8: learn: 0.6519140 total: 10.7ms remaining: 108ms
9: learn: 0.6475038 total: 12.2ms remaining: 110ms
10: learn: 0.6444133 total: 13.1ms remaining: 106ms
11: learn: 0.6400321 total: 14.2ms remaining: 104ms
12: learn: 0.6361821 total: 15.5ms remaining: 104ms
13: learn: 0.6324678 total: 16.8ms remaining: 103ms
14: learn: 0.6284970 total: 18.4ms remaining: 104ms
15: learn: 0.6241548 total: 19.4ms remaining: 102ms
16: learn: 0.6212588 total: 20.6ms remaining: 101ms
17: learn: 0.6181208 total: 22.1ms remaining: 101ms
18: learn: 0.6149727 total: 23.8ms remaining: 102ms
19: learn: 0.6112675 total: 25ms remaining: 100ms
20: learn: 0.6076084 total: 26.2ms remaining: 98.6ms
21: learn: 0.6038614 total: 27ms remaining: 95.7ms
22: learn: 0.5996882 total: 28.5ms remaining: 95.3ms
23: learn: 0.5962705 total: 29.7ms remaining: 94.1ms
24: learn: 0.5923546 total: 30.6ms remaining: 91.8ms
25: learn: 0.5894756 total: 31.2ms remaining: 88.8ms
26: learn: 0.5864478 total: 31.9ms remaining: 86.2ms
27: learn: 0.5833874 total: 32.9ms remaining: 84.7ms
28: learn: 0.5804593 total: 34.4ms remaining: 84.2ms
29: learn: 0.5777455 total: 35.4ms remaining: 82.6ms
30: learn: 0.5751136 total: 36.5ms remaining: 81.3ms
31: learn: 0.5718058 total: 37.3ms remaining: 79.2ms
32: learn: 0.5689155 total: 38.7ms remaining: 78.5ms
33: learn: 0.5658437 total: 39.9ms remaining: 77.4ms
34: learn: 0.5625370 total: 40.8ms remaining: 75.7ms
35: learn: 0.5600906 total: 41.9ms remaining: 74.4ms
36: learn: 0.5574840 total: 42.9ms remaining: 73.1ms
37: learn: 0.5548446 total: 44.1ms remaining: 71.9ms
38: learn: 0.5521241 total: 45.2ms remaining: 70.6ms
39: learn: 0.5493534 total: 46.1ms remaining: 69.2ms
40: learn: 0.5464643 total: 47ms remaining: 67.6ms
41: learn: 0.5439402 total: 47.8ms remaining: 66ms
42: learn: 0.5407503 total: 48.5ms remaining: 64.4ms
43: learn: 0.5383021 total: 49.4ms remaining: 62.8ms
44: learn: 0.5357105 total: 50.2ms remaining: 61.4ms
45: learn: 0.5332736 total: 51ms remaining: 59.9ms
46: learn: 0.5308820 total: 51.8ms remaining: 58.4ms
47: learn: 0.5280463 total: 52.5ms remaining: 56.9ms
48: learn: 0.5251939 total: 53.4ms remaining: 55.6ms
49: learn: 0.5226601 total: 54.2ms remaining: 54.2ms
50: learn: 0.5200151 total: 54.6ms remaining: 52.5ms
51: learn: 0.5174533 total: 55.4ms remaining: 51.1ms
52: learn: 0.5148804 total: 56.3ms remaining: 49.9ms
53: learn: 0.5125037 total: 56.7ms remaining: 48.3ms
54: learn: 0.5103436 total: 57.3ms remaining: 46.9ms
55: learn: 0.5079663 total: 57.8ms remaining: 45.5ms
56: learn: 0.5058975 total: 58.6ms remaining: 44.2ms
57: learn: 0.5036559 total: 59.1ms remaining: 42.8ms
58: learn: 0.5016062 total: 59.6ms remaining: 41.4ms
59: learn: 0.4994963 total: 60.2ms remaining: 40.1ms
60: learn: 0.4971523 total: 60.8ms remaining: 38.9ms
61: learn: 0.4950265 total: 61.2ms remaining: 37.5ms
62: learn: 0.4927465 total: 61.8ms remaining: 36.3ms
63: learn: 0.4903560 total: 62.3ms remaining: 35ms
64: learn: 0.4881810 total: 62.9ms remaining: 33.9ms
65: learn: 0.4861520 total: 63.2ms remaining: 32.6ms
66: learn: 0.4841875 total: 63.9ms remaining: 31.5ms
67: learn: 0.4821350 total: 64.2ms remaining: 30.2ms
68: learn: 0.4798477 total: 64.8ms remaining: 29.1ms
69: learn: 0.4774413 total: 65.3ms remaining: 28ms
70: learn: 0.4754794 total: 65.8ms remaining: 26.9ms
71: learn: 0.4733676 total: 66.2ms remaining: 25.7ms
72: learn: 0.4710711 total: 67ms remaining: 24.8ms
73: learn: 0.4693374 total: 67.5ms remaining: 23.7ms
74: learn: 0.4675932 total: 68ms remaining: 22.7ms
75: learn: 0.4657995 total: 68.7ms remaining: 21.7ms
76: learn: 0.4640184 total: 69.2ms remaining: 20.7ms
77: learn: 0.4623541 total: 69.6ms remaining: 19.6ms
78: learn: 0.4605428 total: 70.3ms remaining: 18.7ms
79: learn: 0.4584769 total: 70.8ms remaining: 17.7ms
80: learn: 0.4566638 total: 71.1ms remaining: 16.7ms
81: learn: 0.4548536 total: 71.7ms remaining: 15.7ms
82: learn: 0.4530710 total: 72.2ms remaining: 14.8ms
83: learn: 0.4510168 total: 72.5ms remaining: 13.8ms
84: learn: 0.4493297 total: 73ms remaining: 12.9ms
85: learn: 0.4474483 total: 73.3ms remaining: 11.9ms
86: learn: 0.4457039 total: 73.9ms remaining: 11ms
87: learn: 0.4440623 total: 74.3ms remaining: 10.1ms
88: learn: 0.4424507 total: 74.8ms remaining: 9.24ms
89: learn: 0.4406137 total: 75.2ms remaining: 8.36ms
90: learn: 0.4388783 total: 75.6ms remaining: 7.48ms
91: learn: 0.4372664 total: 76ms remaining: 6.61ms
92: learn: 0.4356385 total: 76.5ms remaining: 5.76ms
93: learn: 0.4339450 total: 77.1ms remaining: 4.92ms
94: learn: 0.4321958 total: 77.6ms remaining: 4.08ms
95: learn: 0.4305977 total: 77.9ms remaining: 3.25ms
96: learn: 0.4292602 total: 78.6ms remaining: 2.43ms
97: learn: 0.4277594 total: 79ms remaining: 1.61ms
98: learn: 0.4264082 total: 79.9ms remaining: 807us
99: learn: 0.4250075 total: 80.3ms remaining: 0us
0: learn: 0.6885339 total: 1.06ms remaining: 105ms
1: learn: 0.6834227 total: 2.77ms remaining: 136ms
2: learn: 0.6786232 total: 3.65ms remaining: 118ms
3: learn: 0.6739003 total: 4.77ms remaining: 115ms
4: learn: 0.6693001 total: 5.68ms remaining: 108ms
5: learn: 0.6644451 total: 7.3ms remaining: 114ms
6: learn: 0.6598747 total: 8.19ms remaining: 109ms
7: learn: 0.6558602 total: 9.3ms remaining: 107ms
8: learn: 0.6512912 total: 10.2ms remaining: 103ms
9: learn: 0.6467205 total: 11.8ms remaining: 106ms
10: learn: 0.6428079 total: 12.7ms remaining: 103ms
11: learn: 0.6384207 total: 13.7ms remaining: 100ms
12: learn: 0.6344698 total: 14.9ms remaining: 99.5ms
13: learn: 0.6304828 total: 16.3ms remaining: 100ms
14: learn: 0.6266173 total: 17.7ms remaining: 100ms
15: learn: 0.6223299 total: 18.6ms remaining: 97.5ms
16: learn: 0.6186546 total: 19.7ms remaining: 96.3ms
17: learn: 0.6147792 total: 21ms remaining: 95.5ms
18: learn: 0.6110091 total: 21.6ms remaining: 92ms
19: learn: 0.6073387 total: 22.6ms remaining: 90.5ms
20: learn: 0.6038272 total: 24.1ms remaining: 90.8ms
21: learn: 0.6001991 total: 25.4ms remaining: 90ms
22: learn: 0.5964357 total: 26.3ms remaining: 88.1ms
23: learn: 0.5929511 total: 27ms remaining: 85.5ms
24: learn: 0.5893719 total: 28.3ms remaining: 84.8ms
25: learn: 0.5864672 total: 29.3ms remaining: 83.3ms
26: learn: 0.5837256 total: 30.5ms remaining: 82.4ms
27: learn: 0.5805511 total: 31.8ms remaining: 81.7ms
28: learn: 0.5772513 total: 33.5ms remaining: 81.9ms
29: learn: 0.5745393 total: 34.4ms remaining: 80.2ms
30: learn: 0.5717751 total: 35.5ms remaining: 79.1ms
31: learn: 0.5685652 total: 36.4ms remaining: 77.4ms
32: learn: 0.5654609 total: 37.8ms remaining: 76.8ms
33: learn: 0.5622746 total: 38.8ms remaining: 75.4ms
34: learn: 0.5588724 total: 39.9ms remaining: 74ms
35: learn: 0.5555821 total: 41ms remaining: 72.9ms
36: learn: 0.5527355 total: 41.7ms remaining: 71ms
37: learn: 0.5500608 total: 42.7ms remaining: 69.6ms
38: learn: 0.5467145 total: 43.7ms remaining: 68.3ms
39: learn: 0.5441104 total: 45.3ms remaining: 68ms
40: learn: 0.5417590 total: 46.3ms remaining: 66.6ms
41: learn: 0.5389472 total: 46.8ms remaining: 64.7ms
42: learn: 0.5365060 total: 48ms remaining: 63.6ms
43: learn: 0.5333535 total: 48.8ms remaining: 62.1ms
44: learn: 0.5309882 total: 49.8ms remaining: 60.9ms
45: learn: 0.5286618 total: 50.6ms remaining: 59.4ms
46: learn: 0.5259226 total: 51.5ms remaining: 58ms
47: learn: 0.5235263 total: 52.3ms remaining: 56.6ms
48: learn: 0.5210188 total: 53.1ms remaining: 55.2ms
49: learn: 0.5185356 total: 54ms remaining: 54ms
50: learn: 0.5160011 total: 55ms remaining: 52.8ms
51: learn: 0.5139627 total: 55.5ms remaining: 51.3ms
52: learn: 0.5115609 total: 56.3ms remaining: 49.9ms
53: learn: 0.5092764 total: 57ms remaining: 48.5ms
54: learn: 0.5076697 total: 57.7ms remaining: 47.2ms
55: learn: 0.5051063 total: 58.3ms remaining: 45.8ms
56: learn: 0.5025289 total: 58.8ms remaining: 44.3ms
57: learn: 0.5004309 total: 59.5ms remaining: 43.1ms
58: learn: 0.4982484 total: 60.1ms remaining: 41.8ms
59: learn: 0.4958368 total: 60.7ms remaining: 40.5ms
60: learn: 0.4935093 total: 61.2ms remaining: 39.1ms
61: learn: 0.4909244 total: 61.7ms remaining: 37.8ms
62: learn: 0.4885892 total: 62.4ms remaining: 36.6ms
63: learn: 0.4863330 total: 62.8ms remaining: 35.3ms
64: learn: 0.4840421 total: 63.3ms remaining: 34.1ms
65: learn: 0.4819510 total: 63.9ms remaining: 32.9ms
66: learn: 0.4799421 total: 64.5ms remaining: 31.7ms
67: learn: 0.4779539 total: 65ms remaining: 30.6ms
68: learn: 0.4757741 total: 65.4ms remaining: 29.4ms
69: learn: 0.4734062 total: 66ms remaining: 28.3ms
70: learn: 0.4718209 total: 66.5ms remaining: 27.2ms
71: learn: 0.4698129 total: 67ms remaining: 26ms
72: learn: 0.4678973 total: 67.5ms remaining: 25ms
73: learn: 0.4660518 total: 68ms remaining: 23.9ms
74: learn: 0.4641532 total: 68.5ms remaining: 22.8ms
75: learn: 0.4623182 total: 68.9ms remaining: 21.8ms
76: learn: 0.4606424 total: 69.4ms remaining: 20.7ms
77: learn: 0.4589035 total: 69.9ms remaining: 19.7ms
78: learn: 0.4572527 total: 70.2ms remaining: 18.7ms
79: learn: 0.4554347 total: 70.5ms remaining: 17.6ms
80: learn: 0.4534556 total: 70.9ms remaining: 16.6ms
81: learn: 0.4516094 total: 71.3ms remaining: 15.6ms
82: learn: 0.4501233 total: 71.7ms remaining: 14.7ms
83: learn: 0.4489080 total: 72.1ms remaining: 13.7ms
84: learn: 0.4471864 total: 72.5ms remaining: 12.8ms
85: learn: 0.4452958 total: 73ms remaining: 11.9ms
86: learn: 0.4433208 total: 73.3ms remaining: 10.9ms
87: learn: 0.4415841 total: 73.6ms remaining: 10ms
88: learn: 0.4399392 total: 74ms remaining: 9.15ms
89: learn: 0.4381792 total: 74.5ms remaining: 8.28ms
90: learn: 0.4363048 total: 74.8ms remaining: 7.4ms
91: learn: 0.4346445 total: 75.2ms remaining: 6.54ms
92: learn: 0.4330773 total: 75.6ms remaining: 5.69ms
93: learn: 0.4313780 total: 76ms remaining: 4.85ms
94: learn: 0.4299629 total: 76.4ms remaining: 4.02ms
95: learn: 0.4284760 total: 76.7ms remaining: 3.2ms
96: learn: 0.4269777 total: 77.3ms remaining: 2.39ms
97: learn: 0.4254477 total: 77.6ms remaining: 1.58ms
98: learn: 0.4240495 total: 77.9ms remaining: 787us
99: learn: 0.4224510 total: 78.3ms remaining: 0us
0: learn: 0.6883550 total: 1.16ms remaining: 115ms
1: learn: 0.6836109 total: 1.77ms remaining: 86.9ms
2: learn: 0.6787323 total: 2.54ms remaining: 82.2ms
3: learn: 0.6738335 total: 3.54ms remaining: 85.1ms
4: learn: 0.6693032 total: 4.33ms remaining: 82.2ms
5: learn: 0.6644562 total: 5.37ms remaining: 84.2ms
6: learn: 0.6602992 total: 6.29ms remaining: 83.6ms
7: learn: 0.6565796 total: 7.21ms remaining: 82.9ms
8: learn: 0.6517652 total: 7.84ms remaining: 79.3ms
9: learn: 0.6474219 total: 9.26ms remaining: 83.4ms
10: learn: 0.6444322 total: 10.1ms remaining: 81.7ms
11: learn: 0.6397700 total: 11.1ms remaining: 81.2ms
12: learn: 0.6359370 total: 12ms remaining: 80.3ms
13: learn: 0.6324723 total: 12.9ms remaining: 79.4ms
14: learn: 0.6285007 total: 14ms remaining: 79.3ms
15: learn: 0.6241371 total: 15ms remaining: 78.8ms
16: learn: 0.6205886 total: 16ms remaining: 78.2ms
17: learn: 0.6165610 total: 16.9ms remaining: 76.8ms
18: learn: 0.6127567 total: 17.3ms remaining: 73.8ms
19: learn: 0.6088093 total: 18.5ms remaining: 74ms
20: learn: 0.6051968 total: 19.4ms remaining: 72.8ms
21: learn: 0.6014306 total: 20.2ms remaining: 71.5ms
22: learn: 0.5974358 total: 21ms remaining: 70.3ms
23: learn: 0.5939927 total: 21.6ms remaining: 68.3ms
24: learn: 0.5909375 total: 22.5ms remaining: 67.4ms
25: learn: 0.5873963 total: 23.2ms remaining: 66.1ms
26: learn: 0.5844064 total: 24.3ms remaining: 65.6ms
27: learn: 0.5813962 total: 25ms remaining: 64.2ms
28: learn: 0.5785490 total: 25.7ms remaining: 63ms
29: learn: 0.5758908 total: 26.3ms remaining: 61.3ms
30: learn: 0.5732910 total: 27.3ms remaining: 60.8ms
31: learn: 0.5700190 total: 27.7ms remaining: 59ms
32: learn: 0.5669492 total: 28.4ms remaining: 57.7ms
33: learn: 0.5635924 total: 29.1ms remaining: 56.5ms
34: learn: 0.5600908 total: 29.8ms remaining: 55.3ms
35: learn: 0.5572768 total: 30.6ms remaining: 54.4ms
36: learn: 0.5542501 total: 31.2ms remaining: 53.2ms
37: learn: 0.5515929 total: 31.8ms remaining: 51.9ms
38: learn: 0.5480918 total: 32.5ms remaining: 50.8ms
39: learn: 0.5449589 total: 33.2ms remaining: 49.7ms
40: learn: 0.5420133 total: 33.7ms remaining: 48.5ms
41: learn: 0.5389790 total: 34.2ms remaining: 47.2ms
42: learn: 0.5360152 total: 35ms remaining: 46.4ms
43: learn: 0.5336202 total: 35.6ms remaining: 45.4ms
44: learn: 0.5308643 total: 36.3ms remaining: 44.3ms
45: learn: 0.5285430 total: 36.7ms remaining: 43.1ms
46: learn: 0.5260419 total: 37.4ms remaining: 42.1ms
47: learn: 0.5236528 total: 37.8ms remaining: 41ms
48: learn: 0.5206597 total: 38.4ms remaining: 40ms
49: learn: 0.5182717 total: 39.2ms remaining: 39.2ms
50: learn: 0.5159021 total: 39.6ms remaining: 38.1ms
51: learn: 0.5142012 total: 40ms remaining: 36.9ms
52: learn: 0.5118869 total: 40.5ms remaining: 36ms
53: learn: 0.5092385 total: 41.2ms remaining: 35.1ms
54: learn: 0.5074476 total: 41.6ms remaining: 34.1ms
55: learn: 0.5049388 total: 42.1ms remaining: 33.1ms
56: learn: 0.5023647 total: 42.6ms remaining: 32.2ms
57: learn: 0.5003344 total: 43.1ms remaining: 31.2ms
58: learn: 0.4976284 total: 43.5ms remaining: 30.3ms
59: learn: 0.4952153 total: 44ms remaining: 29.3ms
60: learn: 0.4925942 total: 44.5ms remaining: 28.5ms
61: learn: 0.4899387 total: 44.8ms remaining: 27.5ms
62: learn: 0.4879824 total: 45.3ms remaining: 26.6ms
63: learn: 0.4857219 total: 45.7ms remaining: 25.7ms
64: learn: 0.4833536 total: 46.2ms remaining: 24.9ms
65: learn: 0.4812883 total: 46.6ms remaining: 24ms
66: learn: 0.4790204 total: 47.1ms remaining: 23.2ms
67: learn: 0.4769817 total: 47.4ms remaining: 22.3ms
68: learn: 0.4746428 total: 48ms remaining: 21.6ms
69: learn: 0.4722181 total: 48.4ms remaining: 20.7ms
70: learn: 0.4704845 total: 48.8ms remaining: 19.9ms
71: learn: 0.4681145 total: 49.2ms remaining: 19.1ms
72: learn: 0.4659689 total: 49.6ms remaining: 18.4ms
73: learn: 0.4638687 total: 50.1ms remaining: 17.6ms
74: learn: 0.4614369 total: 50.5ms remaining: 16.8ms
75: learn: 0.4592970 total: 50.9ms remaining: 16.1ms
76: learn: 0.4577747 total: 51.4ms remaining: 15.3ms
77: learn: 0.4553532 total: 51.8ms remaining: 14.6ms
78: learn: 0.4536303 total: 52.3ms remaining: 13.9ms
79: learn: 0.4514065 total: 52.7ms remaining: 13.2ms
80: learn: 0.4494436 total: 53ms remaining: 12.4ms
81: learn: 0.4477547 total: 53.3ms remaining: 11.7ms
82: learn: 0.4461889 total: 53.9ms remaining: 11ms
83: learn: 0.4445026 total: 54.2ms remaining: 10.3ms
84: learn: 0.4426023 total: 54.6ms remaining: 9.63ms
85: learn: 0.4405358 total: 55.1ms remaining: 8.97ms
86: learn: 0.4386933 total: 55.5ms remaining: 8.3ms
87: learn: 0.4368974 total: 55.9ms remaining: 7.62ms
88: learn: 0.4351774 total: 56.3ms remaining: 6.96ms
89: learn: 0.4331364 total: 56.6ms remaining: 6.29ms
90: learn: 0.4315189 total: 57ms remaining: 5.64ms
91: learn: 0.4297491 total: 57.3ms remaining: 4.99ms
92: learn: 0.4282855 total: 57.9ms remaining: 4.36ms
93: learn: 0.4269431 total: 58.2ms remaining: 3.72ms
94: learn: 0.4256240 total: 58.6ms remaining: 3.08ms
95: learn: 0.4240214 total: 59ms remaining: 2.46ms
96: learn: 0.4226613 total: 59.4ms remaining: 1.83ms
97: learn: 0.4208426 total: 59.7ms remaining: 1.22ms
98: learn: 0.4195020 total: 60ms remaining: 606us
99: learn: 0.4177747 total: 60.4ms remaining: 0us
0: learn: 0.6889933 total: 862us remaining: 85.4ms
1: learn: 0.6836164 total: 1.6ms remaining: 78.3ms
2: learn: 0.6784902 total: 2.25ms remaining: 72.7ms
3: learn: 0.6738344 total: 3.13ms remaining: 75.2ms
4: learn: 0.6690759 total: 3.8ms remaining: 72.2ms
5: learn: 0.6641179 total: 4.57ms remaining: 71.6ms
6: learn: 0.6609183 total: 5.33ms remaining: 70.8ms
7: learn: 0.6564302 total: 5.9ms remaining: 67.9ms
8: learn: 0.6518274 total: 6.73ms remaining: 68ms
9: learn: 0.6472279 total: 7.27ms remaining: 65.4ms
10: learn: 0.6439145 total: 7.81ms remaining: 63.2ms
11: learn: 0.6400426 total: 8.58ms remaining: 62.9ms
12: learn: 0.6356838 total: 9.33ms remaining: 62.4ms
13: learn: 0.6314445 total: 10.3ms remaining: 63.5ms
14: learn: 0.6280712 total: 11.1ms remaining: 63.2ms
15: learn: 0.6240877 total: 11.9ms remaining: 62.6ms
16: learn: 0.6200849 total: 13.1ms remaining: 63.7ms
17: learn: 0.6168229 total: 13.7ms remaining: 62.2ms
18: learn: 0.6129056 total: 14.5ms remaining: 61.7ms
19: learn: 0.6101052 total: 15.5ms remaining: 61.8ms
20: learn: 0.6064010 total: 16.3ms remaining: 61.2ms
21: learn: 0.6022683 total: 17ms remaining: 60.3ms
22: learn: 0.5980306 total: 17.6ms remaining: 59.1ms
23: learn: 0.5949120 total: 18.2ms remaining: 57.7ms
24: learn: 0.5912501 total: 19.1ms remaining: 57.2ms
25: learn: 0.5879653 total: 20.1ms remaining: 57.1ms
26: learn: 0.5845873 total: 20.8ms remaining: 56.3ms
27: learn: 0.5814684 total: 21.5ms remaining: 55.3ms
28: learn: 0.5788440 total: 22.2ms remaining: 54.3ms
29: learn: 0.5759054 total: 22.8ms remaining: 53.2ms
30: learn: 0.5723048 total: 23.4ms remaining: 52ms
31: learn: 0.5690872 total: 24.1ms remaining: 51.3ms
32: learn: 0.5660702 total: 24.9ms remaining: 50.6ms
33: learn: 0.5628655 total: 25.9ms remaining: 50.3ms
34: learn: 0.5599171 total: 26.7ms remaining: 49.6ms
35: learn: 0.5573603 total: 27.4ms remaining: 48.8ms
36: learn: 0.5551515 total: 28.4ms remaining: 48.4ms
37: learn: 0.5521042 total: 29.3ms remaining: 47.8ms
38: learn: 0.5486314 total: 30.3ms remaining: 47.4ms
39: learn: 0.5453812 total: 31.1ms remaining: 46.7ms
40: learn: 0.5422646 total: 32.3ms remaining: 46.5ms
41: learn: 0.5396314 total: 32.9ms remaining: 45.5ms
42: learn: 0.5369313 total: 33.5ms remaining: 44.5ms
43: learn: 0.5341681 total: 34.5ms remaining: 43.9ms
44: learn: 0.5311134 total: 35.6ms remaining: 43.5ms
45: learn: 0.5279754 total: 36.5ms remaining: 42.8ms
46: learn: 0.5252587 total: 37.3ms remaining: 42.1ms
47: learn: 0.5224699 total: 38.2ms remaining: 41.4ms
48: learn: 0.5196144 total: 39.1ms remaining: 40.7ms
49: learn: 0.5168594 total: 39.7ms remaining: 39.7ms
50: learn: 0.5145349 total: 42ms remaining: 40.4ms
51: learn: 0.5120538 total: 43ms remaining: 39.7ms
52: learn: 0.5094141 total: 43.9ms remaining: 38.9ms
53: learn: 0.5067369 total: 44.7ms remaining: 38.1ms
54: learn: 0.5046851 total: 45.5ms remaining: 37.2ms
55: learn: 0.5021771 total: 46.3ms remaining: 36.3ms
56: learn: 0.5001128 total: 46.6ms remaining: 35.2ms
57: learn: 0.4976841 total: 47.3ms remaining: 34.2ms
58: learn: 0.4952075 total: 48ms remaining: 33.4ms
59: learn: 0.4934166 total: 48.7ms remaining: 32.5ms
60: learn: 0.4908600 total: 49.3ms remaining: 31.5ms
61: learn: 0.4884676 total: 50.1ms remaining: 30.7ms
62: learn: 0.4865485 total: 50.5ms remaining: 29.7ms
63: learn: 0.4844235 total: 50.9ms remaining: 28.6ms
64: learn: 0.4824806 total: 51.1ms remaining: 27.5ms
65: learn: 0.4798479 total: 51.5ms remaining: 26.5ms
66: learn: 0.4773723 total: 52ms remaining: 25.6ms
67: learn: 0.4753494 total: 52.5ms remaining: 24.7ms
68: learn: 0.4732775 total: 53.1ms remaining: 23.9ms
69: learn: 0.4708137 total: 53.8ms remaining: 23.1ms
70: learn: 0.4690268 total: 54.4ms remaining: 22.2ms
71: learn: 0.4671688 total: 55ms remaining: 21.4ms
72: learn: 0.4652406 total: 55.5ms remaining: 20.5ms
73: learn: 0.4632691 total: 56.2ms remaining: 19.7ms
74: learn: 0.4610572 total: 56.7ms remaining: 18.9ms
75: learn: 0.4593666 total: 57.1ms remaining: 18ms
76: learn: 0.4575386 total: 57.8ms remaining: 17.3ms
77: learn: 0.4554613 total: 58.3ms remaining: 16.4ms
78: learn: 0.4531759 total: 58.8ms remaining: 15.6ms
79: learn: 0.4516175 total: 59.2ms remaining: 14.8ms
80: learn: 0.4497692 total: 59.7ms remaining: 14ms
81: learn: 0.4477158 total: 60ms remaining: 13.2ms
82: learn: 0.4458186 total: 60.6ms remaining: 12.4ms
83: learn: 0.4440212 total: 61ms remaining: 11.6ms
84: learn: 0.4421802 total: 61.4ms remaining: 10.8ms
85: learn: 0.4406457 total: 61.8ms remaining: 10.1ms
86: learn: 0.4386856 total: 62.5ms remaining: 9.34ms
87: learn: 0.4367161 total: 62.9ms remaining: 8.58ms
88: learn: 0.4348507 total: 63.3ms remaining: 7.83ms
89: learn: 0.4332635 total: 63.9ms remaining: 7.1ms
90: learn: 0.4317573 total: 64.3ms remaining: 6.36ms
91: learn: 0.4299747 total: 64.7ms remaining: 5.63ms
92: learn: 0.4284364 total: 65.1ms remaining: 4.9ms
93: learn: 0.4272723 total: 65.5ms remaining: 4.18ms
94: learn: 0.4254917 total: 65.9ms remaining: 3.47ms
95: learn: 0.4237938 total: 66.3ms remaining: 2.76ms
96: learn: 0.4223160 total: 66.7ms remaining: 2.06ms
97: learn: 0.4205233 total: 67.1ms remaining: 1.37ms
98: learn: 0.4187850 total: 67.5ms remaining: 681us
99: learn: 0.4171327 total: 67.8ms remaining: 0us
0: learn: 0.6890031 total: 1.45ms remaining: 144ms
1: learn: 0.6839941 total: 2.9ms remaining: 142ms
2: learn: 0.6790950 total: 3.57ms remaining: 116ms
3: learn: 0.6741892 total: 4.36ms remaining: 105ms
4: learn: 0.6699145 total: 4.94ms remaining: 93.9ms
5: learn: 0.6651736 total: 6.38ms remaining: 100ms
6: learn: 0.6605127 total: 6.99ms remaining: 92.9ms
7: learn: 0.6563352 total: 7.83ms remaining: 90ms
8: learn: 0.6525859 total: 8.2ms remaining: 83ms
9: learn: 0.6483100 total: 8.93ms remaining: 80.4ms
10: learn: 0.6443013 total: 9.43ms remaining: 76.3ms
11: learn: 0.6404553 total: 10.6ms remaining: 77.7ms
12: learn: 0.6363750 total: 11.2ms remaining: 75ms
13: learn: 0.6320174 total: 11.8ms remaining: 72.4ms
14: learn: 0.6288432 total: 12.5ms remaining: 70.9ms
15: learn: 0.6252655 total: 13.2ms remaining: 69.5ms
16: learn: 0.6214597 total: 13.8ms remaining: 67.4ms
17: learn: 0.6174034 total: 14.3ms remaining: 65.1ms
18: learn: 0.6143807 total: 15.1ms remaining: 64.5ms
19: learn: 0.6108119 total: 15.7ms remaining: 62.6ms
20: learn: 0.6071982 total: 16.2ms remaining: 60.9ms
21: learn: 0.6035162 total: 16.8ms remaining: 59.5ms
22: learn: 0.5995561 total: 17.3ms remaining: 58ms
23: learn: 0.5967885 total: 17.9ms remaining: 56.8ms
24: learn: 0.5928977 total: 18.2ms remaining: 54.6ms
25: learn: 0.5896768 total: 18.7ms remaining: 53.1ms
26: learn: 0.5864794 total: 19.2ms remaining: 51.9ms
27: learn: 0.5831156 total: 19.8ms remaining: 50.8ms
28: learn: 0.5804258 total: 20.2ms remaining: 49.4ms
29: learn: 0.5780639 total: 20.6ms remaining: 48ms
30: learn: 0.5757587 total: 21.1ms remaining: 47ms
31: learn: 0.5725405 total: 21.8ms remaining: 46.4ms
32: learn: 0.5693987 total: 22.7ms remaining: 46.2ms
33: learn: 0.5660367 total: 23.5ms remaining: 45.7ms
34: learn: 0.5626815 total: 24.5ms remaining: 45.5ms
35: learn: 0.5601347 total: 25.2ms remaining: 44.8ms
36: learn: 0.5570503 total: 25.9ms remaining: 44.1ms
37: learn: 0.5542878 total: 26.6ms remaining: 43.4ms
38: learn: 0.5511386 total: 27.6ms remaining: 43.2ms
39: learn: 0.5481846 total: 28.3ms remaining: 42.5ms
40: learn: 0.5450162 total: 28.8ms remaining: 41.5ms
41: learn: 0.5426760 total: 29.4ms remaining: 40.6ms
42: learn: 0.5395370 total: 30.1ms remaining: 39.9ms
43: learn: 0.5373680 total: 30.6ms remaining: 38.9ms
44: learn: 0.5345015 total: 31ms remaining: 37.8ms
45: learn: 0.5318743 total: 32ms remaining: 37.6ms
46: learn: 0.5292160 total: 32.8ms remaining: 37ms
47: learn: 0.5267250 total: 33.2ms remaining: 35.9ms
48: learn: 0.5243110 total: 33.6ms remaining: 35ms
49: learn: 0.5214591 total: 34ms remaining: 34ms
50: learn: 0.5186970 total: 34.6ms remaining: 33.3ms
51: learn: 0.5158365 total: 35ms remaining: 32.3ms
52: learn: 0.5136888 total: 35.4ms remaining: 31.4ms
53: learn: 0.5112052 total: 36.1ms remaining: 30.8ms
54: learn: 0.5089564 total: 36.7ms remaining: 30ms
55: learn: 0.5065765 total: 37.1ms remaining: 29.2ms
56: learn: 0.5040682 total: 37.4ms remaining: 28.2ms
57: learn: 0.5015641 total: 37.9ms remaining: 27.5ms
58: learn: 0.4991199 total: 38.3ms remaining: 26.6ms
59: learn: 0.4967366 total: 38.8ms remaining: 25.9ms
60: learn: 0.4942500 total: 39.3ms remaining: 25.1ms
61: learn: 0.4921693 total: 39.8ms remaining: 24.4ms
62: learn: 0.4901854 total: 40.4ms remaining: 23.7ms
63: learn: 0.4879617 total: 40.9ms remaining: 23ms
64: learn: 0.4857070 total: 41.4ms remaining: 22.3ms
65: learn: 0.4837889 total: 41.9ms remaining: 21.6ms
66: learn: 0.4814533 total: 42.3ms remaining: 20.8ms
67: learn: 0.4798315 total: 42.7ms remaining: 20.1ms
68: learn: 0.4775769 total: 43.1ms remaining: 19.4ms
69: learn: 0.4754393 total: 43.5ms remaining: 18.6ms
70: learn: 0.4739529 total: 43.9ms remaining: 17.9ms
71: learn: 0.4717598 total: 44.5ms remaining: 17.3ms
72: learn: 0.4696536 total: 45ms remaining: 16.6ms
73: learn: 0.4676443 total: 45.6ms remaining: 16ms
74: learn: 0.4655447 total: 46.3ms remaining: 15.4ms
75: learn: 0.4634157 total: 46.7ms remaining: 14.7ms
76: learn: 0.4614728 total: 47.1ms remaining: 14.1ms
77: learn: 0.4593235 total: 47.5ms remaining: 13.4ms
78: learn: 0.4575239 total: 48ms remaining: 12.8ms
79: learn: 0.4556075 total: 48.3ms remaining: 12.1ms
80: learn: 0.4535342 total: 48.6ms remaining: 11.4ms
81: learn: 0.4515864 total: 49ms remaining: 10.8ms
82: learn: 0.4496692 total: 49.5ms remaining: 10.1ms
83: learn: 0.4484274 total: 49.8ms remaining: 9.49ms
84: learn: 0.4469008 total: 50.1ms remaining: 8.84ms
85: learn: 0.4455647 total: 50.5ms remaining: 8.22ms
86: learn: 0.4436224 total: 51ms remaining: 7.61ms
87: learn: 0.4421467 total: 51.3ms remaining: 6.99ms
88: learn: 0.4405282 total: 51.7ms remaining: 6.39ms
89: learn: 0.4391787 total: 52.2ms remaining: 5.8ms
90: learn: 0.4374349 total: 52.6ms remaining: 5.2ms
91: learn: 0.4359123 total: 53ms remaining: 4.61ms
92: learn: 0.4344414 total: 53.3ms remaining: 4.01ms
93: learn: 0.4327461 total: 53.8ms remaining: 3.43ms
94: learn: 0.4311738 total: 54.1ms remaining: 2.85ms
95: learn: 0.4295184 total: 54.5ms remaining: 2.27ms
96: learn: 0.4276992 total: 55ms remaining: 1.7ms
97: learn: 0.4264722 total: 55.3ms remaining: 1.13ms
98: learn: 0.4249954 total: 55.6ms remaining: 561us
99: learn: 0.4234596 total: 56ms remaining: 0us
0: learn: 0.6884444 total: 1.22ms remaining: 121ms
1: learn: 0.6833777 total: 2.29ms remaining: 112ms
2: learn: 0.6789418 total: 2.99ms remaining: 96.7ms
3: learn: 0.6740883 total: 3.4ms remaining: 81.6ms
4: learn: 0.6695991 total: 4.07ms remaining: 77.3ms
5: learn: 0.6649560 total: 5.37ms remaining: 84.2ms
6: learn: 0.6607099 total: 6.81ms remaining: 90.5ms
7: learn: 0.6568861 total: 8.11ms remaining: 93.2ms
8: learn: 0.6526495 total: 8.77ms remaining: 88.7ms
9: learn: 0.6486747 total: 9.5ms remaining: 85.5ms
10: learn: 0.6446790 total: 10.7ms remaining: 86.9ms
11: learn: 0.6408419 total: 11.7ms remaining: 85.9ms
12: learn: 0.6366769 total: 12.8ms remaining: 85.6ms
13: learn: 0.6327758 total: 14ms remaining: 86ms
14: learn: 0.6289897 total: 15.3ms remaining: 86.5ms
15: learn: 0.6254446 total: 16.3ms remaining: 85.4ms
16: learn: 0.6214642 total: 17.5ms remaining: 85.5ms
17: learn: 0.6179222 total: 18.7ms remaining: 85.3ms
18: learn: 0.6145156 total: 19.4ms remaining: 82.8ms
19: learn: 0.6111632 total: 20.1ms remaining: 80.5ms
20: learn: 0.6075448 total: 21.2ms remaining: 79.8ms
21: learn: 0.6041593 total: 22.1ms remaining: 78.4ms
22: learn: 0.6005087 total: 23.3ms remaining: 78.1ms
23: learn: 0.5967117 total: 24.3ms remaining: 76.9ms
24: learn: 0.5937134 total: 25.2ms remaining: 75.7ms
25: learn: 0.5902819 total: 26ms remaining: 74.1ms
26: learn: 0.5876279 total: 27.3ms remaining: 73.9ms
27: learn: 0.5847773 total: 28.7ms remaining: 73.9ms
28: learn: 0.5817003 total: 30ms remaining: 73.4ms
29: learn: 0.5787036 total: 31ms remaining: 72.3ms
30: learn: 0.5760609 total: 32.2ms remaining: 71.6ms
31: learn: 0.5734735 total: 33.4ms remaining: 71ms
32: learn: 0.5707002 total: 34.1ms remaining: 69.2ms
33: learn: 0.5675329 total: 35.3ms remaining: 68.5ms
34: learn: 0.5643493 total: 36.4ms remaining: 67.6ms
35: learn: 0.5610712 total: 37.5ms remaining: 66.6ms
36: learn: 0.5582207 total: 38.1ms remaining: 64.8ms
37: learn: 0.5559611 total: 39.4ms remaining: 64.3ms
38: learn: 0.5537050 total: 40.4ms remaining: 63.2ms
39: learn: 0.5503936 total: 41.3ms remaining: 62ms
40: learn: 0.5478983 total: 42.5ms remaining: 61.2ms
41: learn: 0.5451844 total: 43.4ms remaining: 59.9ms
42: learn: 0.5418663 total: 43.7ms remaining: 57.9ms
43: learn: 0.5390675 total: 44.7ms remaining: 56.8ms
44: learn: 0.5366144 total: 45.8ms remaining: 56ms
45: learn: 0.5341339 total: 47ms remaining: 55.2ms
46: learn: 0.5320371 total: 47.9ms remaining: 54ms
47: learn: 0.5297100 total: 49ms remaining: 53ms
48: learn: 0.5275120 total: 49.6ms remaining: 51.7ms
49: learn: 0.5249431 total: 50.4ms remaining: 50.4ms
50: learn: 0.5222452 total: 51.1ms remaining: 49.1ms
51: learn: 0.5196150 total: 52ms remaining: 48ms
52: learn: 0.5174188 total: 52.6ms remaining: 46.7ms
53: learn: 0.5150613 total: 53.3ms remaining: 45.4ms
54: learn: 0.5126465 total: 54.2ms remaining: 44.3ms
55: learn: 0.5104492 total: 55ms remaining: 43.2ms
56: learn: 0.5080920 total: 55.6ms remaining: 42ms
57: learn: 0.5056335 total: 56.3ms remaining: 40.8ms
58: learn: 0.5035476 total: 56.6ms remaining: 39.3ms
59: learn: 0.5013622 total: 57ms remaining: 38ms
60: learn: 0.4988686 total: 57.8ms remaining: 37ms
61: learn: 0.4972610 total: 58.5ms remaining: 35.8ms
62: learn: 0.4953017 total: 58.8ms remaining: 34.6ms
63: learn: 0.4927863 total: 59.6ms remaining: 33.5ms
64: learn: 0.4907355 total: 60.2ms remaining: 32.4ms
65: learn: 0.4887132 total: 60.7ms remaining: 31.2ms
66: learn: 0.4864048 total: 60.9ms remaining: 30ms
67: learn: 0.4841681 total: 61.4ms remaining: 28.9ms
68: learn: 0.4822257 total: 62.1ms remaining: 27.9ms
69: learn: 0.4802451 total: 62.6ms remaining: 26.8ms
70: learn: 0.4782052 total: 63.2ms remaining: 25.8ms
71: learn: 0.4761668 total: 63.8ms remaining: 24.8ms
72: learn: 0.4740465 total: 64.5ms remaining: 23.8ms
73: learn: 0.4721290 total: 65.3ms remaining: 22.9ms
74: learn: 0.4701994 total: 65.7ms remaining: 21.9ms
75: learn: 0.4681550 total: 66ms remaining: 20.8ms
76: learn: 0.4665667 total: 66.4ms remaining: 19.8ms
77: learn: 0.4645251 total: 67.2ms remaining: 18.9ms
78: learn: 0.4625883 total: 67.8ms remaining: 18ms
79: learn: 0.4605779 total: 68.3ms remaining: 17.1ms
80: learn: 0.4587713 total: 68.8ms remaining: 16.1ms
81: learn: 0.4568585 total: 69.1ms remaining: 15.2ms
82: learn: 0.4551630 total: 69.6ms remaining: 14.2ms
83: learn: 0.4534023 total: 70ms remaining: 13.3ms
84: learn: 0.4519195 total: 70.7ms remaining: 12.5ms
85: learn: 0.4503519 total: 71.1ms remaining: 11.6ms
86: learn: 0.4485513 total: 71.6ms remaining: 10.7ms
87: learn: 0.4469570 total: 72.1ms remaining: 9.83ms
88: learn: 0.4453554 total: 72.8ms remaining: 8.99ms
89: learn: 0.4437199 total: 73.3ms remaining: 8.15ms
90: learn: 0.4418570 total: 73.8ms remaining: 7.29ms
91: learn: 0.4406584 total: 74.3ms remaining: 6.46ms
92: learn: 0.4389602 total: 74.7ms remaining: 5.62ms
93: learn: 0.4371791 total: 75.1ms remaining: 4.79ms
94: learn: 0.4354748 total: 75.7ms remaining: 3.98ms
95: learn: 0.4339965 total: 76.1ms remaining: 3.17ms
96: learn: 0.4323266 total: 76.6ms remaining: 2.37ms
97: learn: 0.4307838 total: 77.1ms remaining: 1.57ms
98: learn: 0.4293552 total: 77.7ms remaining: 784us
99: learn: 0.4278470 total: 78ms remaining: 0us
0: learn: 0.6885566 total: 1.4ms remaining: 138ms
1: learn: 0.6836071 total: 2.25ms remaining: 110ms
2: learn: 0.6782460 total: 3.22ms remaining: 104ms
3: learn: 0.6743132 total: 4.62ms remaining: 111ms
4: learn: 0.6702237 total: 5.73ms remaining: 109ms
5: learn: 0.6660752 total: 6.86ms remaining: 107ms
6: learn: 0.6616702 total: 8.28ms remaining: 110ms
7: learn: 0.6566263 total: 11ms remaining: 126ms
8: learn: 0.6518664 total: 12.5ms remaining: 126ms
9: learn: 0.6490527 total: 13.6ms remaining: 123ms
10: learn: 0.6444509 total: 14.7ms remaining: 119ms
11: learn: 0.6403671 total: 15.7ms remaining: 115ms
12: learn: 0.6363304 total: 17.2ms remaining: 115ms
13: learn: 0.6327678 total: 18.5ms remaining: 114ms
14: learn: 0.6282780 total: 19.5ms remaining: 110ms
15: learn: 0.6244872 total: 20.4ms remaining: 107ms
16: learn: 0.6203105 total: 21.6ms remaining: 106ms
17: learn: 0.6165432 total: 22.3ms remaining: 101ms
18: learn: 0.6128502 total: 23.6ms remaining: 101ms
19: learn: 0.6090656 total: 24.6ms remaining: 98.5ms
20: learn: 0.6050572 total: 26.5ms remaining: 99.8ms
21: learn: 0.6007740 total: 27.5ms remaining: 97.6ms
22: learn: 0.5970249 total: 28.5ms remaining: 95.3ms
23: learn: 0.5935481 total: 29.2ms remaining: 92.6ms
24: learn: 0.5906288 total: 30.4ms remaining: 91.3ms
25: learn: 0.5877041 total: 31.4ms remaining: 89.5ms
26: learn: 0.5839478 total: 32.5ms remaining: 87.9ms
27: learn: 0.5811081 total: 33.5ms remaining: 86ms
28: learn: 0.5773239 total: 34.8ms remaining: 85.2ms
29: learn: 0.5742893 total: 36.3ms remaining: 84.7ms
30: learn: 0.5706335 total: 37.3ms remaining: 83.1ms
31: learn: 0.5674696 total: 38.2ms remaining: 81.1ms
32: learn: 0.5645132 total: 38.8ms remaining: 78.8ms
33: learn: 0.5608553 total: 39.5ms remaining: 76.7ms
34: learn: 0.5579249 total: 40.9ms remaining: 75.9ms
35: learn: 0.5548740 total: 42.2ms remaining: 74.9ms
36: learn: 0.5521033 total: 43.1ms remaining: 73.4ms
37: learn: 0.5487798 total: 44ms remaining: 71.7ms
38: learn: 0.5458292 total: 45.1ms remaining: 70.6ms
39: learn: 0.5434128 total: 46.2ms remaining: 69.3ms
40: learn: 0.5398872 total: 47ms remaining: 67.6ms
41: learn: 0.5372327 total: 48.2ms remaining: 66.5ms
42: learn: 0.5348076 total: 49.2ms remaining: 65.2ms
43: learn: 0.5319729 total: 50.5ms remaining: 64.3ms
44: learn: 0.5296587 total: 51.5ms remaining: 63ms
45: learn: 0.5271298 total: 52.6ms remaining: 61.8ms
46: learn: 0.5243558 total: 53.7ms remaining: 60.5ms
47: learn: 0.5216964 total: 54.9ms remaining: 59.5ms
48: learn: 0.5191863 total: 55.9ms remaining: 58.2ms
49: learn: 0.5165567 total: 56.5ms remaining: 56.5ms
50: learn: 0.5141248 total: 56.9ms remaining: 54.7ms
51: learn: 0.5116138 total: 57.4ms remaining: 53ms
52: learn: 0.5092609 total: 58.6ms remaining: 52ms
53: learn: 0.5070869 total: 59.5ms remaining: 50.7ms
54: learn: 0.5046551 total: 60.3ms remaining: 49.3ms
55: learn: 0.5022862 total: 60.7ms remaining: 47.7ms
56: learn: 0.4996988 total: 61.5ms remaining: 46.4ms
57: learn: 0.4972926 total: 62ms remaining: 44.9ms
58: learn: 0.4957858 total: 62.4ms remaining: 43.4ms
59: learn: 0.4937636 total: 63.1ms remaining: 42ms
60: learn: 0.4914307 total: 63.6ms remaining: 40.7ms
61: learn: 0.4892601 total: 64.3ms remaining: 39.4ms
62: learn: 0.4870957 total: 65ms remaining: 38.2ms
63: learn: 0.4849294 total: 65.4ms remaining: 36.8ms
64: learn: 0.4828281 total: 66ms remaining: 35.6ms
65: learn: 0.4801656 total: 66.7ms remaining: 34.4ms
66: learn: 0.4779788 total: 67.2ms remaining: 33.1ms
67: learn: 0.4757880 total: 67.7ms remaining: 31.9ms
68: learn: 0.4741170 total: 68.1ms remaining: 30.6ms
69: learn: 0.4721005 total: 68.8ms remaining: 29.5ms
70: learn: 0.4703547 total: 69.5ms remaining: 28.4ms
71: learn: 0.4682511 total: 70.1ms remaining: 27.3ms
72: learn: 0.4666105 total: 70.7ms remaining: 26.2ms
73: learn: 0.4643244 total: 71.3ms remaining: 25.1ms
74: learn: 0.4620866 total: 71.8ms remaining: 23.9ms
75: learn: 0.4599344 total: 72.3ms remaining: 22.8ms
76: learn: 0.4578568 total: 72.9ms remaining: 21.8ms
77: learn: 0.4562268 total: 73.3ms remaining: 20.7ms
78: learn: 0.4540555 total: 73.8ms remaining: 19.6ms
79: learn: 0.4518526 total: 74.3ms remaining: 18.6ms
80: learn: 0.4498825 total: 74.8ms remaining: 17.5ms
81: learn: 0.4476596 total: 75.2ms remaining: 16.5ms
82: learn: 0.4463192 total: 75.7ms remaining: 15.5ms
83: learn: 0.4445282 total: 76ms remaining: 14.5ms
84: learn: 0.4429586 total: 76.7ms remaining: 13.5ms
85: learn: 0.4412175 total: 77.1ms remaining: 12.6ms
86: learn: 0.4394992 total: 77.6ms remaining: 11.6ms
87: learn: 0.4378353 total: 78.1ms remaining: 10.7ms
88: learn: 0.4362414 total: 78.8ms remaining: 9.73ms
89: learn: 0.4346269 total: 79.3ms remaining: 8.81ms
90: learn: 0.4330993 total: 79.7ms remaining: 7.88ms
91: learn: 0.4314341 total: 80.2ms remaining: 6.97ms
92: learn: 0.4296886 total: 80.5ms remaining: 6.06ms
93: learn: 0.4281258 total: 81ms remaining: 5.17ms
94: learn: 0.4266854 total: 81.3ms remaining: 4.28ms
95: learn: 0.4247964 total: 81.9ms remaining: 3.41ms
96: learn: 0.4232147 total: 82.3ms remaining: 2.55ms
97: learn: 0.4217981 total: 82.8ms remaining: 1.69ms
98: learn: 0.4198929 total: 83.1ms remaining: 839us
99: learn: 0.4181065 total: 83.5ms remaining: 0us
0: learn: 0.6896203 total: 1.56ms remaining: 154ms
1: learn: 0.6850714 total: 2.65ms remaining: 130ms
2: learn: 0.6803862 total: 3.87ms remaining: 125ms
3: learn: 0.6758516 total: 5.03ms remaining: 121ms
4: learn: 0.6716479 total: 5.9ms remaining: 112ms
5: learn: 0.6663871 total: 7.39ms remaining: 116ms
6: learn: 0.6619637 total: 8.73ms remaining: 116ms
7: learn: 0.6576289 total: 9.51ms remaining: 109ms
8: learn: 0.6535165 total: 10.1ms remaining: 102ms
9: learn: 0.6494451 total: 10.7ms remaining: 96ms
10: learn: 0.6454696 total: 11.8ms remaining: 95.4ms
11: learn: 0.6411167 total: 12.9ms remaining: 94.4ms
12: learn: 0.6375680 total: 14.1ms remaining: 94.4ms
13: learn: 0.6336478 total: 15.4ms remaining: 94.6ms
14: learn: 0.6296995 total: 16.6ms remaining: 94.1ms
15: learn: 0.6252287 total: 18ms remaining: 94.4ms
16: learn: 0.6216951 total: 19.8ms remaining: 96.8ms
17: learn: 0.6177813 total: 21.1ms remaining: 96.1ms
18: learn: 0.6140884 total: 22.1ms remaining: 94.2ms
19: learn: 0.6108468 total: 23.4ms remaining: 93.6ms
20: learn: 0.6072031 total: 24.5ms remaining: 92.1ms
21: learn: 0.6030750 total: 25.7ms remaining: 91ms
22: learn: 0.5992488 total: 27ms remaining: 90.4ms
23: learn: 0.5955617 total: 28ms remaining: 88.8ms
24: learn: 0.5922429 total: 29ms remaining: 86.9ms
25: learn: 0.5884927 total: 30.1ms remaining: 85.7ms
26: learn: 0.5856491 total: 31.3ms remaining: 84.7ms
27: learn: 0.5820865 total: 32.7ms remaining: 84ms
28: learn: 0.5795715 total: 33.8ms remaining: 82.7ms
29: learn: 0.5761366 total: 35.4ms remaining: 82.5ms
30: learn: 0.5728400 total: 36.5ms remaining: 81.2ms
31: learn: 0.5697248 total: 37.6ms remaining: 79.8ms
32: learn: 0.5668872 total: 38.6ms remaining: 78.5ms
33: learn: 0.5633521 total: 40.4ms remaining: 78.3ms
34: learn: 0.5601400 total: 41.3ms remaining: 76.8ms
35: learn: 0.5574012 total: 42.4ms remaining: 75.4ms
36: learn: 0.5543751 total: 43.7ms remaining: 74.4ms
37: learn: 0.5520800 total: 44.2ms remaining: 72.2ms
38: learn: 0.5495015 total: 45.1ms remaining: 70.6ms
39: learn: 0.5462074 total: 46ms remaining: 69ms
40: learn: 0.5434947 total: 46.8ms remaining: 67.3ms
41: learn: 0.5412224 total: 47.7ms remaining: 65.8ms
42: learn: 0.5385021 total: 48.4ms remaining: 64.2ms
43: learn: 0.5355544 total: 49.3ms remaining: 62.7ms
44: learn: 0.5328038 total: 50ms remaining: 61.1ms
45: learn: 0.5305187 total: 50.8ms remaining: 59.6ms
46: learn: 0.5281554 total: 51.4ms remaining: 58ms
47: learn: 0.5251024 total: 52.5ms remaining: 56.8ms
48: learn: 0.5225982 total: 53ms remaining: 55.2ms
49: learn: 0.5203373 total: 53.8ms remaining: 53.8ms
50: learn: 0.5181042 total: 54.6ms remaining: 52.4ms
51: learn: 0.5153619 total: 55.6ms remaining: 51.3ms
52: learn: 0.5130063 total: 56.2ms remaining: 49.8ms
53: learn: 0.5106676 total: 56.8ms remaining: 48.4ms
54: learn: 0.5085983 total: 57.4ms remaining: 46.9ms
55: learn: 0.5064403 total: 57.9ms remaining: 45.5ms
56: learn: 0.5042995 total: 58.3ms remaining: 44ms
57: learn: 0.5018453 total: 59.1ms remaining: 42.8ms
58: learn: 0.4999030 total: 59.6ms remaining: 41.4ms
59: learn: 0.4970939 total: 60.2ms remaining: 40.2ms
60: learn: 0.4956242 total: 60.7ms remaining: 38.8ms
61: learn: 0.4936966 total: 61.4ms remaining: 37.6ms
62: learn: 0.4915250 total: 62.2ms remaining: 36.5ms
63: learn: 0.4894404 total: 63ms remaining: 35.4ms
64: learn: 0.4869456 total: 63.7ms remaining: 34.3ms
65: learn: 0.4846882 total: 64.2ms remaining: 33.1ms
66: learn: 0.4827632 total: 64.9ms remaining: 32ms
67: learn: 0.4802655 total: 65.4ms remaining: 30.8ms
68: learn: 0.4782428 total: 66ms remaining: 29.7ms
69: learn: 0.4760303 total: 66.6ms remaining: 28.6ms
70: learn: 0.4745876 total: 67.3ms remaining: 27.5ms
71: learn: 0.4721742 total: 67.8ms remaining: 26.4ms
72: learn: 0.4706441 total: 68.4ms remaining: 25.3ms
73: learn: 0.4686661 total: 69ms remaining: 24.2ms
74: learn: 0.4671855 total: 69.6ms remaining: 23.2ms
75: learn: 0.4653315 total: 70.1ms remaining: 22.1ms
76: learn: 0.4632093 total: 70.6ms remaining: 21.1ms
77: learn: 0.4609629 total: 71.5ms remaining: 20.2ms
78: learn: 0.4590973 total: 71.9ms remaining: 19.1ms
79: learn: 0.4575248 total: 72.5ms remaining: 18.1ms
80: learn: 0.4557242 total: 72.9ms remaining: 17.1ms
81: learn: 0.4537280 total: 73.5ms remaining: 16.1ms
82: learn: 0.4518387 total: 74.1ms remaining: 15.2ms
83: learn: 0.4497253 total: 74.5ms remaining: 14.2ms
84: learn: 0.4482405 total: 75.1ms remaining: 13.3ms
85: learn: 0.4465275 total: 75.5ms remaining: 12.3ms
86: learn: 0.4450708 total: 76ms remaining: 11.4ms
87: learn: 0.4429419 total: 76.5ms remaining: 10.4ms
88: learn: 0.4413283 total: 77ms remaining: 9.52ms
89: learn: 0.4397207 total: 77.5ms remaining: 8.61ms
90: learn: 0.4384493 total: 78.1ms remaining: 7.72ms
91: learn: 0.4367745 total: 78.5ms remaining: 6.83ms
92: learn: 0.4353104 total: 79.2ms remaining: 5.96ms
93: learn: 0.4333906 total: 79.7ms remaining: 5.09ms
94: learn: 0.4317244 total: 80.3ms remaining: 4.22ms
95: learn: 0.4302170 total: 80.7ms remaining: 3.36ms
96: learn: 0.4286522 total: 81.2ms remaining: 2.51ms
97: learn: 0.4268209 total: 81.7ms remaining: 1.67ms
98: learn: 0.4253835 total: 82.1ms remaining: 829us
99: learn: 0.4237831 total: 82.5ms remaining: 0us
0: learn: 0.6894871 total: 1.99ms remaining: 197ms
1: learn: 0.6842252 total: 2.8ms remaining: 137ms
2: learn: 0.6798605 total: 4.19ms remaining: 135ms
3: learn: 0.6746510 total: 5.17ms remaining: 124ms
4: learn: 0.6695916 total: 6.58ms remaining: 125ms
5: learn: 0.6646934 total: 8.19ms remaining: 128ms
6: learn: 0.6608368 total: 9.33ms remaining: 124ms
7: learn: 0.6560198 total: 10.3ms remaining: 119ms
8: learn: 0.6514894 total: 11.4ms remaining: 115ms
9: learn: 0.6484876 total: 12.5ms remaining: 113ms
10: learn: 0.6438394 total: 13.5ms remaining: 109ms
11: learn: 0.6400430 total: 14.5ms remaining: 107ms
12: learn: 0.6364403 total: 16.2ms remaining: 108ms
13: learn: 0.6328070 total: 17.2ms remaining: 106ms
14: learn: 0.6285253 total: 18.3ms remaining: 103ms
15: learn: 0.6244703 total: 19.5ms remaining: 102ms
16: learn: 0.6203785 total: 20.9ms remaining: 102ms
17: learn: 0.6170779 total: 21.5ms remaining: 97.9ms
18: learn: 0.6134879 total: 21.9ms remaining: 93.3ms
19: learn: 0.6098244 total: 23.1ms remaining: 92.4ms
20: learn: 0.6058554 total: 24.4ms remaining: 91.7ms
21: learn: 0.6020761 total: 25.1ms remaining: 89ms
22: learn: 0.5989954 total: 25.8ms remaining: 86.2ms
23: learn: 0.5950739 total: 27ms remaining: 85.3ms
24: learn: 0.5921883 total: 28.6ms remaining: 85.8ms
25: learn: 0.5889009 total: 29.9ms remaining: 85ms
26: learn: 0.5860139 total: 31.1ms remaining: 84.1ms
27: learn: 0.5833418 total: 33.6ms remaining: 86.4ms
28: learn: 0.5810032 total: 35ms remaining: 85.8ms
29: learn: 0.5784543 total: 36.5ms remaining: 85.2ms
30: learn: 0.5750472 total: 37.2ms remaining: 82.8ms
31: learn: 0.5717814 total: 38.4ms remaining: 81.7ms
32: learn: 0.5686437 total: 39.4ms remaining: 80ms
33: learn: 0.5653167 total: 40.6ms remaining: 78.7ms
34: learn: 0.5630180 total: 42.2ms remaining: 78.4ms
35: learn: 0.5603828 total: 43.6ms remaining: 77.5ms
36: learn: 0.5575529 total: 44.9ms remaining: 76.4ms
37: learn: 0.5553556 total: 46.2ms remaining: 75.4ms
38: learn: 0.5525472 total: 47.4ms remaining: 74.1ms
39: learn: 0.5495731 total: 48ms remaining: 72ms
40: learn: 0.5473859 total: 48.9ms remaining: 70.4ms
41: learn: 0.5444052 total: 50.1ms remaining: 69.2ms
42: learn: 0.5422100 total: 50.9ms remaining: 67.5ms
43: learn: 0.5393793 total: 51.9ms remaining: 66.1ms
44: learn: 0.5367392 total: 53ms remaining: 64.8ms
45: learn: 0.5339108 total: 54ms remaining: 63.3ms
46: learn: 0.5312177 total: 55ms remaining: 62.1ms
47: learn: 0.5287355 total: 55.7ms remaining: 60.4ms
48: learn: 0.5265024 total: 56.1ms remaining: 58.4ms
49: learn: 0.5235980 total: 57.1ms remaining: 57.1ms
50: learn: 0.5207756 total: 58.2ms remaining: 55.9ms
51: learn: 0.5177716 total: 59.2ms remaining: 54.6ms
52: learn: 0.5154983 total: 60.1ms remaining: 53.3ms
53: learn: 0.5126700 total: 60.7ms remaining: 51.7ms
54: learn: 0.5101043 total: 61.2ms remaining: 50.1ms
55: learn: 0.5078914 total: 62.2ms remaining: 48.9ms
56: learn: 0.5052694 total: 63.3ms remaining: 47.7ms
57: learn: 0.5027824 total: 64ms remaining: 46.4ms
58: learn: 0.5004041 total: 64.9ms remaining: 45.1ms
59: learn: 0.4979322 total: 65.7ms remaining: 43.8ms
60: learn: 0.4954362 total: 66.4ms remaining: 42.5ms
61: learn: 0.4929859 total: 67ms remaining: 41.1ms
62: learn: 0.4911638 total: 67.7ms remaining: 39.8ms
63: learn: 0.4890122 total: 68.6ms remaining: 38.6ms
64: learn: 0.4866211 total: 69.4ms remaining: 37.4ms
65: learn: 0.4845082 total: 70ms remaining: 36.1ms
66: learn: 0.4821234 total: 70.8ms remaining: 34.9ms
67: learn: 0.4798660 total: 71.2ms remaining: 33.5ms
68: learn: 0.4779862 total: 72.1ms remaining: 32.4ms
69: learn: 0.4762877 total: 73ms remaining: 31.3ms
70: learn: 0.4741045 total: 73.3ms remaining: 29.9ms
71: learn: 0.4722938 total: 74.1ms remaining: 28.8ms
72: learn: 0.4700146 total: 74.7ms remaining: 27.6ms
73: learn: 0.4680951 total: 75.4ms remaining: 26.5ms
74: learn: 0.4659906 total: 76ms remaining: 25.3ms
75: learn: 0.4640971 total: 76.6ms remaining: 24.2ms
76: learn: 0.4620922 total: 77.3ms remaining: 23.1ms
77: learn: 0.4600803 total: 77.8ms remaining: 21.9ms
78: learn: 0.4586681 total: 78.3ms remaining: 20.8ms
79: learn: 0.4566174 total: 78.8ms remaining: 19.7ms
80: learn: 0.4545173 total: 79.3ms remaining: 18.6ms
81: learn: 0.4524986 total: 79.7ms remaining: 17.5ms
82: learn: 0.4505864 total: 80.2ms remaining: 16.4ms
83: learn: 0.4489424 total: 80.6ms remaining: 15.4ms
84: learn: 0.4474078 total: 81.1ms remaining: 14.3ms
85: learn: 0.4460705 total: 81.6ms remaining: 13.3ms
86: learn: 0.4445821 total: 82ms remaining: 12.3ms
87: learn: 0.4428882 total: 82.5ms remaining: 11.2ms
88: learn: 0.4411782 total: 83ms remaining: 10.3ms
89: learn: 0.4392787 total: 83.5ms remaining: 9.28ms
90: learn: 0.4375469 total: 84ms remaining: 8.31ms
91: learn: 0.4358815 total: 84.5ms remaining: 7.35ms
92: learn: 0.4342353 total: 84.9ms remaining: 6.39ms
93: learn: 0.4325697 total: 85.3ms remaining: 5.45ms
94: learn: 0.4309381 total: 86ms remaining: 4.52ms
95: learn: 0.4293767 total: 86.4ms remaining: 3.6ms
96: learn: 0.4278734 total: 86.9ms remaining: 2.69ms
97: learn: 0.4261342 total: 87.3ms remaining: 1.78ms
98: learn: 0.4245995 total: 87.7ms remaining: 886us
99: learn: 0.4228280 total: 88.1ms remaining: 0us
0: learn: 0.6895315 total: 1.28ms remaining: 127ms
1: learn: 0.6844180 total: 2.46ms remaining: 121ms
2: learn: 0.6797461 total: 3.48ms remaining: 113ms
3: learn: 0.6750861 total: 5.01ms remaining: 120ms
4: learn: 0.6707944 total: 6.62ms remaining: 126ms
5: learn: 0.6661817 total: 7.46ms remaining: 117ms
6: learn: 0.6616283 total: 8.72ms remaining: 116ms
7: learn: 0.6576498 total: 10.1ms remaining: 117ms
8: learn: 0.6531931 total: 11.2ms remaining: 113ms
9: learn: 0.6487036 total: 12.5ms remaining: 112ms
10: learn: 0.6449099 total: 13.3ms remaining: 107ms
11: learn: 0.6406385 total: 14.3ms remaining: 105ms
12: learn: 0.6364890 total: 15.6ms remaining: 105ms
13: learn: 0.6327426 total: 17.3ms remaining: 106ms
14: learn: 0.6293871 total: 18.5ms remaining: 105ms
15: learn: 0.6251434 total: 19.5ms remaining: 103ms
16: learn: 0.6214766 total: 20.7ms remaining: 101ms
17: learn: 0.6178709 total: 21.3ms remaining: 97.2ms
18: learn: 0.6143560 total: 21.6ms remaining: 92.1ms
19: learn: 0.6108690 total: 22.8ms remaining: 91.4ms
20: learn: 0.6073974 total: 24ms remaining: 90.4ms
21: learn: 0.6036627 total: 24.7ms remaining: 87.4ms
22: learn: 0.5997537 total: 25.3ms remaining: 84.6ms
23: learn: 0.5964076 total: 26.2ms remaining: 83.1ms
24: learn: 0.5934630 total: 27.3ms remaining: 81.9ms
25: learn: 0.5903772 total: 28ms remaining: 79.7ms
26: learn: 0.5873292 total: 28.6ms remaining: 77.4ms
27: learn: 0.5843486 total: 29.8ms remaining: 76.6ms
28: learn: 0.5815989 total: 31.9ms remaining: 78.1ms
29: learn: 0.5790148 total: 33.2ms remaining: 77.5ms
30: learn: 0.5764376 total: 34.4ms remaining: 76.6ms
31: learn: 0.5734280 total: 35.3ms remaining: 75ms
32: learn: 0.5703529 total: 36.4ms remaining: 73.9ms
33: learn: 0.5671999 total: 37.4ms remaining: 72.7ms
34: learn: 0.5637969 total: 38.5ms remaining: 71.5ms
35: learn: 0.5609537 total: 39.9ms remaining: 70.9ms
36: learn: 0.5583443 total: 41ms remaining: 69.7ms
37: learn: 0.5554906 total: 42.3ms remaining: 69ms
38: learn: 0.5523799 total: 43.4ms remaining: 67.8ms
39: learn: 0.5496638 total: 44.1ms remaining: 66.2ms
40: learn: 0.5467338 total: 44.8ms remaining: 64.5ms
41: learn: 0.5437414 total: 45.4ms remaining: 62.7ms
42: learn: 0.5410515 total: 46.5ms remaining: 61.6ms
43: learn: 0.5385706 total: 47.5ms remaining: 60.4ms
44: learn: 0.5359205 total: 48.6ms remaining: 59.4ms
45: learn: 0.5335821 total: 49ms remaining: 57.5ms
46: learn: 0.5311304 total: 49.5ms remaining: 55.8ms
47: learn: 0.5287105 total: 50.2ms remaining: 54.3ms
48: learn: 0.5262265 total: 50.9ms remaining: 52.9ms
49: learn: 0.5235670 total: 51.7ms remaining: 51.7ms
50: learn: 0.5209126 total: 52.4ms remaining: 50.4ms
51: learn: 0.5187759 total: 53.2ms remaining: 49.1ms
52: learn: 0.5164240 total: 53.7ms remaining: 47.6ms
53: learn: 0.5139137 total: 54.6ms remaining: 46.5ms
54: learn: 0.5115121 total: 55.4ms remaining: 45.3ms
55: learn: 0.5090840 total: 56.3ms remaining: 44.3ms
56: learn: 0.5065030 total: 57ms remaining: 43ms
57: learn: 0.5039316 total: 58ms remaining: 42ms
58: learn: 0.5014690 total: 58.5ms remaining: 40.7ms
59: learn: 0.4988399 total: 59.2ms remaining: 39.5ms
60: learn: 0.4965204 total: 59.8ms remaining: 38.2ms
61: learn: 0.4942442 total: 60.4ms remaining: 37ms
62: learn: 0.4920413 total: 60.9ms remaining: 35.8ms
63: learn: 0.4900332 total: 61.3ms remaining: 34.5ms
64: learn: 0.4879750 total: 61.8ms remaining: 33.3ms
65: learn: 0.4859368 total: 62.3ms remaining: 32.1ms
66: learn: 0.4841339 total: 62.7ms remaining: 30.9ms
67: learn: 0.4817868 total: 63.2ms remaining: 29.7ms
68: learn: 0.4798315 total: 63.7ms remaining: 28.6ms
69: learn: 0.4776966 total: 64.5ms remaining: 27.6ms
70: learn: 0.4754465 total: 65ms remaining: 26.6ms
71: learn: 0.4733520 total: 65.6ms remaining: 25.5ms
72: learn: 0.4714535 total: 66.2ms remaining: 24.5ms
73: learn: 0.4695847 total: 66.7ms remaining: 23.4ms
74: learn: 0.4679735 total: 67.1ms remaining: 22.4ms
75: learn: 0.4661721 total: 67.6ms remaining: 21.4ms
76: learn: 0.4641415 total: 68.2ms remaining: 20.4ms
77: learn: 0.4622109 total: 68.6ms remaining: 19.3ms
78: learn: 0.4602079 total: 69ms remaining: 18.3ms
79: learn: 0.4584789 total: 69.3ms remaining: 17.3ms
80: learn: 0.4564984 total: 69.7ms remaining: 16.4ms
81: learn: 0.4546838 total: 70.1ms remaining: 15.4ms
82: learn: 0.4528079 total: 70.5ms remaining: 14.4ms
83: learn: 0.4513652 total: 71ms remaining: 13.5ms
84: learn: 0.4497571 total: 71.5ms remaining: 12.6ms
85: learn: 0.4479051 total: 71.8ms remaining: 11.7ms
86: learn: 0.4458026 total: 72.1ms remaining: 10.8ms
87: learn: 0.4441070 total: 72.6ms remaining: 9.89ms
88: learn: 0.4425226 total: 73ms remaining: 9.03ms
89: learn: 0.4407601 total: 73.4ms remaining: 8.16ms
90: learn: 0.4391970 total: 73.8ms remaining: 7.3ms
91: learn: 0.4375876 total: 74.3ms remaining: 6.46ms
92: learn: 0.4359720 total: 74.7ms remaining: 5.62ms
93: learn: 0.4342904 total: 75.1ms remaining: 4.79ms
94: learn: 0.4329354 total: 75.6ms remaining: 3.98ms
95: learn: 0.4312499 total: 76ms remaining: 3.17ms
96: learn: 0.4294859 total: 76.4ms remaining: 2.36ms
97: learn: 0.4281853 total: 76.7ms remaining: 1.57ms
98: learn: 0.4265789 total: 77.3ms remaining: 780us
99: learn: 0.4249783 total: 77.7ms remaining: 0us
0: learn: 0.6884251 total: 1.36ms remaining: 135ms
1: learn: 0.6837882 total: 2.38ms remaining: 116ms
2: learn: 0.6788893 total: 3.53ms remaining: 114ms
3: learn: 0.6742531 total: 4.37ms remaining: 105ms
4: learn: 0.6694941 total: 5.43ms remaining: 103ms
5: learn: 0.6644974 total: 6.68ms remaining: 105ms
6: learn: 0.6602396 total: 8.12ms remaining: 108ms
7: learn: 0.6546575 total: 9.43ms remaining: 108ms
8: learn: 0.6495477 total: 11.1ms remaining: 113ms
9: learn: 0.6460980 total: 12ms remaining: 108ms
10: learn: 0.6407818 total: 13ms remaining: 105ms
11: learn: 0.6358886 total: 14.1ms remaining: 103ms
12: learn: 0.6319770 total: 15.7ms remaining: 105ms
13: learn: 0.6280995 total: 16.8ms remaining: 103ms
14: learn: 0.6233202 total: 17.7ms remaining: 100ms
15: learn: 0.6192099 total: 18.9ms remaining: 99.2ms
16: learn: 0.6147762 total: 20.3ms remaining: 99.3ms
17: learn: 0.6103238 total: 21.1ms remaining: 96.2ms
18: learn: 0.6060995 total: 22.4ms remaining: 95.6ms
19: learn: 0.6020556 total: 23.8ms remaining: 95.1ms
20: learn: 0.5977037 total: 25.4ms remaining: 95.6ms
21: learn: 0.5931581 total: 26.3ms remaining: 93.4ms
22: learn: 0.5890359 total: 27.2ms remaining: 91.1ms
23: learn: 0.5846377 total: 28.2ms remaining: 89.4ms
24: learn: 0.5814286 total: 29.6ms remaining: 88.7ms
25: learn: 0.5780560 total: 30.8ms remaining: 87.7ms
26: learn: 0.5739783 total: 31.8ms remaining: 85.9ms
27: learn: 0.5709059 total: 32.8ms remaining: 84.4ms
28: learn: 0.5681418 total: 33.8ms remaining: 82.7ms
29: learn: 0.5650585 total: 35ms remaining: 81.7ms
30: learn: 0.5612944 total: 35.7ms remaining: 79.4ms
31: learn: 0.5579837 total: 36.9ms remaining: 78.4ms
32: learn: 0.5545437 total: 37.6ms remaining: 76.3ms
33: learn: 0.5508624 total: 38.4ms remaining: 74.6ms
34: learn: 0.5476947 total: 39.4ms remaining: 73.2ms
35: learn: 0.5443277 total: 41ms remaining: 72.9ms
36: learn: 0.5410425 total: 41.8ms remaining: 71.2ms
37: learn: 0.5381051 total: 42.7ms remaining: 69.7ms
38: learn: 0.5349005 total: 44ms remaining: 68.8ms
39: learn: 0.5320597 total: 46ms remaining: 69ms
40: learn: 0.5288238 total: 47ms remaining: 67.7ms
41: learn: 0.5252659 total: 48ms remaining: 66.3ms
42: learn: 0.5227562 total: 49.1ms remaining: 65.1ms
43: learn: 0.5199700 total: 49.9ms remaining: 63.5ms
44: learn: 0.5171837 total: 50.9ms remaining: 62.2ms
45: learn: 0.5143990 total: 51.6ms remaining: 60.5ms
46: learn: 0.5116150 total: 52.4ms remaining: 59.1ms
47: learn: 0.5085206 total: 53.6ms remaining: 58ms
48: learn: 0.5052346 total: 54.3ms remaining: 56.5ms
49: learn: 0.5021518 total: 55.2ms remaining: 55.2ms
50: learn: 0.4995699 total: 55.7ms remaining: 53.5ms
51: learn: 0.4966323 total: 56.6ms remaining: 52.2ms
52: learn: 0.4942124 total: 57.3ms remaining: 50.8ms
53: learn: 0.4918257 total: 58ms remaining: 49.4ms
54: learn: 0.4890696 total: 58.5ms remaining: 47.9ms
55: learn: 0.4870031 total: 59.3ms remaining: 46.6ms
56: learn: 0.4843219 total: 59.9ms remaining: 45.2ms
57: learn: 0.4816078 total: 60.5ms remaining: 43.8ms
58: learn: 0.4785999 total: 61.1ms remaining: 42.5ms
59: learn: 0.4760945 total: 61.8ms remaining: 41.2ms
60: learn: 0.4733829 total: 62.1ms remaining: 39.7ms
61: learn: 0.4708449 total: 62.7ms remaining: 38.4ms
62: learn: 0.4687759 total: 63.1ms remaining: 37.1ms
63: learn: 0.4666086 total: 63.8ms remaining: 35.9ms
64: learn: 0.4640723 total: 64.4ms remaining: 34.7ms
65: learn: 0.4615086 total: 64.9ms remaining: 33.4ms
66: learn: 0.4593938 total: 65.7ms remaining: 32.3ms
67: learn: 0.4573500 total: 66.4ms remaining: 31.2ms
68: learn: 0.4549034 total: 66.9ms remaining: 30.1ms
69: learn: 0.4531617 total: 67.4ms remaining: 28.9ms
70: learn: 0.4508314 total: 67.9ms remaining: 27.7ms
71: learn: 0.4484736 total: 68.5ms remaining: 26.6ms
72: learn: 0.4462847 total: 68.8ms remaining: 25.5ms
73: learn: 0.4440720 total: 69.3ms remaining: 24.4ms
74: learn: 0.4418016 total: 70ms remaining: 23.3ms
75: learn: 0.4398148 total: 70.7ms remaining: 22.3ms
76: learn: 0.4376869 total: 71.1ms remaining: 21.3ms
77: learn: 0.4353386 total: 71.7ms remaining: 20.2ms
78: learn: 0.4331077 total: 72.1ms remaining: 19.2ms
79: learn: 0.4309541 total: 72.4ms remaining: 18.1ms
80: learn: 0.4292700 total: 72.9ms remaining: 17.1ms
81: learn: 0.4273715 total: 73.4ms remaining: 16.1ms
82: learn: 0.4253411 total: 73.8ms remaining: 15.1ms
83: learn: 0.4232837 total: 74.1ms remaining: 14.1ms
84: learn: 0.4217062 total: 74.6ms remaining: 13.2ms
85: learn: 0.4202087 total: 75ms remaining: 12.2ms
86: learn: 0.4181963 total: 75.4ms remaining: 11.3ms
87: learn: 0.4162303 total: 75.8ms remaining: 10.3ms
88: learn: 0.4145317 total: 76.3ms remaining: 9.43ms
89: learn: 0.4126805 total: 76.7ms remaining: 8.52ms
90: learn: 0.4109249 total: 77.1ms remaining: 7.62ms
91: learn: 0.4090370 total: 77.5ms remaining: 6.74ms
92: learn: 0.4075740 total: 77.9ms remaining: 5.86ms
93: learn: 0.4056473 total: 78.2ms remaining: 4.99ms
94: learn: 0.4043221 total: 78.5ms remaining: 4.13ms
95: learn: 0.4023332 total: 79.1ms remaining: 3.29ms
96: learn: 0.4003395 total: 79.5ms remaining: 2.46ms
97: learn: 0.3984095 total: 79.7ms remaining: 1.63ms
98: learn: 0.3966427 total: 80.1ms remaining: 808us
99: learn: 0.3947914 total: 80.5ms remaining: 0us
0: learn: 0.6878984 total: 1.26ms remaining: 125ms
1: learn: 0.6825637 total: 1.69ms remaining: 82.9ms
2: learn: 0.6778136 total: 2.01ms remaining: 65ms
3: learn: 0.6730922 total: 2.42ms remaining: 58.1ms
4: learn: 0.6683979 total: 2.72ms remaining: 51.7ms
5: learn: 0.6639298 total: 3.21ms remaining: 50.3ms
6: learn: 0.6588845 total: 3.54ms remaining: 47ms
7: learn: 0.6547090 total: 3.87ms remaining: 44.4ms
8: learn: 0.6501215 total: 4.16ms remaining: 42ms
9: learn: 0.6455656 total: 4.72ms remaining: 42.5ms
10: learn: 0.6411502 total: 5.03ms remaining: 40.7ms
11: learn: 0.6367748 total: 5.33ms remaining: 39.1ms
12: learn: 0.6324957 total: 5.71ms remaining: 38.2ms
13: learn: 0.6282916 total: 6.31ms remaining: 38.8ms
14: learn: 0.6244643 total: 6.71ms remaining: 38ms
15: learn: 0.6197827 total: 7.02ms remaining: 36.9ms
16: learn: 0.6162918 total: 7.39ms remaining: 36.1ms
17: learn: 0.6117805 total: 8.46ms remaining: 38.5ms
18: learn: 0.6076094 total: 9.01ms remaining: 38.4ms
19: learn: 0.6039850 total: 9.98ms remaining: 39.9ms
20: learn: 0.6001150 total: 11.3ms remaining: 42.6ms
21: learn: 0.5961289 total: 13ms remaining: 46ms
22: learn: 0.5919174 total: 13.9ms remaining: 46.6ms
23: learn: 0.5880261 total: 14.8ms remaining: 46.9ms
24: learn: 0.5845686 total: 16.2ms remaining: 48.6ms
25: learn: 0.5814502 total: 17.9ms remaining: 51ms
26: learn: 0.5783653 total: 19.5ms remaining: 52.7ms
27: learn: 0.5749444 total: 21ms remaining: 54ms
28: learn: 0.5717356 total: 22ms remaining: 54ms
29: learn: 0.5686947 total: 23.4ms remaining: 54.6ms
30: learn: 0.5658574 total: 24.7ms remaining: 55ms
31: learn: 0.5627563 total: 25.3ms remaining: 53.8ms
32: learn: 0.5594251 total: 26.6ms remaining: 54ms
33: learn: 0.5561764 total: 27.9ms remaining: 54.2ms
34: learn: 0.5529952 total: 29.2ms remaining: 54.3ms
35: learn: 0.5508336 total: 30.1ms remaining: 53.5ms
36: learn: 0.5478067 total: 31.3ms remaining: 53.4ms
37: learn: 0.5449646 total: 32.7ms remaining: 53.3ms
38: learn: 0.5420459 total: 33.9ms remaining: 53ms
39: learn: 0.5388165 total: 35.2ms remaining: 52.8ms
40: learn: 0.5358141 total: 36.2ms remaining: 52.1ms
41: learn: 0.5331245 total: 37.8ms remaining: 52.2ms
42: learn: 0.5300696 total: 38.6ms remaining: 51.2ms
43: learn: 0.5273135 total: 39.6ms remaining: 50.5ms
44: learn: 0.5247408 total: 40.7ms remaining: 49.8ms
45: learn: 0.5220816 total: 42.1ms remaining: 49.4ms
46: learn: 0.5198094 total: 43.1ms remaining: 48.6ms
47: learn: 0.5168347 total: 44.4ms remaining: 48.1ms
48: learn: 0.5144139 total: 45.7ms remaining: 47.6ms
49: learn: 0.5116065 total: 46.6ms remaining: 46.6ms
50: learn: 0.5090462 total: 47.6ms remaining: 45.8ms
51: learn: 0.5064657 total: 48.8ms remaining: 45ms
52: learn: 0.5045712 total: 50.2ms remaining: 44.5ms
53: learn: 0.5023182 total: 51.3ms remaining: 43.7ms
54: learn: 0.4998693 total: 52.6ms remaining: 43ms
55: learn: 0.4978502 total: 53.6ms remaining: 42.1ms
56: learn: 0.4956555 total: 54.6ms remaining: 41.2ms
57: learn: 0.4933164 total: 56ms remaining: 40.5ms
58: learn: 0.4907979 total: 57ms remaining: 39.6ms
59: learn: 0.4880570 total: 58.4ms remaining: 38.9ms
60: learn: 0.4857546 total: 59.7ms remaining: 38.2ms
61: learn: 0.4835209 total: 60.8ms remaining: 37.3ms
62: learn: 0.4807113 total: 61.8ms remaining: 36.3ms
63: learn: 0.4785444 total: 63ms remaining: 35.4ms
64: learn: 0.4762250 total: 64.2ms remaining: 34.6ms
65: learn: 0.4739229 total: 65.5ms remaining: 33.7ms
66: learn: 0.4715654 total: 66.3ms remaining: 32.6ms
67: learn: 0.4690922 total: 67.4ms remaining: 31.7ms
68: learn: 0.4664602 total: 68.9ms remaining: 30.9ms
69: learn: 0.4642567 total: 70.1ms remaining: 30ms
70: learn: 0.4621358 total: 71ms remaining: 29ms
71: learn: 0.4598338 total: 72.2ms remaining: 28.1ms
72: learn: 0.4581320 total: 73.1ms remaining: 27ms
73: learn: 0.4560829 total: 73.9ms remaining: 26ms
74: learn: 0.4539861 total: 75.1ms remaining: 25ms
75: learn: 0.4522336 total: 76ms remaining: 24ms
76: learn: 0.4502632 total: 77.3ms remaining: 23.1ms
77: learn: 0.4484178 total: 78.6ms remaining: 22.2ms
78: learn: 0.4465482 total: 79.8ms remaining: 21.2ms
79: learn: 0.4445991 total: 80.8ms remaining: 20.2ms
80: learn: 0.4427816 total: 81.4ms remaining: 19.1ms
81: learn: 0.4407815 total: 81.7ms remaining: 17.9ms
82: learn: 0.4389686 total: 82.7ms remaining: 16.9ms
83: learn: 0.4371507 total: 83.8ms remaining: 16ms
84: learn: 0.4352346 total: 84.6ms remaining: 14.9ms
85: learn: 0.4339660 total: 85.8ms remaining: 14ms
86: learn: 0.4323103 total: 86.9ms remaining: 13ms
87: learn: 0.4302934 total: 88.1ms remaining: 12ms
88: learn: 0.4281793 total: 88.9ms remaining: 11ms
89: learn: 0.4264841 total: 90ms remaining: 9.99ms
90: learn: 0.4245832 total: 91.1ms remaining: 9.01ms
91: learn: 0.4228376 total: 92.2ms remaining: 8.02ms
92: learn: 0.4210649 total: 92.9ms remaining: 7ms
93: learn: 0.4194267 total: 93.7ms remaining: 5.98ms
94: learn: 0.4178059 total: 94.6ms remaining: 4.98ms
95: learn: 0.4160512 total: 95.6ms remaining: 3.98ms
96: learn: 0.4147059 total: 96.4ms remaining: 2.98ms
97: learn: 0.4131006 total: 96.8ms remaining: 1.97ms
98: learn: 0.4116573 total: 97.5ms remaining: 985us
99: learn: 0.4098773 total: 98.4ms remaining: 0us
0: learn: 0.6882230 total: 1.28ms remaining: 127ms
1: learn: 0.6830130 total: 2.04ms remaining: 100ms
2: learn: 0.6777502 total: 2.4ms remaining: 77.7ms
3: learn: 0.6725869 total: 3.69ms remaining: 88.6ms
4: learn: 0.6678779 total: 4.92ms remaining: 93.5ms
5: learn: 0.6627342 total: 6.22ms remaining: 97.4ms
6: learn: 0.6578700 total: 7.47ms remaining: 99.3ms
7: learn: 0.6539456 total: 8.51ms remaining: 97.9ms
8: learn: 0.6489295 total: 9.4ms remaining: 95ms
9: learn: 0.6447199 total: 10.7ms remaining: 96.2ms
10: learn: 0.6410565 total: 11.7ms remaining: 95ms
11: learn: 0.6370334 total: 12.9ms remaining: 94.7ms
12: learn: 0.6323019 total: 14ms remaining: 93.6ms
13: learn: 0.6278074 total: 14.8ms remaining: 91ms
14: learn: 0.6244961 total: 15.6ms remaining: 88.6ms
15: learn: 0.6205365 total: 17ms remaining: 89.4ms
16: learn: 0.6164157 total: 18.5ms remaining: 90.2ms
17: learn: 0.6125643 total: 19.5ms remaining: 88.8ms
18: learn: 0.6087596 total: 20.6ms remaining: 87.7ms
19: learn: 0.6051407 total: 21.8ms remaining: 87.2ms
20: learn: 0.6012506 total: 23ms remaining: 86.7ms
21: learn: 0.5969953 total: 23.9ms remaining: 84.7ms
22: learn: 0.5926725 total: 25ms remaining: 83.6ms
23: learn: 0.5892457 total: 25.7ms remaining: 81.4ms
24: learn: 0.5861760 total: 26.9ms remaining: 80.7ms
25: learn: 0.5827887 total: 28.2ms remaining: 80.3ms
26: learn: 0.5792932 total: 29.3ms remaining: 79.3ms
27: learn: 0.5751741 total: 30.4ms remaining: 78.1ms
28: learn: 0.5722502 total: 31.2ms remaining: 76.5ms
29: learn: 0.5696262 total: 31.6ms remaining: 73.7ms
30: learn: 0.5670091 total: 32.5ms remaining: 72.4ms
31: learn: 0.5635108 total: 33.1ms remaining: 70.3ms
32: learn: 0.5600613 total: 33.8ms remaining: 68.6ms
33: learn: 0.5568582 total: 34.1ms remaining: 66.2ms
34: learn: 0.5533880 total: 35ms remaining: 65ms
35: learn: 0.5503392 total: 35.8ms remaining: 63.6ms
36: learn: 0.5472394 total: 36.4ms remaining: 62ms
37: learn: 0.5443595 total: 37.1ms remaining: 60.6ms
38: learn: 0.5412247 total: 38ms remaining: 59.4ms
39: learn: 0.5382340 total: 38.7ms remaining: 58ms
40: learn: 0.5349641 total: 39.8ms remaining: 57.2ms
41: learn: 0.5321799 total: 40.6ms remaining: 56.1ms
42: learn: 0.5289777 total: 41.3ms remaining: 54.8ms
43: learn: 0.5259768 total: 42.1ms remaining: 53.6ms
44: learn: 0.5230374 total: 42.9ms remaining: 52.4ms
45: learn: 0.5207025 total: 43.4ms remaining: 51ms
46: learn: 0.5181586 total: 44.2ms remaining: 49.8ms
47: learn: 0.5151432 total: 44.8ms remaining: 48.6ms
48: learn: 0.5125814 total: 45.5ms remaining: 47.3ms
49: learn: 0.5098965 total: 46.2ms remaining: 46.2ms
50: learn: 0.5069506 total: 46.9ms remaining: 45.1ms
51: learn: 0.5041681 total: 47.7ms remaining: 44ms
52: learn: 0.5016870 total: 48.5ms remaining: 43ms
53: learn: 0.4994037 total: 49.1ms remaining: 41.8ms
54: learn: 0.4966429 total: 49.8ms remaining: 40.7ms
55: learn: 0.4946069 total: 50.5ms remaining: 39.6ms
56: learn: 0.4919759 total: 51.2ms remaining: 38.6ms
57: learn: 0.4890635 total: 51.6ms remaining: 37.4ms
58: learn: 0.4867096 total: 52.4ms remaining: 36.4ms
59: learn: 0.4841801 total: 53ms remaining: 35.4ms
60: learn: 0.4816752 total: 53.8ms remaining: 34.4ms
61: learn: 0.4793108 total: 54.5ms remaining: 33.4ms
62: learn: 0.4767157 total: 55ms remaining: 32.3ms
63: learn: 0.4742852 total: 55.7ms remaining: 31.3ms
64: learn: 0.4723054 total: 56.1ms remaining: 30.2ms
65: learn: 0.4700578 total: 56.7ms remaining: 29.2ms
66: learn: 0.4679812 total: 57ms remaining: 28.1ms
67: learn: 0.4653752 total: 57.6ms remaining: 27.1ms
68: learn: 0.4629021 total: 58.1ms remaining: 26.1ms
69: learn: 0.4608863 total: 58.6ms remaining: 25.1ms
70: learn: 0.4589026 total: 59.1ms remaining: 24.2ms
71: learn: 0.4564408 total: 59.7ms remaining: 23.2ms
72: learn: 0.4540118 total: 60.2ms remaining: 22.3ms
73: learn: 0.4517610 total: 60.8ms remaining: 21.4ms
74: learn: 0.4497752 total: 61.3ms remaining: 20.4ms
75: learn: 0.4478640 total: 61.8ms remaining: 19.5ms
76: learn: 0.4457896 total: 62.4ms remaining: 18.6ms
77: learn: 0.4437100 total: 62.9ms remaining: 17.8ms
78: learn: 0.4415177 total: 63.4ms remaining: 16.9ms
79: learn: 0.4393996 total: 63.9ms remaining: 16ms
80: learn: 0.4373214 total: 64.4ms remaining: 15.1ms
81: learn: 0.4352059 total: 64.9ms remaining: 14.3ms
82: learn: 0.4336374 total: 65.2ms remaining: 13.4ms
83: learn: 0.4320289 total: 65.7ms remaining: 12.5ms
84: learn: 0.4300997 total: 66.3ms remaining: 11.7ms
85: learn: 0.4281833 total: 66.7ms remaining: 10.9ms
86: learn: 0.4263781 total: 67.1ms remaining: 10ms
87: learn: 0.4250742 total: 67.5ms remaining: 9.21ms
88: learn: 0.4231440 total: 68.1ms remaining: 8.42ms
89: learn: 0.4210817 total: 68.5ms remaining: 7.62ms
90: learn: 0.4192910 total: 68.9ms remaining: 6.81ms
91: learn: 0.4175057 total: 69.3ms remaining: 6.02ms
92: learn: 0.4157031 total: 69.9ms remaining: 5.26ms
93: learn: 0.4137212 total: 70.3ms remaining: 4.49ms
94: learn: 0.4120097 total: 70.7ms remaining: 3.72ms
95: learn: 0.4104197 total: 71.2ms remaining: 2.97ms
96: learn: 0.4086656 total: 71.6ms remaining: 2.21ms
97: learn: 0.4068281 total: 72ms remaining: 1.47ms
98: learn: 0.4053149 total: 72.4ms remaining: 731us
99: learn: 0.4035909 total: 72.9ms remaining: 0us
0: learn: 0.6886159 total: 866us remaining: 85.8ms
1: learn: 0.6833054 total: 2.1ms remaining: 103ms
2: learn: 0.6784831 total: 3.21ms remaining: 104ms
3: learn: 0.6744239 total: 4.92ms remaining: 118ms
4: learn: 0.6703678 total: 5.83ms remaining: 111ms
5: learn: 0.6657698 total: 7.09ms remaining: 111ms
6: learn: 0.6613446 total: 8.14ms remaining: 108ms
7: learn: 0.6575104 total: 9.07ms remaining: 104ms
8: learn: 0.6529390 total: 9.9ms remaining: 100ms
9: learn: 0.6485341 total: 11.1ms remaining: 99.5ms
10: learn: 0.6445752 total: 12ms remaining: 97.5ms
11: learn: 0.6403021 total: 12.7ms remaining: 93.2ms
12: learn: 0.6365289 total: 13.6ms remaining: 91ms
13: learn: 0.6324526 total: 14.9ms remaining: 91.8ms
14: learn: 0.6290089 total: 16.5ms remaining: 93.4ms
15: learn: 0.6245414 total: 17.6ms remaining: 92.4ms
16: learn: 0.6210188 total: 19.4ms remaining: 94.8ms
17: learn: 0.6174320 total: 20.8ms remaining: 94.7ms
18: learn: 0.6140296 total: 21.4ms remaining: 91.1ms
19: learn: 0.6105599 total: 22.5ms remaining: 90ms
20: learn: 0.6069129 total: 23.6ms remaining: 88.9ms
21: learn: 0.6030373 total: 24.6ms remaining: 87.2ms
22: learn: 0.5995989 total: 25.8ms remaining: 86.4ms
23: learn: 0.5963684 total: 27.1ms remaining: 85.8ms
24: learn: 0.5922892 total: 27.9ms remaining: 83.8ms
25: learn: 0.5894788 total: 29.2ms remaining: 83.1ms
26: learn: 0.5863141 total: 30.1ms remaining: 81.3ms
27: learn: 0.5833325 total: 31.1ms remaining: 80.1ms
28: learn: 0.5804530 total: 32.2ms remaining: 78.8ms
29: learn: 0.5778179 total: 33.1ms remaining: 77.3ms
30: learn: 0.5753587 total: 34ms remaining: 75.8ms
31: learn: 0.5724407 total: 34.9ms remaining: 74.1ms
32: learn: 0.5690258 total: 35.9ms remaining: 72.8ms
33: learn: 0.5657888 total: 37ms remaining: 71.9ms
34: learn: 0.5621902 total: 37.6ms remaining: 69.8ms
35: learn: 0.5593933 total: 38.4ms remaining: 68.3ms
36: learn: 0.5564423 total: 39.6ms remaining: 67.5ms
37: learn: 0.5537762 total: 40.8ms remaining: 66.6ms
38: learn: 0.5504946 total: 42.1ms remaining: 65.9ms
39: learn: 0.5481516 total: 43.5ms remaining: 65.2ms
40: learn: 0.5452933 total: 44.7ms remaining: 64.4ms
41: learn: 0.5425361 total: 45.5ms remaining: 62.8ms
42: learn: 0.5398155 total: 46.3ms remaining: 61.3ms
43: learn: 0.5365296 total: 47.3ms remaining: 60.2ms
44: learn: 0.5338784 total: 48.3ms remaining: 59.1ms
45: learn: 0.5312372 total: 49.4ms remaining: 58ms
46: learn: 0.5285292 total: 50.3ms remaining: 56.7ms
47: learn: 0.5257266 total: 51ms remaining: 55.2ms
48: learn: 0.5232061 total: 51.7ms remaining: 53.8ms
49: learn: 0.5203838 total: 53.1ms remaining: 53.1ms
50: learn: 0.5178191 total: 54.2ms remaining: 52.1ms
51: learn: 0.5153748 total: 55.2ms remaining: 51ms
52: learn: 0.5132619 total: 56.1ms remaining: 49.7ms
53: learn: 0.5111186 total: 56.7ms remaining: 48.3ms
54: learn: 0.5089255 total: 57.5ms remaining: 47.1ms
55: learn: 0.5064440 total: 58.4ms remaining: 45.9ms
56: learn: 0.5038217 total: 59ms remaining: 44.5ms
57: learn: 0.5015302 total: 59.7ms remaining: 43.2ms
58: learn: 0.4997467 total: 60.6ms remaining: 42.1ms
59: learn: 0.4974990 total: 61.7ms remaining: 41.1ms
60: learn: 0.4953613 total: 62.4ms remaining: 39.9ms
61: learn: 0.4926778 total: 62.9ms remaining: 38.6ms
62: learn: 0.4903292 total: 63.5ms remaining: 37.3ms
63: learn: 0.4884905 total: 64.3ms remaining: 36.2ms
64: learn: 0.4859898 total: 65.1ms remaining: 35.1ms
65: learn: 0.4837754 total: 66ms remaining: 34ms
66: learn: 0.4818255 total: 66.8ms remaining: 32.9ms
67: learn: 0.4798123 total: 67.5ms remaining: 31.8ms
68: learn: 0.4778911 total: 68.3ms remaining: 30.7ms
69: learn: 0.4760780 total: 69ms remaining: 29.6ms
70: learn: 0.4738256 total: 69.6ms remaining: 28.4ms
71: learn: 0.4720699 total: 70.1ms remaining: 27.3ms
72: learn: 0.4699009 total: 70.7ms remaining: 26.2ms
73: learn: 0.4679986 total: 71.5ms remaining: 25.1ms
74: learn: 0.4660636 total: 72.1ms remaining: 24ms
75: learn: 0.4639535 total: 72.9ms remaining: 23ms
76: learn: 0.4622288 total: 73.8ms remaining: 22ms
77: learn: 0.4603284 total: 74.3ms remaining: 21ms
78: learn: 0.4585539 total: 74.9ms remaining: 19.9ms
79: learn: 0.4570284 total: 75.2ms remaining: 18.8ms
80: learn: 0.4549886 total: 75.7ms remaining: 17.8ms
81: learn: 0.4530752 total: 76.2ms remaining: 16.7ms
82: learn: 0.4514332 total: 76.8ms remaining: 15.7ms
83: learn: 0.4498867 total: 77.4ms remaining: 14.7ms
84: learn: 0.4478527 total: 77.8ms remaining: 13.7ms
85: learn: 0.4457519 total: 78.3ms remaining: 12.8ms
86: learn: 0.4439592 total: 78.7ms remaining: 11.8ms
87: learn: 0.4423116 total: 79.4ms remaining: 10.8ms
88: learn: 0.4404342 total: 79.8ms remaining: 9.86ms
89: learn: 0.4388356 total: 80.1ms remaining: 8.9ms
90: learn: 0.4372264 total: 80.6ms remaining: 7.97ms
91: learn: 0.4357569 total: 81.1ms remaining: 7.05ms
92: learn: 0.4341066 total: 81.5ms remaining: 6.14ms
93: learn: 0.4328156 total: 82.1ms remaining: 5.24ms
94: learn: 0.4311393 total: 82.5ms remaining: 4.34ms
95: learn: 0.4296042 total: 82.9ms remaining: 3.45ms
96: learn: 0.4279636 total: 83.3ms remaining: 2.58ms
97: learn: 0.4267022 total: 83.8ms remaining: 1.71ms
98: learn: 0.4249715 total: 84.2ms remaining: 850us
99: learn: 0.4236659 total: 84.5ms remaining: 0us
Evaluating param combo 11/16: {'boosted_cb_model__iterations': 100, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 6, 'boosted_cb_model__num_leaves': 8}
Duplicate found for this combo (hash=3f34ed272e80fbe03fd44105e587bcef). Removing old run: a9ae36d1081444369b42179b44b2b3d2
0: learn: 0.6882547 total: 2ms remaining: 198ms
1: learn: 0.6826413 total: 3.42ms remaining: 168ms
2: learn: 0.6787574 total: 5.09ms remaining: 165ms
3: learn: 0.6737900 total: 6.84ms remaining: 164ms
4: learn: 0.6686791 total: 8.69ms remaining: 165ms
5: learn: 0.6643458 total: 10.2ms remaining: 160ms
6: learn: 0.6596252 total: 12.3ms remaining: 164ms
7: learn: 0.6554987 total: 13.5ms remaining: 155ms
8: learn: 0.6509583 total: 15.6ms remaining: 157ms
9: learn: 0.6469695 total: 17.1ms remaining: 154ms
10: learn: 0.6423582 total: 18.7ms remaining: 152ms
11: learn: 0.6374666 total: 20.4ms remaining: 150ms
12: learn: 0.6326421 total: 21.2ms remaining: 142ms
13: learn: 0.6295545 total: 23.1ms remaining: 142ms
14: learn: 0.6256949 total: 25.2ms remaining: 143ms
15: learn: 0.6213727 total: 26.8ms remaining: 141ms
16: learn: 0.6171218 total: 28.4ms remaining: 138ms
17: learn: 0.6139615 total: 30.1ms remaining: 137ms
18: learn: 0.6105782 total: 32ms remaining: 136ms
19: learn: 0.6064788 total: 33.8ms remaining: 135ms
20: learn: 0.6038663 total: 35.6ms remaining: 134ms
21: learn: 0.6003718 total: 37.2ms remaining: 132ms
22: learn: 0.5963038 total: 38.2ms remaining: 128ms
23: learn: 0.5942131 total: 40.4ms remaining: 128ms
24: learn: 0.5901593 total: 42.1ms remaining: 126ms
25: learn: 0.5866114 total: 44ms remaining: 125ms
26: learn: 0.5830859 total: 45ms remaining: 122ms
27: learn: 0.5797169 total: 46.6ms remaining: 120ms
28: learn: 0.5765771 total: 48.5ms remaining: 119ms
29: learn: 0.5731998 total: 49.3ms remaining: 115ms
30: learn: 0.5698716 total: 51.2ms remaining: 114ms
31: learn: 0.5662749 total: 52.9ms remaining: 113ms
32: learn: 0.5633926 total: 54.5ms remaining: 111ms
33: learn: 0.5596447 total: 56.2ms remaining: 109ms
34: learn: 0.5563727 total: 57.8ms remaining: 107ms
35: learn: 0.5536910 total: 60.6ms remaining: 108ms
36: learn: 0.5511013 total: 62.8ms remaining: 107ms
37: learn: 0.5473601 total: 64.5ms remaining: 105ms
38: learn: 0.5440328 total: 66ms remaining: 103ms
39: learn: 0.5413512 total: 67.3ms remaining: 101ms
40: learn: 0.5382235 total: 68.5ms remaining: 98.6ms
41: learn: 0.5355798 total: 69.5ms remaining: 96ms
42: learn: 0.5320209 total: 70.7ms remaining: 93.7ms
43: learn: 0.5292923 total: 71.9ms remaining: 91.5ms
44: learn: 0.5264982 total: 72.3ms remaining: 88.4ms
45: learn: 0.5236175 total: 73.5ms remaining: 86.3ms
46: learn: 0.5205947 total: 74.4ms remaining: 83.9ms
47: learn: 0.5178331 total: 75.5ms remaining: 81.8ms
48: learn: 0.5149838 total: 76.4ms remaining: 79.5ms
49: learn: 0.5120349 total: 77.2ms remaining: 77.2ms
50: learn: 0.5094310 total: 78ms remaining: 75ms
51: learn: 0.5063628 total: 78.8ms remaining: 72.7ms
52: learn: 0.5041036 total: 79.8ms remaining: 70.8ms
53: learn: 0.5018156 total: 80.4ms remaining: 68.5ms
54: learn: 0.4990495 total: 81ms remaining: 66.3ms
55: learn: 0.4970765 total: 81.7ms remaining: 64.2ms
56: learn: 0.4946661 total: 82.5ms remaining: 62.2ms
57: learn: 0.4919709 total: 83.2ms remaining: 60.2ms
58: learn: 0.4897959 total: 83.7ms remaining: 58.2ms
59: learn: 0.4874367 total: 84ms remaining: 56ms
60: learn: 0.4851208 total: 84.5ms remaining: 54ms
61: learn: 0.4833134 total: 85.2ms remaining: 52.2ms
62: learn: 0.4807927 total: 85.7ms remaining: 50.3ms
63: learn: 0.4790429 total: 86.3ms remaining: 48.6ms
64: learn: 0.4767107 total: 87.1ms remaining: 46.9ms
65: learn: 0.4741546 total: 87.6ms remaining: 45.1ms
66: learn: 0.4717371 total: 88.1ms remaining: 43.4ms
67: learn: 0.4697934 total: 88.7ms remaining: 41.8ms
68: learn: 0.4675539 total: 89.2ms remaining: 40.1ms
69: learn: 0.4651952 total: 89.7ms remaining: 38.4ms
70: learn: 0.4629150 total: 90.4ms remaining: 36.9ms
71: learn: 0.4608229 total: 91ms remaining: 35.4ms
72: learn: 0.4586819 total: 91.4ms remaining: 33.8ms
73: learn: 0.4566460 total: 92ms remaining: 32.3ms
74: learn: 0.4546153 total: 92.5ms remaining: 30.8ms
75: learn: 0.4527045 total: 93ms remaining: 29.4ms
76: learn: 0.4509724 total: 93.7ms remaining: 28ms
77: learn: 0.4490774 total: 94.2ms remaining: 26.6ms
78: learn: 0.4472504 total: 94.8ms remaining: 25.2ms
79: learn: 0.4453785 total: 95.5ms remaining: 23.9ms
80: learn: 0.4436674 total: 96.1ms remaining: 22.5ms
81: learn: 0.4418352 total: 96.3ms remaining: 21.1ms
82: learn: 0.4398787 total: 96.9ms remaining: 19.8ms
83: learn: 0.4378228 total: 97.4ms remaining: 18.5ms
84: learn: 0.4360598 total: 97.9ms remaining: 17.3ms
85: learn: 0.4343564 total: 98.6ms remaining: 16.1ms
86: learn: 0.4332180 total: 99.2ms remaining: 14.8ms
87: learn: 0.4314023 total: 99.8ms remaining: 13.6ms
88: learn: 0.4299614 total: 100ms remaining: 12.4ms
89: learn: 0.4284609 total: 101ms remaining: 11.2ms
90: learn: 0.4268117 total: 102ms remaining: 10ms
91: learn: 0.4250903 total: 102ms remaining: 8.87ms
92: learn: 0.4235067 total: 103ms remaining: 7.72ms
93: learn: 0.4215657 total: 103ms remaining: 6.58ms
94: learn: 0.4202177 total: 104ms remaining: 5.46ms
95: learn: 0.4185972 total: 104ms remaining: 4.34ms
96: learn: 0.4170424 total: 105ms remaining: 3.24ms
97: learn: 0.4153982 total: 105ms remaining: 2.15ms
98: learn: 0.4139034 total: 106ms remaining: 1.07ms
99: learn: 0.4122535 total: 106ms remaining: 0us
0: learn: 0.6879855 total: 1.93ms remaining: 191ms
1: learn: 0.6817990 total: 3.75ms remaining: 184ms
2: learn: 0.6768301 total: 5.13ms remaining: 166ms
3: learn: 0.6725467 total: 6.51ms remaining: 156ms
4: learn: 0.6677060 total: 8.21ms remaining: 156ms
5: learn: 0.6633421 total: 9.77ms remaining: 153ms
6: learn: 0.6587639 total: 11.5ms remaining: 153ms
7: learn: 0.6545091 total: 13.3ms remaining: 153ms
8: learn: 0.6492749 total: 14.6ms remaining: 148ms
9: learn: 0.6451569 total: 16.3ms remaining: 147ms
10: learn: 0.6404551 total: 18.1ms remaining: 146ms
11: learn: 0.6362056 total: 19.9ms remaining: 146ms
12: learn: 0.6319192 total: 21.3ms remaining: 143ms
13: learn: 0.6281266 total: 23.2ms remaining: 143ms
14: learn: 0.6229575 total: 24.9ms remaining: 141ms
15: learn: 0.6189410 total: 26.2ms remaining: 138ms
16: learn: 0.6150797 total: 27.6ms remaining: 135ms
17: learn: 0.6112747 total: 28.5ms remaining: 130ms
18: learn: 0.6075500 total: 30.4ms remaining: 130ms
19: learn: 0.6030214 total: 32.1ms remaining: 128ms
20: learn: 0.5989044 total: 33.9ms remaining: 128ms
21: learn: 0.5945766 total: 35.3ms remaining: 125ms
22: learn: 0.5899263 total: 36.8ms remaining: 123ms
23: learn: 0.5859453 total: 37.7ms remaining: 119ms
24: learn: 0.5817255 total: 39.7ms remaining: 119ms
25: learn: 0.5787281 total: 41.6ms remaining: 119ms
26: learn: 0.5750515 total: 43.2ms remaining: 117ms
27: learn: 0.5722523 total: 44.8ms remaining: 115ms
28: learn: 0.5688370 total: 46.6ms remaining: 114ms
29: learn: 0.5650730 total: 47.3ms remaining: 110ms
30: learn: 0.5614533 total: 48.6ms remaining: 108ms
31: learn: 0.5578750 total: 50.6ms remaining: 108ms
32: learn: 0.5542367 total: 52ms remaining: 106ms
33: learn: 0.5516665 total: 53.5ms remaining: 104ms
34: learn: 0.5487423 total: 55.2ms remaining: 102ms
35: learn: 0.5451976 total: 56.5ms remaining: 101ms
36: learn: 0.5415315 total: 57.8ms remaining: 98.4ms
37: learn: 0.5385923 total: 59.1ms remaining: 96.4ms
38: learn: 0.5357241 total: 59.9ms remaining: 93.7ms
39: learn: 0.5327391 total: 60.6ms remaining: 90.9ms
40: learn: 0.5294844 total: 61.6ms remaining: 88.6ms
41: learn: 0.5267391 total: 62.5ms remaining: 86.3ms
42: learn: 0.5243481 total: 63.6ms remaining: 84.3ms
43: learn: 0.5215191 total: 65ms remaining: 82.7ms
44: learn: 0.5183146 total: 66.4ms remaining: 81.1ms
45: learn: 0.5150006 total: 67.6ms remaining: 79.3ms
46: learn: 0.5117049 total: 68.8ms remaining: 77.6ms
47: learn: 0.5084576 total: 69.9ms remaining: 75.7ms
48: learn: 0.5058160 total: 70.8ms remaining: 73.7ms
49: learn: 0.5032421 total: 71.9ms remaining: 71.9ms
50: learn: 0.5007677 total: 72.6ms remaining: 69.7ms
51: learn: 0.4979306 total: 73.5ms remaining: 67.9ms
52: learn: 0.4954326 total: 74.5ms remaining: 66ms
53: learn: 0.4925781 total: 75.2ms remaining: 64.1ms
54: learn: 0.4898815 total: 76ms remaining: 62.2ms
55: learn: 0.4870882 total: 76.7ms remaining: 60.3ms
56: learn: 0.4845337 total: 77.5ms remaining: 58.4ms
57: learn: 0.4820966 total: 78.2ms remaining: 56.7ms
58: learn: 0.4796332 total: 79ms remaining: 54.9ms
59: learn: 0.4776606 total: 79.8ms remaining: 53.2ms
60: learn: 0.4749781 total: 80.3ms remaining: 51.3ms
61: learn: 0.4720528 total: 80.9ms remaining: 49.6ms
62: learn: 0.4701890 total: 81.4ms remaining: 47.8ms
63: learn: 0.4678341 total: 81.9ms remaining: 46ms
64: learn: 0.4661527 total: 82.6ms remaining: 44.5ms
65: learn: 0.4638934 total: 83.2ms remaining: 42.9ms
66: learn: 0.4615825 total: 83.8ms remaining: 41.3ms
67: learn: 0.4589831 total: 84.3ms remaining: 39.7ms
68: learn: 0.4570743 total: 84.9ms remaining: 38.2ms
69: learn: 0.4543068 total: 85.6ms remaining: 36.7ms
70: learn: 0.4522936 total: 86.2ms remaining: 35.2ms
71: learn: 0.4501520 total: 86.7ms remaining: 33.7ms
72: learn: 0.4477899 total: 87.5ms remaining: 32.4ms
73: learn: 0.4459078 total: 88ms remaining: 30.9ms
74: learn: 0.4440784 total: 88.6ms remaining: 29.5ms
75: learn: 0.4421673 total: 89.2ms remaining: 28.2ms
76: learn: 0.4399315 total: 89.8ms remaining: 26.8ms
77: learn: 0.4376167 total: 90.4ms remaining: 25.5ms
78: learn: 0.4359490 total: 90.9ms remaining: 24.2ms
79: learn: 0.4339178 total: 91.4ms remaining: 22.9ms
80: learn: 0.4316639 total: 92.1ms remaining: 21.6ms
81: learn: 0.4294796 total: 92.6ms remaining: 20.3ms
82: learn: 0.4278307 total: 93.1ms remaining: 19.1ms
83: learn: 0.4258583 total: 93.7ms remaining: 17.9ms
84: learn: 0.4240872 total: 94.2ms remaining: 16.6ms
85: learn: 0.4224970 total: 94.8ms remaining: 15.4ms
86: learn: 0.4213246 total: 95.4ms remaining: 14.3ms
87: learn: 0.4193919 total: 95.9ms remaining: 13.1ms
88: learn: 0.4170500 total: 96.6ms remaining: 11.9ms
89: learn: 0.4152032 total: 97.2ms remaining: 10.8ms
90: learn: 0.4134219 total: 97.7ms remaining: 9.66ms
91: learn: 0.4114825 total: 98.2ms remaining: 8.54ms
92: learn: 0.4094175 total: 98.6ms remaining: 7.42ms
93: learn: 0.4081632 total: 99.1ms remaining: 6.33ms
94: learn: 0.4066762 total: 99.7ms remaining: 5.25ms
95: learn: 0.4048239 total: 100ms remaining: 4.17ms
96: learn: 0.4030484 total: 101ms remaining: 3.11ms
97: learn: 0.4014973 total: 101ms remaining: 2.07ms
98: learn: 0.3998837 total: 102ms remaining: 1.03ms
99: learn: 0.3981028 total: 102ms remaining: 0us
0: learn: 0.6878694 total: 2.48ms remaining: 245ms
1: learn: 0.6827595 total: 4.33ms remaining: 212ms
2: learn: 0.6778842 total: 5.9ms remaining: 191ms
3: learn: 0.6740388 total: 7.26ms remaining: 174ms
4: learn: 0.6691401 total: 8.88ms remaining: 169ms
5: learn: 0.6648926 total: 10.5ms remaining: 164ms
6: learn: 0.6611328 total: 12ms remaining: 159ms
7: learn: 0.6560550 total: 13.3ms remaining: 153ms
8: learn: 0.6511893 total: 15ms remaining: 151ms
9: learn: 0.6473611 total: 16.5ms remaining: 149ms
10: learn: 0.6425144 total: 18.7ms remaining: 151ms
11: learn: 0.6383460 total: 21.6ms remaining: 158ms
12: learn: 0.6332335 total: 23.5ms remaining: 157ms
13: learn: 0.6286910 total: 25.3ms remaining: 156ms
14: learn: 0.6242482 total: 27.4ms remaining: 155ms
15: learn: 0.6204319 total: 29ms remaining: 152ms
16: learn: 0.6162046 total: 30.6ms remaining: 149ms
17: learn: 0.6121904 total: 32ms remaining: 146ms
18: learn: 0.6077890 total: 33.5ms remaining: 143ms
19: learn: 0.6045111 total: 35ms remaining: 140ms
20: learn: 0.6007578 total: 36.8ms remaining: 138ms
21: learn: 0.5985328 total: 38.7ms remaining: 137ms
22: learn: 0.5943166 total: 40.4ms remaining: 135ms
23: learn: 0.5910600 total: 41.7ms remaining: 132ms
24: learn: 0.5876570 total: 43.3ms remaining: 130ms
25: learn: 0.5843096 total: 45.1ms remaining: 128ms
26: learn: 0.5808736 total: 46.2ms remaining: 125ms
27: learn: 0.5782591 total: 47.6ms remaining: 122ms
28: learn: 0.5752312 total: 49.4ms remaining: 121ms
29: learn: 0.5713676 total: 50.8ms remaining: 119ms
30: learn: 0.5680353 total: 52.3ms remaining: 116ms
31: learn: 0.5645367 total: 55.2ms remaining: 117ms
32: learn: 0.5611002 total: 56.6ms remaining: 115ms
33: learn: 0.5572774 total: 57.9ms remaining: 112ms
34: learn: 0.5541482 total: 59.3ms remaining: 110ms
35: learn: 0.5509010 total: 60.5ms remaining: 107ms
36: learn: 0.5473483 total: 61.9ms remaining: 105ms
37: learn: 0.5442582 total: 63.2ms remaining: 103ms
38: learn: 0.5409799 total: 64.2ms remaining: 100ms
39: learn: 0.5380030 total: 64.8ms remaining: 97.2ms
40: learn: 0.5347076 total: 65.9ms remaining: 94.8ms
41: learn: 0.5320555 total: 67.1ms remaining: 92.6ms
42: learn: 0.5289706 total: 67.7ms remaining: 89.7ms
43: learn: 0.5261645 total: 68.3ms remaining: 87ms
44: learn: 0.5233001 total: 68.9ms remaining: 84.2ms
45: learn: 0.5201561 total: 69.6ms remaining: 81.6ms
46: learn: 0.5175403 total: 70.1ms remaining: 79.1ms
47: learn: 0.5144315 total: 70.6ms remaining: 76.4ms
48: learn: 0.5114985 total: 71.3ms remaining: 74.3ms
49: learn: 0.5095739 total: 72ms remaining: 72ms
50: learn: 0.5064724 total: 72.8ms remaining: 69.9ms
51: learn: 0.5035834 total: 73.5ms remaining: 67.8ms
52: learn: 0.5015171 total: 74.1ms remaining: 65.7ms
53: learn: 0.4983474 total: 74.8ms remaining: 63.7ms
54: learn: 0.4957118 total: 75.5ms remaining: 61.8ms
55: learn: 0.4935364 total: 76.1ms remaining: 59.8ms
56: learn: 0.4911946 total: 76.9ms remaining: 58ms
57: learn: 0.4884428 total: 77.4ms remaining: 56ms
58: learn: 0.4862087 total: 78ms remaining: 54.2ms
59: learn: 0.4840193 total: 78.6ms remaining: 52.4ms
60: learn: 0.4817624 total: 79.1ms remaining: 50.6ms
61: learn: 0.4791168 total: 79.6ms remaining: 48.8ms
62: learn: 0.4761302 total: 80.2ms remaining: 47.1ms
63: learn: 0.4738800 total: 80.7ms remaining: 45.4ms
64: learn: 0.4715675 total: 81.3ms remaining: 43.8ms
65: learn: 0.4692935 total: 81.9ms remaining: 42.2ms
66: learn: 0.4667453 total: 82.4ms remaining: 40.6ms
67: learn: 0.4646977 total: 83ms remaining: 39.1ms
68: learn: 0.4623724 total: 83.5ms remaining: 37.5ms
69: learn: 0.4601356 total: 84.1ms remaining: 36.1ms
70: learn: 0.4576502 total: 84.7ms remaining: 34.6ms
71: learn: 0.4559931 total: 85.2ms remaining: 33.1ms
72: learn: 0.4540637 total: 85.9ms remaining: 31.8ms
73: learn: 0.4519782 total: 86.4ms remaining: 30.4ms
74: learn: 0.4497355 total: 86.8ms remaining: 28.9ms
75: learn: 0.4473424 total: 87.4ms remaining: 27.6ms
76: learn: 0.4458190 total: 87.9ms remaining: 26.2ms
77: learn: 0.4437176 total: 88.4ms remaining: 24.9ms
78: learn: 0.4424880 total: 88.9ms remaining: 23.6ms
79: learn: 0.4403117 total: 89.4ms remaining: 22.4ms
80: learn: 0.4380973 total: 90ms remaining: 21.1ms
81: learn: 0.4362197 total: 91.1ms remaining: 20ms
82: learn: 0.4341420 total: 91.8ms remaining: 18.8ms
83: learn: 0.4322704 total: 92.4ms remaining: 17.6ms
84: learn: 0.4301970 total: 93.1ms remaining: 16.4ms
85: learn: 0.4284361 total: 93.6ms remaining: 15.2ms
86: learn: 0.4265964 total: 94.2ms remaining: 14.1ms
87: learn: 0.4246779 total: 94.8ms remaining: 12.9ms
88: learn: 0.4226993 total: 95.4ms remaining: 11.8ms
89: learn: 0.4209481 total: 95.9ms remaining: 10.7ms
90: learn: 0.4191342 total: 96.6ms remaining: 9.56ms
91: learn: 0.4172939 total: 97.2ms remaining: 8.45ms
92: learn: 0.4156818 total: 97.7ms remaining: 7.36ms
93: learn: 0.4140700 total: 98.5ms remaining: 6.29ms
94: learn: 0.4127733 total: 99ms remaining: 5.21ms
95: learn: 0.4113930 total: 99.6ms remaining: 4.15ms
96: learn: 0.4098590 total: 100ms remaining: 3.1ms
97: learn: 0.4080056 total: 101ms remaining: 2.06ms
98: learn: 0.4063266 total: 101ms remaining: 1.02ms
99: learn: 0.4045030 total: 102ms remaining: 0us
0: learn: 0.6878111 total: 2.97ms remaining: 294ms
1: learn: 0.6830677 total: 5.89ms remaining: 288ms
2: learn: 0.6786716 total: 7.99ms remaining: 258ms
3: learn: 0.6730396 total: 9.02ms remaining: 216ms
4: learn: 0.6684266 total: 9.92ms remaining: 189ms
5: learn: 0.6638953 total: 12.1ms remaining: 189ms
6: learn: 0.6589777 total: 14.7ms remaining: 195ms
7: learn: 0.6538375 total: 17.2ms remaining: 197ms
8: learn: 0.6500576 total: 19.5ms remaining: 197ms
9: learn: 0.6454226 total: 21.7ms remaining: 195ms
10: learn: 0.6414998 total: 23.9ms remaining: 193ms
11: learn: 0.6373951 total: 25.5ms remaining: 187ms
12: learn: 0.6336399 total: 27.2ms remaining: 182ms
13: learn: 0.6301762 total: 29ms remaining: 178ms
14: learn: 0.6265799 total: 31.2ms remaining: 177ms
15: learn: 0.6228602 total: 33.3ms remaining: 175ms
16: learn: 0.6182332 total: 35.1ms remaining: 171ms
17: learn: 0.6135086 total: 36.9ms remaining: 168ms
18: learn: 0.6104827 total: 38.4ms remaining: 164ms
19: learn: 0.6071668 total: 40.1ms remaining: 160ms
20: learn: 0.6035631 total: 41.4ms remaining: 156ms
21: learn: 0.5992836 total: 43.2ms remaining: 153ms
22: learn: 0.5954905 total: 45.3ms remaining: 152ms
23: learn: 0.5915550 total: 47ms remaining: 149ms
24: learn: 0.5876077 total: 48.3ms remaining: 145ms
25: learn: 0.5835114 total: 49.8ms remaining: 142ms
26: learn: 0.5796600 total: 51.2ms remaining: 139ms
27: learn: 0.5766405 total: 52.2ms remaining: 134ms
28: learn: 0.5731260 total: 53.8ms remaining: 132ms
29: learn: 0.5697091 total: 55.1ms remaining: 128ms
30: learn: 0.5658183 total: 56.1ms remaining: 125ms
31: learn: 0.5626938 total: 57.4ms remaining: 122ms
32: learn: 0.5596719 total: 59.4ms remaining: 121ms
33: learn: 0.5562376 total: 60.4ms remaining: 117ms
34: learn: 0.5526401 total: 61.6ms remaining: 114ms
35: learn: 0.5492243 total: 62.5ms remaining: 111ms
36: learn: 0.5457598 total: 63.7ms remaining: 109ms
37: learn: 0.5420782 total: 64.7ms remaining: 106ms
38: learn: 0.5390517 total: 65.5ms remaining: 102ms
39: learn: 0.5357773 total: 66.3ms remaining: 99.4ms
40: learn: 0.5328531 total: 67.1ms remaining: 96.6ms
41: learn: 0.5300062 total: 68ms remaining: 93.9ms
42: learn: 0.5272410 total: 68.7ms remaining: 91.1ms
43: learn: 0.5240909 total: 69.4ms remaining: 88.3ms
44: learn: 0.5214789 total: 70.2ms remaining: 85.8ms
45: learn: 0.5191970 total: 70.9ms remaining: 83.2ms
46: learn: 0.5161765 total: 71.6ms remaining: 80.7ms
47: learn: 0.5133697 total: 71.9ms remaining: 77.9ms
48: learn: 0.5104980 total: 72.5ms remaining: 75.5ms
49: learn: 0.5075510 total: 73.3ms remaining: 73.3ms
50: learn: 0.5048457 total: 74ms remaining: 71.1ms
51: learn: 0.5021142 total: 74.8ms remaining: 69ms
52: learn: 0.4995906 total: 75.1ms remaining: 66.6ms
53: learn: 0.4966072 total: 75.7ms remaining: 64.5ms
54: learn: 0.4938185 total: 76.5ms remaining: 62.6ms
55: learn: 0.4910611 total: 77.1ms remaining: 60.6ms
56: learn: 0.4889639 total: 77.6ms remaining: 58.6ms
57: learn: 0.4863851 total: 78.3ms remaining: 56.7ms
58: learn: 0.4837062 total: 78.9ms remaining: 54.9ms
59: learn: 0.4810830 total: 79.6ms remaining: 53.1ms
60: learn: 0.4785150 total: 80.2ms remaining: 51.2ms
61: learn: 0.4759963 total: 80.6ms remaining: 49.4ms
62: learn: 0.4738162 total: 81.4ms remaining: 47.8ms
63: learn: 0.4720720 total: 81.9ms remaining: 46.1ms
64: learn: 0.4700173 total: 82.7ms remaining: 44.5ms
65: learn: 0.4676757 total: 83.2ms remaining: 42.9ms
66: learn: 0.4654652 total: 83.7ms remaining: 41.2ms
67: learn: 0.4631012 total: 84.2ms remaining: 39.6ms
68: learn: 0.4607469 total: 84.7ms remaining: 38.1ms
69: learn: 0.4583304 total: 85.3ms remaining: 36.5ms
70: learn: 0.4559507 total: 85.8ms remaining: 35.1ms
71: learn: 0.4535407 total: 86.4ms remaining: 33.6ms
72: learn: 0.4512914 total: 86.8ms remaining: 32.1ms
73: learn: 0.4492981 total: 87.5ms remaining: 30.7ms
74: learn: 0.4472471 total: 88ms remaining: 29.3ms
75: learn: 0.4455808 total: 88.6ms remaining: 28ms
76: learn: 0.4435213 total: 89.1ms remaining: 26.6ms
77: learn: 0.4411004 total: 89.6ms remaining: 25.3ms
78: learn: 0.4392804 total: 90.1ms remaining: 24ms
79: learn: 0.4372280 total: 90.8ms remaining: 22.7ms
80: learn: 0.4351228 total: 91.3ms remaining: 21.4ms
81: learn: 0.4335048 total: 92ms remaining: 20.2ms
82: learn: 0.4316939 total: 92.6ms remaining: 19ms
83: learn: 0.4295739 total: 93ms remaining: 17.7ms
84: learn: 0.4279941 total: 93.7ms remaining: 16.5ms
85: learn: 0.4261938 total: 94.2ms remaining: 15.3ms
86: learn: 0.4243969 total: 94.4ms remaining: 14.1ms
87: learn: 0.4223284 total: 95ms remaining: 13ms
88: learn: 0.4202255 total: 95.4ms remaining: 11.8ms
89: learn: 0.4182940 total: 96.2ms remaining: 10.7ms
90: learn: 0.4163520 total: 96.7ms remaining: 9.57ms
91: learn: 0.4143460 total: 97.3ms remaining: 8.46ms
92: learn: 0.4126951 total: 97.8ms remaining: 7.36ms
93: learn: 0.4109849 total: 98.3ms remaining: 6.28ms
94: learn: 0.4090185 total: 99ms remaining: 5.21ms
95: learn: 0.4071831 total: 99.5ms remaining: 4.14ms
96: learn: 0.4056752 total: 100ms remaining: 3.1ms
97: learn: 0.4040005 total: 101ms remaining: 2.05ms
98: learn: 0.4025078 total: 101ms remaining: 1.02ms
99: learn: 0.4008263 total: 102ms remaining: 0us
0: learn: 0.6881984 total: 2.17ms remaining: 215ms
1: learn: 0.6837389 total: 3.64ms remaining: 179ms
2: learn: 0.6789850 total: 4.3ms remaining: 139ms
3: learn: 0.6740545 total: 6.48ms remaining: 156ms
4: learn: 0.6700245 total: 8.6ms remaining: 163ms
5: learn: 0.6664139 total: 10.3ms remaining: 161ms
6: learn: 0.6617050 total: 12.4ms remaining: 165ms
7: learn: 0.6579000 total: 14.2ms remaining: 163ms
8: learn: 0.6528585 total: 16.1ms remaining: 163ms
9: learn: 0.6487785 total: 17.6ms remaining: 158ms
10: learn: 0.6439531 total: 19.5ms remaining: 158ms
11: learn: 0.6395388 total: 21.3ms remaining: 156ms
12: learn: 0.6351148 total: 22.9ms remaining: 153ms
13: learn: 0.6311540 total: 24.9ms remaining: 153ms
14: learn: 0.6266968 total: 26.6ms remaining: 151ms
15: learn: 0.6225980 total: 28.2ms remaining: 148ms
16: learn: 0.6190533 total: 29.3ms remaining: 143ms
17: learn: 0.6146239 total: 31ms remaining: 141ms
18: learn: 0.6109725 total: 32.9ms remaining: 140ms
19: learn: 0.6069686 total: 34.5ms remaining: 138ms
20: learn: 0.6025956 total: 36.2ms remaining: 136ms
21: learn: 0.5986945 total: 37.8ms remaining: 134ms
22: learn: 0.5944782 total: 39.6ms remaining: 132ms
23: learn: 0.5917354 total: 40.9ms remaining: 130ms
24: learn: 0.5882377 total: 41.8ms remaining: 125ms
25: learn: 0.5847297 total: 43.3ms remaining: 123ms
26: learn: 0.5815059 total: 44.3ms remaining: 120ms
27: learn: 0.5781487 total: 45.4ms remaining: 117ms
28: learn: 0.5743941 total: 46.7ms remaining: 114ms
29: learn: 0.5714101 total: 48ms remaining: 112ms
30: learn: 0.5680936 total: 49.3ms remaining: 110ms
31: learn: 0.5645795 total: 50.1ms remaining: 106ms
32: learn: 0.5609279 total: 51.2ms remaining: 104ms
33: learn: 0.5577243 total: 52.4ms remaining: 102ms
34: learn: 0.5546932 total: 53.4ms remaining: 99.1ms
35: learn: 0.5516329 total: 54.2ms remaining: 96.4ms
36: learn: 0.5484561 total: 55.7ms remaining: 94.8ms
37: learn: 0.5458463 total: 56.7ms remaining: 92.5ms
38: learn: 0.5429156 total: 57.7ms remaining: 90.2ms
39: learn: 0.5401707 total: 58.6ms remaining: 87.8ms
40: learn: 0.5371278 total: 59.6ms remaining: 85.8ms
41: learn: 0.5348207 total: 61.3ms remaining: 84.7ms
42: learn: 0.5322859 total: 62.4ms remaining: 82.7ms
43: learn: 0.5295026 total: 63.2ms remaining: 80.4ms
44: learn: 0.5272640 total: 64.1ms remaining: 78.4ms
45: learn: 0.5245788 total: 64.4ms remaining: 75.6ms
46: learn: 0.5220263 total: 65.1ms remaining: 73.5ms
47: learn: 0.5199606 total: 66ms remaining: 71.5ms
48: learn: 0.5177430 total: 66.7ms remaining: 69.4ms
49: learn: 0.5155567 total: 67.3ms remaining: 67.3ms
50: learn: 0.5134795 total: 68ms remaining: 65.3ms
51: learn: 0.5106185 total: 68.6ms remaining: 63.4ms
52: learn: 0.5078960 total: 69.3ms remaining: 61.4ms
53: learn: 0.5052445 total: 70ms remaining: 59.6ms
54: learn: 0.5025090 total: 70.6ms remaining: 57.8ms
55: learn: 0.5002670 total: 71.2ms remaining: 55.9ms
56: learn: 0.4979050 total: 71.6ms remaining: 54ms
57: learn: 0.4953377 total: 72.3ms remaining: 52.3ms
58: learn: 0.4930925 total: 72.8ms remaining: 50.6ms
59: learn: 0.4907548 total: 73.5ms remaining: 49ms
60: learn: 0.4888160 total: 74ms remaining: 47.3ms
61: learn: 0.4866885 total: 74.3ms remaining: 45.5ms
62: learn: 0.4849705 total: 74.8ms remaining: 44ms
63: learn: 0.4828227 total: 75.4ms remaining: 42.4ms
64: learn: 0.4809728 total: 75.8ms remaining: 40.8ms
65: learn: 0.4789222 total: 76.2ms remaining: 39.3ms
66: learn: 0.4765588 total: 76.7ms remaining: 37.8ms
67: learn: 0.4746616 total: 77.2ms remaining: 36.3ms
68: learn: 0.4720778 total: 77.7ms remaining: 34.9ms
69: learn: 0.4698665 total: 78.4ms remaining: 33.6ms
70: learn: 0.4679023 total: 78.9ms remaining: 32.2ms
71: learn: 0.4657162 total: 79.4ms remaining: 30.9ms
72: learn: 0.4637961 total: 80.1ms remaining: 29.6ms
73: learn: 0.4615775 total: 80.5ms remaining: 28.3ms
74: learn: 0.4593678 total: 81.2ms remaining: 27.1ms
75: learn: 0.4578329 total: 81.7ms remaining: 25.8ms
76: learn: 0.4560153 total: 82.3ms remaining: 24.6ms
77: learn: 0.4541984 total: 82.8ms remaining: 23.4ms
78: learn: 0.4523104 total: 83.3ms remaining: 22.2ms
79: learn: 0.4500421 total: 83.8ms remaining: 20.9ms
80: learn: 0.4481602 total: 84.3ms remaining: 19.8ms
81: learn: 0.4464342 total: 84.7ms remaining: 18.6ms
82: learn: 0.4445703 total: 85.3ms remaining: 17.5ms
83: learn: 0.4431809 total: 85.8ms remaining: 16.3ms
84: learn: 0.4417564 total: 86.5ms remaining: 15.3ms
85: learn: 0.4401982 total: 87ms remaining: 14.2ms
86: learn: 0.4384260 total: 87.6ms remaining: 13.1ms
87: learn: 0.4363719 total: 88.1ms remaining: 12ms
88: learn: 0.4347263 total: 88.7ms remaining: 11ms
89: learn: 0.4333496 total: 89.3ms remaining: 9.93ms
90: learn: 0.4315995 total: 89.9ms remaining: 8.89ms
91: learn: 0.4296464 total: 90.4ms remaining: 7.86ms
92: learn: 0.4277424 total: 91ms remaining: 6.85ms
93: learn: 0.4258887 total: 91.6ms remaining: 5.84ms
94: learn: 0.4243223 total: 92.1ms remaining: 4.85ms
95: learn: 0.4227430 total: 92.5ms remaining: 3.86ms
96: learn: 0.4211127 total: 93.1ms remaining: 2.88ms
97: learn: 0.4194151 total: 93.7ms remaining: 1.91ms
98: learn: 0.4178400 total: 94.2ms remaining: 951us
99: learn: 0.4162946 total: 94.8ms remaining: 0us
0: learn: 0.6884902 total: 1.04ms remaining: 103ms
1: learn: 0.6827814 total: 2.16ms remaining: 106ms
2: learn: 0.6775249 total: 2.94ms remaining: 95ms
3: learn: 0.6730922 total: 3.79ms remaining: 90.9ms
4: learn: 0.6678643 total: 4.67ms remaining: 88.6ms
5: learn: 0.6641156 total: 5.36ms remaining: 84ms
6: learn: 0.6591491 total: 6.22ms remaining: 82.6ms
7: learn: 0.6544223 total: 7.36ms remaining: 84.6ms
8: learn: 0.6493682 total: 8.25ms remaining: 83.4ms
9: learn: 0.6451104 total: 9.03ms remaining: 81.3ms
10: learn: 0.6405221 total: 10ms remaining: 81.3ms
11: learn: 0.6370647 total: 10.8ms remaining: 79.4ms
12: learn: 0.6327924 total: 11.5ms remaining: 77ms
13: learn: 0.6287428 total: 12.4ms remaining: 75.9ms
14: learn: 0.6240275 total: 13ms remaining: 73.6ms
15: learn: 0.6208828 total: 13.7ms remaining: 71.8ms
16: learn: 0.6173008 total: 14.4ms remaining: 70.5ms
17: learn: 0.6141311 total: 15ms remaining: 68.5ms
18: learn: 0.6099239 total: 15.8ms remaining: 67.3ms
19: learn: 0.6060360 total: 16.7ms remaining: 66.6ms
20: learn: 0.6018792 total: 17.3ms remaining: 64.9ms
21: learn: 0.5990134 total: 17.8ms remaining: 63.2ms
22: learn: 0.5950741 total: 18.4ms remaining: 61.7ms
23: learn: 0.5913950 total: 19.3ms remaining: 61.1ms
24: learn: 0.5878841 total: 19.9ms remaining: 59.7ms
25: learn: 0.5840340 total: 20.6ms remaining: 58.5ms
26: learn: 0.5802704 total: 21.3ms remaining: 57.5ms
27: learn: 0.5774147 total: 21.9ms remaining: 56.4ms
28: learn: 0.5737299 total: 22.5ms remaining: 55.1ms
29: learn: 0.5707954 total: 22.8ms remaining: 53.3ms
30: learn: 0.5676180 total: 23.5ms remaining: 52.3ms
31: learn: 0.5639358 total: 24.1ms remaining: 51.2ms
32: learn: 0.5602758 total: 24.9ms remaining: 50.5ms
33: learn: 0.5569481 total: 25.4ms remaining: 49.3ms
34: learn: 0.5540790 total: 25.9ms remaining: 48.2ms
35: learn: 0.5507536 total: 26.6ms remaining: 47.3ms
36: learn: 0.5476666 total: 27.1ms remaining: 46.2ms
37: learn: 0.5454448 total: 27.6ms remaining: 45ms
38: learn: 0.5422534 total: 28.3ms remaining: 44.2ms
39: learn: 0.5391597 total: 28.9ms remaining: 43.4ms
40: learn: 0.5361588 total: 29.5ms remaining: 42.5ms
41: learn: 0.5336828 total: 30.1ms remaining: 41.6ms
42: learn: 0.5301573 total: 30.5ms remaining: 40.5ms
43: learn: 0.5268429 total: 31ms remaining: 39.5ms
44: learn: 0.5243637 total: 31.6ms remaining: 38.6ms
45: learn: 0.5215132 total: 32ms remaining: 37.6ms
46: learn: 0.5184502 total: 32.6ms remaining: 36.8ms
47: learn: 0.5157361 total: 33.2ms remaining: 36ms
48: learn: 0.5133939 total: 33.7ms remaining: 35.1ms
49: learn: 0.5110648 total: 34.4ms remaining: 34.4ms
50: learn: 0.5084614 total: 35ms remaining: 33.6ms
51: learn: 0.5064103 total: 35.5ms remaining: 32.8ms
52: learn: 0.5036007 total: 36.2ms remaining: 32.1ms
53: learn: 0.5008922 total: 36.7ms remaining: 31.2ms
54: learn: 0.4980136 total: 37.1ms remaining: 30.4ms
55: learn: 0.4952527 total: 37.8ms remaining: 29.7ms
56: learn: 0.4925733 total: 38.3ms remaining: 28.9ms
57: learn: 0.4906550 total: 38.7ms remaining: 28ms
58: learn: 0.4879110 total: 39.2ms remaining: 27.2ms
59: learn: 0.4855741 total: 39.8ms remaining: 26.5ms
60: learn: 0.4828698 total: 40.1ms remaining: 25.7ms
61: learn: 0.4806057 total: 40.6ms remaining: 24.9ms
62: learn: 0.4782623 total: 41.2ms remaining: 24.2ms
63: learn: 0.4762935 total: 41.4ms remaining: 23.3ms
64: learn: 0.4737258 total: 42.1ms remaining: 22.7ms
65: learn: 0.4714130 total: 42.6ms remaining: 21.9ms
66: learn: 0.4692257 total: 43.1ms remaining: 21.2ms
67: learn: 0.4670355 total: 43.8ms remaining: 20.6ms
68: learn: 0.4647485 total: 44.3ms remaining: 19.9ms
69: learn: 0.4627915 total: 44.9ms remaining: 19.2ms
70: learn: 0.4607737 total: 45.5ms remaining: 18.6ms
71: learn: 0.4587871 total: 46ms remaining: 17.9ms
72: learn: 0.4565595 total: 46.5ms remaining: 17.2ms
73: learn: 0.4541840 total: 47.1ms remaining: 16.6ms
74: learn: 0.4522133 total: 47.6ms remaining: 15.9ms
75: learn: 0.4499972 total: 48.1ms remaining: 15.2ms
76: learn: 0.4477514 total: 48.8ms remaining: 14.6ms
77: learn: 0.4463573 total: 49.3ms remaining: 13.9ms
78: learn: 0.4449833 total: 49.8ms remaining: 13.2ms
79: learn: 0.4430477 total: 50.5ms remaining: 12.6ms
80: learn: 0.4414868 total: 51.1ms remaining: 12ms
81: learn: 0.4399330 total: 51.8ms remaining: 11.4ms
82: learn: 0.4378218 total: 52.4ms remaining: 10.7ms
83: learn: 0.4357907 total: 52.9ms remaining: 10.1ms
84: learn: 0.4342854 total: 53.5ms remaining: 9.44ms
85: learn: 0.4329505 total: 54.2ms remaining: 8.83ms
86: learn: 0.4314940 total: 54.7ms remaining: 8.18ms
87: learn: 0.4298428 total: 55.3ms remaining: 7.55ms
88: learn: 0.4281492 total: 56ms remaining: 6.92ms
89: learn: 0.4259604 total: 56.6ms remaining: 6.29ms
90: learn: 0.4239700 total: 57.2ms remaining: 5.65ms
91: learn: 0.4221725 total: 57.9ms remaining: 5.03ms
92: learn: 0.4204447 total: 58.4ms remaining: 4.4ms
93: learn: 0.4184858 total: 58.9ms remaining: 3.76ms
94: learn: 0.4168876 total: 59.6ms remaining: 3.13ms
95: learn: 0.4149810 total: 60.1ms remaining: 2.5ms
96: learn: 0.4134731 total: 60.7ms remaining: 1.88ms
97: learn: 0.4119213 total: 61.4ms remaining: 1.25ms
98: learn: 0.4101695 total: 61.9ms remaining: 625us
99: learn: 0.4083226 total: 62.5ms remaining: 0us
0: learn: 0.6882498 total: 1.05ms remaining: 104ms
1: learn: 0.6828704 total: 1.85ms remaining: 90.8ms
2: learn: 0.6783092 total: 2.8ms remaining: 90.6ms
3: learn: 0.6728309 total: 3.62ms remaining: 86.9ms
4: learn: 0.6682349 total: 3.93ms remaining: 74.6ms
5: learn: 0.6635672 total: 4.75ms remaining: 74.4ms
6: learn: 0.6588318 total: 5.5ms remaining: 73ms
7: learn: 0.6534401 total: 6.13ms remaining: 70.5ms
8: learn: 0.6493226 total: 7.08ms remaining: 71.6ms
9: learn: 0.6449812 total: 7.95ms remaining: 71.5ms
10: learn: 0.6405017 total: 8.67ms remaining: 70.2ms
11: learn: 0.6369216 total: 9.38ms remaining: 68.8ms
12: learn: 0.6332908 total: 10ms remaining: 66.9ms
13: learn: 0.6289660 total: 10.7ms remaining: 65.7ms
14: learn: 0.6249958 total: 11.5ms remaining: 65.3ms
15: learn: 0.6212372 total: 11.9ms remaining: 62.3ms
16: learn: 0.6166556 total: 12.5ms remaining: 61ms
17: learn: 0.6131139 total: 13.2ms remaining: 60.3ms
18: learn: 0.6101849 total: 13.8ms remaining: 59ms
19: learn: 0.6060846 total: 14.5ms remaining: 57.9ms
20: learn: 0.6024470 total: 15.2ms remaining: 57.2ms
21: learn: 0.5990534 total: 15.7ms remaining: 55.7ms
22: learn: 0.5953439 total: 16.3ms remaining: 54.6ms
23: learn: 0.5917527 total: 17.1ms remaining: 54.2ms
24: learn: 0.5879160 total: 17.7ms remaining: 53.1ms
25: learn: 0.5843541 total: 18.3ms remaining: 52ms
26: learn: 0.5802603 total: 18.9ms remaining: 51.2ms
27: learn: 0.5771810 total: 19.5ms remaining: 50.1ms
28: learn: 0.5734008 total: 20.2ms remaining: 49.4ms
29: learn: 0.5700816 total: 20.8ms remaining: 48.4ms
30: learn: 0.5669234 total: 21ms remaining: 46.8ms
31: learn: 0.5643094 total: 21.6ms remaining: 45.8ms
32: learn: 0.5608718 total: 22.3ms remaining: 45.3ms
33: learn: 0.5573620 total: 22.8ms remaining: 44.2ms
34: learn: 0.5536805 total: 23.3ms remaining: 43.3ms
35: learn: 0.5506021 total: 23.9ms remaining: 42.4ms
36: learn: 0.5478504 total: 24.4ms remaining: 41.5ms
37: learn: 0.5445196 total: 25ms remaining: 40.8ms
38: learn: 0.5421972 total: 25.5ms remaining: 39.9ms
39: learn: 0.5397971 total: 26ms remaining: 39.1ms
40: learn: 0.5368651 total: 26.6ms remaining: 38.3ms
41: learn: 0.5341032 total: 27.1ms remaining: 37.4ms
42: learn: 0.5313339 total: 27.7ms remaining: 36.7ms
43: learn: 0.5282895 total: 28.2ms remaining: 35.9ms
44: learn: 0.5253477 total: 28.7ms remaining: 35ms
45: learn: 0.5226034 total: 29.4ms remaining: 34.5ms
46: learn: 0.5197743 total: 29.9ms remaining: 33.7ms
47: learn: 0.5174824 total: 30.4ms remaining: 33ms
48: learn: 0.5148874 total: 31.1ms remaining: 32.4ms
49: learn: 0.5125514 total: 31.5ms remaining: 31.5ms
50: learn: 0.5100936 total: 32.1ms remaining: 30.8ms
51: learn: 0.5080227 total: 32.7ms remaining: 30.2ms
52: learn: 0.5058244 total: 33.2ms remaining: 29.4ms
53: learn: 0.5029077 total: 33.6ms remaining: 28.6ms
54: learn: 0.5010657 total: 34.3ms remaining: 28.1ms
55: learn: 0.4982809 total: 34.8ms remaining: 27.3ms
56: learn: 0.4955976 total: 35.3ms remaining: 26.6ms
57: learn: 0.4928007 total: 35.9ms remaining: 26ms
58: learn: 0.4902985 total: 36.5ms remaining: 25.4ms
59: learn: 0.4879107 total: 36.7ms remaining: 24.5ms
60: learn: 0.4852656 total: 37.4ms remaining: 23.9ms
61: learn: 0.4826753 total: 37.9ms remaining: 23.2ms
62: learn: 0.4803594 total: 38.4ms remaining: 22.6ms
63: learn: 0.4779874 total: 39.1ms remaining: 22ms
64: learn: 0.4757642 total: 39.6ms remaining: 21.3ms
65: learn: 0.4734295 total: 40.2ms remaining: 20.7ms
66: learn: 0.4709168 total: 40.8ms remaining: 20.1ms
67: learn: 0.4686218 total: 41.3ms remaining: 19.4ms
68: learn: 0.4663017 total: 41.9ms remaining: 18.8ms
69: learn: 0.4641567 total: 42.5ms remaining: 18.2ms
70: learn: 0.4622478 total: 43ms remaining: 17.6ms
71: learn: 0.4599877 total: 43.6ms remaining: 17ms
72: learn: 0.4577739 total: 44.2ms remaining: 16.3ms
73: learn: 0.4554528 total: 44.7ms remaining: 15.7ms
74: learn: 0.4537921 total: 45.3ms remaining: 15.1ms
75: learn: 0.4520921 total: 45.8ms remaining: 14.5ms
76: learn: 0.4503531 total: 46.6ms remaining: 13.9ms
77: learn: 0.4482864 total: 47.1ms remaining: 13.3ms
78: learn: 0.4461399 total: 47.6ms remaining: 12.7ms
79: learn: 0.4442617 total: 48.2ms remaining: 12ms
80: learn: 0.4422994 total: 48.8ms remaining: 11.4ms
81: learn: 0.4407674 total: 49.2ms remaining: 10.8ms
82: learn: 0.4390264 total: 49.6ms remaining: 10.2ms
83: learn: 0.4369447 total: 50.3ms remaining: 9.57ms
84: learn: 0.4351189 total: 50.5ms remaining: 8.91ms
85: learn: 0.4339728 total: 51ms remaining: 8.3ms
86: learn: 0.4320184 total: 51.6ms remaining: 7.71ms
87: learn: 0.4303732 total: 52ms remaining: 7.09ms
88: learn: 0.4284251 total: 52.5ms remaining: 6.49ms
89: learn: 0.4264471 total: 53.2ms remaining: 5.91ms
90: learn: 0.4251425 total: 53.7ms remaining: 5.31ms
91: learn: 0.4231876 total: 54.3ms remaining: 4.72ms
92: learn: 0.4216493 total: 54.9ms remaining: 4.13ms
93: learn: 0.4201183 total: 55.4ms remaining: 3.53ms
94: learn: 0.4186990 total: 56ms remaining: 2.95ms
95: learn: 0.4174029 total: 56.6ms remaining: 2.36ms
96: learn: 0.4158460 total: 56.9ms remaining: 1.76ms
97: learn: 0.4142413 total: 57.5ms remaining: 1.17ms
98: learn: 0.4128025 total: 58.2ms remaining: 587us
99: learn: 0.4109770 total: 58.7ms remaining: 0us
0: learn: 0.6884755 total: 1.93ms remaining: 191ms
1: learn: 0.6840549 total: 2.83ms remaining: 139ms
2: learn: 0.6800972 total: 4.22ms remaining: 136ms
3: learn: 0.6768483 total: 5.84ms remaining: 140ms
4: learn: 0.6715695 total: 7.57ms remaining: 144ms
5: learn: 0.6672446 total: 9.39ms remaining: 147ms
6: learn: 0.6625330 total: 11.4ms remaining: 151ms
7: learn: 0.6589198 total: 13.1ms remaining: 151ms
8: learn: 0.6546621 total: 14.8ms remaining: 149ms
9: learn: 0.6501226 total: 16.5ms remaining: 149ms
10: learn: 0.6460963 total: 17.8ms remaining: 144ms
11: learn: 0.6423398 total: 18.8ms remaining: 138ms
12: learn: 0.6379660 total: 20.4ms remaining: 137ms
13: learn: 0.6342444 total: 21.8ms remaining: 134ms
14: learn: 0.6305205 total: 22.8ms remaining: 129ms
15: learn: 0.6264874 total: 24.1ms remaining: 127ms
16: learn: 0.6230416 total: 25.6ms remaining: 125ms
17: learn: 0.6197055 total: 26.8ms remaining: 122ms
18: learn: 0.6157189 total: 28ms remaining: 119ms
19: learn: 0.6125868 total: 29ms remaining: 116ms
20: learn: 0.6083547 total: 29.9ms remaining: 112ms
21: learn: 0.6046788 total: 31.3ms remaining: 111ms
22: learn: 0.6015356 total: 32.6ms remaining: 109ms
23: learn: 0.5973653 total: 33.6ms remaining: 106ms
24: learn: 0.5941012 total: 34.7ms remaining: 104ms
25: learn: 0.5900899 total: 35.7ms remaining: 102ms
26: learn: 0.5864145 total: 36.6ms remaining: 98.9ms
27: learn: 0.5826043 total: 37.8ms remaining: 97.2ms
28: learn: 0.5791785 total: 39.2ms remaining: 96ms
29: learn: 0.5761813 total: 40.3ms remaining: 94ms
30: learn: 0.5732834 total: 41.2ms remaining: 91.7ms
31: learn: 0.5699500 total: 42.1ms remaining: 89.5ms
32: learn: 0.5664212 total: 43.2ms remaining: 87.6ms
33: learn: 0.5634228 total: 44.2ms remaining: 85.8ms
34: learn: 0.5609187 total: 45ms remaining: 83.6ms
35: learn: 0.5576450 total: 45.8ms remaining: 81.5ms
36: learn: 0.5546704 total: 46.5ms remaining: 79.3ms
37: learn: 0.5519747 total: 48ms remaining: 78.3ms
38: learn: 0.5486644 total: 49ms remaining: 76.6ms
39: learn: 0.5462319 total: 49.9ms remaining: 74.9ms
40: learn: 0.5433373 total: 50.8ms remaining: 73.1ms
41: learn: 0.5405130 total: 51.7ms remaining: 71.4ms
42: learn: 0.5383285 total: 52.4ms remaining: 69.5ms
43: learn: 0.5363352 total: 53.2ms remaining: 67.7ms
44: learn: 0.5333441 total: 53.9ms remaining: 65.9ms
45: learn: 0.5304898 total: 54.2ms remaining: 63.7ms
46: learn: 0.5277943 total: 55ms remaining: 62ms
47: learn: 0.5254893 total: 55.6ms remaining: 60.3ms
48: learn: 0.5225329 total: 56.3ms remaining: 58.6ms
49: learn: 0.5198716 total: 57ms remaining: 57ms
50: learn: 0.5169834 total: 57.7ms remaining: 55.4ms
51: learn: 0.5144067 total: 58.4ms remaining: 53.9ms
52: learn: 0.5118940 total: 59.1ms remaining: 52.4ms
53: learn: 0.5096997 total: 59.7ms remaining: 50.8ms
54: learn: 0.5070050 total: 60ms remaining: 49.1ms
55: learn: 0.5045958 total: 60.5ms remaining: 47.5ms
56: learn: 0.5020338 total: 61.1ms remaining: 46.1ms
57: learn: 0.5000771 total: 61.6ms remaining: 44.6ms
58: learn: 0.4981839 total: 62.4ms remaining: 43.3ms
59: learn: 0.4960523 total: 62.6ms remaining: 41.7ms
60: learn: 0.4935454 total: 63.2ms remaining: 40.4ms
61: learn: 0.4916064 total: 63.7ms remaining: 39ms
62: learn: 0.4892696 total: 64.3ms remaining: 37.8ms
63: learn: 0.4870125 total: 64.9ms remaining: 36.5ms
64: learn: 0.4848185 total: 65.5ms remaining: 35.3ms
65: learn: 0.4826492 total: 66.1ms remaining: 34.1ms
66: learn: 0.4802407 total: 66.6ms remaining: 32.8ms
67: learn: 0.4779000 total: 67.3ms remaining: 31.7ms
68: learn: 0.4753992 total: 67.8ms remaining: 30.5ms
69: learn: 0.4739093 total: 68.4ms remaining: 29.3ms
70: learn: 0.4714523 total: 69.1ms remaining: 28.2ms
71: learn: 0.4695382 total: 69.6ms remaining: 27.1ms
72: learn: 0.4674394 total: 70.3ms remaining: 26ms
73: learn: 0.4652941 total: 70.8ms remaining: 24.9ms
74: learn: 0.4636822 total: 71.2ms remaining: 23.7ms
75: learn: 0.4618156 total: 71.9ms remaining: 22.7ms
76: learn: 0.4594714 total: 72.4ms remaining: 21.6ms
77: learn: 0.4575470 total: 72.9ms remaining: 20.6ms
78: learn: 0.4558903 total: 73.4ms remaining: 19.5ms
79: learn: 0.4543526 total: 74ms remaining: 18.5ms
80: learn: 0.4527884 total: 74.6ms remaining: 17.5ms
81: learn: 0.4508507 total: 75.2ms remaining: 16.5ms
82: learn: 0.4489548 total: 75.7ms remaining: 15.5ms
83: learn: 0.4473538 total: 76.3ms remaining: 14.5ms
84: learn: 0.4461317 total: 76.9ms remaining: 13.6ms
85: learn: 0.4447457 total: 77.5ms remaining: 12.6ms
86: learn: 0.4426733 total: 78.2ms remaining: 11.7ms
87: learn: 0.4411311 total: 78.7ms remaining: 10.7ms
88: learn: 0.4395233 total: 79.3ms remaining: 9.79ms
89: learn: 0.4382878 total: 79.6ms remaining: 8.85ms
90: learn: 0.4363979 total: 80.1ms remaining: 7.92ms
91: learn: 0.4345390 total: 80.6ms remaining: 7.01ms
92: learn: 0.4330218 total: 81.3ms remaining: 6.12ms
93: learn: 0.4312193 total: 81.8ms remaining: 5.22ms
94: learn: 0.4296027 total: 82.3ms remaining: 4.33ms
95: learn: 0.4279780 total: 82.8ms remaining: 3.45ms
96: learn: 0.4263970 total: 83.4ms remaining: 2.58ms
97: learn: 0.4246456 total: 83.9ms remaining: 1.71ms
98: learn: 0.4231325 total: 84.6ms remaining: 854us
99: learn: 0.4215310 total: 85.1ms remaining: 0us
0: learn: 0.6880341 total: 2.22ms remaining: 220ms
1: learn: 0.6821741 total: 3.66ms remaining: 179ms
2: learn: 0.6772413 total: 5.32ms remaining: 172ms
3: learn: 0.6719167 total: 6.31ms remaining: 152ms
4: learn: 0.6673770 total: 7.34ms remaining: 139ms
5: learn: 0.6631005 total: 8.48ms remaining: 133ms
6: learn: 0.6583423 total: 9.44ms remaining: 125ms
7: learn: 0.6541579 total: 11.1ms remaining: 128ms
8: learn: 0.6500454 total: 12.4ms remaining: 126ms
9: learn: 0.6451953 total: 13.8ms remaining: 125ms
10: learn: 0.6408795 total: 15.1ms remaining: 122ms
11: learn: 0.6365962 total: 15.6ms remaining: 114ms
12: learn: 0.6326614 total: 16.8ms remaining: 112ms
13: learn: 0.6286050 total: 18ms remaining: 110ms
14: learn: 0.6244195 total: 19.4ms remaining: 110ms
15: learn: 0.6206224 total: 20.4ms remaining: 107ms
16: learn: 0.6166255 total: 21.3ms remaining: 104ms
17: learn: 0.6129454 total: 22.3ms remaining: 101ms
18: learn: 0.6091903 total: 23.2ms remaining: 99.1ms
19: learn: 0.6051157 total: 24ms remaining: 95.8ms
20: learn: 0.6008821 total: 25.1ms remaining: 94.4ms
21: learn: 0.5965324 total: 25.8ms remaining: 91.6ms
22: learn: 0.5923261 total: 26.9ms remaining: 90.2ms
23: learn: 0.5880464 total: 27.8ms remaining: 88.1ms
24: learn: 0.5839825 total: 28.8ms remaining: 86.5ms
25: learn: 0.5796767 total: 29.7ms remaining: 84.5ms
26: learn: 0.5762351 total: 30.5ms remaining: 82.6ms
27: learn: 0.5730085 total: 31.4ms remaining: 80.6ms
28: learn: 0.5698990 total: 32.4ms remaining: 79.4ms
29: learn: 0.5668733 total: 33.1ms remaining: 77.2ms
30: learn: 0.5631105 total: 33.7ms remaining: 75ms
31: learn: 0.5594105 total: 34.6ms remaining: 73.4ms
32: learn: 0.5556754 total: 35.2ms remaining: 71.5ms
33: learn: 0.5528355 total: 35.9ms remaining: 69.7ms
34: learn: 0.5497371 total: 36.6ms remaining: 68ms
35: learn: 0.5463105 total: 37.2ms remaining: 66.1ms
36: learn: 0.5429736 total: 38ms remaining: 64.7ms
37: learn: 0.5404733 total: 38.7ms remaining: 63.1ms
38: learn: 0.5372550 total: 39.3ms remaining: 61.5ms
39: learn: 0.5341701 total: 40.1ms remaining: 60.1ms
40: learn: 0.5312312 total: 40.3ms remaining: 58ms
41: learn: 0.5278372 total: 41ms remaining: 56.6ms
42: learn: 0.5251373 total: 41.8ms remaining: 55.4ms
43: learn: 0.5228357 total: 42.4ms remaining: 54ms
44: learn: 0.5200075 total: 43ms remaining: 52.6ms
45: learn: 0.5178263 total: 43.7ms remaining: 51.3ms
46: learn: 0.5150005 total: 44.2ms remaining: 49.8ms
47: learn: 0.5119695 total: 44.9ms remaining: 48.6ms
48: learn: 0.5091177 total: 45.4ms remaining: 47.3ms
49: learn: 0.5060475 total: 46ms remaining: 46ms
50: learn: 0.5036737 total: 46.6ms remaining: 44.8ms
51: learn: 0.5012514 total: 47.2ms remaining: 43.5ms
52: learn: 0.4987633 total: 47.7ms remaining: 42.3ms
53: learn: 0.4960564 total: 48.3ms remaining: 41.2ms
54: learn: 0.4936300 total: 48.8ms remaining: 39.9ms
55: learn: 0.4909804 total: 49.4ms remaining: 38.8ms
56: learn: 0.4884961 total: 50.1ms remaining: 37.8ms
57: learn: 0.4860099 total: 50.3ms remaining: 36.4ms
58: learn: 0.4833494 total: 50.8ms remaining: 35.3ms
59: learn: 0.4809202 total: 51.4ms remaining: 34.3ms
60: learn: 0.4785807 total: 51.9ms remaining: 33.2ms
61: learn: 0.4757666 total: 52.4ms remaining: 32.1ms
62: learn: 0.4735325 total: 53.1ms remaining: 31.2ms
63: learn: 0.4709927 total: 53.6ms remaining: 30.1ms
64: learn: 0.4683417 total: 54.2ms remaining: 29.2ms
65: learn: 0.4657681 total: 54.7ms remaining: 28.2ms
66: learn: 0.4637961 total: 55.5ms remaining: 27.3ms
67: learn: 0.4614103 total: 56.1ms remaining: 26.4ms
68: learn: 0.4599264 total: 56.6ms remaining: 25.4ms
69: learn: 0.4577802 total: 57.4ms remaining: 24.6ms
70: learn: 0.4558063 total: 57.9ms remaining: 23.7ms
71: learn: 0.4536017 total: 58.5ms remaining: 22.8ms
72: learn: 0.4511466 total: 59.1ms remaining: 21.8ms
73: learn: 0.4490527 total: 59.5ms remaining: 20.9ms
74: learn: 0.4476742 total: 60.3ms remaining: 20.1ms
75: learn: 0.4455227 total: 60.9ms remaining: 19.2ms
76: learn: 0.4439339 total: 61.6ms remaining: 18.4ms
77: learn: 0.4417317 total: 62ms remaining: 17.5ms
78: learn: 0.4398648 total: 62.7ms remaining: 16.7ms
79: learn: 0.4383028 total: 63.2ms remaining: 15.8ms
80: learn: 0.4363247 total: 63.8ms remaining: 15ms
81: learn: 0.4343102 total: 64.4ms remaining: 14.1ms
82: learn: 0.4325206 total: 65ms remaining: 13.3ms
83: learn: 0.4310468 total: 65.5ms remaining: 12.5ms
84: learn: 0.4290302 total: 66.1ms remaining: 11.7ms
85: learn: 0.4271689 total: 66.3ms remaining: 10.8ms
86: learn: 0.4250570 total: 66.7ms remaining: 9.97ms
87: learn: 0.4228321 total: 67.4ms remaining: 9.19ms
88: learn: 0.4209581 total: 67.9ms remaining: 8.39ms
89: learn: 0.4191468 total: 68.4ms remaining: 7.6ms
90: learn: 0.4172830 total: 69.1ms remaining: 6.84ms
91: learn: 0.4152090 total: 69.7ms remaining: 6.06ms
92: learn: 0.4135675 total: 70.3ms remaining: 5.29ms
93: learn: 0.4119928 total: 70.7ms remaining: 4.51ms
94: learn: 0.4105038 total: 71.9ms remaining: 3.78ms
95: learn: 0.4090142 total: 72.8ms remaining: 3.03ms
96: learn: 0.4073067 total: 73.4ms remaining: 2.27ms
97: learn: 0.4054502 total: 74.1ms remaining: 1.51ms
98: learn: 0.4037744 total: 74.7ms remaining: 754us
99: learn: 0.4027672 total: 75.3ms remaining: 0us
0: learn: 0.6886303 total: 1.93ms remaining: 191ms
1: learn: 0.6848097 total: 3.33ms remaining: 163ms
2: learn: 0.6801248 total: 4.87ms remaining: 157ms
3: learn: 0.6770560 total: 6.76ms remaining: 162ms
4: learn: 0.6719868 total: 8.23ms remaining: 156ms
5: learn: 0.6677443 total: 8.78ms remaining: 138ms
6: learn: 0.6637682 total: 10.2ms remaining: 136ms
7: learn: 0.6595154 total: 11.5ms remaining: 132ms
8: learn: 0.6558830 total: 12.4ms remaining: 125ms
9: learn: 0.6517791 total: 13.7ms remaining: 123ms
10: learn: 0.6471647 total: 15ms remaining: 122ms
11: learn: 0.6430895 total: 16.1ms remaining: 118ms
12: learn: 0.6393340 total: 17.2ms remaining: 115ms
13: learn: 0.6354497 total: 18.5ms remaining: 114ms
14: learn: 0.6311215 total: 19.7ms remaining: 112ms
15: learn: 0.6275276 total: 20.7ms remaining: 109ms
16: learn: 0.6236488 total: 21.7ms remaining: 106ms
17: learn: 0.6202546 total: 22.4ms remaining: 102ms
18: learn: 0.6166884 total: 23.4ms remaining: 99.6ms
19: learn: 0.6129113 total: 24.6ms remaining: 98.5ms
20: learn: 0.6093491 total: 25.4ms remaining: 95.7ms
21: learn: 0.6065245 total: 26.4ms remaining: 93.7ms
22: learn: 0.6034042 total: 27.4ms remaining: 91.6ms
23: learn: 0.5997530 total: 28.3ms remaining: 89.7ms
24: learn: 0.5959880 total: 29.5ms remaining: 88.4ms
25: learn: 0.5922939 total: 30.4ms remaining: 86.6ms
26: learn: 0.5897792 total: 31.2ms remaining: 84.4ms
27: learn: 0.5861889 total: 32.1ms remaining: 82.6ms
28: learn: 0.5835273 total: 33.4ms remaining: 81.8ms
29: learn: 0.5808634 total: 34.2ms remaining: 79.9ms
30: learn: 0.5772724 total: 35ms remaining: 78ms
31: learn: 0.5740322 total: 36ms remaining: 76.4ms
32: learn: 0.5710547 total: 36.7ms remaining: 74.6ms
33: learn: 0.5677978 total: 37.7ms remaining: 73.3ms
34: learn: 0.5645430 total: 38.7ms remaining: 71.9ms
35: learn: 0.5610716 total: 39.6ms remaining: 70.3ms
36: learn: 0.5578719 total: 40.4ms remaining: 68.8ms
37: learn: 0.5552181 total: 41.2ms remaining: 67.1ms
38: learn: 0.5524750 total: 41.5ms remaining: 65ms
39: learn: 0.5500543 total: 42.4ms remaining: 63.6ms
40: learn: 0.5472859 total: 43.4ms remaining: 62.4ms
41: learn: 0.5445638 total: 44.2ms remaining: 61.1ms
42: learn: 0.5419454 total: 44.9ms remaining: 59.5ms
43: learn: 0.5391157 total: 45.6ms remaining: 58ms
44: learn: 0.5367238 total: 46.3ms remaining: 56.6ms
45: learn: 0.5340405 total: 47.1ms remaining: 55.3ms
46: learn: 0.5314801 total: 47.9ms remaining: 54ms
47: learn: 0.5288782 total: 48.6ms remaining: 52.6ms
48: learn: 0.5259124 total: 49.3ms remaining: 51.3ms
49: learn: 0.5231423 total: 49.9ms remaining: 49.9ms
50: learn: 0.5207599 total: 50.5ms remaining: 48.6ms
51: learn: 0.5187159 total: 51.2ms remaining: 47.3ms
52: learn: 0.5162719 total: 51.9ms remaining: 46ms
53: learn: 0.5139486 total: 52.6ms remaining: 44.8ms
54: learn: 0.5117177 total: 53.2ms remaining: 43.5ms
55: learn: 0.5092999 total: 53.7ms remaining: 42.2ms
56: learn: 0.5067726 total: 54.3ms remaining: 41ms
57: learn: 0.5043010 total: 54.8ms remaining: 39.7ms
58: learn: 0.5021702 total: 55.3ms remaining: 38.5ms
59: learn: 0.4997819 total: 56ms remaining: 37.3ms
60: learn: 0.4973616 total: 56.5ms remaining: 36.1ms
61: learn: 0.4949667 total: 57ms remaining: 35ms
62: learn: 0.4928978 total: 57.4ms remaining: 33.7ms
63: learn: 0.4908787 total: 58ms remaining: 32.6ms
64: learn: 0.4883756 total: 58.4ms remaining: 31.5ms
65: learn: 0.4867250 total: 59.1ms remaining: 30.4ms
66: learn: 0.4845296 total: 59.6ms remaining: 29.4ms
67: learn: 0.4821400 total: 60.2ms remaining: 28.3ms
68: learn: 0.4800275 total: 60.7ms remaining: 27.3ms
69: learn: 0.4781675 total: 61.2ms remaining: 26.2ms
70: learn: 0.4760829 total: 61.9ms remaining: 25.3ms
71: learn: 0.4741379 total: 62.4ms remaining: 24.3ms
72: learn: 0.4726351 total: 62.9ms remaining: 23.3ms
73: learn: 0.4705210 total: 63.8ms remaining: 22.4ms
74: learn: 0.4683300 total: 64.4ms remaining: 21.5ms
75: learn: 0.4664747 total: 65ms remaining: 20.5ms
76: learn: 0.4645237 total: 65.7ms remaining: 19.6ms
77: learn: 0.4627381 total: 66.3ms remaining: 18.7ms
78: learn: 0.4609883 total: 66.9ms remaining: 17.8ms
79: learn: 0.4591362 total: 67.5ms remaining: 16.9ms
80: learn: 0.4575176 total: 68ms remaining: 15.9ms
81: learn: 0.4556861 total: 68.5ms remaining: 15ms
82: learn: 0.4542708 total: 69ms remaining: 14.1ms
83: learn: 0.4526115 total: 69.5ms remaining: 13.2ms
84: learn: 0.4509860 total: 70.2ms remaining: 12.4ms
85: learn: 0.4491232 total: 70.7ms remaining: 11.5ms
86: learn: 0.4475120 total: 71.2ms remaining: 10.6ms
87: learn: 0.4456911 total: 71.9ms remaining: 9.8ms
88: learn: 0.4442656 total: 72.3ms remaining: 8.94ms
89: learn: 0.4427431 total: 72.5ms remaining: 8.06ms
90: learn: 0.4412647 total: 73.2ms remaining: 7.24ms
91: learn: 0.4395599 total: 73.6ms remaining: 6.4ms
92: learn: 0.4379309 total: 74.1ms remaining: 5.58ms
93: learn: 0.4361173 total: 74.7ms remaining: 4.77ms
94: learn: 0.4343678 total: 75.2ms remaining: 3.96ms
95: learn: 0.4326400 total: 75.7ms remaining: 3.15ms
96: learn: 0.4308858 total: 76.3ms remaining: 2.36ms
97: learn: 0.4296773 total: 76.8ms remaining: 1.57ms
98: learn: 0.4280932 total: 77.4ms remaining: 781us
99: learn: 0.4265640 total: 78.1ms remaining: 0us
0: learn: 0.6888083 total: 1.16ms remaining: 115ms
1: learn: 0.6840100 total: 2.19ms remaining: 108ms
2: learn: 0.6794042 total: 3.32ms remaining: 107ms
3: learn: 0.6755347 total: 4.3ms remaining: 103ms
4: learn: 0.6710576 total: 5.22ms remaining: 99.2ms
5: learn: 0.6663095 total: 6.2ms remaining: 97.1ms
6: learn: 0.6626385 total: 7.11ms remaining: 94.4ms
7: learn: 0.6575897 total: 8.09ms remaining: 93.1ms
8: learn: 0.6527714 total: 8.84ms remaining: 89.3ms
9: learn: 0.6479300 total: 9.57ms remaining: 86.1ms
10: learn: 0.6447237 total: 10.5ms remaining: 85.3ms
11: learn: 0.6410750 total: 11.5ms remaining: 84.2ms
12: learn: 0.6368673 total: 12.3ms remaining: 82.6ms
13: learn: 0.6331436 total: 13.2ms remaining: 81ms
14: learn: 0.6287820 total: 13.9ms remaining: 78.6ms
15: learn: 0.6257715 total: 14.6ms remaining: 76.7ms
16: learn: 0.6217684 total: 15.6ms remaining: 76.4ms
17: learn: 0.6173308 total: 16.4ms remaining: 74.7ms
18: learn: 0.6138749 total: 17ms remaining: 72.4ms
19: learn: 0.6098700 total: 17.7ms remaining: 70.9ms
20: learn: 0.6058963 total: 18.4ms remaining: 69.3ms
21: learn: 0.6024329 total: 19.1ms remaining: 67.7ms
22: learn: 0.5992812 total: 19.9ms remaining: 66.6ms
23: learn: 0.5963980 total: 20.5ms remaining: 64.8ms
24: learn: 0.5932907 total: 21ms remaining: 63ms
25: learn: 0.5906091 total: 21.6ms remaining: 61.4ms
26: learn: 0.5863905 total: 22.1ms remaining: 59.7ms
27: learn: 0.5837556 total: 22.6ms remaining: 58ms
28: learn: 0.5809663 total: 23.1ms remaining: 56.7ms
29: learn: 0.5773555 total: 23.9ms remaining: 55.8ms
30: learn: 0.5738775 total: 24.4ms remaining: 54.4ms
31: learn: 0.5705333 total: 25.1ms remaining: 53.3ms
32: learn: 0.5675880 total: 25.7ms remaining: 52.1ms
33: learn: 0.5643922 total: 26.2ms remaining: 50.9ms
34: learn: 0.5619492 total: 26.7ms remaining: 49.6ms
35: learn: 0.5596123 total: 27.2ms remaining: 48.4ms
36: learn: 0.5563666 total: 27.7ms remaining: 47.1ms
37: learn: 0.5530458 total: 28.4ms remaining: 46.4ms
38: learn: 0.5505357 total: 28.7ms remaining: 45ms
39: learn: 0.5473357 total: 29.3ms remaining: 44ms
40: learn: 0.5448640 total: 30ms remaining: 43.2ms
41: learn: 0.5414981 total: 30.6ms remaining: 42.2ms
42: learn: 0.5392637 total: 31ms remaining: 41.1ms
43: learn: 0.5362985 total: 31.7ms remaining: 40.4ms
44: learn: 0.5339898 total: 32.2ms remaining: 39.4ms
45: learn: 0.5305500 total: 32.7ms remaining: 38.4ms
46: learn: 0.5278988 total: 33.4ms remaining: 37.6ms
47: learn: 0.5249098 total: 34ms remaining: 36.8ms
48: learn: 0.5224657 total: 34.5ms remaining: 35.9ms
49: learn: 0.5203313 total: 35.1ms remaining: 35.1ms
50: learn: 0.5183110 total: 35.8ms remaining: 34.4ms
51: learn: 0.5156346 total: 36.4ms remaining: 33.6ms
52: learn: 0.5132979 total: 36.9ms remaining: 32.7ms
53: learn: 0.5110004 total: 37.5ms remaining: 31.9ms
54: learn: 0.5087149 total: 38ms remaining: 31.1ms
55: learn: 0.5059530 total: 38.5ms remaining: 30.3ms
56: learn: 0.5035587 total: 39ms remaining: 29.4ms
57: learn: 0.5009002 total: 39.6ms remaining: 28.7ms
58: learn: 0.4984494 total: 40.1ms remaining: 27.9ms
59: learn: 0.4967913 total: 40.7ms remaining: 27.1ms
60: learn: 0.4942841 total: 41.2ms remaining: 26.3ms
61: learn: 0.4920158 total: 41.8ms remaining: 25.6ms
62: learn: 0.4894883 total: 42.4ms remaining: 24.9ms
63: learn: 0.4871693 total: 42.9ms remaining: 24.1ms
64: learn: 0.4845915 total: 43.5ms remaining: 23.4ms
65: learn: 0.4821749 total: 44.1ms remaining: 22.7ms
66: learn: 0.4796513 total: 44.6ms remaining: 22ms
67: learn: 0.4776679 total: 45.3ms remaining: 21.3ms
68: learn: 0.4754361 total: 45.9ms remaining: 20.6ms
69: learn: 0.4732956 total: 46.4ms remaining: 19.9ms
70: learn: 0.4707850 total: 47.1ms remaining: 19.2ms
71: learn: 0.4685379 total: 47.5ms remaining: 18.5ms
72: learn: 0.4666592 total: 48.2ms remaining: 17.8ms
73: learn: 0.4641637 total: 48.8ms remaining: 17.2ms
74: learn: 0.4619504 total: 49.3ms remaining: 16.4ms
75: learn: 0.4595146 total: 49.9ms remaining: 15.8ms
76: learn: 0.4577572 total: 50.4ms remaining: 15.1ms
77: learn: 0.4556302 total: 51ms remaining: 14.4ms
78: learn: 0.4541389 total: 51.5ms remaining: 13.7ms
79: learn: 0.4518847 total: 51.9ms remaining: 13ms
80: learn: 0.4496552 total: 52.4ms remaining: 12.3ms
81: learn: 0.4474852 total: 53ms remaining: 11.6ms
82: learn: 0.4450534 total: 53.5ms remaining: 11ms
83: learn: 0.4432547 total: 54.4ms remaining: 10.4ms
84: learn: 0.4412831 total: 54.8ms remaining: 9.67ms
85: learn: 0.4398874 total: 55.4ms remaining: 9.01ms
86: learn: 0.4381570 total: 56ms remaining: 8.37ms
87: learn: 0.4362008 total: 56.6ms remaining: 7.72ms
88: learn: 0.4342811 total: 57.3ms remaining: 7.08ms
89: learn: 0.4325549 total: 57.8ms remaining: 6.42ms
90: learn: 0.4310816 total: 58.3ms remaining: 5.77ms
91: learn: 0.4289986 total: 59ms remaining: 5.13ms
92: learn: 0.4270305 total: 59.6ms remaining: 4.48ms
93: learn: 0.4251537 total: 60ms remaining: 3.83ms
94: learn: 0.4233687 total: 60.6ms remaining: 3.19ms
95: learn: 0.4214113 total: 61.1ms remaining: 2.54ms
96: learn: 0.4196621 total: 61.5ms remaining: 1.9ms
97: learn: 0.4176539 total: 62ms remaining: 1.27ms
98: learn: 0.4158150 total: 62.7ms remaining: 633us
99: learn: 0.4144446 total: 63.2ms remaining: 0us
0: learn: 0.6885484 total: 1.66ms remaining: 164ms
1: learn: 0.6832990 total: 3.13ms remaining: 153ms
2: learn: 0.6784705 total: 4.36ms remaining: 141ms
3: learn: 0.6740281 total: 5.64ms remaining: 135ms
4: learn: 0.6689528 total: 7.07ms remaining: 134ms
5: learn: 0.6651681 total: 8.48ms remaining: 133ms
6: learn: 0.6602428 total: 9.82ms remaining: 130ms
7: learn: 0.6565745 total: 11.3ms remaining: 130ms
8: learn: 0.6526943 total: 12.4ms remaining: 125ms
9: learn: 0.6481739 total: 13.6ms remaining: 122ms
10: learn: 0.6445998 total: 14.5ms remaining: 118ms
11: learn: 0.6406037 total: 15.1ms remaining: 111ms
12: learn: 0.6369031 total: 16.4ms remaining: 110ms
13: learn: 0.6331369 total: 17.7ms remaining: 109ms
14: learn: 0.6293530 total: 18.8ms remaining: 107ms
15: learn: 0.6256101 total: 19.8ms remaining: 104ms
16: learn: 0.6223479 total: 20.8ms remaining: 102ms
17: learn: 0.6191322 total: 21.7ms remaining: 98.8ms
18: learn: 0.6159982 total: 22.6ms remaining: 96.2ms
19: learn: 0.6125919 total: 23.5ms remaining: 93.8ms
20: learn: 0.6087055 total: 24.6ms remaining: 92.6ms
21: learn: 0.6053173 total: 25.5ms remaining: 90.5ms
22: learn: 0.6016938 total: 26.3ms remaining: 88ms
23: learn: 0.5979304 total: 27.3ms remaining: 86.4ms
24: learn: 0.5951165 total: 28ms remaining: 84.1ms
25: learn: 0.5919794 total: 28.8ms remaining: 82.1ms
26: learn: 0.5887875 total: 29.6ms remaining: 80ms
27: learn: 0.5852175 total: 30.3ms remaining: 77.9ms
28: learn: 0.5813837 total: 30.7ms remaining: 75.1ms
29: learn: 0.5779479 total: 31.4ms remaining: 73.4ms
30: learn: 0.5747208 total: 32.2ms remaining: 71.8ms
31: learn: 0.5711797 total: 33ms remaining: 70.1ms
32: learn: 0.5692595 total: 33.6ms remaining: 68.3ms
33: learn: 0.5661201 total: 34.4ms remaining: 66.8ms
34: learn: 0.5634923 total: 35.2ms remaining: 65.3ms
35: learn: 0.5607615 total: 35.8ms remaining: 63.6ms
36: learn: 0.5580848 total: 36.5ms remaining: 62.1ms
37: learn: 0.5548941 total: 37.2ms remaining: 60.7ms
38: learn: 0.5520169 total: 37.8ms remaining: 59.2ms
39: learn: 0.5491247 total: 38.6ms remaining: 57.9ms
40: learn: 0.5471747 total: 39.3ms remaining: 56.5ms
41: learn: 0.5441979 total: 39.9ms remaining: 55.1ms
42: learn: 0.5408542 total: 40.5ms remaining: 53.7ms
43: learn: 0.5378777 total: 41.1ms remaining: 52.3ms
44: learn: 0.5352505 total: 41.8ms remaining: 51.1ms
45: learn: 0.5326579 total: 42.2ms remaining: 49.6ms
46: learn: 0.5301107 total: 42.8ms remaining: 48.2ms
47: learn: 0.5270970 total: 43.4ms remaining: 47ms
48: learn: 0.5243750 total: 43.9ms remaining: 45.6ms
49: learn: 0.5221046 total: 44.4ms remaining: 44.4ms
50: learn: 0.5197129 total: 44.9ms remaining: 43.1ms
51: learn: 0.5172321 total: 45.5ms remaining: 42ms
52: learn: 0.5147543 total: 46ms remaining: 40.8ms
53: learn: 0.5125193 total: 46.5ms remaining: 39.6ms
54: learn: 0.5105005 total: 47.1ms remaining: 38.6ms
55: learn: 0.5079834 total: 47.6ms remaining: 37.4ms
56: learn: 0.5055446 total: 48.2ms remaining: 36.4ms
57: learn: 0.5029891 total: 48.7ms remaining: 35.3ms
58: learn: 0.5005422 total: 49.3ms remaining: 34.2ms
59: learn: 0.4981034 total: 49.7ms remaining: 33.1ms
60: learn: 0.4957149 total: 50.2ms remaining: 32.1ms
61: learn: 0.4934286 total: 50.7ms remaining: 31.1ms
62: learn: 0.4916000 total: 51.4ms remaining: 30.2ms
63: learn: 0.4891685 total: 51.9ms remaining: 29.2ms
64: learn: 0.4868845 total: 52.4ms remaining: 28.2ms
65: learn: 0.4852369 total: 52.9ms remaining: 27.3ms
66: learn: 0.4831013 total: 53.4ms remaining: 26.3ms
67: learn: 0.4806681 total: 54.1ms remaining: 25.5ms
68: learn: 0.4781447 total: 54.5ms remaining: 24.5ms
69: learn: 0.4759038 total: 55ms remaining: 23.6ms
70: learn: 0.4740721 total: 55.6ms remaining: 22.7ms
71: learn: 0.4720991 total: 56.1ms remaining: 21.8ms
72: learn: 0.4697717 total: 56.8ms remaining: 21ms
73: learn: 0.4678502 total: 57.3ms remaining: 20.1ms
74: learn: 0.4662440 total: 57.8ms remaining: 19.3ms
75: learn: 0.4644569 total: 58.4ms remaining: 18.4ms
76: learn: 0.4624719 total: 59ms remaining: 17.6ms
77: learn: 0.4607071 total: 59.6ms remaining: 16.8ms
78: learn: 0.4586385 total: 60.1ms remaining: 16ms
79: learn: 0.4569248 total: 60.5ms remaining: 15.1ms
80: learn: 0.4549812 total: 61ms remaining: 14.3ms
81: learn: 0.4530521 total: 61.7ms remaining: 13.5ms
82: learn: 0.4510327 total: 62.2ms remaining: 12.7ms
83: learn: 0.4492313 total: 62.7ms remaining: 11.9ms
84: learn: 0.4477621 total: 63.3ms remaining: 11.2ms
85: learn: 0.4458929 total: 63.8ms remaining: 10.4ms
86: learn: 0.4441596 total: 64.3ms remaining: 9.6ms
87: learn: 0.4423847 total: 65ms remaining: 8.86ms
88: learn: 0.4405744 total: 65.5ms remaining: 8.1ms
89: learn: 0.4389385 total: 66ms remaining: 7.33ms
90: learn: 0.4370101 total: 66.5ms remaining: 6.58ms
91: learn: 0.4351913 total: 67ms remaining: 5.83ms
92: learn: 0.4333438 total: 67.7ms remaining: 5.09ms
93: learn: 0.4318603 total: 68.2ms remaining: 4.35ms
94: learn: 0.4302856 total: 68.7ms remaining: 3.61ms
95: learn: 0.4287658 total: 69.4ms remaining: 2.89ms
96: learn: 0.4273389 total: 69.9ms remaining: 2.16ms
97: learn: 0.4258300 total: 70.4ms remaining: 1.44ms
98: learn: 0.4241782 total: 71ms remaining: 717us
99: learn: 0.4228510 total: 71.5ms remaining: 0us
0: learn: 0.6884063 total: 2.04ms remaining: 202ms
1: learn: 0.6836852 total: 3.07ms remaining: 150ms
2: learn: 0.6790473 total: 3.94ms remaining: 127ms
3: learn: 0.6744778 total: 5.54ms remaining: 133ms
4: learn: 0.6704643 total: 7.48ms remaining: 142ms
5: learn: 0.6659800 total: 8.92ms remaining: 140ms
6: learn: 0.6620535 total: 10.6ms remaining: 141ms
7: learn: 0.6583088 total: 12ms remaining: 138ms
8: learn: 0.6545450 total: 13.4ms remaining: 135ms
9: learn: 0.6505076 total: 15ms remaining: 135ms
10: learn: 0.6461325 total: 16.2ms remaining: 131ms
11: learn: 0.6423199 total: 17.4ms remaining: 128ms
12: learn: 0.6383978 total: 18.8ms remaining: 126ms
13: learn: 0.6350323 total: 19.7ms remaining: 121ms
14: learn: 0.6308430 total: 21ms remaining: 119ms
15: learn: 0.6269783 total: 22.4ms remaining: 117ms
16: learn: 0.6229115 total: 23.3ms remaining: 114ms
17: learn: 0.6187152 total: 24.2ms remaining: 110ms
18: learn: 0.6157313 total: 25.4ms remaining: 108ms
19: learn: 0.6119853 total: 26.6ms remaining: 106ms
20: learn: 0.6076172 total: 27.5ms remaining: 103ms
21: learn: 0.6042139 total: 28.7ms remaining: 102ms
22: learn: 0.6002465 total: 29.8ms remaining: 99.7ms
23: learn: 0.5968348 total: 30.7ms remaining: 97.1ms
24: learn: 0.5937729 total: 31.6ms remaining: 94.7ms
25: learn: 0.5904123 total: 32.3ms remaining: 92ms
26: learn: 0.5879717 total: 32.9ms remaining: 89ms
27: learn: 0.5843604 total: 34.5ms remaining: 88.8ms
28: learn: 0.5809902 total: 35.2ms remaining: 86.2ms
29: learn: 0.5778176 total: 35.9ms remaining: 83.7ms
30: learn: 0.5746172 total: 36.5ms remaining: 81.3ms
31: learn: 0.5717418 total: 37.4ms remaining: 79.4ms
32: learn: 0.5683009 total: 38.1ms remaining: 77.5ms
33: learn: 0.5651867 total: 38.9ms remaining: 75.4ms
34: learn: 0.5622274 total: 39.5ms remaining: 73.4ms
35: learn: 0.5593118 total: 40.3ms remaining: 71.6ms
36: learn: 0.5561789 total: 40.9ms remaining: 69.7ms
37: learn: 0.5528329 total: 41.6ms remaining: 67.8ms
38: learn: 0.5501205 total: 42.2ms remaining: 66ms
39: learn: 0.5471151 total: 43ms remaining: 64.5ms
40: learn: 0.5442372 total: 43.6ms remaining: 62.8ms
41: learn: 0.5415876 total: 44.2ms remaining: 61.1ms
42: learn: 0.5386989 total: 44.9ms remaining: 59.5ms
43: learn: 0.5359933 total: 45.5ms remaining: 57.9ms
44: learn: 0.5330209 total: 46.2ms remaining: 56.5ms
45: learn: 0.5301928 total: 46.7ms remaining: 54.9ms
46: learn: 0.5281150 total: 47.3ms remaining: 53.4ms
47: learn: 0.5254504 total: 47.9ms remaining: 51.9ms
48: learn: 0.5232090 total: 48.5ms remaining: 50.5ms
49: learn: 0.5201480 total: 49ms remaining: 49ms
50: learn: 0.5178021 total: 49.7ms remaining: 47.8ms
51: learn: 0.5149536 total: 50.3ms remaining: 46.4ms
52: learn: 0.5122542 total: 50.9ms remaining: 45.1ms
53: learn: 0.5095375 total: 51.5ms remaining: 43.8ms
54: learn: 0.5070496 total: 51.7ms remaining: 42.3ms
55: learn: 0.5045973 total: 51.9ms remaining: 40.8ms
56: learn: 0.5026500 total: 52.6ms remaining: 39.7ms
57: learn: 0.5000555 total: 53ms remaining: 38.4ms
58: learn: 0.4980693 total: 53.6ms remaining: 37.2ms
59: learn: 0.4960656 total: 54.2ms remaining: 36.2ms
60: learn: 0.4934948 total: 54.8ms remaining: 35ms
61: learn: 0.4910221 total: 55.3ms remaining: 33.9ms
62: learn: 0.4885432 total: 55.9ms remaining: 32.8ms
63: learn: 0.4861779 total: 56.4ms remaining: 31.7ms
64: learn: 0.4842216 total: 56.9ms remaining: 30.7ms
65: learn: 0.4822751 total: 57.4ms remaining: 29.6ms
66: learn: 0.4801862 total: 57.9ms remaining: 28.5ms
67: learn: 0.4777808 total: 58.4ms remaining: 27.5ms
68: learn: 0.4756700 total: 58.7ms remaining: 26.4ms
69: learn: 0.4734203 total: 59.2ms remaining: 25.4ms
70: learn: 0.4710669 total: 59.7ms remaining: 24.4ms
71: learn: 0.4687042 total: 60.4ms remaining: 23.5ms
72: learn: 0.4670045 total: 60.9ms remaining: 22.5ms
73: learn: 0.4647839 total: 61.5ms remaining: 21.6ms
74: learn: 0.4624318 total: 62.2ms remaining: 20.7ms
75: learn: 0.4606327 total: 62.6ms remaining: 19.8ms
76: learn: 0.4588003 total: 63.1ms remaining: 18.9ms
77: learn: 0.4565626 total: 63.8ms remaining: 18ms
78: learn: 0.4550084 total: 64.3ms remaining: 17.1ms
79: learn: 0.4533107 total: 65ms remaining: 16.2ms
80: learn: 0.4514708 total: 65.6ms remaining: 15.4ms
81: learn: 0.4496577 total: 66.1ms remaining: 14.5ms
82: learn: 0.4483382 total: 66.7ms remaining: 13.7ms
83: learn: 0.4467164 total: 67.2ms remaining: 12.8ms
84: learn: 0.4448025 total: 67.6ms remaining: 11.9ms
85: learn: 0.4434261 total: 68.3ms remaining: 11.1ms
86: learn: 0.4417151 total: 68.8ms remaining: 10.3ms
87: learn: 0.4400095 total: 69ms remaining: 9.41ms
88: learn: 0.4385232 total: 69.6ms remaining: 8.6ms
89: learn: 0.4364587 total: 70.1ms remaining: 7.78ms
90: learn: 0.4347775 total: 70.6ms remaining: 6.98ms
91: learn: 0.4331790 total: 71.1ms remaining: 6.18ms
92: learn: 0.4313283 total: 71.6ms remaining: 5.39ms
93: learn: 0.4297055 total: 72.2ms remaining: 4.61ms
94: learn: 0.4278951 total: 72.8ms remaining: 3.83ms
95: learn: 0.4262358 total: 73.3ms remaining: 3.06ms
96: learn: 0.4247683 total: 74ms remaining: 2.29ms
97: learn: 0.4230988 total: 74.5ms remaining: 1.52ms
98: learn: 0.4215698 total: 75.4ms remaining: 761us
99: learn: 0.4201487 total: 76ms remaining: 0us
0: learn: 0.6884317 total: 1.09ms remaining: 108ms
1: learn: 0.6842219 total: 2.17ms remaining: 106ms
2: learn: 0.6788840 total: 3.1ms remaining: 100ms
3: learn: 0.6745675 total: 3.9ms remaining: 93.5ms
4: learn: 0.6701844 total: 4.18ms remaining: 79.3ms
5: learn: 0.6658574 total: 4.95ms remaining: 77.6ms
6: learn: 0.6613603 total: 5.93ms remaining: 78.8ms
7: learn: 0.6565105 total: 6.92ms remaining: 79.6ms
8: learn: 0.6527962 total: 7.92ms remaining: 80.1ms
9: learn: 0.6488140 total: 8.96ms remaining: 80.7ms
10: learn: 0.6442703 total: 9.92ms remaining: 80.3ms
11: learn: 0.6409121 total: 10.8ms remaining: 79.2ms
12: learn: 0.6361741 total: 11.5ms remaining: 77.1ms
13: learn: 0.6320954 total: 12.4ms remaining: 76.4ms
14: learn: 0.6277527 total: 13.5ms remaining: 76.2ms
15: learn: 0.6234568 total: 14.2ms remaining: 74.4ms
16: learn: 0.6197360 total: 14.9ms remaining: 72.6ms
17: learn: 0.6161622 total: 15.8ms remaining: 72.1ms
18: learn: 0.6126012 total: 16.6ms remaining: 70.8ms
19: learn: 0.6093254 total: 17.3ms remaining: 69.3ms
20: learn: 0.6050689 total: 18.2ms remaining: 68.5ms
21: learn: 0.6013927 total: 19ms remaining: 67.5ms
22: learn: 0.5975716 total: 19.8ms remaining: 66.4ms
23: learn: 0.5938500 total: 20.7ms remaining: 65.6ms
24: learn: 0.5904517 total: 21.5ms remaining: 64.4ms
25: learn: 0.5872580 total: 22.2ms remaining: 63.1ms
26: learn: 0.5841571 total: 22.8ms remaining: 61.7ms
27: learn: 0.5809397 total: 23.3ms remaining: 60ms
28: learn: 0.5776263 total: 24ms remaining: 58.9ms
29: learn: 0.5740388 total: 24.8ms remaining: 57.8ms
30: learn: 0.5708005 total: 25.2ms remaining: 56.2ms
31: learn: 0.5674383 total: 25.9ms remaining: 55ms
32: learn: 0.5643648 total: 26.6ms remaining: 54.1ms
33: learn: 0.5609892 total: 27.3ms remaining: 53ms
34: learn: 0.5574797 total: 28.1ms remaining: 52.1ms
35: learn: 0.5546357 total: 28.8ms remaining: 51.2ms
36: learn: 0.5519836 total: 29.4ms remaining: 50ms
37: learn: 0.5496245 total: 29.9ms remaining: 48.9ms
38: learn: 0.5462709 total: 30.4ms remaining: 47.6ms
39: learn: 0.5438816 total: 31.1ms remaining: 46.6ms
40: learn: 0.5412759 total: 31.7ms remaining: 45.7ms
41: learn: 0.5381534 total: 32.3ms remaining: 44.7ms
42: learn: 0.5350876 total: 32.8ms remaining: 43.5ms
43: learn: 0.5324341 total: 33.5ms remaining: 42.7ms
44: learn: 0.5297221 total: 34.1ms remaining: 41.6ms
45: learn: 0.5269718 total: 34.6ms remaining: 40.7ms
46: learn: 0.5241960 total: 34.9ms remaining: 39.3ms
47: learn: 0.5216740 total: 35.4ms remaining: 38.3ms
48: learn: 0.5188038 total: 35.9ms remaining: 37.4ms
49: learn: 0.5159962 total: 36.6ms remaining: 36.6ms
50: learn: 0.5134614 total: 37.1ms remaining: 35.7ms
51: learn: 0.5114242 total: 37.8ms remaining: 34.9ms
52: learn: 0.5089879 total: 38.3ms remaining: 33.9ms
53: learn: 0.5066030 total: 38.7ms remaining: 33ms
54: learn: 0.5038953 total: 39.5ms remaining: 32.3ms
55: learn: 0.5010538 total: 40ms remaining: 31.4ms
56: learn: 0.4986885 total: 40.5ms remaining: 30.5ms
57: learn: 0.4961412 total: 41.1ms remaining: 29.8ms
58: learn: 0.4942195 total: 41.5ms remaining: 28.8ms
59: learn: 0.4921222 total: 42ms remaining: 28ms
60: learn: 0.4893808 total: 42.7ms remaining: 27.3ms
61: learn: 0.4874546 total: 43.1ms remaining: 26.4ms
62: learn: 0.4855385 total: 43.7ms remaining: 25.7ms
63: learn: 0.4831312 total: 44.4ms remaining: 25ms
64: learn: 0.4809026 total: 45ms remaining: 24.2ms
65: learn: 0.4785241 total: 45.6ms remaining: 23.5ms
66: learn: 0.4758036 total: 46.1ms remaining: 22.7ms
67: learn: 0.4734743 total: 46.6ms remaining: 21.9ms
68: learn: 0.4709811 total: 47.3ms remaining: 21.3ms
69: learn: 0.4688838 total: 47.8ms remaining: 20.5ms
70: learn: 0.4667464 total: 48.5ms remaining: 19.8ms
71: learn: 0.4651299 total: 48.7ms remaining: 18.9ms
72: learn: 0.4632938 total: 49.3ms remaining: 18.2ms
73: learn: 0.4609648 total: 49.8ms remaining: 17.5ms
74: learn: 0.4590642 total: 50.5ms remaining: 16.8ms
75: learn: 0.4568245 total: 50.9ms remaining: 16.1ms
76: learn: 0.4547441 total: 51.5ms remaining: 15.4ms
77: learn: 0.4526380 total: 52.2ms remaining: 14.7ms
78: learn: 0.4505788 total: 52.8ms remaining: 14ms
79: learn: 0.4488851 total: 53.2ms remaining: 13.3ms
80: learn: 0.4468512 total: 53.9ms remaining: 12.6ms
81: learn: 0.4449372 total: 54.4ms remaining: 11.9ms
82: learn: 0.4430671 total: 54.9ms remaining: 11.2ms
83: learn: 0.4410632 total: 55.3ms remaining: 10.5ms
84: learn: 0.4388456 total: 55.8ms remaining: 9.85ms
85: learn: 0.4370682 total: 56.2ms remaining: 9.14ms
86: learn: 0.4359577 total: 56.7ms remaining: 8.48ms
87: learn: 0.4339157 total: 57.2ms remaining: 7.8ms
88: learn: 0.4321642 total: 57.9ms remaining: 7.16ms
89: learn: 0.4306499 total: 58.4ms remaining: 6.49ms
90: learn: 0.4290748 total: 58.9ms remaining: 5.83ms
91: learn: 0.4277414 total: 59.7ms remaining: 5.19ms
92: learn: 0.4257343 total: 60.1ms remaining: 4.53ms
93: learn: 0.4240245 total: 60.6ms remaining: 3.87ms
94: learn: 0.4224567 total: 61.1ms remaining: 3.22ms
95: learn: 0.4204940 total: 61.6ms remaining: 2.57ms
96: learn: 0.4186419 total: 62.1ms remaining: 1.92ms
97: learn: 0.4170743 total: 62.8ms remaining: 1.28ms
98: learn: 0.4158949 total: 63.2ms remaining: 638us
99: learn: 0.4145162 total: 63.7ms remaining: 0us
0: learn: 0.6889045 total: 1.58ms remaining: 156ms
1: learn: 0.6837910 total: 3.07ms remaining: 150ms
2: learn: 0.6796625 total: 4.25ms remaining: 137ms
3: learn: 0.6755106 total: 5.27ms remaining: 127ms
4: learn: 0.6704930 total: 6.12ms remaining: 116ms
5: learn: 0.6659863 total: 7.21ms remaining: 113ms
6: learn: 0.6611236 total: 7.74ms remaining: 103ms
7: learn: 0.6570025 total: 8.9ms remaining: 102ms
8: learn: 0.6525141 total: 10.8ms remaining: 109ms
9: learn: 0.6488466 total: 11.9ms remaining: 107ms
10: learn: 0.6439622 total: 13.1ms remaining: 106ms
11: learn: 0.6399417 total: 14.3ms remaining: 105ms
12: learn: 0.6355915 total: 15.7ms remaining: 105ms
13: learn: 0.6311927 total: 17ms remaining: 104ms
14: learn: 0.6264151 total: 18ms remaining: 102ms
15: learn: 0.6224202 total: 19.3ms remaining: 101ms
16: learn: 0.6179388 total: 20.7ms remaining: 101ms
17: learn: 0.6142164 total: 21.6ms remaining: 98.2ms
18: learn: 0.6103310 total: 22.8ms remaining: 97.1ms
19: learn: 0.6061065 total: 23.9ms remaining: 95.7ms
20: learn: 0.6024065 total: 25.8ms remaining: 97.2ms
21: learn: 0.5988295 total: 26.7ms remaining: 94.7ms
22: learn: 0.5947404 total: 27.6ms remaining: 92.4ms
23: learn: 0.5927571 total: 28.7ms remaining: 91ms
24: learn: 0.5893932 total: 29.6ms remaining: 88.8ms
25: learn: 0.5859566 total: 30.3ms remaining: 86.2ms
26: learn: 0.5824083 total: 31ms remaining: 83.8ms
27: learn: 0.5785285 total: 31.9ms remaining: 82ms
28: learn: 0.5759217 total: 32.7ms remaining: 80.1ms
29: learn: 0.5725713 total: 33.1ms remaining: 77.3ms
30: learn: 0.5701297 total: 33.7ms remaining: 74.9ms
31: learn: 0.5669196 total: 34.2ms remaining: 72.7ms
32: learn: 0.5633647 total: 35ms remaining: 71.1ms
33: learn: 0.5600124 total: 35.6ms remaining: 69ms
34: learn: 0.5570166 total: 36.4ms remaining: 67.7ms
35: learn: 0.5536585 total: 37ms remaining: 65.8ms
36: learn: 0.5501919 total: 37.7ms remaining: 64.1ms
37: learn: 0.5476065 total: 38.2ms remaining: 62.3ms
38: learn: 0.5450105 total: 38.9ms remaining: 60.8ms
39: learn: 0.5427028 total: 39.4ms remaining: 59ms
40: learn: 0.5399803 total: 39.9ms remaining: 57.4ms
41: learn: 0.5374643 total: 40.7ms remaining: 56.2ms
42: learn: 0.5349718 total: 41.1ms remaining: 54.5ms
43: learn: 0.5319629 total: 41.9ms remaining: 53.3ms
44: learn: 0.5290022 total: 42.5ms remaining: 52ms
45: learn: 0.5263070 total: 43.1ms remaining: 50.6ms
46: learn: 0.5235474 total: 43.7ms remaining: 49.3ms
47: learn: 0.5206222 total: 43.9ms remaining: 47.6ms
48: learn: 0.5179628 total: 44.5ms remaining: 46.4ms
49: learn: 0.5158435 total: 45.3ms remaining: 45.3ms
50: learn: 0.5129976 total: 46.3ms remaining: 44.5ms
51: learn: 0.5103865 total: 46.9ms remaining: 43.3ms
52: learn: 0.5075014 total: 47.6ms remaining: 42.2ms
53: learn: 0.5047716 total: 48.1ms remaining: 41ms
54: learn: 0.5022449 total: 48.4ms remaining: 39.6ms
55: learn: 0.4995605 total: 49ms remaining: 38.5ms
56: learn: 0.4978050 total: 49.7ms remaining: 37.5ms
57: learn: 0.4950124 total: 50.5ms remaining: 36.5ms
58: learn: 0.4930733 total: 51.2ms remaining: 35.6ms
59: learn: 0.4904718 total: 51.7ms remaining: 34.5ms
60: learn: 0.4887344 total: 52.3ms remaining: 33.5ms
61: learn: 0.4865266 total: 53.2ms remaining: 32.6ms
62: learn: 0.4841574 total: 54ms remaining: 31.7ms
63: learn: 0.4819785 total: 54.6ms remaining: 30.7ms
64: learn: 0.4804902 total: 55.2ms remaining: 29.7ms
65: learn: 0.4783669 total: 55.9ms remaining: 28.8ms
66: learn: 0.4763882 total: 56.6ms remaining: 27.9ms
67: learn: 0.4736970 total: 57.2ms remaining: 26.9ms
68: learn: 0.4719603 total: 58ms remaining: 26.1ms
69: learn: 0.4694067 total: 58.7ms remaining: 25.1ms
70: learn: 0.4674972 total: 59.2ms remaining: 24.2ms
71: learn: 0.4653817 total: 59.9ms remaining: 23.3ms
72: learn: 0.4635985 total: 60.5ms remaining: 22.4ms
73: learn: 0.4621394 total: 60.9ms remaining: 21.4ms
74: learn: 0.4604022 total: 61.6ms remaining: 20.5ms
75: learn: 0.4587158 total: 62.2ms remaining: 19.6ms
76: learn: 0.4569038 total: 62.9ms remaining: 18.8ms
77: learn: 0.4550002 total: 63.4ms remaining: 17.9ms
78: learn: 0.4529074 total: 64.3ms remaining: 17.1ms
79: learn: 0.4509757 total: 65ms remaining: 16.3ms
80: learn: 0.4487447 total: 65.6ms remaining: 15.4ms
81: learn: 0.4465551 total: 66.1ms remaining: 14.5ms
82: learn: 0.4446271 total: 66.9ms remaining: 13.7ms
83: learn: 0.4426076 total: 67.7ms remaining: 12.9ms
84: learn: 0.4404503 total: 68.2ms remaining: 12ms
85: learn: 0.4387267 total: 68.9ms remaining: 11.2ms
86: learn: 0.4373792 total: 69.3ms remaining: 10.4ms
87: learn: 0.4353366 total: 69.9ms remaining: 9.53ms
88: learn: 0.4331099 total: 70.6ms remaining: 8.72ms
89: learn: 0.4309840 total: 71.3ms remaining: 7.92ms
90: learn: 0.4289889 total: 72.1ms remaining: 7.13ms
91: learn: 0.4270319 total: 72.6ms remaining: 6.31ms
92: learn: 0.4252756 total: 73.3ms remaining: 5.51ms
93: learn: 0.4234406 total: 73.7ms remaining: 4.71ms
94: learn: 0.4218919 total: 74.3ms remaining: 3.91ms
95: learn: 0.4204512 total: 75ms remaining: 3.13ms
96: learn: 0.4189420 total: 75.6ms remaining: 2.34ms
97: learn: 0.4176835 total: 76.3ms remaining: 1.56ms
98: learn: 0.4156564 total: 76.9ms remaining: 776us
99: learn: 0.4141609 total: 77.4ms remaining: 0us
0: learn: 0.6881669 total: 911us remaining: 90.2ms
1: learn: 0.6833468 total: 1.57ms remaining: 76.8ms
2: learn: 0.6791169 total: 3.59ms remaining: 116ms
3: learn: 0.6758376 total: 4.42ms remaining: 106ms
4: learn: 0.6712291 total: 5.11ms remaining: 97.2ms
5: learn: 0.6669118 total: 8.96ms remaining: 140ms
6: learn: 0.6632769 total: 10.3ms remaining: 137ms
7: learn: 0.6589892 total: 11.5ms remaining: 133ms
8: learn: 0.6545352 total: 12.4ms remaining: 125ms
9: learn: 0.6497433 total: 13.6ms remaining: 122ms
10: learn: 0.6459731 total: 14.1ms remaining: 114ms
11: learn: 0.6423070 total: 14.7ms remaining: 108ms
12: learn: 0.6378382 total: 15.4ms remaining: 103ms
13: learn: 0.6342830 total: 15.9ms remaining: 97.9ms
14: learn: 0.6305179 total: 17.3ms remaining: 98.1ms
15: learn: 0.6273236 total: 18ms remaining: 94.5ms
16: learn: 0.6233593 total: 18.9ms remaining: 92.1ms
17: learn: 0.6202737 total: 19.5ms remaining: 89ms
18: learn: 0.6164205 total: 20.2ms remaining: 85.9ms
19: learn: 0.6125395 total: 21.5ms remaining: 86ms
20: learn: 0.6093116 total: 22.7ms remaining: 85.3ms
21: learn: 0.6057181 total: 23.2ms remaining: 82.4ms
22: learn: 0.6021154 total: 23.9ms remaining: 80.2ms
23: learn: 0.5986431 total: 24.4ms remaining: 77.3ms
24: learn: 0.5948586 total: 25.1ms remaining: 75.3ms
25: learn: 0.5912135 total: 26.1ms remaining: 74.2ms
26: learn: 0.5876534 total: 26.7ms remaining: 72.1ms
27: learn: 0.5839998 total: 27.2ms remaining: 69.9ms
28: learn: 0.5802642 total: 27.7ms remaining: 67.8ms
29: learn: 0.5771353 total: 28.5ms remaining: 66.5ms
30: learn: 0.5739135 total: 29.1ms remaining: 64.7ms
31: learn: 0.5708365 total: 30.5ms remaining: 64.9ms
32: learn: 0.5672966 total: 31.3ms remaining: 63.6ms
33: learn: 0.5646367 total: 32.3ms remaining: 62.7ms
34: learn: 0.5611941 total: 32.9ms remaining: 61.1ms
35: learn: 0.5586881 total: 33.6ms remaining: 59.8ms
36: learn: 0.5552103 total: 34.5ms remaining: 58.7ms
37: learn: 0.5529219 total: 35.3ms remaining: 57.5ms
38: learn: 0.5496374 total: 35.8ms remaining: 55.9ms
39: learn: 0.5473225 total: 36.2ms remaining: 54.3ms
40: learn: 0.5442109 total: 37ms remaining: 53.3ms
41: learn: 0.5409920 total: 37.6ms remaining: 51.9ms
42: learn: 0.5386969 total: 38.2ms remaining: 50.6ms
43: learn: 0.5358866 total: 38.9ms remaining: 49.5ms
44: learn: 0.5330785 total: 39.4ms remaining: 48.2ms
45: learn: 0.5305521 total: 40.1ms remaining: 47.1ms
46: learn: 0.5278588 total: 40.6ms remaining: 45.8ms
47: learn: 0.5249340 total: 41.3ms remaining: 44.8ms
48: learn: 0.5217724 total: 41.9ms remaining: 43.6ms
49: learn: 0.5202045 total: 42.5ms remaining: 42.5ms
50: learn: 0.5176788 total: 43.6ms remaining: 41.9ms
51: learn: 0.5153876 total: 44.6ms remaining: 41.2ms
52: learn: 0.5134912 total: 45.1ms remaining: 40ms
53: learn: 0.5109341 total: 45.8ms remaining: 39ms
54: learn: 0.5087370 total: 46.4ms remaining: 37.9ms
55: learn: 0.5064716 total: 46.9ms remaining: 36.9ms
56: learn: 0.5044754 total: 47.8ms remaining: 36.1ms
57: learn: 0.5018445 total: 48.8ms remaining: 35.3ms
58: learn: 0.5001153 total: 49.5ms remaining: 34.4ms
59: learn: 0.4983187 total: 50.1ms remaining: 33.4ms
60: learn: 0.4963267 total: 50.6ms remaining: 32.4ms
61: learn: 0.4940461 total: 51.2ms remaining: 31.4ms
62: learn: 0.4919278 total: 51.8ms remaining: 30.4ms
63: learn: 0.4896749 total: 52.8ms remaining: 29.7ms
64: learn: 0.4871274 total: 53.3ms remaining: 28.7ms
65: learn: 0.4849744 total: 53.9ms remaining: 27.8ms
66: learn: 0.4826793 total: 54.5ms remaining: 26.8ms
67: learn: 0.4805057 total: 55ms remaining: 25.9ms
68: learn: 0.4780862 total: 55.5ms remaining: 25ms
69: learn: 0.4767565 total: 56ms remaining: 24ms
70: learn: 0.4748172 total: 56.8ms remaining: 23.2ms
71: learn: 0.4731001 total: 57.4ms remaining: 22.3ms
72: learn: 0.4707187 total: 58.3ms remaining: 21.6ms
73: learn: 0.4686882 total: 58.9ms remaining: 20.7ms
74: learn: 0.4667510 total: 59.4ms remaining: 19.8ms
75: learn: 0.4646721 total: 60.2ms remaining: 19ms
76: learn: 0.4628371 total: 60.8ms remaining: 18.2ms
77: learn: 0.4609072 total: 61.5ms remaining: 17.3ms
78: learn: 0.4590288 total: 62ms remaining: 16.5ms
79: learn: 0.4570339 total: 62.5ms remaining: 15.6ms
80: learn: 0.4548811 total: 63ms remaining: 14.8ms
81: learn: 0.4532088 total: 63.6ms remaining: 14ms
82: learn: 0.4512085 total: 64.3ms remaining: 13.2ms
83: learn: 0.4492292 total: 64.7ms remaining: 12.3ms
84: learn: 0.4471357 total: 65.2ms remaining: 11.5ms
85: learn: 0.4453762 total: 66.1ms remaining: 10.8ms
86: learn: 0.4435967 total: 66.8ms remaining: 9.98ms
87: learn: 0.4418189 total: 67.3ms remaining: 9.17ms
88: learn: 0.4400782 total: 67.8ms remaining: 8.38ms
89: learn: 0.4380934 total: 68.4ms remaining: 7.6ms
90: learn: 0.4362762 total: 69ms remaining: 6.83ms
91: learn: 0.4348678 total: 69.4ms remaining: 6.04ms
92: learn: 0.4329523 total: 70.3ms remaining: 5.29ms
93: learn: 0.4312429 total: 71ms remaining: 4.53ms
94: learn: 0.4295643 total: 71.6ms remaining: 3.77ms
95: learn: 0.4275956 total: 72.1ms remaining: 3ms
96: learn: 0.4261009 total: 72.5ms remaining: 2.24ms
97: learn: 0.4245874 total: 73.1ms remaining: 1.49ms
98: learn: 0.4235023 total: 73.6ms remaining: 743us
99: learn: 0.4217160 total: 74.3ms remaining: 0us
0: learn: 0.6881313 total: 719us remaining: 71.3ms
1: learn: 0.6834396 total: 1.23ms remaining: 60.5ms
2: learn: 0.6788678 total: 1.84ms remaining: 59.6ms
3: learn: 0.6745282 total: 2.35ms remaining: 56.4ms
4: learn: 0.6705313 total: 3.15ms remaining: 59.8ms
5: learn: 0.6664501 total: 3.88ms remaining: 60.8ms
6: learn: 0.6625251 total: 4.48ms remaining: 59.5ms
7: learn: 0.6589340 total: 5.26ms remaining: 60.5ms
8: learn: 0.6541913 total: 5.98ms remaining: 60.4ms
9: learn: 0.6498026 total: 6.46ms remaining: 58.2ms
10: learn: 0.6453986 total: 7.02ms remaining: 56.8ms
11: learn: 0.6421787 total: 7.65ms remaining: 56.1ms
12: learn: 0.6387500 total: 8.22ms remaining: 55ms
13: learn: 0.6352154 total: 8.76ms remaining: 53.8ms
14: learn: 0.6312877 total: 9.55ms remaining: 54.1ms
15: learn: 0.6279324 total: 10.2ms remaining: 53.4ms
16: learn: 0.6247067 total: 10.6ms remaining: 51.8ms
17: learn: 0.6210395 total: 11.3ms remaining: 51.5ms
18: learn: 0.6179746 total: 11.9ms remaining: 50.7ms
19: learn: 0.6143485 total: 12.5ms remaining: 49.9ms
20: learn: 0.6106524 total: 13.2ms remaining: 49.5ms
21: learn: 0.6066544 total: 13.7ms remaining: 48.4ms
22: learn: 0.6042317 total: 14.3ms remaining: 47.8ms
23: learn: 0.6005581 total: 15.1ms remaining: 47.7ms
24: learn: 0.5968835 total: 15.6ms remaining: 46.8ms
25: learn: 0.5931447 total: 16.1ms remaining: 45.8ms
26: learn: 0.5890090 total: 16.7ms remaining: 45.1ms
27: learn: 0.5856988 total: 17.2ms remaining: 44.3ms
28: learn: 0.5825885 total: 17.8ms remaining: 43.5ms
29: learn: 0.5794557 total: 18.5ms remaining: 43.1ms
30: learn: 0.5759834 total: 19ms remaining: 42.4ms
31: learn: 0.5738438 total: 19.6ms remaining: 41.7ms
32: learn: 0.5708319 total: 20.3ms remaining: 41.2ms
33: learn: 0.5679970 total: 20.9ms remaining: 40.5ms
34: learn: 0.5651570 total: 21.4ms remaining: 39.8ms
35: learn: 0.5631887 total: 21.9ms remaining: 39ms
36: learn: 0.5605275 total: 22.4ms remaining: 38.1ms
37: learn: 0.5577177 total: 23ms remaining: 37.6ms
38: learn: 0.5547583 total: 24ms remaining: 37.5ms
39: learn: 0.5524270 total: 24.6ms remaining: 36.9ms
40: learn: 0.5500806 total: 25.1ms remaining: 36.2ms
41: learn: 0.5475216 total: 25.7ms remaining: 35.5ms
42: learn: 0.5446629 total: 26.3ms remaining: 34.9ms
43: learn: 0.5419058 total: 27ms remaining: 34.3ms
44: learn: 0.5387193 total: 27.5ms remaining: 33.6ms
45: learn: 0.5362276 total: 28.1ms remaining: 33ms
46: learn: 0.5335108 total: 28.6ms remaining: 32.2ms
47: learn: 0.5313886 total: 29.2ms remaining: 31.7ms
48: learn: 0.5288603 total: 29.6ms remaining: 30.8ms
49: learn: 0.5261072 total: 30.2ms remaining: 30.2ms
50: learn: 0.5233041 total: 30.9ms remaining: 29.7ms
51: learn: 0.5202423 total: 31.4ms remaining: 29ms
52: learn: 0.5173466 total: 31.9ms remaining: 28.3ms
53: learn: 0.5144277 total: 32.7ms remaining: 27.8ms
54: learn: 0.5122916 total: 33.1ms remaining: 27.1ms
55: learn: 0.5098521 total: 33.7ms remaining: 26.5ms
56: learn: 0.5074225 total: 34.2ms remaining: 25.8ms
57: learn: 0.5051557 total: 35ms remaining: 25.3ms
58: learn: 0.5028183 total: 35.2ms remaining: 24.5ms
59: learn: 0.5006007 total: 35.8ms remaining: 23.9ms
60: learn: 0.4986387 total: 36.8ms remaining: 23.5ms
61: learn: 0.4959748 total: 37.7ms remaining: 23.1ms
62: learn: 0.4936445 total: 38.3ms remaining: 22.5ms
63: learn: 0.4911664 total: 39ms remaining: 21.9ms
64: learn: 0.4888798 total: 39.5ms remaining: 21.3ms
65: learn: 0.4864808 total: 40ms remaining: 20.6ms
66: learn: 0.4852232 total: 40.5ms remaining: 19.9ms
67: learn: 0.4828485 total: 41ms remaining: 19.3ms
68: learn: 0.4805835 total: 41.6ms remaining: 18.7ms
69: learn: 0.4785631 total: 42.2ms remaining: 18.1ms
70: learn: 0.4760562 total: 42.8ms remaining: 17.5ms
71: learn: 0.4736100 total: 43.4ms remaining: 16.9ms
72: learn: 0.4716507 total: 44ms remaining: 16.3ms
73: learn: 0.4697942 total: 44.7ms remaining: 15.7ms
74: learn: 0.4674482 total: 45.1ms remaining: 15.1ms
75: learn: 0.4658586 total: 45.7ms remaining: 14.4ms
76: learn: 0.4637583 total: 46.1ms remaining: 13.8ms
77: learn: 0.4619726 total: 46.7ms remaining: 13.2ms
78: learn: 0.4600564 total: 47.2ms remaining: 12.5ms
79: learn: 0.4579343 total: 47.7ms remaining: 11.9ms
80: learn: 0.4559391 total: 48.8ms remaining: 11.5ms
81: learn: 0.4543580 total: 49.4ms remaining: 10.8ms
82: learn: 0.4521845 total: 49.9ms remaining: 10.2ms
83: learn: 0.4505865 total: 50.7ms remaining: 9.65ms
84: learn: 0.4484197 total: 51.2ms remaining: 9.04ms
85: learn: 0.4466925 total: 51.7ms remaining: 8.41ms
86: learn: 0.4447067 total: 52.3ms remaining: 7.82ms
87: learn: 0.4426456 total: 53ms remaining: 7.22ms
88: learn: 0.4409240 total: 53.5ms remaining: 6.62ms
89: learn: 0.4392475 total: 54.2ms remaining: 6.02ms
90: learn: 0.4375688 total: 54.7ms remaining: 5.41ms
91: learn: 0.4357843 total: 55.2ms remaining: 4.8ms
92: learn: 0.4340597 total: 55.9ms remaining: 4.21ms
93: learn: 0.4321912 total: 56.5ms remaining: 3.61ms
94: learn: 0.4307065 total: 57ms remaining: 3ms
95: learn: 0.4292643 total: 57.5ms remaining: 2.4ms
96: learn: 0.4275736 total: 58.1ms remaining: 1.8ms
97: learn: 0.4259144 total: 58.6ms remaining: 1.2ms
98: learn: 0.4248509 total: 59.1ms remaining: 596us
99: learn: 0.4234812 total: 59.8ms remaining: 0us
0: learn: 0.6882460 total: 764us remaining: 75.7ms
1: learn: 0.6843912 total: 1.33ms remaining: 65ms
2: learn: 0.6800541 total: 1.92ms remaining: 62.1ms
3: learn: 0.6760891 total: 2.57ms remaining: 61.7ms
4: learn: 0.6712217 total: 3.11ms remaining: 59.1ms
5: learn: 0.6672614 total: 3.83ms remaining: 60ms
6: learn: 0.6625482 total: 4.45ms remaining: 59.2ms
7: learn: 0.6584332 total: 5.09ms remaining: 58.5ms
8: learn: 0.6542506 total: 5.64ms remaining: 57ms
9: learn: 0.6496036 total: 6.26ms remaining: 56.3ms
10: learn: 0.6458211 total: 6.72ms remaining: 54.4ms
11: learn: 0.6409358 total: 7.08ms remaining: 51.9ms
12: learn: 0.6366605 total: 7.77ms remaining: 52ms
13: learn: 0.6327545 total: 8.53ms remaining: 52.4ms
14: learn: 0.6285988 total: 9.21ms remaining: 52.2ms
15: learn: 0.6249008 total: 9.71ms remaining: 51ms
16: learn: 0.6202061 total: 10.3ms remaining: 50.3ms
17: learn: 0.6165730 total: 10.9ms remaining: 49.5ms
18: learn: 0.6143048 total: 11.5ms remaining: 49ms
19: learn: 0.6101026 total: 12.1ms remaining: 48.4ms
20: learn: 0.6062540 total: 12.6ms remaining: 47.4ms
21: learn: 0.6024226 total: 13.2ms remaining: 46.9ms
22: learn: 0.5992711 total: 14ms remaining: 46.8ms
23: learn: 0.5962842 total: 14.4ms remaining: 45.7ms
24: learn: 0.5930634 total: 15ms remaining: 45ms
25: learn: 0.5896212 total: 15.7ms remaining: 44.7ms
26: learn: 0.5862146 total: 16.2ms remaining: 43.9ms
27: learn: 0.5836004 total: 16.8ms remaining: 43.2ms
28: learn: 0.5806826 total: 17.5ms remaining: 42.9ms
29: learn: 0.5778009 total: 18.2ms remaining: 42.5ms
30: learn: 0.5747073 total: 18.7ms remaining: 41.7ms
31: learn: 0.5711173 total: 19.4ms remaining: 41.1ms
32: learn: 0.5676276 total: 20ms remaining: 40.6ms
33: learn: 0.5649525 total: 20.5ms remaining: 39.8ms
34: learn: 0.5614809 total: 21ms remaining: 38.9ms
35: learn: 0.5586365 total: 21.5ms remaining: 38.2ms
36: learn: 0.5555984 total: 22.2ms remaining: 37.8ms
37: learn: 0.5521355 total: 22.9ms remaining: 37.4ms
38: learn: 0.5491231 total: 23.6ms remaining: 36.9ms
39: learn: 0.5461905 total: 24.2ms remaining: 36.3ms
40: learn: 0.5435150 total: 24.7ms remaining: 35.6ms
41: learn: 0.5400794 total: 25.8ms remaining: 35.6ms
42: learn: 0.5369342 total: 26.8ms remaining: 35.6ms
43: learn: 0.5338750 total: 28.3ms remaining: 36ms
44: learn: 0.5307068 total: 29.8ms remaining: 36.4ms
45: learn: 0.5278970 total: 31.7ms remaining: 37.2ms
46: learn: 0.5253307 total: 33.2ms remaining: 37.4ms
47: learn: 0.5227516 total: 35.4ms remaining: 38.3ms
48: learn: 0.5203989 total: 37.2ms remaining: 38.7ms
49: learn: 0.5173267 total: 39ms remaining: 39ms
50: learn: 0.5145928 total: 40.6ms remaining: 39ms
51: learn: 0.5118905 total: 42.3ms remaining: 39.1ms
52: learn: 0.5091929 total: 44.2ms remaining: 39.2ms
53: learn: 0.5063144 total: 45.6ms remaining: 38.8ms
54: learn: 0.5039401 total: 46.3ms remaining: 37.9ms
55: learn: 0.5018232 total: 48.3ms remaining: 38ms
56: learn: 0.4995300 total: 49.8ms remaining: 37.6ms
57: learn: 0.4967105 total: 51.3ms remaining: 37.2ms
58: learn: 0.4942444 total: 53.5ms remaining: 37.2ms
59: learn: 0.4914428 total: 55.2ms remaining: 36.8ms
60: learn: 0.4890672 total: 56.4ms remaining: 36ms
61: learn: 0.4865293 total: 58.2ms remaining: 35.7ms
62: learn: 0.4842583 total: 60.3ms remaining: 35.4ms
63: learn: 0.4821064 total: 61.8ms remaining: 34.8ms
64: learn: 0.4797389 total: 63.4ms remaining: 34.1ms
65: learn: 0.4770447 total: 65ms remaining: 33.5ms
66: learn: 0.4746364 total: 66.7ms remaining: 32.8ms
67: learn: 0.4720423 total: 68.6ms remaining: 32.3ms
68: learn: 0.4695496 total: 70.3ms remaining: 31.6ms
69: learn: 0.4675379 total: 72.4ms remaining: 31ms
70: learn: 0.4651009 total: 74.5ms remaining: 30.4ms
71: learn: 0.4625320 total: 76.3ms remaining: 29.7ms
72: learn: 0.4602102 total: 78ms remaining: 28.8ms
73: learn: 0.4587625 total: 78.8ms remaining: 27.7ms
74: learn: 0.4569151 total: 80.5ms remaining: 26.8ms
75: learn: 0.4547828 total: 82.4ms remaining: 26ms
76: learn: 0.4529937 total: 84.3ms remaining: 25.2ms
77: learn: 0.4508657 total: 86.4ms remaining: 24.4ms
78: learn: 0.4491949 total: 88.3ms remaining: 23.5ms
79: learn: 0.4468518 total: 89.6ms remaining: 22.4ms
80: learn: 0.4447912 total: 91.2ms remaining: 21.4ms
81: learn: 0.4434849 total: 93.1ms remaining: 20.4ms
82: learn: 0.4417231 total: 94.4ms remaining: 19.3ms
83: learn: 0.4396984 total: 96.3ms remaining: 18.4ms
84: learn: 0.4379999 total: 97.1ms remaining: 17.1ms
85: learn: 0.4360282 total: 99ms remaining: 16.1ms
86: learn: 0.4341880 total: 101ms remaining: 15.1ms
87: learn: 0.4319460 total: 102ms remaining: 14ms
88: learn: 0.4299689 total: 104ms remaining: 12.9ms
89: learn: 0.4280744 total: 106ms remaining: 11.8ms
90: learn: 0.4261640 total: 108ms remaining: 10.7ms
91: learn: 0.4248069 total: 109ms remaining: 9.51ms
92: learn: 0.4230969 total: 110ms remaining: 8.29ms
93: learn: 0.4215900 total: 111ms remaining: 7.1ms
94: learn: 0.4198851 total: 113ms remaining: 5.93ms
95: learn: 0.4179986 total: 114ms remaining: 4.76ms
96: learn: 0.4166283 total: 116ms remaining: 3.59ms
97: learn: 0.4152687 total: 117ms remaining: 2.4ms
98: learn: 0.4135474 total: 119ms remaining: 1.2ms
99: learn: 0.4116823 total: 120ms remaining: 0us
0: learn: 0.6893152 total: 3.63ms remaining: 360ms
1: learn: 0.6844835 total: 5.97ms remaining: 293ms
2: learn: 0.6800974 total: 7.31ms remaining: 236ms
3: learn: 0.6760208 total: 9.65ms remaining: 232ms
4: learn: 0.6719295 total: 11.3ms remaining: 215ms
5: learn: 0.6673758 total: 12.8ms remaining: 201ms
6: learn: 0.6621889 total: 14.8ms remaining: 197ms
7: learn: 0.6576188 total: 16.1ms remaining: 185ms
8: learn: 0.6534662 total: 17.8ms remaining: 180ms
9: learn: 0.6495321 total: 19.9ms remaining: 179ms
10: learn: 0.6456604 total: 21ms remaining: 170ms
11: learn: 0.6406508 total: 22.2ms remaining: 162ms
12: learn: 0.6362949 total: 24.1ms remaining: 161ms
13: learn: 0.6323097 total: 25.3ms remaining: 155ms
14: learn: 0.6280563 total: 26.3ms remaining: 149ms
15: learn: 0.6241475 total: 26.9ms remaining: 141ms
16: learn: 0.6197190 total: 28.2ms remaining: 138ms
17: learn: 0.6159291 total: 28.6ms remaining: 130ms
18: learn: 0.6123843 total: 29.5ms remaining: 126ms
19: learn: 0.6095165 total: 30.6ms remaining: 122ms
20: learn: 0.6066874 total: 31.6ms remaining: 119ms
21: learn: 0.6032681 total: 32.2ms remaining: 114ms
22: learn: 0.6000013 total: 33.2ms remaining: 111ms
23: learn: 0.5968636 total: 33.8ms remaining: 107ms
24: learn: 0.5936499 total: 34.4ms remaining: 103ms
25: learn: 0.5902258 total: 35ms remaining: 99.7ms
26: learn: 0.5872464 total: 36ms remaining: 97.2ms
27: learn: 0.5840496 total: 36.8ms remaining: 94.6ms
28: learn: 0.5803844 total: 37.7ms remaining: 92.4ms
29: learn: 0.5765966 total: 38.3ms remaining: 89.3ms
30: learn: 0.5729305 total: 38.9ms remaining: 86.6ms
31: learn: 0.5693852 total: 39.8ms remaining: 84.5ms
32: learn: 0.5662854 total: 40.4ms remaining: 82.1ms
33: learn: 0.5633348 total: 41.3ms remaining: 80.1ms
34: learn: 0.5603130 total: 42ms remaining: 78.1ms
35: learn: 0.5570380 total: 42.7ms remaining: 75.9ms
36: learn: 0.5535011 total: 43.4ms remaining: 73.9ms
37: learn: 0.5506970 total: 43.9ms remaining: 71.7ms
38: learn: 0.5475887 total: 44.6ms remaining: 69.8ms
39: learn: 0.5448545 total: 45.2ms remaining: 67.8ms
40: learn: 0.5416395 total: 46ms remaining: 66.1ms
41: learn: 0.5385460 total: 46.4ms remaining: 64.1ms
42: learn: 0.5358333 total: 47ms remaining: 62.3ms
43: learn: 0.5329987 total: 47.7ms remaining: 60.7ms
44: learn: 0.5306597 total: 48.3ms remaining: 59ms
45: learn: 0.5274482 total: 48.9ms remaining: 57.4ms
46: learn: 0.5249510 total: 49.4ms remaining: 55.7ms
47: learn: 0.5218486 total: 49.9ms remaining: 54.1ms
48: learn: 0.5192859 total: 50.6ms remaining: 52.6ms
49: learn: 0.5163437 total: 51.1ms remaining: 51.1ms
50: learn: 0.5135689 total: 51.6ms remaining: 49.6ms
51: learn: 0.5116517 total: 52.3ms remaining: 48.3ms
52: learn: 0.5093598 total: 52.8ms remaining: 46.8ms
53: learn: 0.5070659 total: 53.2ms remaining: 45.3ms
54: learn: 0.5050415 total: 53.7ms remaining: 44ms
55: learn: 0.5028938 total: 54.4ms remaining: 42.8ms
56: learn: 0.5009031 total: 55.1ms remaining: 41.5ms
57: learn: 0.4986966 total: 55.6ms remaining: 40.3ms
58: learn: 0.4965786 total: 56.2ms remaining: 39ms
59: learn: 0.4941129 total: 56.6ms remaining: 37.8ms
60: learn: 0.4915667 total: 57.3ms remaining: 36.6ms
61: learn: 0.4892120 total: 57.8ms remaining: 35.4ms
62: learn: 0.4869451 total: 58.1ms remaining: 34.1ms
63: learn: 0.4852238 total: 58.7ms remaining: 33ms
64: learn: 0.4831940 total: 59.2ms remaining: 31.9ms
65: learn: 0.4807932 total: 59.8ms remaining: 30.8ms
66: learn: 0.4788962 total: 60.2ms remaining: 29.6ms
67: learn: 0.4765024 total: 60.7ms remaining: 28.6ms
68: learn: 0.4741526 total: 61.3ms remaining: 27.5ms
69: learn: 0.4715532 total: 61.8ms remaining: 26.5ms
70: learn: 0.4699507 total: 62.5ms remaining: 25.5ms
71: learn: 0.4676305 total: 63ms remaining: 24.5ms
72: learn: 0.4656121 total: 63.6ms remaining: 23.5ms
73: learn: 0.4637803 total: 64.3ms remaining: 22.6ms
74: learn: 0.4620908 total: 64.7ms remaining: 21.6ms
75: learn: 0.4602817 total: 65.3ms remaining: 20.6ms
76: learn: 0.4583853 total: 65.9ms remaining: 19.7ms
77: learn: 0.4563427 total: 66.4ms remaining: 18.7ms
78: learn: 0.4543921 total: 67.1ms remaining: 17.8ms
79: learn: 0.4526397 total: 67.7ms remaining: 16.9ms
80: learn: 0.4505288 total: 68.2ms remaining: 16ms
81: learn: 0.4492302 total: 68.7ms remaining: 15.1ms
82: learn: 0.4476243 total: 69.2ms remaining: 14.2ms
83: learn: 0.4458867 total: 69.7ms remaining: 13.3ms
84: learn: 0.4440911 total: 70.4ms remaining: 12.4ms
85: learn: 0.4425387 total: 70.9ms remaining: 11.5ms
86: learn: 0.4410423 total: 71.4ms remaining: 10.7ms
87: learn: 0.4391265 total: 71.8ms remaining: 9.8ms
88: learn: 0.4379439 total: 72.4ms remaining: 8.94ms
89: learn: 0.4361668 total: 72.9ms remaining: 8.1ms
90: learn: 0.4345088 total: 73.6ms remaining: 7.28ms
91: learn: 0.4325576 total: 74.2ms remaining: 6.45ms
92: learn: 0.4307234 total: 74.7ms remaining: 5.63ms
93: learn: 0.4288036 total: 75.3ms remaining: 4.8ms
94: learn: 0.4272949 total: 75.7ms remaining: 3.99ms
95: learn: 0.4257101 total: 76.3ms remaining: 3.18ms
96: learn: 0.4241792 total: 76.8ms remaining: 2.38ms
97: learn: 0.4225862 total: 77.4ms remaining: 1.58ms
98: learn: 0.4210460 total: 77.9ms remaining: 787us
99: learn: 0.4196173 total: 78.6ms remaining: 0us
0: learn: 0.6898834 total: 1.58ms remaining: 157ms
1: learn: 0.6842462 total: 2.19ms remaining: 108ms
2: learn: 0.6792858 total: 2.69ms remaining: 86.9ms
3: learn: 0.6746738 total: 3.33ms remaining: 79.9ms
4: learn: 0.6695928 total: 4.52ms remaining: 85.9ms
5: learn: 0.6661403 total: 6.01ms remaining: 94.1ms
6: learn: 0.6611720 total: 7.32ms remaining: 97.3ms
7: learn: 0.6566346 total: 8.03ms remaining: 92.3ms
8: learn: 0.6521007 total: 8.76ms remaining: 88.5ms
9: learn: 0.6483892 total: 9.84ms remaining: 88.5ms
10: learn: 0.6445378 total: 11ms remaining: 89.3ms
11: learn: 0.6402623 total: 11.5ms remaining: 84.4ms
12: learn: 0.6356433 total: 12.5ms remaining: 83.3ms
13: learn: 0.6320741 total: 13.7ms remaining: 84.1ms
14: learn: 0.6293125 total: 15ms remaining: 85ms
15: learn: 0.6252919 total: 16ms remaining: 84ms
16: learn: 0.6215804 total: 16.8ms remaining: 82.1ms
17: learn: 0.6181259 total: 17.4ms remaining: 79.5ms
18: learn: 0.6150490 total: 18.7ms remaining: 79.5ms
19: learn: 0.6107875 total: 19.9ms remaining: 79.5ms
20: learn: 0.6067928 total: 20.4ms remaining: 76.6ms
21: learn: 0.6026918 total: 20.9ms remaining: 74ms
22: learn: 0.5992985 total: 21.5ms remaining: 72.1ms
23: learn: 0.5966905 total: 22.5ms remaining: 71.3ms
24: learn: 0.5929736 total: 23.8ms remaining: 71.4ms
25: learn: 0.5903067 total: 24.7ms remaining: 70.4ms
26: learn: 0.5869536 total: 25.7ms remaining: 69.4ms
27: learn: 0.5834395 total: 26.8ms remaining: 68.9ms
28: learn: 0.5804730 total: 27.9ms remaining: 68.4ms
29: learn: 0.5774620 total: 28.9ms remaining: 67.5ms
30: learn: 0.5742536 total: 30.1ms remaining: 67ms
31: learn: 0.5709241 total: 30.8ms remaining: 65.4ms
32: learn: 0.5676292 total: 31.6ms remaining: 64.2ms
33: learn: 0.5644774 total: 32.6ms remaining: 63.3ms
34: learn: 0.5607723 total: 33.4ms remaining: 62ms
35: learn: 0.5575395 total: 34.7ms remaining: 61.8ms
36: learn: 0.5550180 total: 35.5ms remaining: 60.5ms
37: learn: 0.5519310 total: 36.4ms remaining: 59.3ms
38: learn: 0.5485639 total: 37.1ms remaining: 58.1ms
39: learn: 0.5453633 total: 37.9ms remaining: 56.8ms
40: learn: 0.5423500 total: 38.7ms remaining: 55.6ms
41: learn: 0.5392707 total: 39.2ms remaining: 54.2ms
42: learn: 0.5363255 total: 39.7ms remaining: 52.7ms
43: learn: 0.5335525 total: 40.6ms remaining: 51.7ms
44: learn: 0.5315166 total: 41.3ms remaining: 50.5ms
45: learn: 0.5284654 total: 42.1ms remaining: 49.4ms
46: learn: 0.5254547 total: 42.6ms remaining: 48.1ms
47: learn: 0.5226909 total: 42.9ms remaining: 46.5ms
48: learn: 0.5204454 total: 43.7ms remaining: 45.5ms
49: learn: 0.5174313 total: 44.3ms remaining: 44.3ms
50: learn: 0.5152630 total: 44.7ms remaining: 43ms
51: learn: 0.5123739 total: 45.5ms remaining: 42ms
52: learn: 0.5097095 total: 46.1ms remaining: 40.9ms
53: learn: 0.5073269 total: 46.7ms remaining: 39.8ms
54: learn: 0.5046066 total: 47.3ms remaining: 38.7ms
55: learn: 0.5022936 total: 47.9ms remaining: 37.6ms
56: learn: 0.5000231 total: 48.5ms remaining: 36.6ms
57: learn: 0.4977305 total: 49ms remaining: 35.5ms
58: learn: 0.4960952 total: 49.5ms remaining: 34.4ms
59: learn: 0.4938946 total: 50.3ms remaining: 33.6ms
60: learn: 0.4915094 total: 50.8ms remaining: 32.5ms
61: learn: 0.4893572 total: 51.4ms remaining: 31.5ms
62: learn: 0.4875191 total: 51.8ms remaining: 30.4ms
63: learn: 0.4851330 total: 52.3ms remaining: 29.4ms
64: learn: 0.4826003 total: 52.8ms remaining: 28.4ms
65: learn: 0.4804328 total: 53.3ms remaining: 27.5ms
66: learn: 0.4784701 total: 53.9ms remaining: 26.5ms
67: learn: 0.4765333 total: 56.3ms remaining: 26.5ms
68: learn: 0.4744135 total: 56.9ms remaining: 25.6ms
69: learn: 0.4719490 total: 57.4ms remaining: 24.6ms
70: learn: 0.4697763 total: 58ms remaining: 23.7ms
71: learn: 0.4674152 total: 58.6ms remaining: 22.8ms
72: learn: 0.4649742 total: 61.9ms remaining: 22.9ms
73: learn: 0.4626694 total: 62.4ms remaining: 21.9ms
74: learn: 0.4604859 total: 63ms remaining: 21ms
75: learn: 0.4588696 total: 63.5ms remaining: 20.1ms
76: learn: 0.4570569 total: 64.5ms remaining: 19.3ms
77: learn: 0.4549811 total: 65.3ms remaining: 18.4ms
78: learn: 0.4529830 total: 66.6ms remaining: 17.7ms
79: learn: 0.4513307 total: 67.2ms remaining: 16.8ms
80: learn: 0.4494753 total: 67.7ms remaining: 15.9ms
81: learn: 0.4477859 total: 68.3ms remaining: 15ms
82: learn: 0.4465069 total: 68.9ms remaining: 14.1ms
83: learn: 0.4444738 total: 69.1ms remaining: 13.2ms
84: learn: 0.4428198 total: 70.9ms remaining: 12.5ms
85: learn: 0.4412138 total: 71.5ms remaining: 11.6ms
86: learn: 0.4394230 total: 72ms remaining: 10.8ms
87: learn: 0.4382404 total: 72.8ms remaining: 9.92ms
88: learn: 0.4362849 total: 73.3ms remaining: 9.06ms
89: learn: 0.4344100 total: 74ms remaining: 8.22ms
90: learn: 0.4324197 total: 74.6ms remaining: 7.38ms
91: learn: 0.4308327 total: 75.1ms remaining: 6.53ms
92: learn: 0.4288311 total: 75.7ms remaining: 5.7ms
93: learn: 0.4269936 total: 76.4ms remaining: 4.88ms
94: learn: 0.4254260 total: 77ms remaining: 4.05ms
95: learn: 0.4237694 total: 79ms remaining: 3.29ms
96: learn: 0.4224569 total: 79.5ms remaining: 2.46ms
97: learn: 0.4206735 total: 80ms remaining: 1.63ms
98: learn: 0.4189745 total: 80.9ms remaining: 816us
99: learn: 0.4175085 total: 81.8ms remaining: 0us
0: learn: 0.6896049 total: 1.13ms remaining: 112ms
1: learn: 0.6849136 total: 2.1ms remaining: 103ms
2: learn: 0.6803576 total: 2.89ms remaining: 93.3ms
3: learn: 0.6768148 total: 3.77ms remaining: 90.4ms
4: learn: 0.6727358 total: 4.6ms remaining: 87.5ms
5: learn: 0.6684443 total: 5.5ms remaining: 86.2ms
6: learn: 0.6645143 total: 6.14ms remaining: 81.6ms
7: learn: 0.6608152 total: 7.1ms remaining: 81.7ms
8: learn: 0.6561977 total: 7.76ms remaining: 78.5ms
9: learn: 0.6516975 total: 8.48ms remaining: 76.3ms
10: learn: 0.6481278 total: 9.08ms remaining: 73.5ms
11: learn: 0.6440783 total: 9.84ms remaining: 72.2ms
12: learn: 0.6404162 total: 10.5ms remaining: 70.2ms
13: learn: 0.6370865 total: 11.1ms remaining: 68.1ms
14: learn: 0.6330448 total: 11.8ms remaining: 66.6ms
15: learn: 0.6293072 total: 12.4ms remaining: 65.2ms
16: learn: 0.6255331 total: 13.1ms remaining: 63.9ms
17: learn: 0.6212560 total: 13.7ms remaining: 62.3ms
18: learn: 0.6170654 total: 14.4ms remaining: 61.5ms
19: learn: 0.6131645 total: 15ms remaining: 59.9ms
20: learn: 0.6095121 total: 15.6ms remaining: 58.6ms
21: learn: 0.6071570 total: 16.2ms remaining: 57.5ms
22: learn: 0.6038082 total: 16.8ms remaining: 56.3ms
23: learn: 0.6003898 total: 17.5ms remaining: 55.4ms
24: learn: 0.5968834 total: 18.1ms remaining: 54.3ms
25: learn: 0.5930002 total: 18.7ms remaining: 53.1ms
26: learn: 0.5898503 total: 19.3ms remaining: 52.2ms
27: learn: 0.5866331 total: 19.9ms remaining: 51.2ms
28: learn: 0.5835247 total: 20.5ms remaining: 50.2ms
29: learn: 0.5802822 total: 21.2ms remaining: 49.4ms
30: learn: 0.5774329 total: 21.7ms remaining: 48.2ms
31: learn: 0.5738669 total: 22.2ms remaining: 47.2ms
32: learn: 0.5708731 total: 22.8ms remaining: 46.4ms
33: learn: 0.5672658 total: 23.3ms remaining: 45.3ms
34: learn: 0.5635881 total: 24ms remaining: 44.6ms
35: learn: 0.5611327 total: 24.5ms remaining: 43.5ms
36: learn: 0.5586970 total: 25ms remaining: 42.6ms
37: learn: 0.5557206 total: 25.7ms remaining: 41.9ms
38: learn: 0.5533724 total: 26.2ms remaining: 41ms
39: learn: 0.5506490 total: 26.5ms remaining: 39.8ms
40: learn: 0.5481978 total: 27.1ms remaining: 39ms
41: learn: 0.5449705 total: 27.7ms remaining: 38.2ms
42: learn: 0.5425293 total: 28.1ms remaining: 37.3ms
43: learn: 0.5401083 total: 28.9ms remaining: 36.7ms
44: learn: 0.5378922 total: 29.4ms remaining: 35.9ms
45: learn: 0.5347885 total: 30.1ms remaining: 35.3ms
46: learn: 0.5320211 total: 30.6ms remaining: 34.5ms
47: learn: 0.5299872 total: 31.1ms remaining: 33.7ms
48: learn: 0.5269199 total: 31.6ms remaining: 32.9ms
49: learn: 0.5241069 total: 32.1ms remaining: 32.1ms
50: learn: 0.5215167 total: 32.6ms remaining: 31.4ms
51: learn: 0.5188360 total: 33.2ms remaining: 30.6ms
52: learn: 0.5160112 total: 33.8ms remaining: 29.9ms
53: learn: 0.5135812 total: 34.5ms remaining: 29.4ms
54: learn: 0.5108101 total: 35ms remaining: 28.7ms
55: learn: 0.5083556 total: 35.7ms remaining: 28ms
56: learn: 0.5059947 total: 36.2ms remaining: 27.3ms
57: learn: 0.5035406 total: 36.7ms remaining: 26.6ms
58: learn: 0.5016698 total: 37.4ms remaining: 26ms
59: learn: 0.4994005 total: 37.9ms remaining: 25.2ms
60: learn: 0.4972218 total: 38.3ms remaining: 24.5ms
61: learn: 0.4951677 total: 38.8ms remaining: 23.8ms
62: learn: 0.4930645 total: 39.5ms remaining: 23.2ms
63: learn: 0.4904773 total: 40ms remaining: 22.5ms
64: learn: 0.4881770 total: 40.6ms remaining: 21.9ms
65: learn: 0.4861932 total: 41.3ms remaining: 21.3ms
66: learn: 0.4837416 total: 41.8ms remaining: 20.6ms
67: learn: 0.4817091 total: 42.5ms remaining: 20ms
68: learn: 0.4793937 total: 43.1ms remaining: 19.4ms
69: learn: 0.4773097 total: 43.6ms remaining: 18.7ms
70: learn: 0.4750601 total: 44.3ms remaining: 18.1ms
71: learn: 0.4727539 total: 44.8ms remaining: 17.4ms
72: learn: 0.4706514 total: 45.5ms remaining: 16.8ms
73: learn: 0.4687804 total: 46.1ms remaining: 16.2ms
74: learn: 0.4664653 total: 46.7ms remaining: 15.6ms
75: learn: 0.4646016 total: 47.3ms remaining: 15ms
76: learn: 0.4625819 total: 47.9ms remaining: 14.3ms
77: learn: 0.4604296 total: 48.4ms remaining: 13.7ms
78: learn: 0.4582116 total: 49.1ms remaining: 13ms
79: learn: 0.4564384 total: 49.6ms remaining: 12.4ms
80: learn: 0.4544613 total: 50.1ms remaining: 11.8ms
81: learn: 0.4523994 total: 50.6ms remaining: 11.1ms
82: learn: 0.4506000 total: 51.1ms remaining: 10.5ms
83: learn: 0.4485890 total: 51.8ms remaining: 9.87ms
84: learn: 0.4469787 total: 52.3ms remaining: 9.22ms
85: learn: 0.4453135 total: 52.8ms remaining: 8.6ms
86: learn: 0.4435503 total: 53.5ms remaining: 7.99ms
87: learn: 0.4420554 total: 53.9ms remaining: 7.36ms
88: learn: 0.4403759 total: 54.6ms remaining: 6.75ms
89: learn: 0.4387024 total: 55.1ms remaining: 6.12ms
90: learn: 0.4370041 total: 55.5ms remaining: 5.49ms
91: learn: 0.4354101 total: 56.1ms remaining: 4.88ms
92: learn: 0.4341167 total: 56.6ms remaining: 4.26ms
93: learn: 0.4328660 total: 57.1ms remaining: 3.64ms
94: learn: 0.4313935 total: 57.7ms remaining: 3.04ms
95: learn: 0.4296744 total: 58.2ms remaining: 2.42ms
96: learn: 0.4280102 total: 59.2ms remaining: 1.83ms
97: learn: 0.4264289 total: 59.8ms remaining: 1.22ms
98: learn: 0.4245965 total: 60.4ms remaining: 609us
99: learn: 0.4226310 total: 60.9ms remaining: 0us
0: learn: 0.6881868 total: 1.02ms remaining: 101ms
1: learn: 0.6829738 total: 2.49ms remaining: 122ms
2: learn: 0.6776030 total: 3.81ms remaining: 123ms
3: learn: 0.6738265 total: 5.03ms remaining: 121ms
4: learn: 0.6695172 total: 5.88ms remaining: 112ms
5: learn: 0.6648879 total: 7.49ms remaining: 117ms
6: learn: 0.6603464 total: 8.8ms remaining: 117ms
7: learn: 0.6554638 total: 10.3ms remaining: 118ms
8: learn: 0.6500920 total: 11.7ms remaining: 119ms
9: learn: 0.6452815 total: 13ms remaining: 117ms
10: learn: 0.6408534 total: 13.8ms remaining: 112ms
11: learn: 0.6366700 total: 15.3ms remaining: 112ms
12: learn: 0.6325433 total: 16.7ms remaining: 112ms
13: learn: 0.6274845 total: 18ms remaining: 110ms
14: learn: 0.6231921 total: 19.1ms remaining: 108ms
15: learn: 0.6185671 total: 20.5ms remaining: 108ms
16: learn: 0.6144366 total: 21.8ms remaining: 106ms
17: learn: 0.6104962 total: 23.2ms remaining: 106ms
18: learn: 0.6058526 total: 24.5ms remaining: 105ms
19: learn: 0.6011075 total: 25.8ms remaining: 103ms
20: learn: 0.5967877 total: 27ms remaining: 102ms
21: learn: 0.5921092 total: 28.4ms remaining: 101ms
22: learn: 0.5891360 total: 29.2ms remaining: 97.9ms
23: learn: 0.5852300 total: 30.2ms remaining: 95.6ms
24: learn: 0.5813717 total: 31ms remaining: 93ms
25: learn: 0.5776055 total: 31.9ms remaining: 90.7ms
26: learn: 0.5742867 total: 32.7ms remaining: 88.5ms
27: learn: 0.5700137 total: 33.6ms remaining: 86.5ms
28: learn: 0.5669218 total: 34.4ms remaining: 84.2ms
29: learn: 0.5633576 total: 35.5ms remaining: 82.8ms
30: learn: 0.5598367 total: 36.4ms remaining: 81.1ms
31: learn: 0.5569285 total: 37.4ms remaining: 79.4ms
32: learn: 0.5533664 total: 38.2ms remaining: 77.6ms
33: learn: 0.5500605 total: 39.1ms remaining: 75.9ms
34: learn: 0.5466335 total: 39.8ms remaining: 73.9ms
35: learn: 0.5439169 total: 40.4ms remaining: 71.8ms
36: learn: 0.5401360 total: 41.1ms remaining: 70ms
37: learn: 0.5374215 total: 42ms remaining: 68.6ms
38: learn: 0.5337420 total: 42.7ms remaining: 66.8ms
39: learn: 0.5306709 total: 43.6ms remaining: 65.5ms
40: learn: 0.5274138 total: 44.4ms remaining: 63.8ms
41: learn: 0.5238286 total: 45.1ms remaining: 62.3ms
42: learn: 0.5208717 total: 46ms remaining: 61ms
43: learn: 0.5179086 total: 46.9ms remaining: 59.6ms
44: learn: 0.5147624 total: 47.4ms remaining: 58ms
45: learn: 0.5118639 total: 48.1ms remaining: 56.5ms
46: learn: 0.5087510 total: 48.8ms remaining: 55.1ms
47: learn: 0.5062515 total: 49.5ms remaining: 53.7ms
48: learn: 0.5031440 total: 50.3ms remaining: 52.3ms
49: learn: 0.4998412 total: 50.9ms remaining: 50.9ms
50: learn: 0.4977029 total: 51.4ms remaining: 49.4ms
51: learn: 0.4950439 total: 52.1ms remaining: 48.1ms
52: learn: 0.4920730 total: 52.7ms remaining: 46.7ms
53: learn: 0.4893246 total: 53.5ms remaining: 45.5ms
54: learn: 0.4867335 total: 54.1ms remaining: 44.3ms
55: learn: 0.4847649 total: 54.7ms remaining: 43ms
56: learn: 0.4825782 total: 55.5ms remaining: 41.9ms
57: learn: 0.4798402 total: 56.1ms remaining: 40.6ms
58: learn: 0.4770542 total: 56.7ms remaining: 39.4ms
59: learn: 0.4743373 total: 57.3ms remaining: 38.2ms
60: learn: 0.4715615 total: 58ms remaining: 37.1ms
61: learn: 0.4688409 total: 58.4ms remaining: 35.8ms
62: learn: 0.4667864 total: 58.8ms remaining: 34.5ms
63: learn: 0.4642890 total: 59.4ms remaining: 33.4ms
64: learn: 0.4615690 total: 60ms remaining: 32.3ms
65: learn: 0.4593597 total: 60.6ms remaining: 31.2ms
66: learn: 0.4568570 total: 61.3ms remaining: 30.2ms
67: learn: 0.4547259 total: 61.9ms remaining: 29.1ms
68: learn: 0.4522637 total: 62.5ms remaining: 28.1ms
69: learn: 0.4500281 total: 62.9ms remaining: 27ms
70: learn: 0.4476853 total: 63.4ms remaining: 25.9ms
71: learn: 0.4459522 total: 63.9ms remaining: 24.9ms
72: learn: 0.4441427 total: 64.4ms remaining: 23.8ms
73: learn: 0.4419336 total: 65ms remaining: 22.8ms
74: learn: 0.4402443 total: 65.5ms remaining: 21.8ms
75: learn: 0.4380476 total: 66ms remaining: 20.8ms
76: learn: 0.4357243 total: 66.7ms remaining: 19.9ms
77: learn: 0.4332320 total: 67.2ms remaining: 19ms
78: learn: 0.4309344 total: 67.9ms remaining: 18.1ms
79: learn: 0.4289794 total: 68.5ms remaining: 17.1ms
80: learn: 0.4268334 total: 69ms remaining: 16.2ms
81: learn: 0.4250378 total: 69.6ms remaining: 15.3ms
82: learn: 0.4233009 total: 69.9ms remaining: 14.3ms
83: learn: 0.4210661 total: 70.4ms remaining: 13.4ms
84: learn: 0.4191080 total: 71.1ms remaining: 12.5ms
85: learn: 0.4174551 total: 71.7ms remaining: 11.7ms
86: learn: 0.4155001 total: 72.2ms remaining: 10.8ms
87: learn: 0.4139865 total: 72.8ms remaining: 9.92ms
88: learn: 0.4126125 total: 73.2ms remaining: 9.05ms
89: learn: 0.4107598 total: 74ms remaining: 8.22ms
90: learn: 0.4088837 total: 74.5ms remaining: 7.37ms
91: learn: 0.4071511 total: 75.1ms remaining: 6.53ms
92: learn: 0.4053648 total: 75.7ms remaining: 5.7ms
93: learn: 0.4032347 total: 76.2ms remaining: 4.86ms
94: learn: 0.4016395 total: 76.8ms remaining: 4.04ms
95: learn: 0.3999227 total: 77.3ms remaining: 3.22ms
96: learn: 0.3979591 total: 77.8ms remaining: 2.41ms
97: learn: 0.3967040 total: 78.4ms remaining: 1.6ms
98: learn: 0.3953452 total: 78.8ms remaining: 795us
99: learn: 0.3935704 total: 79.4ms remaining: 0us
0: learn: 0.6877711 total: 2.04ms remaining: 202ms
1: learn: 0.6818905 total: 2.83ms remaining: 139ms
2: learn: 0.6769580 total: 3.38ms remaining: 109ms
3: learn: 0.6719819 total: 4.79ms remaining: 115ms
4: learn: 0.6675527 total: 5.68ms remaining: 108ms
5: learn: 0.6629202 total: 6.85ms remaining: 107ms
6: learn: 0.6587006 total: 8.1ms remaining: 108ms
7: learn: 0.6530943 total: 9.72ms remaining: 112ms
8: learn: 0.6487981 total: 11.4ms remaining: 116ms
9: learn: 0.6443788 total: 13.2ms remaining: 119ms
10: learn: 0.6410016 total: 14.7ms remaining: 119ms
11: learn: 0.6366268 total: 16.2ms remaining: 119ms
12: learn: 0.6316172 total: 17.1ms remaining: 115ms
13: learn: 0.6281314 total: 19.2ms remaining: 118ms
14: learn: 0.6239560 total: 20.8ms remaining: 118ms
15: learn: 0.6193631 total: 21.8ms remaining: 114ms
16: learn: 0.6150314 total: 23.3ms remaining: 114ms
17: learn: 0.6115652 total: 24.6ms remaining: 112ms
18: learn: 0.6084149 total: 26.5ms remaining: 113ms
19: learn: 0.6048026 total: 28.3ms remaining: 113ms
20: learn: 0.6008272 total: 29.9ms remaining: 112ms
21: learn: 0.5968209 total: 31.5ms remaining: 112ms
22: learn: 0.5930977 total: 33ms remaining: 110ms
23: learn: 0.5888537 total: 34.2ms remaining: 108ms
24: learn: 0.5851596 total: 35.7ms remaining: 107ms
25: learn: 0.5814760 total: 37ms remaining: 105ms
26: learn: 0.5787512 total: 37.9ms remaining: 102ms
27: learn: 0.5757786 total: 39.1ms remaining: 101ms
28: learn: 0.5730349 total: 41ms remaining: 100ms
29: learn: 0.5696539 total: 42.2ms remaining: 98.4ms
30: learn: 0.5661847 total: 43.3ms remaining: 96.4ms
31: learn: 0.5623431 total: 44.6ms remaining: 94.7ms
32: learn: 0.5595707 total: 45.8ms remaining: 93ms
33: learn: 0.5563657 total: 46.8ms remaining: 90.8ms
34: learn: 0.5532213 total: 47.8ms remaining: 88.8ms
35: learn: 0.5496803 total: 49.1ms remaining: 87.3ms
36: learn: 0.5464033 total: 50.1ms remaining: 85.3ms
37: learn: 0.5437436 total: 51.1ms remaining: 83.4ms
38: learn: 0.5404662 total: 51.7ms remaining: 80.9ms
39: learn: 0.5371853 total: 52.6ms remaining: 78.8ms
40: learn: 0.5340354 total: 53.4ms remaining: 76.9ms
41: learn: 0.5313063 total: 54.6ms remaining: 75.4ms
42: learn: 0.5287670 total: 55.6ms remaining: 73.7ms
43: learn: 0.5257274 total: 56.4ms remaining: 71.7ms
44: learn: 0.5226840 total: 57.1ms remaining: 69.8ms
45: learn: 0.5192317 total: 58ms remaining: 68.1ms
46: learn: 0.5162763 total: 58.3ms remaining: 65.8ms
47: learn: 0.5130371 total: 59ms remaining: 63.9ms
48: learn: 0.5109865 total: 59.7ms remaining: 62.1ms
49: learn: 0.5086913 total: 60.3ms remaining: 60.3ms
50: learn: 0.5059362 total: 61.1ms remaining: 58.7ms
51: learn: 0.5036115 total: 61.9ms remaining: 57.1ms
52: learn: 0.5006941 total: 62.6ms remaining: 55.5ms
53: learn: 0.4985983 total: 63.2ms remaining: 53.8ms
54: learn: 0.4962046 total: 63.9ms remaining: 52.3ms
55: learn: 0.4937513 total: 64.5ms remaining: 50.7ms
56: learn: 0.4912702 total: 64.9ms remaining: 49ms
57: learn: 0.4891981 total: 65.4ms remaining: 47.4ms
58: learn: 0.4869499 total: 66ms remaining: 45.9ms
59: learn: 0.4851253 total: 66.5ms remaining: 44.3ms
60: learn: 0.4829265 total: 67.2ms remaining: 42.9ms
61: learn: 0.4800915 total: 67.7ms remaining: 41.5ms
62: learn: 0.4777068 total: 68.2ms remaining: 40.1ms
63: learn: 0.4754654 total: 68.8ms remaining: 38.7ms
64: learn: 0.4730451 total: 69ms remaining: 37.2ms
65: learn: 0.4709285 total: 69.7ms remaining: 35.9ms
66: learn: 0.4688354 total: 70.2ms remaining: 34.6ms
67: learn: 0.4667783 total: 70.6ms remaining: 33.2ms
68: learn: 0.4641239 total: 71.3ms remaining: 32ms
69: learn: 0.4617831 total: 71.8ms remaining: 30.8ms
70: learn: 0.4591348 total: 72.5ms remaining: 29.6ms
71: learn: 0.4568819 total: 73.2ms remaining: 28.5ms
72: learn: 0.4551343 total: 73.9ms remaining: 27.3ms
73: learn: 0.4531559 total: 74.4ms remaining: 26.2ms
74: learn: 0.4512009 total: 75.1ms remaining: 25ms
75: learn: 0.4489969 total: 75.7ms remaining: 23.9ms
76: learn: 0.4468744 total: 76.4ms remaining: 22.8ms
77: learn: 0.4451108 total: 77ms remaining: 21.7ms
78: learn: 0.4426581 total: 77.5ms remaining: 20.6ms
79: learn: 0.4408923 total: 78.1ms remaining: 19.5ms
80: learn: 0.4395120 total: 78.8ms remaining: 18.5ms
81: learn: 0.4374392 total: 79.4ms remaining: 17.4ms
82: learn: 0.4352890 total: 79.9ms remaining: 16.4ms
83: learn: 0.4335777 total: 80.5ms remaining: 15.3ms
84: learn: 0.4318931 total: 80.7ms remaining: 14.2ms
85: learn: 0.4298907 total: 81.2ms remaining: 13.2ms
86: learn: 0.4278883 total: 82ms remaining: 12.3ms
87: learn: 0.4263150 total: 82.7ms remaining: 11.3ms
88: learn: 0.4250235 total: 83.1ms remaining: 10.3ms
89: learn: 0.4232212 total: 83.6ms remaining: 9.29ms
90: learn: 0.4216586 total: 84.2ms remaining: 8.33ms
91: learn: 0.4198502 total: 84.7ms remaining: 7.37ms
92: learn: 0.4190742 total: 85.2ms remaining: 6.41ms
93: learn: 0.4172805 total: 85.6ms remaining: 5.46ms
94: learn: 0.4152617 total: 86ms remaining: 4.53ms
95: learn: 0.4136929 total: 86.6ms remaining: 3.61ms
96: learn: 0.4118276 total: 87.1ms remaining: 2.69ms
97: learn: 0.4099107 total: 87.7ms remaining: 1.79ms
98: learn: 0.4082152 total: 88.3ms remaining: 892us
99: learn: 0.4065726 total: 88.7ms remaining: 0us
0: learn: 0.6881719 total: 2.2ms remaining: 218ms
1: learn: 0.6822875 total: 3.56ms remaining: 174ms
2: learn: 0.6772719 total: 4.93ms remaining: 159ms
3: learn: 0.6722092 total: 7.33ms remaining: 176ms
4: learn: 0.6668137 total: 8.9ms remaining: 169ms
5: learn: 0.6627716 total: 10.4ms remaining: 164ms
6: learn: 0.6577771 total: 11.3ms remaining: 150ms
7: learn: 0.6539072 total: 13ms remaining: 150ms
8: learn: 0.6498281 total: 14.6ms remaining: 148ms
9: learn: 0.6449070 total: 17ms remaining: 153ms
10: learn: 0.6409257 total: 18.4ms remaining: 149ms
11: learn: 0.6361254 total: 20.5ms remaining: 151ms
12: learn: 0.6330183 total: 22ms remaining: 148ms
13: learn: 0.6287459 total: 23.8ms remaining: 146ms
14: learn: 0.6242019 total: 25.2ms remaining: 143ms
15: learn: 0.6199916 total: 25.8ms remaining: 135ms
16: learn: 0.6158487 total: 27.4ms remaining: 134ms
17: learn: 0.6123479 total: 29.1ms remaining: 133ms
18: learn: 0.6079944 total: 30.7ms remaining: 131ms
19: learn: 0.6040598 total: 32.3ms remaining: 129ms
20: learn: 0.6006360 total: 33.5ms remaining: 126ms
21: learn: 0.5968636 total: 34.8ms remaining: 123ms
22: learn: 0.5938489 total: 36.4ms remaining: 122ms
23: learn: 0.5899764 total: 37.9ms remaining: 120ms
24: learn: 0.5861951 total: 39.1ms remaining: 117ms
25: learn: 0.5821266 total: 40.2ms remaining: 114ms
26: learn: 0.5785535 total: 41.2ms remaining: 112ms
27: learn: 0.5753401 total: 42.2ms remaining: 108ms
28: learn: 0.5713715 total: 43ms remaining: 105ms
29: learn: 0.5679069 total: 44ms remaining: 103ms
30: learn: 0.5639393 total: 45.1ms remaining: 100ms
31: learn: 0.5609833 total: 46.2ms remaining: 98.2ms
32: learn: 0.5575458 total: 47ms remaining: 95.5ms
33: learn: 0.5542170 total: 47.9ms remaining: 93ms
34: learn: 0.5514027 total: 48.8ms remaining: 90.6ms
35: learn: 0.5492300 total: 49.5ms remaining: 87.9ms
36: learn: 0.5457673 total: 50.1ms remaining: 85.4ms
37: learn: 0.5426376 total: 51.1ms remaining: 83.3ms
38: learn: 0.5399211 total: 51.9ms remaining: 81.2ms
39: learn: 0.5364066 total: 52.7ms remaining: 79.1ms
40: learn: 0.5339090 total: 53.4ms remaining: 76.9ms
41: learn: 0.5304076 total: 54.1ms remaining: 74.7ms
42: learn: 0.5277597 total: 54.8ms remaining: 72.7ms
43: learn: 0.5246306 total: 55.6ms remaining: 70.8ms
44: learn: 0.5211611 total: 56.5ms remaining: 69ms
45: learn: 0.5181130 total: 57.2ms remaining: 67.1ms
46: learn: 0.5151812 total: 57.9ms remaining: 65.3ms
47: learn: 0.5121221 total: 58.6ms remaining: 63.5ms
48: learn: 0.5090498 total: 59.4ms remaining: 61.8ms
49: learn: 0.5066203 total: 60ms remaining: 60ms
50: learn: 0.5036575 total: 60.7ms remaining: 58.3ms
51: learn: 0.5007699 total: 61.4ms remaining: 56.7ms
52: learn: 0.4979053 total: 62.1ms remaining: 55.1ms
53: learn: 0.4951215 total: 62.7ms remaining: 53.4ms
54: learn: 0.4925006 total: 63ms remaining: 51.5ms
55: learn: 0.4905055 total: 63.7ms remaining: 50ms
56: learn: 0.4880459 total: 64.2ms remaining: 48.5ms
57: learn: 0.4850937 total: 64.8ms remaining: 47ms
58: learn: 0.4833149 total: 65.5ms remaining: 45.5ms
59: learn: 0.4810325 total: 66ms remaining: 44ms
60: learn: 0.4786283 total: 66.6ms remaining: 42.6ms
61: learn: 0.4758581 total: 67.3ms remaining: 41.2ms
62: learn: 0.4735562 total: 67.8ms remaining: 39.8ms
63: learn: 0.4708692 total: 68.4ms remaining: 38.5ms
64: learn: 0.4683422 total: 68.9ms remaining: 37.1ms
65: learn: 0.4664656 total: 69.4ms remaining: 35.7ms
66: learn: 0.4643844 total: 70ms remaining: 34.5ms
67: learn: 0.4617902 total: 70.7ms remaining: 33.2ms
68: learn: 0.4593479 total: 71.2ms remaining: 32ms
69: learn: 0.4568910 total: 71.7ms remaining: 30.7ms
70: learn: 0.4547430 total: 72.5ms remaining: 29.6ms
71: learn: 0.4523202 total: 73.1ms remaining: 28.4ms
72: learn: 0.4502394 total: 73.9ms remaining: 27.3ms
73: learn: 0.4479709 total: 74.4ms remaining: 26.2ms
74: learn: 0.4460912 total: 75ms remaining: 25ms
75: learn: 0.4441840 total: 75.5ms remaining: 23.8ms
76: learn: 0.4421638 total: 76.1ms remaining: 22.7ms
77: learn: 0.4401003 total: 76.6ms remaining: 21.6ms
78: learn: 0.4380573 total: 77.1ms remaining: 20.5ms
79: learn: 0.4358753 total: 77.9ms remaining: 19.5ms
80: learn: 0.4341868 total: 78.6ms remaining: 18.4ms
81: learn: 0.4319715 total: 79.3ms remaining: 17.4ms
82: learn: 0.4299201 total: 79.8ms remaining: 16.3ms
83: learn: 0.4280455 total: 80.4ms remaining: 15.3ms
84: learn: 0.4258657 total: 81ms remaining: 14.3ms
85: learn: 0.4240289 total: 81.2ms remaining: 13.2ms
86: learn: 0.4223571 total: 81.9ms remaining: 12.2ms
87: learn: 0.4204075 total: 82.5ms remaining: 11.2ms
88: learn: 0.4183153 total: 83.2ms remaining: 10.3ms
89: learn: 0.4164581 total: 83.9ms remaining: 9.32ms
90: learn: 0.4149003 total: 86.6ms remaining: 8.57ms
91: learn: 0.4130581 total: 87.3ms remaining: 7.59ms
92: learn: 0.4112342 total: 88ms remaining: 6.62ms
93: learn: 0.4094360 total: 88.6ms remaining: 5.66ms
94: learn: 0.4079925 total: 91ms remaining: 4.79ms
95: learn: 0.4062496 total: 91.5ms remaining: 3.81ms
96: learn: 0.4046321 total: 92ms remaining: 2.85ms
97: learn: 0.4025814 total: 92.7ms remaining: 1.89ms
98: learn: 0.4009699 total: 93.4ms remaining: 943us
99: learn: 0.3990377 total: 95.9ms remaining: 0us
0: learn: 0.6885583 total: 816us remaining: 80.8ms
1: learn: 0.6834753 total: 1.71ms remaining: 84ms
2: learn: 0.6789470 total: 2.9ms remaining: 93.9ms
3: learn: 0.6755916 total: 4.25ms remaining: 102ms
4: learn: 0.6715284 total: 5.3ms remaining: 101ms
5: learn: 0.6673140 total: 6.41ms remaining: 100ms
6: learn: 0.6627801 total: 7.76ms remaining: 103ms
7: learn: 0.6575259 total: 8.7ms remaining: 100ms
8: learn: 0.6536172 total: 9.97ms remaining: 101ms
9: learn: 0.6497181 total: 10.9ms remaining: 97.7ms
10: learn: 0.6457477 total: 11.8ms remaining: 95.9ms
11: learn: 0.6413658 total: 13.4ms remaining: 98.1ms
12: learn: 0.6373549 total: 14.5ms remaining: 97.1ms
13: learn: 0.6336955 total: 15.6ms remaining: 95.8ms
14: learn: 0.6307611 total: 16.5ms remaining: 93.5ms
15: learn: 0.6264181 total: 17.4ms remaining: 91.5ms
16: learn: 0.6234381 total: 18.3ms remaining: 89.3ms
17: learn: 0.6204603 total: 19ms remaining: 86.6ms
18: learn: 0.6166111 total: 20ms remaining: 85.2ms
19: learn: 0.6133843 total: 20.8ms remaining: 83.1ms
20: learn: 0.6095574 total: 21.6ms remaining: 81.4ms
21: learn: 0.6058448 total: 22.5ms remaining: 79.7ms
22: learn: 0.6034212 total: 23.4ms remaining: 78.4ms
23: learn: 0.6010433 total: 24.2ms remaining: 76.7ms
24: learn: 0.5977298 total: 25ms remaining: 74.9ms
25: learn: 0.5951096 total: 25.9ms remaining: 73.6ms
26: learn: 0.5909807 total: 26.9ms remaining: 72.7ms
27: learn: 0.5869012 total: 27.7ms remaining: 71.3ms
28: learn: 0.5835725 total: 28.5ms remaining: 69.8ms
29: learn: 0.5798497 total: 29.4ms remaining: 68.5ms
30: learn: 0.5763549 total: 30.1ms remaining: 67ms
31: learn: 0.5730186 total: 30.9ms remaining: 65.6ms
32: learn: 0.5708685 total: 31.5ms remaining: 64ms
33: learn: 0.5670709 total: 32.1ms remaining: 62.4ms
34: learn: 0.5640302 total: 32.8ms remaining: 60.9ms
35: learn: 0.5606153 total: 33.5ms remaining: 59.5ms
36: learn: 0.5584798 total: 34.1ms remaining: 58ms
37: learn: 0.5551818 total: 34.7ms remaining: 56.7ms
38: learn: 0.5516796 total: 35.3ms remaining: 55.3ms
39: learn: 0.5484479 total: 35.9ms remaining: 53.9ms
40: learn: 0.5452117 total: 36.6ms remaining: 52.7ms
41: learn: 0.5417497 total: 37.2ms remaining: 51.4ms
42: learn: 0.5394314 total: 37.7ms remaining: 50ms
43: learn: 0.5371108 total: 38.2ms remaining: 48.6ms
44: learn: 0.5346540 total: 38.9ms remaining: 47.5ms
45: learn: 0.5318031 total: 39.4ms remaining: 46.2ms
46: learn: 0.5296798 total: 40ms remaining: 45.1ms
47: learn: 0.5272447 total: 40.8ms remaining: 44.2ms
48: learn: 0.5249427 total: 41.3ms remaining: 42.9ms
49: learn: 0.5225004 total: 41.6ms remaining: 41.6ms
50: learn: 0.5198720 total: 42.3ms remaining: 40.6ms
51: learn: 0.5171538 total: 42.9ms remaining: 39.6ms
52: learn: 0.5146010 total: 43.5ms remaining: 38.6ms
53: learn: 0.5120055 total: 44.2ms remaining: 37.7ms
54: learn: 0.5092152 total: 44.8ms remaining: 36.7ms
55: learn: 0.5062983 total: 45.4ms remaining: 35.7ms
56: learn: 0.5041371 total: 46.2ms remaining: 34.9ms
57: learn: 0.5019974 total: 46.9ms remaining: 34ms
58: learn: 0.4997200 total: 47.3ms remaining: 32.9ms
59: learn: 0.4971815 total: 48ms remaining: 32ms
60: learn: 0.4953989 total: 48.6ms remaining: 31.1ms
61: learn: 0.4931661 total: 49.2ms remaining: 30.2ms
62: learn: 0.4907795 total: 49.8ms remaining: 29.2ms
63: learn: 0.4881739 total: 50.3ms remaining: 28.3ms
64: learn: 0.4856616 total: 50.9ms remaining: 27.4ms
65: learn: 0.4836175 total: 51.6ms remaining: 26.6ms
66: learn: 0.4813518 total: 52.3ms remaining: 25.7ms
67: learn: 0.4793712 total: 52.8ms remaining: 24.9ms
68: learn: 0.4772529 total: 53.6ms remaining: 24.1ms
69: learn: 0.4751869 total: 54.1ms remaining: 23.2ms
70: learn: 0.4735373 total: 54.8ms remaining: 22.4ms
71: learn: 0.4715085 total: 55.4ms remaining: 21.6ms
72: learn: 0.4697620 total: 56ms remaining: 20.7ms
73: learn: 0.4678527 total: 56.6ms remaining: 19.9ms
74: learn: 0.4663328 total: 57.4ms remaining: 19.1ms
75: learn: 0.4639681 total: 57.8ms remaining: 18.3ms
76: learn: 0.4618942 total: 58.4ms remaining: 17.4ms
77: learn: 0.4599932 total: 59.2ms remaining: 16.7ms
78: learn: 0.4577392 total: 59.7ms remaining: 15.9ms
79: learn: 0.4556219 total: 60.3ms remaining: 15.1ms
80: learn: 0.4537197 total: 60.8ms remaining: 14.3ms
81: learn: 0.4519369 total: 61.3ms remaining: 13.5ms
82: learn: 0.4505394 total: 62.3ms remaining: 12.8ms
83: learn: 0.4490933 total: 62.8ms remaining: 12ms
84: learn: 0.4475676 total: 63.3ms remaining: 11.2ms
85: learn: 0.4455236 total: 63.9ms remaining: 10.4ms
86: learn: 0.4431977 total: 64.4ms remaining: 9.62ms
87: learn: 0.4411327 total: 65.1ms remaining: 8.88ms
88: learn: 0.4393624 total: 66ms remaining: 8.15ms
89: learn: 0.4378524 total: 66.6ms remaining: 7.39ms
90: learn: 0.4363968 total: 67.3ms remaining: 6.65ms
91: learn: 0.4347528 total: 67.8ms remaining: 5.9ms
92: learn: 0.4329627 total: 68.4ms remaining: 5.15ms
93: learn: 0.4307560 total: 69.2ms remaining: 4.41ms
94: learn: 0.4290811 total: 69.7ms remaining: 3.67ms
95: learn: 0.4274502 total: 70.4ms remaining: 2.93ms
96: learn: 0.4258416 total: 71.3ms remaining: 2.2ms
97: learn: 0.4245272 total: 71.9ms remaining: 1.47ms
98: learn: 0.4229103 total: 72.5ms remaining: 732us
99: learn: 0.4212071 total: 73.3ms remaining: 0us
Evaluating param combo 12/16: {'boosted_cb_model__iterations': 100, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 6, 'boosted_cb_model__num_leaves': 16}
Duplicate found for this combo (hash=6de1910ecc338ba01cb5204f802a7109). Removing old run: 21b89a03688c40fda614e136bbdc74db
0: learn: 0.6882686 total: 1.77ms remaining: 175ms
1: learn: 0.6842287 total: 3.66ms remaining: 180ms
2: learn: 0.6795568 total: 5.08ms remaining: 164ms
3: learn: 0.6751320 total: 6.47ms remaining: 155ms
4: learn: 0.6706698 total: 7.95ms remaining: 151ms
5: learn: 0.6664309 total: 9.09ms remaining: 142ms
6: learn: 0.6621163 total: 10.6ms remaining: 140ms
7: learn: 0.6580139 total: 11.6ms remaining: 133ms
8: learn: 0.6528908 total: 13.2ms remaining: 134ms
9: learn: 0.6489797 total: 14ms remaining: 126ms
10: learn: 0.6451126 total: 15.5ms remaining: 126ms
11: learn: 0.6408115 total: 15.8ms remaining: 116ms
12: learn: 0.6371277 total: 16.9ms remaining: 113ms
13: learn: 0.6335652 total: 17.9ms remaining: 110ms
14: learn: 0.6295871 total: 19.2ms remaining: 109ms
15: learn: 0.6263508 total: 20.3ms remaining: 107ms
16: learn: 0.6230431 total: 21.3ms remaining: 104ms
17: learn: 0.6195287 total: 22.4ms remaining: 102ms
18: learn: 0.6158131 total: 23.3ms remaining: 99.5ms
19: learn: 0.6118211 total: 24.5ms remaining: 98.1ms
20: learn: 0.6079949 total: 24.8ms remaining: 93.3ms
21: learn: 0.6045961 total: 25.8ms remaining: 91.6ms
22: learn: 0.6018823 total: 26.8ms remaining: 89.6ms
23: learn: 0.5984668 total: 27.9ms remaining: 88.3ms
24: learn: 0.5950315 total: 28.6ms remaining: 85.9ms
25: learn: 0.5916049 total: 29.5ms remaining: 84.1ms
26: learn: 0.5879565 total: 30.4ms remaining: 82.3ms
27: learn: 0.5845201 total: 31.4ms remaining: 80.8ms
28: learn: 0.5810843 total: 32ms remaining: 78.4ms
29: learn: 0.5783354 total: 32.9ms remaining: 76.9ms
30: learn: 0.5757722 total: 33.6ms remaining: 74.9ms
31: learn: 0.5724279 total: 33.9ms remaining: 72ms
32: learn: 0.5683808 total: 34.7ms remaining: 70.5ms
33: learn: 0.5646839 total: 35.6ms remaining: 69.1ms
34: learn: 0.5617004 total: 36.6ms remaining: 68ms
35: learn: 0.5581773 total: 37.1ms remaining: 65.9ms
36: learn: 0.5554810 total: 37.9ms remaining: 64.5ms
37: learn: 0.5519617 total: 38.6ms remaining: 63ms
38: learn: 0.5486472 total: 39.4ms remaining: 61.7ms
39: learn: 0.5453312 total: 40.2ms remaining: 60.3ms
40: learn: 0.5429028 total: 40.8ms remaining: 58.8ms
41: learn: 0.5397943 total: 41.5ms remaining: 57.3ms
42: learn: 0.5365728 total: 42.4ms remaining: 56.2ms
43: learn: 0.5339146 total: 43ms remaining: 54.7ms
44: learn: 0.5313950 total: 43.9ms remaining: 53.7ms
45: learn: 0.5285409 total: 44.6ms remaining: 52.3ms
46: learn: 0.5255187 total: 45.3ms remaining: 51.1ms
47: learn: 0.5227771 total: 45.5ms remaining: 49.3ms
48: learn: 0.5197882 total: 46.4ms remaining: 48.3ms
49: learn: 0.5169313 total: 47.3ms remaining: 47.3ms
50: learn: 0.5139815 total: 48.1ms remaining: 46.2ms
51: learn: 0.5115501 total: 48.9ms remaining: 45.1ms
52: learn: 0.5088597 total: 49.7ms remaining: 44ms
53: learn: 0.5064938 total: 50.5ms remaining: 43ms
54: learn: 0.5038095 total: 51.4ms remaining: 42ms
55: learn: 0.5014791 total: 52.2ms remaining: 41ms
56: learn: 0.4992139 total: 52.7ms remaining: 39.8ms
57: learn: 0.4970104 total: 53.3ms remaining: 38.6ms
58: learn: 0.4949760 total: 54ms remaining: 37.5ms
59: learn: 0.4929837 total: 54.8ms remaining: 36.5ms
60: learn: 0.4907196 total: 55.6ms remaining: 35.6ms
61: learn: 0.4880442 total: 56.4ms remaining: 34.5ms
62: learn: 0.4856791 total: 57.5ms remaining: 33.8ms
63: learn: 0.4835952 total: 58.1ms remaining: 32.7ms
64: learn: 0.4812582 total: 59ms remaining: 31.8ms
65: learn: 0.4789467 total: 59.5ms remaining: 30.7ms
66: learn: 0.4764567 total: 60.6ms remaining: 29.8ms
67: learn: 0.4742982 total: 61.6ms remaining: 29ms
68: learn: 0.4721584 total: 62.1ms remaining: 27.9ms
69: learn: 0.4697551 total: 62.6ms remaining: 26.8ms
70: learn: 0.4676067 total: 63.4ms remaining: 25.9ms
71: learn: 0.4651015 total: 63.9ms remaining: 24.8ms
72: learn: 0.4630352 total: 64.2ms remaining: 23.7ms
73: learn: 0.4607316 total: 65ms remaining: 22.8ms
74: learn: 0.4588498 total: 65.5ms remaining: 21.8ms
75: learn: 0.4566244 total: 66.2ms remaining: 20.9ms
76: learn: 0.4546299 total: 67.3ms remaining: 20.1ms
77: learn: 0.4524906 total: 68.1ms remaining: 19.2ms
78: learn: 0.4504720 total: 69ms remaining: 18.3ms
79: learn: 0.4485331 total: 69.7ms remaining: 17.4ms
80: learn: 0.4462447 total: 70.6ms remaining: 16.6ms
81: learn: 0.4443546 total: 71.3ms remaining: 15.7ms
82: learn: 0.4422947 total: 72.2ms remaining: 14.8ms
83: learn: 0.4406292 total: 73ms remaining: 13.9ms
84: learn: 0.4388911 total: 73.7ms remaining: 13ms
85: learn: 0.4370487 total: 74.7ms remaining: 12.2ms
86: learn: 0.4350158 total: 75.4ms remaining: 11.3ms
87: learn: 0.4332813 total: 75.6ms remaining: 10.3ms
88: learn: 0.4316803 total: 76.7ms remaining: 9.48ms
89: learn: 0.4303429 total: 77.5ms remaining: 8.61ms
90: learn: 0.4287259 total: 78.4ms remaining: 7.76ms
91: learn: 0.4269499 total: 79ms remaining: 6.87ms
92: learn: 0.4256529 total: 80ms remaining: 6.02ms
93: learn: 0.4236397 total: 80.6ms remaining: 5.14ms
94: learn: 0.4222452 total: 81.3ms remaining: 4.28ms
95: learn: 0.4202560 total: 82.1ms remaining: 3.42ms
96: learn: 0.4186381 total: 82.9ms remaining: 2.56ms
97: learn: 0.4167344 total: 83.6ms remaining: 1.71ms
98: learn: 0.4154784 total: 84.6ms remaining: 854us
99: learn: 0.4135975 total: 85.1ms remaining: 0us
0: learn: 0.6879001 total: 1.17ms remaining: 116ms
1: learn: 0.6825664 total: 1.93ms remaining: 94.4ms
2: learn: 0.6778624 total: 2.58ms remaining: 83.4ms
3: learn: 0.6728604 total: 2.85ms remaining: 68.4ms
4: learn: 0.6685503 total: 3.58ms remaining: 67.9ms
5: learn: 0.6637032 total: 3.8ms remaining: 59.6ms
6: learn: 0.6594021 total: 4.91ms remaining: 65.2ms
7: learn: 0.6552368 total: 5.73ms remaining: 65.9ms
8: learn: 0.6509857 total: 6.68ms remaining: 67.6ms
9: learn: 0.6465138 total: 7.54ms remaining: 67.9ms
10: learn: 0.6427414 total: 8.62ms remaining: 69.7ms
11: learn: 0.6380331 total: 9.46ms remaining: 69.4ms
12: learn: 0.6340114 total: 10.3ms remaining: 69ms
13: learn: 0.6297944 total: 11.1ms remaining: 68.3ms
14: learn: 0.6258446 total: 11.5ms remaining: 64.9ms
15: learn: 0.6215928 total: 12.3ms remaining: 64.5ms
16: learn: 0.6177414 total: 13.4ms remaining: 65.3ms
17: learn: 0.6139682 total: 14.5ms remaining: 65.8ms
18: learn: 0.6104304 total: 15.1ms remaining: 64.3ms
19: learn: 0.6064870 total: 15.8ms remaining: 63ms
20: learn: 0.6031981 total: 16.6ms remaining: 62.6ms
21: learn: 0.5993083 total: 17.8ms remaining: 62.9ms
22: learn: 0.5955826 total: 18.6ms remaining: 62.2ms
23: learn: 0.5917941 total: 19.3ms remaining: 61.2ms
24: learn: 0.5885132 total: 20.4ms remaining: 61.2ms
25: learn: 0.5856755 total: 21.3ms remaining: 60.5ms
26: learn: 0.5819673 total: 21.9ms remaining: 59.2ms
27: learn: 0.5785210 total: 22.6ms remaining: 58.2ms
28: learn: 0.5748315 total: 23.6ms remaining: 57.9ms
29: learn: 0.5708227 total: 24.3ms remaining: 56.7ms
30: learn: 0.5676059 total: 24.6ms remaining: 54.7ms
31: learn: 0.5649729 total: 25.5ms remaining: 54.2ms
32: learn: 0.5616275 total: 26.5ms remaining: 53.7ms
33: learn: 0.5587318 total: 27.3ms remaining: 53.1ms
34: learn: 0.5551036 total: 28.1ms remaining: 52.2ms
35: learn: 0.5516173 total: 28.4ms remaining: 50.5ms
36: learn: 0.5482520 total: 29.4ms remaining: 50.1ms
37: learn: 0.5449372 total: 30ms remaining: 49ms
38: learn: 0.5416244 total: 30.8ms remaining: 48.2ms
39: learn: 0.5384832 total: 31.6ms remaining: 47.4ms
40: learn: 0.5356229 total: 31.8ms remaining: 45.8ms
41: learn: 0.5325318 total: 32.7ms remaining: 45.2ms
42: learn: 0.5291555 total: 33.5ms remaining: 44.4ms
43: learn: 0.5267413 total: 34.4ms remaining: 43.7ms
44: learn: 0.5239407 total: 35.3ms remaining: 43.1ms
45: learn: 0.5205549 total: 36ms remaining: 42.3ms
46: learn: 0.5179840 total: 36.7ms remaining: 41.4ms
47: learn: 0.5152208 total: 37.7ms remaining: 40.9ms
48: learn: 0.5124560 total: 38.6ms remaining: 40.2ms
49: learn: 0.5101575 total: 39.4ms remaining: 39.4ms
50: learn: 0.5072371 total: 40.4ms remaining: 38.8ms
51: learn: 0.5044088 total: 41ms remaining: 37.9ms
52: learn: 0.5014327 total: 42ms remaining: 37.2ms
53: learn: 0.4991858 total: 42.9ms remaining: 36.5ms
54: learn: 0.4966112 total: 43.6ms remaining: 35.7ms
55: learn: 0.4940952 total: 44.4ms remaining: 34.9ms
56: learn: 0.4920825 total: 45.4ms remaining: 34.3ms
57: learn: 0.4894764 total: 46.3ms remaining: 33.5ms
58: learn: 0.4870588 total: 47.6ms remaining: 33.1ms
59: learn: 0.4847156 total: 48ms remaining: 32ms
60: learn: 0.4825264 total: 48.8ms remaining: 31.2ms
61: learn: 0.4802567 total: 49.6ms remaining: 30.4ms
62: learn: 0.4775950 total: 50.3ms remaining: 29.6ms
63: learn: 0.4749240 total: 51ms remaining: 28.7ms
64: learn: 0.4724742 total: 51.8ms remaining: 27.9ms
65: learn: 0.4702017 total: 52.4ms remaining: 27ms
66: learn: 0.4677725 total: 53.1ms remaining: 26.2ms
67: learn: 0.4655453 total: 54ms remaining: 25.4ms
68: learn: 0.4627189 total: 54.7ms remaining: 24.6ms
69: learn: 0.4601613 total: 55.6ms remaining: 23.8ms
70: learn: 0.4580353 total: 56.5ms remaining: 23.1ms
71: learn: 0.4562184 total: 57ms remaining: 22.2ms
72: learn: 0.4543514 total: 57.8ms remaining: 21.4ms
73: learn: 0.4523479 total: 58.6ms remaining: 20.6ms
74: learn: 0.4504560 total: 59.1ms remaining: 19.7ms
75: learn: 0.4485326 total: 60.1ms remaining: 19ms
76: learn: 0.4470981 total: 61.2ms remaining: 18.3ms
77: learn: 0.4448518 total: 61.9ms remaining: 17.5ms
78: learn: 0.4429101 total: 62.8ms remaining: 16.7ms
79: learn: 0.4410034 total: 63.6ms remaining: 15.9ms
80: learn: 0.4393089 total: 64.4ms remaining: 15.1ms
81: learn: 0.4374687 total: 65.3ms remaining: 14.3ms
82: learn: 0.4354235 total: 65.9ms remaining: 13.5ms
83: learn: 0.4336120 total: 66.6ms remaining: 12.7ms
84: learn: 0.4316595 total: 67.5ms remaining: 11.9ms
85: learn: 0.4298922 total: 68.4ms remaining: 11.1ms
86: learn: 0.4283712 total: 69.3ms remaining: 10.4ms
87: learn: 0.4261745 total: 70.1ms remaining: 9.56ms
88: learn: 0.4241850 total: 70.5ms remaining: 8.72ms
89: learn: 0.4227062 total: 71.5ms remaining: 7.94ms
90: learn: 0.4208794 total: 72.3ms remaining: 7.15ms
91: learn: 0.4195547 total: 72.7ms remaining: 6.32ms
92: learn: 0.4172180 total: 73.1ms remaining: 5.5ms
93: learn: 0.4156820 total: 74ms remaining: 4.72ms
94: learn: 0.4140858 total: 74.8ms remaining: 3.94ms
95: learn: 0.4124925 total: 75.4ms remaining: 3.14ms
96: learn: 0.4101614 total: 76.1ms remaining: 2.35ms
97: learn: 0.4086172 total: 76.8ms remaining: 1.57ms
98: learn: 0.4066562 total: 77.7ms remaining: 784us
99: learn: 0.4046721 total: 78.5ms remaining: 0us
0: learn: 0.6879038 total: 1.83ms remaining: 181ms
1: learn: 0.6833723 total: 3.56ms remaining: 175ms
2: learn: 0.6793155 total: 4.81ms remaining: 156ms
3: learn: 0.6755335 total: 6.62ms remaining: 159ms
4: learn: 0.6704722 total: 8.2ms remaining: 156ms
5: learn: 0.6661947 total: 9.98ms remaining: 156ms
6: learn: 0.6614249 total: 11.3ms remaining: 150ms
7: learn: 0.6571407 total: 12.9ms remaining: 149ms
8: learn: 0.6534196 total: 14.6ms remaining: 148ms
9: learn: 0.6486063 total: 16.2ms remaining: 146ms
10: learn: 0.6447700 total: 17.5ms remaining: 142ms
11: learn: 0.6408974 total: 19ms remaining: 140ms
12: learn: 0.6366198 total: 19.7ms remaining: 132ms
13: learn: 0.6325800 total: 21.2ms remaining: 130ms
14: learn: 0.6277010 total: 22.1ms remaining: 125ms
15: learn: 0.6239838 total: 23.2ms remaining: 122ms
16: learn: 0.6196373 total: 24.6ms remaining: 120ms
17: learn: 0.6153141 total: 25.5ms remaining: 116ms
18: learn: 0.6108362 total: 26.6ms remaining: 113ms
19: learn: 0.6073908 total: 27.4ms remaining: 110ms
20: learn: 0.6036408 total: 28.5ms remaining: 107ms
21: learn: 0.5996456 total: 29.6ms remaining: 105ms
22: learn: 0.5964015 total: 30.7ms remaining: 103ms
23: learn: 0.5925342 total: 31.8ms remaining: 101ms
24: learn: 0.5889169 total: 33ms remaining: 99ms
25: learn: 0.5853063 total: 34.1ms remaining: 97ms
26: learn: 0.5814669 total: 35ms remaining: 94.7ms
27: learn: 0.5781858 total: 36.1ms remaining: 92.8ms
28: learn: 0.5749039 total: 37.8ms remaining: 92.7ms
29: learn: 0.5712272 total: 38.7ms remaining: 90.4ms
30: learn: 0.5686921 total: 39.8ms remaining: 88.5ms
31: learn: 0.5653936 total: 40.8ms remaining: 86.7ms
32: learn: 0.5625110 total: 42ms remaining: 85.2ms
33: learn: 0.5589728 total: 42.9ms remaining: 83.3ms
34: learn: 0.5561277 total: 43.9ms remaining: 81.6ms
35: learn: 0.5527337 total: 44.7ms remaining: 79.5ms
36: learn: 0.5493706 total: 45.7ms remaining: 77.8ms
37: learn: 0.5462561 total: 46.4ms remaining: 75.7ms
38: learn: 0.5436280 total: 47ms remaining: 73.4ms
39: learn: 0.5413929 total: 47.6ms remaining: 71.5ms
40: learn: 0.5384565 total: 48.4ms remaining: 69.7ms
41: learn: 0.5354266 total: 49.4ms remaining: 68.3ms
42: learn: 0.5322718 total: 50.3ms remaining: 66.7ms
43: learn: 0.5294400 total: 51.3ms remaining: 65.3ms
44: learn: 0.5265466 total: 51.9ms remaining: 63.4ms
45: learn: 0.5235887 total: 52.7ms remaining: 61.8ms
46: learn: 0.5208130 total: 53.5ms remaining: 60.3ms
47: learn: 0.5181899 total: 54.4ms remaining: 58.9ms
48: learn: 0.5153383 total: 55.1ms remaining: 57.3ms
49: learn: 0.5125170 total: 56ms remaining: 56ms
50: learn: 0.5095813 total: 56.7ms remaining: 54.5ms
51: learn: 0.5069522 total: 57.4ms remaining: 53ms
52: learn: 0.5042881 total: 58ms remaining: 51.4ms
53: learn: 0.5016992 total: 58.9ms remaining: 50.2ms
54: learn: 0.4993265 total: 59.2ms remaining: 48.4ms
55: learn: 0.4968023 total: 60ms remaining: 47.1ms
56: learn: 0.4944942 total: 60.8ms remaining: 45.9ms
57: learn: 0.4922157 total: 61.8ms remaining: 44.7ms
58: learn: 0.4897362 total: 62.5ms remaining: 43.5ms
59: learn: 0.4871803 total: 63.6ms remaining: 42.4ms
60: learn: 0.4852816 total: 64.3ms remaining: 41.1ms
61: learn: 0.4824799 total: 64.6ms remaining: 39.6ms
62: learn: 0.4802973 total: 65.5ms remaining: 38.5ms
63: learn: 0.4780778 total: 66.3ms remaining: 37.3ms
64: learn: 0.4755148 total: 67.1ms remaining: 36.1ms
65: learn: 0.4729485 total: 67.5ms remaining: 34.8ms
66: learn: 0.4710472 total: 68.4ms remaining: 33.7ms
67: learn: 0.4687673 total: 69ms remaining: 32.5ms
68: learn: 0.4663062 total: 69.6ms remaining: 31.3ms
69: learn: 0.4648952 total: 69.8ms remaining: 29.9ms
70: learn: 0.4629342 total: 70.6ms remaining: 28.9ms
71: learn: 0.4610131 total: 71.7ms remaining: 27.9ms
72: learn: 0.4589731 total: 72.5ms remaining: 26.8ms
73: learn: 0.4572571 total: 73.4ms remaining: 25.8ms
74: learn: 0.4551794 total: 73.8ms remaining: 24.6ms
75: learn: 0.4534776 total: 74.5ms remaining: 23.5ms
76: learn: 0.4514043 total: 75.2ms remaining: 22.5ms
77: learn: 0.4490440 total: 76.1ms remaining: 21.5ms
78: learn: 0.4467232 total: 76.9ms remaining: 20.4ms
79: learn: 0.4447211 total: 77.5ms remaining: 19.4ms
80: learn: 0.4431372 total: 78.4ms remaining: 18.4ms
81: learn: 0.4413268 total: 79ms remaining: 17.3ms
82: learn: 0.4395766 total: 79.9ms remaining: 16.4ms
83: learn: 0.4375347 total: 80.7ms remaining: 15.4ms
84: learn: 0.4359025 total: 81.6ms remaining: 14.4ms
85: learn: 0.4340720 total: 82.5ms remaining: 13.4ms
86: learn: 0.4322101 total: 83.4ms remaining: 12.5ms
87: learn: 0.4301023 total: 84.1ms remaining: 11.5ms
88: learn: 0.4283585 total: 85ms remaining: 10.5ms
89: learn: 0.4271138 total: 85.9ms remaining: 9.55ms
90: learn: 0.4253296 total: 87ms remaining: 8.6ms
91: learn: 0.4237699 total: 87.7ms remaining: 7.63ms
92: learn: 0.4216927 total: 88.5ms remaining: 6.66ms
93: learn: 0.4195077 total: 89.4ms remaining: 5.71ms
94: learn: 0.4176092 total: 90.2ms remaining: 4.75ms
95: learn: 0.4156904 total: 90.9ms remaining: 3.79ms
96: learn: 0.4138553 total: 91.6ms remaining: 2.83ms
97: learn: 0.4123674 total: 92.5ms remaining: 1.89ms
98: learn: 0.4105459 total: 93ms remaining: 939us
99: learn: 0.4088539 total: 94.1ms remaining: 0us
0: learn: 0.6877830 total: 2.85ms remaining: 283ms
1: learn: 0.6834925 total: 5.52ms remaining: 270ms
2: learn: 0.6789587 total: 8.42ms remaining: 272ms
3: learn: 0.6742087 total: 10.3ms remaining: 247ms
4: learn: 0.6697901 total: 12.6ms remaining: 240ms
5: learn: 0.6651971 total: 14.5ms remaining: 226ms
6: learn: 0.6608724 total: 16.5ms remaining: 219ms
7: learn: 0.6570621 total: 18.4ms remaining: 211ms
8: learn: 0.6525786 total: 20.2ms remaining: 204ms
9: learn: 0.6485319 total: 21.9ms remaining: 197ms
10: learn: 0.6438388 total: 23.5ms remaining: 191ms
11: learn: 0.6399976 total: 25.3ms remaining: 186ms
12: learn: 0.6354262 total: 26.9ms remaining: 180ms
13: learn: 0.6318842 total: 28.6ms remaining: 176ms
14: learn: 0.6283602 total: 30.1ms remaining: 171ms
15: learn: 0.6239873 total: 31.2ms remaining: 164ms
16: learn: 0.6197663 total: 32ms remaining: 156ms
17: learn: 0.6160495 total: 33.5ms remaining: 153ms
18: learn: 0.6121943 total: 34.8ms remaining: 148ms
19: learn: 0.6087217 total: 36.2ms remaining: 145ms
20: learn: 0.6050074 total: 37.3ms remaining: 140ms
21: learn: 0.6011373 total: 38.8ms remaining: 138ms
22: learn: 0.5971935 total: 40ms remaining: 134ms
23: learn: 0.5927861 total: 40.7ms remaining: 129ms
24: learn: 0.5895579 total: 41.9ms remaining: 126ms
25: learn: 0.5859251 total: 42.6ms remaining: 121ms
26: learn: 0.5830791 total: 43.8ms remaining: 119ms
27: learn: 0.5796411 total: 45.1ms remaining: 116ms
28: learn: 0.5763626 total: 46.1ms remaining: 113ms
29: learn: 0.5729057 total: 47.1ms remaining: 110ms
30: learn: 0.5696363 total: 48.1ms remaining: 107ms
31: learn: 0.5660485 total: 49ms remaining: 104ms
32: learn: 0.5629348 total: 50ms remaining: 102ms
33: learn: 0.5591928 total: 51ms remaining: 99ms
34: learn: 0.5556857 total: 51.9ms remaining: 96.3ms
35: learn: 0.5529631 total: 52.6ms remaining: 93.4ms
36: learn: 0.5495135 total: 53.3ms remaining: 90.7ms
37: learn: 0.5466279 total: 54.3ms remaining: 88.5ms
38: learn: 0.5434318 total: 55.8ms remaining: 87.2ms
39: learn: 0.5405497 total: 56.5ms remaining: 84.8ms
40: learn: 0.5371825 total: 57.3ms remaining: 82.5ms
41: learn: 0.5341038 total: 58ms remaining: 80.1ms
42: learn: 0.5308408 total: 58.7ms remaining: 77.7ms
43: learn: 0.5280881 total: 59.7ms remaining: 76ms
44: learn: 0.5251138 total: 60.2ms remaining: 73.5ms
45: learn: 0.5217302 total: 61.1ms remaining: 71.7ms
46: learn: 0.5193920 total: 61.9ms remaining: 69.8ms
47: learn: 0.5161583 total: 62.6ms remaining: 67.8ms
48: learn: 0.5128156 total: 63ms remaining: 65.5ms
49: learn: 0.5101414 total: 63.7ms remaining: 63.7ms
50: learn: 0.5072125 total: 64.6ms remaining: 62.1ms
51: learn: 0.5045709 total: 65.8ms remaining: 60.8ms
52: learn: 0.5019071 total: 66.6ms remaining: 59.1ms
53: learn: 0.4991868 total: 67.6ms remaining: 57.6ms
54: learn: 0.4965691 total: 67.8ms remaining: 55.5ms
55: learn: 0.4937257 total: 68.6ms remaining: 53.9ms
56: learn: 0.4910990 total: 69ms remaining: 52.1ms
57: learn: 0.4886427 total: 70ms remaining: 50.7ms
58: learn: 0.4858006 total: 70.7ms remaining: 49.1ms
59: learn: 0.4831352 total: 71.5ms remaining: 47.7ms
60: learn: 0.4807835 total: 71.9ms remaining: 46ms
61: learn: 0.4787501 total: 72.8ms remaining: 44.6ms
62: learn: 0.4759399 total: 73.7ms remaining: 43.3ms
63: learn: 0.4736924 total: 74.2ms remaining: 41.7ms
64: learn: 0.4712170 total: 75.2ms remaining: 40.5ms
65: learn: 0.4686388 total: 75.9ms remaining: 39.1ms
66: learn: 0.4659725 total: 76.8ms remaining: 37.8ms
67: learn: 0.4634761 total: 77.6ms remaining: 36.5ms
68: learn: 0.4611674 total: 78.7ms remaining: 35.4ms
69: learn: 0.4588186 total: 79.6ms remaining: 34.1ms
70: learn: 0.4568943 total: 80.6ms remaining: 32.9ms
71: learn: 0.4548001 total: 81.5ms remaining: 31.7ms
72: learn: 0.4530243 total: 82.7ms remaining: 30.6ms
73: learn: 0.4507764 total: 83.5ms remaining: 29.3ms
74: learn: 0.4486108 total: 84.1ms remaining: 28ms
75: learn: 0.4463555 total: 84.9ms remaining: 26.8ms
76: learn: 0.4441325 total: 85.7ms remaining: 25.6ms
77: learn: 0.4421374 total: 85.9ms remaining: 24.2ms
78: learn: 0.4401454 total: 86.2ms remaining: 22.9ms
79: learn: 0.4380102 total: 87ms remaining: 21.8ms
80: learn: 0.4361960 total: 87.8ms remaining: 20.6ms
81: learn: 0.4343025 total: 89ms remaining: 19.5ms
82: learn: 0.4325398 total: 89.4ms remaining: 18.3ms
83: learn: 0.4306687 total: 89.6ms remaining: 17.1ms
84: learn: 0.4285668 total: 90.6ms remaining: 16ms
85: learn: 0.4264108 total: 91.3ms remaining: 14.9ms
86: learn: 0.4244385 total: 92.2ms remaining: 13.8ms
87: learn: 0.4225429 total: 93ms remaining: 12.7ms
88: learn: 0.4205017 total: 94ms remaining: 11.6ms
89: learn: 0.4187612 total: 94.8ms remaining: 10.5ms
90: learn: 0.4170778 total: 95.5ms remaining: 9.45ms
91: learn: 0.4156857 total: 96.2ms remaining: 8.37ms
92: learn: 0.4138484 total: 97ms remaining: 7.3ms
93: learn: 0.4121859 total: 97.9ms remaining: 6.25ms
94: learn: 0.4102246 total: 98.8ms remaining: 5.2ms
95: learn: 0.4086265 total: 99.6ms remaining: 4.15ms
96: learn: 0.4069158 total: 100ms remaining: 3.1ms
97: learn: 0.4052973 total: 101ms remaining: 2.06ms
98: learn: 0.4033980 total: 102ms remaining: 1.03ms
99: learn: 0.4018856 total: 103ms remaining: 0us
0: learn: 0.6885024 total: 2.44ms remaining: 241ms
1: learn: 0.6846428 total: 5.16ms remaining: 253ms
2: learn: 0.6800668 total: 7.8ms remaining: 252ms
3: learn: 0.6756460 total: 9.43ms remaining: 226ms
4: learn: 0.6712975 total: 10.9ms remaining: 207ms
5: learn: 0.6676982 total: 11.9ms remaining: 186ms
6: learn: 0.6642036 total: 14.1ms remaining: 187ms
7: learn: 0.6601437 total: 15.6ms remaining: 179ms
8: learn: 0.6557718 total: 17ms remaining: 172ms
9: learn: 0.6518354 total: 19.2ms remaining: 173ms
10: learn: 0.6477087 total: 20.5ms remaining: 166ms
11: learn: 0.6435351 total: 21.3ms remaining: 156ms
12: learn: 0.6394047 total: 23.9ms remaining: 160ms
13: learn: 0.6358291 total: 25.8ms remaining: 158ms
14: learn: 0.6320091 total: 27.9ms remaining: 158ms
15: learn: 0.6279222 total: 29.1ms remaining: 153ms
16: learn: 0.6242789 total: 30.8ms remaining: 151ms
17: learn: 0.6204739 total: 31.6ms remaining: 144ms
18: learn: 0.6170662 total: 33.1ms remaining: 141ms
19: learn: 0.6138748 total: 34.6ms remaining: 138ms
20: learn: 0.6101235 total: 35.2ms remaining: 132ms
21: learn: 0.6064071 total: 36.7ms remaining: 130ms
22: learn: 0.6035068 total: 38.2ms remaining: 128ms
23: learn: 0.5998397 total: 39.1ms remaining: 124ms
24: learn: 0.5964036 total: 40.2ms remaining: 121ms
25: learn: 0.5933263 total: 41.4ms remaining: 118ms
26: learn: 0.5891559 total: 42.3ms remaining: 114ms
27: learn: 0.5862627 total: 43.5ms remaining: 112ms
28: learn: 0.5829233 total: 44.6ms remaining: 109ms
29: learn: 0.5799922 total: 45.6ms remaining: 106ms
30: learn: 0.5764535 total: 46.8ms remaining: 104ms
31: learn: 0.5732734 total: 48ms remaining: 102ms
32: learn: 0.5703912 total: 48.8ms remaining: 99ms
33: learn: 0.5677847 total: 49.8ms remaining: 96.6ms
34: learn: 0.5644089 total: 51ms remaining: 94.7ms
35: learn: 0.5612078 total: 51.9ms remaining: 92.3ms
36: learn: 0.5579158 total: 52.8ms remaining: 90ms
37: learn: 0.5548959 total: 53.9ms remaining: 88ms
38: learn: 0.5517562 total: 54.8ms remaining: 85.8ms
39: learn: 0.5485756 total: 55.8ms remaining: 83.7ms
40: learn: 0.5452331 total: 56.5ms remaining: 81.4ms
41: learn: 0.5428162 total: 57.4ms remaining: 79.3ms
42: learn: 0.5404160 total: 58.3ms remaining: 77.3ms
43: learn: 0.5380578 total: 58.8ms remaining: 74.8ms
44: learn: 0.5355467 total: 59.4ms remaining: 72.6ms
45: learn: 0.5328272 total: 60.4ms remaining: 70.9ms
46: learn: 0.5299021 total: 61ms remaining: 68.8ms
47: learn: 0.5268303 total: 61.8ms remaining: 67ms
48: learn: 0.5242317 total: 62.7ms remaining: 65.3ms
49: learn: 0.5215018 total: 63.5ms remaining: 63.5ms
50: learn: 0.5191895 total: 64.4ms remaining: 61.9ms
51: learn: 0.5169240 total: 65.4ms remaining: 60.3ms
52: learn: 0.5147491 total: 66.3ms remaining: 58.8ms
53: learn: 0.5125515 total: 66.8ms remaining: 56.9ms
54: learn: 0.5092663 total: 67.7ms remaining: 55.4ms
55: learn: 0.5067094 total: 68.5ms remaining: 53.8ms
56: learn: 0.5043970 total: 69.3ms remaining: 52.3ms
57: learn: 0.5020777 total: 71ms remaining: 51.4ms
58: learn: 0.5005792 total: 71.6ms remaining: 49.8ms
59: learn: 0.4977722 total: 72.3ms remaining: 48.2ms
60: learn: 0.4950910 total: 73.1ms remaining: 46.8ms
61: learn: 0.4930249 total: 74ms remaining: 45.4ms
62: learn: 0.4907534 total: 74.6ms remaining: 43.8ms
63: learn: 0.4884731 total: 75.4ms remaining: 42.4ms
64: learn: 0.4858895 total: 76.4ms remaining: 41.1ms
65: learn: 0.4839893 total: 77.1ms remaining: 39.7ms
66: learn: 0.4818980 total: 78ms remaining: 38.4ms
67: learn: 0.4799184 total: 78.7ms remaining: 37ms
68: learn: 0.4774662 total: 79.4ms remaining: 35.7ms
69: learn: 0.4754508 total: 79.6ms remaining: 34.1ms
70: learn: 0.4735379 total: 80.2ms remaining: 32.7ms
71: learn: 0.4719689 total: 80.7ms remaining: 31.4ms
72: learn: 0.4695853 total: 81.6ms remaining: 30.2ms
73: learn: 0.4683372 total: 82.4ms remaining: 29ms
74: learn: 0.4661421 total: 83.1ms remaining: 27.7ms
75: learn: 0.4641302 total: 83.9ms remaining: 26.5ms
76: learn: 0.4621320 total: 84.9ms remaining: 25.4ms
77: learn: 0.4601545 total: 85.5ms remaining: 24.1ms
78: learn: 0.4580497 total: 86.4ms remaining: 23ms
79: learn: 0.4560310 total: 87.1ms remaining: 21.8ms
80: learn: 0.4540271 total: 87.7ms remaining: 20.6ms
81: learn: 0.4522298 total: 88.5ms remaining: 19.4ms
82: learn: 0.4505709 total: 89.1ms remaining: 18.2ms
83: learn: 0.4485105 total: 89.6ms remaining: 17.1ms
84: learn: 0.4469941 total: 90.4ms remaining: 16ms
85: learn: 0.4455582 total: 91.4ms remaining: 14.9ms
86: learn: 0.4434393 total: 92.3ms remaining: 13.8ms
87: learn: 0.4417809 total: 93.1ms remaining: 12.7ms
88: learn: 0.4401956 total: 93.9ms remaining: 11.6ms
89: learn: 0.4387408 total: 94.7ms remaining: 10.5ms
90: learn: 0.4373206 total: 95.5ms remaining: 9.45ms
91: learn: 0.4356324 total: 96.4ms remaining: 8.38ms
92: learn: 0.4338526 total: 97.4ms remaining: 7.33ms
93: learn: 0.4322867 total: 98.3ms remaining: 6.27ms
94: learn: 0.4307339 total: 99.2ms remaining: 5.22ms
95: learn: 0.4287344 total: 100ms remaining: 4.17ms
96: learn: 0.4277387 total: 101ms remaining: 3.12ms
97: learn: 0.4260245 total: 102ms remaining: 2.08ms
98: learn: 0.4245470 total: 103ms remaining: 1.04ms
99: learn: 0.4229554 total: 103ms remaining: 0us
0: learn: 0.6884559 total: 1.86ms remaining: 184ms
1: learn: 0.6843430 total: 3.98ms remaining: 195ms
2: learn: 0.6796675 total: 5.43ms remaining: 176ms
3: learn: 0.6752330 total: 7.06ms remaining: 170ms
4: learn: 0.6710458 total: 8.69ms remaining: 165ms
5: learn: 0.6662186 total: 10.3ms remaining: 162ms
6: learn: 0.6616622 total: 11.6ms remaining: 155ms
7: learn: 0.6570717 total: 13ms remaining: 150ms
8: learn: 0.6531693 total: 13.4ms remaining: 135ms
9: learn: 0.6481402 total: 14.9ms remaining: 134ms
10: learn: 0.6431442 total: 16.2ms remaining: 131ms
11: learn: 0.6392234 total: 17.6ms remaining: 129ms
12: learn: 0.6358571 total: 18.9ms remaining: 126ms
13: learn: 0.6329785 total: 20.1ms remaining: 123ms
14: learn: 0.6288396 total: 21.2ms remaining: 120ms
15: learn: 0.6247382 total: 22.3ms remaining: 117ms
16: learn: 0.6214486 total: 23.3ms remaining: 114ms
17: learn: 0.6172429 total: 24.2ms remaining: 110ms
18: learn: 0.6139519 total: 25.1ms remaining: 107ms
19: learn: 0.6099444 total: 26.1ms remaining: 104ms
20: learn: 0.6062216 total: 26.9ms remaining: 101ms
21: learn: 0.6022794 total: 27.9ms remaining: 99.1ms
22: learn: 0.5979682 total: 28.8ms remaining: 96.4ms
23: learn: 0.5944250 total: 29.4ms remaining: 93.2ms
24: learn: 0.5907559 total: 30.3ms remaining: 90.8ms
25: learn: 0.5870441 total: 30.8ms remaining: 87.8ms
26: learn: 0.5837875 total: 31.8ms remaining: 86ms
27: learn: 0.5807009 total: 32.8ms remaining: 84.4ms
28: learn: 0.5776645 total: 33.6ms remaining: 82.3ms
29: learn: 0.5749488 total: 34.4ms remaining: 80.3ms
30: learn: 0.5714051 total: 35ms remaining: 78ms
31: learn: 0.5682202 total: 35.7ms remaining: 75.9ms
32: learn: 0.5646589 total: 36.3ms remaining: 73.7ms
33: learn: 0.5612332 total: 36.8ms remaining: 71.5ms
34: learn: 0.5580240 total: 37.7ms remaining: 69.9ms
35: learn: 0.5552691 total: 38ms remaining: 67.6ms
36: learn: 0.5523508 total: 38.4ms remaining: 65.5ms
37: learn: 0.5496641 total: 38.9ms remaining: 63.5ms
38: learn: 0.5467321 total: 39.5ms remaining: 61.8ms
39: learn: 0.5439376 total: 40.2ms remaining: 60.3ms
40: learn: 0.5411322 total: 41ms remaining: 58.9ms
41: learn: 0.5381168 total: 41.5ms remaining: 57.2ms
42: learn: 0.5351125 total: 42ms remaining: 55.6ms
43: learn: 0.5321597 total: 42.6ms remaining: 54.2ms
44: learn: 0.5296471 total: 43.2ms remaining: 52.9ms
45: learn: 0.5268437 total: 43.9ms remaining: 51.6ms
46: learn: 0.5236156 total: 44.5ms remaining: 50.1ms
47: learn: 0.5209652 total: 45ms remaining: 48.7ms
48: learn: 0.5180780 total: 45.7ms remaining: 47.5ms
49: learn: 0.5153886 total: 46.2ms remaining: 46.2ms
50: learn: 0.5131090 total: 46.5ms remaining: 44.7ms
51: learn: 0.5104505 total: 47.1ms remaining: 43.5ms
52: learn: 0.5080560 total: 47.8ms remaining: 42.4ms
53: learn: 0.5054340 total: 48.3ms remaining: 41.1ms
54: learn: 0.5028857 total: 49ms remaining: 40.1ms
55: learn: 0.5003848 total: 49.4ms remaining: 38.8ms
56: learn: 0.4982614 total: 50.1ms remaining: 37.8ms
57: learn: 0.4964142 total: 50.5ms remaining: 36.6ms
58: learn: 0.4937670 total: 51ms remaining: 35.5ms
59: learn: 0.4911476 total: 51.2ms remaining: 34.1ms
60: learn: 0.4885780 total: 51.6ms remaining: 33ms
61: learn: 0.4861432 total: 52.2ms remaining: 32ms
62: learn: 0.4836285 total: 52.7ms remaining: 30.9ms
63: learn: 0.4812985 total: 53.4ms remaining: 30.1ms
64: learn: 0.4787342 total: 54.1ms remaining: 29.1ms
65: learn: 0.4768285 total: 54.6ms remaining: 28.1ms
66: learn: 0.4746037 total: 55.1ms remaining: 27.1ms
67: learn: 0.4723183 total: 55.8ms remaining: 26.2ms
68: learn: 0.4701508 total: 56.3ms remaining: 25.3ms
69: learn: 0.4681316 total: 56.9ms remaining: 24.4ms
70: learn: 0.4661125 total: 57.5ms remaining: 23.5ms
71: learn: 0.4643893 total: 58ms remaining: 22.6ms
72: learn: 0.4621581 total: 59ms remaining: 21.8ms
73: learn: 0.4607279 total: 59.6ms remaining: 20.9ms
74: learn: 0.4585197 total: 60.1ms remaining: 20ms
75: learn: 0.4564410 total: 61ms remaining: 19.3ms
76: learn: 0.4545530 total: 61.9ms remaining: 18.5ms
77: learn: 0.4524360 total: 62.5ms remaining: 17.6ms
78: learn: 0.4500605 total: 63.2ms remaining: 16.8ms
79: learn: 0.4484366 total: 64ms remaining: 16ms
80: learn: 0.4466695 total: 64.8ms remaining: 15.2ms
81: learn: 0.4444775 total: 65.5ms remaining: 14.4ms
82: learn: 0.4422397 total: 66.4ms remaining: 13.6ms
83: learn: 0.4405110 total: 67.2ms remaining: 12.8ms
84: learn: 0.4387115 total: 67.9ms remaining: 12ms
85: learn: 0.4367680 total: 68.4ms remaining: 11.1ms
86: learn: 0.4349723 total: 69.1ms remaining: 10.3ms
87: learn: 0.4334843 total: 69.6ms remaining: 9.48ms
88: learn: 0.4314610 total: 70.3ms remaining: 8.68ms
89: learn: 0.4295193 total: 70.8ms remaining: 7.87ms
90: learn: 0.4278404 total: 71.6ms remaining: 7.08ms
91: learn: 0.4261553 total: 72.4ms remaining: 6.3ms
92: learn: 0.4242387 total: 73ms remaining: 5.49ms
93: learn: 0.4221441 total: 73.8ms remaining: 4.71ms
94: learn: 0.4206136 total: 74.5ms remaining: 3.92ms
95: learn: 0.4187796 total: 75.2ms remaining: 3.13ms
96: learn: 0.4170621 total: 75.8ms remaining: 2.35ms
97: learn: 0.4154551 total: 76.6ms remaining: 1.56ms
98: learn: 0.4137332 total: 77.3ms remaining: 780us
99: learn: 0.4123082 total: 77.9ms remaining: 0us
0: learn: 0.6881859 total: 956us remaining: 94.7ms
1: learn: 0.6844111 total: 2.52ms remaining: 124ms
2: learn: 0.6799576 total: 4.25ms remaining: 137ms
3: learn: 0.6755406 total: 5.99ms remaining: 144ms
4: learn: 0.6713634 total: 8.32ms remaining: 158ms
5: learn: 0.6666994 total: 9.67ms remaining: 151ms
6: learn: 0.6625210 total: 11.4ms remaining: 151ms
7: learn: 0.6582164 total: 13.7ms remaining: 157ms
8: learn: 0.6542482 total: 15.2ms remaining: 153ms
9: learn: 0.6492810 total: 16.8ms remaining: 151ms
10: learn: 0.6452562 total: 17.4ms remaining: 141ms
11: learn: 0.6405148 total: 18.5ms remaining: 136ms
12: learn: 0.6367805 total: 19.7ms remaining: 132ms
13: learn: 0.6323872 total: 21.6ms remaining: 133ms
14: learn: 0.6287385 total: 24.3ms remaining: 137ms
15: learn: 0.6245734 total: 25.2ms remaining: 132ms
16: learn: 0.6206634 total: 26.7ms remaining: 130ms
17: learn: 0.6162322 total: 27.3ms remaining: 124ms
18: learn: 0.6130019 total: 28.3ms remaining: 120ms
19: learn: 0.6087847 total: 30.3ms remaining: 121ms
20: learn: 0.6049334 total: 32.3ms remaining: 122ms
21: learn: 0.6013334 total: 34.2ms remaining: 121ms
22: learn: 0.5976211 total: 35.5ms remaining: 119ms
23: learn: 0.5941389 total: 38.2ms remaining: 121ms
24: learn: 0.5904175 total: 41ms remaining: 123ms
25: learn: 0.5872989 total: 44ms remaining: 125ms
26: learn: 0.5843788 total: 44.9ms remaining: 121ms
27: learn: 0.5810017 total: 46.9ms remaining: 121ms
28: learn: 0.5770647 total: 47.8ms remaining: 117ms
29: learn: 0.5734684 total: 49ms remaining: 114ms
30: learn: 0.5696450 total: 49.7ms remaining: 111ms
31: learn: 0.5669904 total: 51.5ms remaining: 109ms
32: learn: 0.5633582 total: 54.1ms remaining: 110ms
33: learn: 0.5602036 total: 56.9ms remaining: 110ms
34: learn: 0.5574471 total: 59.7ms remaining: 111ms
35: learn: 0.5546641 total: 60.8ms remaining: 108ms
36: learn: 0.5520727 total: 62.3ms remaining: 106ms
37: learn: 0.5495315 total: 62.8ms remaining: 102ms
38: learn: 0.5463314 total: 63.3ms remaining: 99ms
39: learn: 0.5434985 total: 64.6ms remaining: 96.8ms
40: learn: 0.5406721 total: 65.5ms remaining: 94.3ms
41: learn: 0.5374358 total: 67.3ms remaining: 92.9ms
42: learn: 0.5343420 total: 68.2ms remaining: 90.4ms
43: learn: 0.5313562 total: 68.7ms remaining: 87.4ms
44: learn: 0.5281717 total: 70.4ms remaining: 86.1ms
45: learn: 0.5257638 total: 72.2ms remaining: 84.8ms
46: learn: 0.5230920 total: 73.7ms remaining: 83.1ms
47: learn: 0.5204169 total: 75.4ms remaining: 81.6ms
48: learn: 0.5177728 total: 76.7ms remaining: 79.8ms
49: learn: 0.5152570 total: 77.9ms remaining: 77.9ms
50: learn: 0.5126190 total: 79.1ms remaining: 76ms
51: learn: 0.5099504 total: 79.5ms remaining: 73.4ms
52: learn: 0.5070353 total: 80.8ms remaining: 71.6ms
53: learn: 0.5042589 total: 82.5ms remaining: 70.3ms
54: learn: 0.5019193 total: 84.4ms remaining: 69ms
55: learn: 0.4993186 total: 85.4ms remaining: 67.1ms
56: learn: 0.4972080 total: 87.1ms remaining: 65.7ms
57: learn: 0.4944874 total: 88.2ms remaining: 63.9ms
58: learn: 0.4915904 total: 89.1ms remaining: 61.9ms
59: learn: 0.4892463 total: 90.5ms remaining: 60.4ms
60: learn: 0.4864875 total: 92.1ms remaining: 58.9ms
61: learn: 0.4841580 total: 92.9ms remaining: 57ms
62: learn: 0.4819972 total: 94.2ms remaining: 55.3ms
63: learn: 0.4794997 total: 95.3ms remaining: 53.6ms
64: learn: 0.4774879 total: 96.6ms remaining: 52ms
65: learn: 0.4758185 total: 98.1ms remaining: 50.5ms
66: learn: 0.4734715 total: 99.3ms remaining: 48.9ms
67: learn: 0.4715649 total: 100ms remaining: 47.3ms
68: learn: 0.4693481 total: 102ms remaining: 45.7ms
69: learn: 0.4673057 total: 102ms remaining: 43.8ms
70: learn: 0.4649696 total: 103ms remaining: 42.2ms
71: learn: 0.4627991 total: 105ms remaining: 40.6ms
72: learn: 0.4609100 total: 106ms remaining: 39.1ms
73: learn: 0.4587119 total: 107ms remaining: 37.5ms
74: learn: 0.4568139 total: 108ms remaining: 35.9ms
75: learn: 0.4548912 total: 109ms remaining: 34.5ms
76: learn: 0.4529579 total: 110ms remaining: 33ms
77: learn: 0.4513188 total: 111ms remaining: 31.4ms
78: learn: 0.4494767 total: 112ms remaining: 29.8ms
79: learn: 0.4476995 total: 113ms remaining: 28.3ms
80: learn: 0.4454494 total: 114ms remaining: 26.8ms
81: learn: 0.4437134 total: 115ms remaining: 25.3ms
82: learn: 0.4417487 total: 116ms remaining: 23.8ms
83: learn: 0.4399505 total: 117ms remaining: 22.3ms
84: learn: 0.4380141 total: 118ms remaining: 20.8ms
85: learn: 0.4363505 total: 119ms remaining: 19.3ms
86: learn: 0.4340481 total: 119ms remaining: 17.8ms
87: learn: 0.4323121 total: 120ms remaining: 16.4ms
88: learn: 0.4308088 total: 121ms remaining: 14.9ms
89: learn: 0.4292199 total: 121ms remaining: 13.5ms
90: learn: 0.4276296 total: 122ms remaining: 12.1ms
91: learn: 0.4262623 total: 123ms remaining: 10.7ms
92: learn: 0.4245822 total: 124ms remaining: 9.3ms
93: learn: 0.4226833 total: 124ms remaining: 7.93ms
94: learn: 0.4210408 total: 125ms remaining: 6.58ms
95: learn: 0.4193460 total: 126ms remaining: 5.24ms
96: learn: 0.4179500 total: 126ms remaining: 3.91ms
97: learn: 0.4161901 total: 127ms remaining: 2.59ms
98: learn: 0.4145113 total: 128ms remaining: 1.29ms
99: learn: 0.4130434 total: 128ms remaining: 0us
0: learn: 0.6884949 total: 478us remaining: 47.4ms
1: learn: 0.6839801 total: 992us remaining: 48.6ms
2: learn: 0.6791613 total: 1.68ms remaining: 54.2ms
3: learn: 0.6742374 total: 2.18ms remaining: 52.3ms
4: learn: 0.6698433 total: 3.11ms remaining: 59.1ms
5: learn: 0.6657356 total: 3.81ms remaining: 59.8ms
6: learn: 0.6612373 total: 4.61ms remaining: 61.3ms
7: learn: 0.6568788 total: 5.17ms remaining: 59.5ms
8: learn: 0.6536179 total: 5.6ms remaining: 56.6ms
9: learn: 0.6493948 total: 5.96ms remaining: 53.6ms
10: learn: 0.6452865 total: 7.17ms remaining: 58ms
11: learn: 0.6416073 total: 7.81ms remaining: 57.3ms
12: learn: 0.6391135 total: 8.97ms remaining: 60ms
13: learn: 0.6346678 total: 9.82ms remaining: 60.3ms
14: learn: 0.6311541 total: 10.6ms remaining: 60ms
15: learn: 0.6275356 total: 11.2ms remaining: 58.7ms
16: learn: 0.6240274 total: 11.9ms remaining: 58ms
17: learn: 0.6205983 total: 12.5ms remaining: 56.8ms
18: learn: 0.6164443 total: 12.9ms remaining: 55.2ms
19: learn: 0.6122633 total: 13.4ms remaining: 53.7ms
20: learn: 0.6092096 total: 13.9ms remaining: 52.3ms
21: learn: 0.6057245 total: 14.4ms remaining: 51.1ms
22: learn: 0.6021422 total: 15.1ms remaining: 50.7ms
23: learn: 0.5981987 total: 15.6ms remaining: 49.4ms
24: learn: 0.5948994 total: 16.2ms remaining: 48.5ms
25: learn: 0.5921669 total: 16.7ms remaining: 47.4ms
26: learn: 0.5890569 total: 17.3ms remaining: 46.7ms
27: learn: 0.5853665 total: 18.1ms remaining: 46.5ms
28: learn: 0.5821117 total: 18.7ms remaining: 45.9ms
29: learn: 0.5789751 total: 19.3ms remaining: 45.1ms
30: learn: 0.5760359 total: 19.9ms remaining: 44.3ms
31: learn: 0.5726499 total: 20.4ms remaining: 43.2ms
32: learn: 0.5692102 total: 20.9ms remaining: 42.4ms
33: learn: 0.5659861 total: 21.4ms remaining: 41.6ms
34: learn: 0.5629366 total: 22.1ms remaining: 41ms
35: learn: 0.5603221 total: 22.8ms remaining: 40.4ms
36: learn: 0.5568870 total: 23.5ms remaining: 39.9ms
37: learn: 0.5537936 total: 24ms remaining: 39.1ms
38: learn: 0.5507915 total: 24.5ms remaining: 38.4ms
39: learn: 0.5481011 total: 25ms remaining: 37.4ms
40: learn: 0.5452670 total: 25.6ms remaining: 36.9ms
41: learn: 0.5417452 total: 26.2ms remaining: 36.2ms
42: learn: 0.5391062 total: 26.8ms remaining: 35.6ms
43: learn: 0.5367240 total: 27.6ms remaining: 35.2ms
44: learn: 0.5338743 total: 28.6ms remaining: 34.9ms
45: learn: 0.5310151 total: 29.3ms remaining: 34.3ms
46: learn: 0.5279937 total: 29.6ms remaining: 33.4ms
47: learn: 0.5251938 total: 30.2ms remaining: 32.7ms
48: learn: 0.5222575 total: 30.5ms remaining: 31.8ms
49: learn: 0.5198283 total: 31.2ms remaining: 31.2ms
50: learn: 0.5176906 total: 32ms remaining: 30.7ms
51: learn: 0.5150305 total: 32.6ms remaining: 30.1ms
52: learn: 0.5124388 total: 33.2ms remaining: 29.5ms
53: learn: 0.5101732 total: 33.8ms remaining: 28.8ms
54: learn: 0.5077479 total: 34.5ms remaining: 28.2ms
55: learn: 0.5056719 total: 35.2ms remaining: 27.6ms
56: learn: 0.5030096 total: 35.7ms remaining: 26.9ms
57: learn: 0.5003582 total: 36.3ms remaining: 26.3ms
58: learn: 0.4983844 total: 36.9ms remaining: 25.6ms
59: learn: 0.4960864 total: 37.5ms remaining: 25ms
60: learn: 0.4935340 total: 37.9ms remaining: 24.2ms
61: learn: 0.4919227 total: 38.5ms remaining: 23.6ms
62: learn: 0.4895177 total: 39.1ms remaining: 23ms
63: learn: 0.4869231 total: 39.6ms remaining: 22.3ms
64: learn: 0.4846806 total: 40.2ms remaining: 21.6ms
65: learn: 0.4819794 total: 40.7ms remaining: 21ms
66: learn: 0.4798768 total: 41.2ms remaining: 20.3ms
67: learn: 0.4778844 total: 41.8ms remaining: 19.7ms
68: learn: 0.4756829 total: 42.4ms remaining: 19ms
69: learn: 0.4733200 total: 43.1ms remaining: 18.5ms
70: learn: 0.4712280 total: 43.7ms remaining: 17.9ms
71: learn: 0.4691641 total: 44.3ms remaining: 17.2ms
72: learn: 0.4670457 total: 44.8ms remaining: 16.6ms
73: learn: 0.4654191 total: 45.5ms remaining: 16ms
74: learn: 0.4634164 total: 45.9ms remaining: 15.3ms
75: learn: 0.4613756 total: 46.5ms remaining: 14.7ms
76: learn: 0.4593780 total: 47.1ms remaining: 14.1ms
77: learn: 0.4570923 total: 47.8ms remaining: 13.5ms
78: learn: 0.4550906 total: 48.5ms remaining: 12.9ms
79: learn: 0.4534051 total: 49.2ms remaining: 12.3ms
80: learn: 0.4517382 total: 49.8ms remaining: 11.7ms
81: learn: 0.4500298 total: 50.4ms remaining: 11.1ms
82: learn: 0.4479994 total: 51ms remaining: 10.4ms
83: learn: 0.4461639 total: 51.7ms remaining: 9.84ms
84: learn: 0.4444105 total: 52.2ms remaining: 9.2ms
85: learn: 0.4431826 total: 52.7ms remaining: 8.58ms
86: learn: 0.4412751 total: 53.4ms remaining: 7.98ms
87: learn: 0.4396587 total: 54ms remaining: 7.36ms
88: learn: 0.4378393 total: 54.7ms remaining: 6.76ms
89: learn: 0.4359180 total: 55.4ms remaining: 6.15ms
90: learn: 0.4345389 total: 56.1ms remaining: 5.54ms
91: learn: 0.4328381 total: 56.7ms remaining: 4.93ms
92: learn: 0.4310307 total: 57.3ms remaining: 4.31ms
93: learn: 0.4292151 total: 57.9ms remaining: 3.69ms
94: learn: 0.4275225 total: 58.6ms remaining: 3.08ms
95: learn: 0.4260639 total: 59.3ms remaining: 2.47ms
96: learn: 0.4243990 total: 59.8ms remaining: 1.85ms
97: learn: 0.4225974 total: 60.5ms remaining: 1.23ms
98: learn: 0.4209589 total: 61.3ms remaining: 618us
99: learn: 0.4194141 total: 61.8ms remaining: 0us
0: learn: 0.6878969 total: 695us remaining: 68.9ms
1: learn: 0.6834371 total: 1.53ms remaining: 74.8ms
2: learn: 0.6793659 total: 2.81ms remaining: 90.9ms
3: learn: 0.6746757 total: 3.99ms remaining: 95.8ms
4: learn: 0.6702315 total: 4.92ms remaining: 93.4ms
5: learn: 0.6660132 total: 6.3ms remaining: 98.8ms
6: learn: 0.6620074 total: 7.26ms remaining: 96.5ms
7: learn: 0.6577334 total: 8.35ms remaining: 96.1ms
8: learn: 0.6533813 total: 9.29ms remaining: 93.9ms
9: learn: 0.6487205 total: 10.2ms remaining: 91.6ms
10: learn: 0.6447954 total: 11.2ms remaining: 90.8ms
11: learn: 0.6401955 total: 12.1ms remaining: 88.9ms
12: learn: 0.6355459 total: 12.8ms remaining: 85.9ms
13: learn: 0.6317789 total: 13.8ms remaining: 84.9ms
14: learn: 0.6274385 total: 15ms remaining: 84.9ms
15: learn: 0.6239107 total: 16ms remaining: 83.9ms
16: learn: 0.6199303 total: 16.8ms remaining: 82.2ms
17: learn: 0.6165104 total: 17.8ms remaining: 81ms
18: learn: 0.6121992 total: 18.4ms remaining: 78.5ms
19: learn: 0.6090521 total: 19.3ms remaining: 77.1ms
20: learn: 0.6049247 total: 20.5ms remaining: 77.2ms
21: learn: 0.6009591 total: 21.5ms remaining: 76.2ms
22: learn: 0.5972106 total: 22.6ms remaining: 75.7ms
23: learn: 0.5934511 total: 22.9ms remaining: 72.6ms
24: learn: 0.5896801 total: 23.5ms remaining: 70.5ms
25: learn: 0.5857917 total: 24.1ms remaining: 68.7ms
26: learn: 0.5824441 total: 25.2ms remaining: 68ms
27: learn: 0.5782137 total: 26.3ms remaining: 67.5ms
28: learn: 0.5745073 total: 27.3ms remaining: 66.7ms
29: learn: 0.5714868 total: 28.3ms remaining: 65.9ms
30: learn: 0.5678615 total: 29.2ms remaining: 64.9ms
31: learn: 0.5640808 total: 29.9ms remaining: 63.6ms
32: learn: 0.5609747 total: 30.9ms remaining: 62.6ms
33: learn: 0.5572637 total: 31.8ms remaining: 61.7ms
34: learn: 0.5541527 total: 32.2ms remaining: 59.8ms
35: learn: 0.5513270 total: 33.1ms remaining: 58.9ms
36: learn: 0.5478448 total: 34ms remaining: 57.9ms
37: learn: 0.5444886 total: 34.7ms remaining: 56.5ms
38: learn: 0.5418822 total: 35.3ms remaining: 55.2ms
39: learn: 0.5388862 total: 36.3ms remaining: 54.4ms
40: learn: 0.5363399 total: 36.9ms remaining: 53.1ms
41: learn: 0.5334605 total: 37.9ms remaining: 52.3ms
42: learn: 0.5303391 total: 38.5ms remaining: 51.1ms
43: learn: 0.5271799 total: 39.2ms remaining: 49.9ms
44: learn: 0.5242581 total: 40ms remaining: 48.9ms
45: learn: 0.5213147 total: 40.8ms remaining: 47.9ms
46: learn: 0.5184782 total: 41.7ms remaining: 47ms
47: learn: 0.5158614 total: 42.4ms remaining: 45.9ms
48: learn: 0.5129389 total: 43.1ms remaining: 44.9ms
49: learn: 0.5103840 total: 43.5ms remaining: 43.5ms
50: learn: 0.5079012 total: 44.2ms remaining: 42.4ms
51: learn: 0.5053963 total: 45.1ms remaining: 41.6ms
52: learn: 0.5027932 total: 45.8ms remaining: 40.6ms
53: learn: 0.5002335 total: 46.6ms remaining: 39.7ms
54: learn: 0.4975191 total: 47.2ms remaining: 38.6ms
55: learn: 0.4947258 total: 48ms remaining: 37.7ms
56: learn: 0.4923294 total: 48.6ms remaining: 36.7ms
57: learn: 0.4901360 total: 49.3ms remaining: 35.7ms
58: learn: 0.4874011 total: 49.9ms remaining: 34.7ms
59: learn: 0.4851628 total: 50.6ms remaining: 33.7ms
60: learn: 0.4822885 total: 51ms remaining: 32.6ms
61: learn: 0.4797427 total: 51.3ms remaining: 31.5ms
62: learn: 0.4771268 total: 51.9ms remaining: 30.5ms
63: learn: 0.4746619 total: 52.3ms remaining: 29.4ms
64: learn: 0.4723054 total: 52.8ms remaining: 28.4ms
65: learn: 0.4698461 total: 53.3ms remaining: 27.4ms
66: learn: 0.4678423 total: 53.8ms remaining: 26.5ms
67: learn: 0.4656479 total: 54.4ms remaining: 25.6ms
68: learn: 0.4636092 total: 54.9ms remaining: 24.7ms
69: learn: 0.4613971 total: 55ms remaining: 23.6ms
70: learn: 0.4590672 total: 55.6ms remaining: 22.7ms
71: learn: 0.4566890 total: 56.3ms remaining: 21.9ms
72: learn: 0.4549533 total: 56.8ms remaining: 21ms
73: learn: 0.4527144 total: 57.2ms remaining: 20.1ms
74: learn: 0.4508319 total: 57.8ms remaining: 19.3ms
75: learn: 0.4485957 total: 58.5ms remaining: 18.5ms
76: learn: 0.4461832 total: 59.3ms remaining: 17.7ms
77: learn: 0.4442718 total: 60.2ms remaining: 17ms
78: learn: 0.4424299 total: 60.9ms remaining: 16.2ms
79: learn: 0.4405656 total: 61.4ms remaining: 15.4ms
80: learn: 0.4386087 total: 62ms remaining: 14.5ms
81: learn: 0.4369478 total: 62.6ms remaining: 13.7ms
82: learn: 0.4350702 total: 63.2ms remaining: 13ms
83: learn: 0.4333680 total: 64ms remaining: 12.2ms
84: learn: 0.4311063 total: 64.7ms remaining: 11.4ms
85: learn: 0.4293329 total: 65.1ms remaining: 10.6ms
86: learn: 0.4275034 total: 65.7ms remaining: 9.82ms
87: learn: 0.4257185 total: 66.4ms remaining: 9.05ms
88: learn: 0.4237555 total: 67ms remaining: 8.29ms
89: learn: 0.4214402 total: 67.5ms remaining: 7.5ms
90: learn: 0.4196571 total: 68.2ms remaining: 6.74ms
91: learn: 0.4179091 total: 68.3ms remaining: 5.94ms
92: learn: 0.4163563 total: 69ms remaining: 5.19ms
93: learn: 0.4144657 total: 69.5ms remaining: 4.44ms
94: learn: 0.4128859 total: 70.5ms remaining: 3.71ms
95: learn: 0.4108897 total: 71ms remaining: 2.96ms
96: learn: 0.4095732 total: 71.5ms remaining: 2.21ms
97: learn: 0.4079204 total: 72.2ms remaining: 1.47ms
98: learn: 0.4063518 total: 72.8ms remaining: 735us
99: learn: 0.4046437 total: 73.5ms remaining: 0us
0: learn: 0.6886303 total: 651us remaining: 64.5ms
1: learn: 0.6848532 total: 1.18ms remaining: 57.8ms
2: learn: 0.6807654 total: 1.77ms remaining: 57.1ms
3: learn: 0.6757109 total: 2.44ms remaining: 58.6ms
4: learn: 0.6712357 total: 3.19ms remaining: 60.6ms
5: learn: 0.6677912 total: 3.81ms remaining: 59.7ms
6: learn: 0.6633335 total: 4.33ms remaining: 57.6ms
7: learn: 0.6595120 total: 4.65ms remaining: 53.5ms
8: learn: 0.6547988 total: 5.09ms remaining: 51.5ms
9: learn: 0.6508939 total: 5.49ms remaining: 49.5ms
10: learn: 0.6474754 total: 6.08ms remaining: 49.2ms
11: learn: 0.6435513 total: 6.6ms remaining: 48.4ms
12: learn: 0.6399169 total: 7.24ms remaining: 48.4ms
13: learn: 0.6360485 total: 7.87ms remaining: 48.3ms
14: learn: 0.6320116 total: 8.02ms remaining: 45.5ms
15: learn: 0.6283337 total: 8.67ms remaining: 45.5ms
16: learn: 0.6250711 total: 9.3ms remaining: 45.4ms
17: learn: 0.6215353 total: 10ms remaining: 45.7ms
18: learn: 0.6181452 total: 10.7ms remaining: 45.6ms
19: learn: 0.6144406 total: 11.3ms remaining: 45.1ms
20: learn: 0.6107661 total: 11.9ms remaining: 44.7ms
21: learn: 0.6074419 total: 12.5ms remaining: 44.3ms
22: learn: 0.6040458 total: 13.1ms remaining: 43.8ms
23: learn: 0.6009696 total: 13.7ms remaining: 43.5ms
24: learn: 0.5974595 total: 14.3ms remaining: 43ms
25: learn: 0.5941549 total: 15ms remaining: 42.7ms
26: learn: 0.5910260 total: 15.6ms remaining: 42.2ms
27: learn: 0.5879450 total: 16ms remaining: 41.1ms
28: learn: 0.5848512 total: 16.6ms remaining: 40.6ms
29: learn: 0.5820150 total: 17.2ms remaining: 40.1ms
30: learn: 0.5785911 total: 17.8ms remaining: 39.7ms
31: learn: 0.5753528 total: 18.3ms remaining: 38.9ms
32: learn: 0.5725451 total: 19.1ms remaining: 38.7ms
33: learn: 0.5700475 total: 19.7ms remaining: 38.2ms
34: learn: 0.5667296 total: 20.4ms remaining: 37.8ms
35: learn: 0.5638499 total: 20.9ms remaining: 37.1ms
36: learn: 0.5609647 total: 21.3ms remaining: 36.3ms
37: learn: 0.5576302 total: 21.7ms remaining: 35.4ms
38: learn: 0.5545089 total: 22.3ms remaining: 34.9ms
39: learn: 0.5522755 total: 22.9ms remaining: 34.3ms
40: learn: 0.5490363 total: 23.6ms remaining: 33.9ms
41: learn: 0.5461510 total: 24ms remaining: 33.1ms
42: learn: 0.5430897 total: 24.4ms remaining: 32.3ms
43: learn: 0.5398943 total: 24.8ms remaining: 31.5ms
44: learn: 0.5372769 total: 25.5ms remaining: 31.2ms
45: learn: 0.5347099 total: 26.4ms remaining: 31ms
46: learn: 0.5322609 total: 26.9ms remaining: 30.4ms
47: learn: 0.5299848 total: 27.6ms remaining: 29.9ms
48: learn: 0.5275044 total: 28.3ms remaining: 29.5ms
49: learn: 0.5247466 total: 28.9ms remaining: 28.9ms
50: learn: 0.5227275 total: 29.6ms remaining: 28.4ms
51: learn: 0.5202591 total: 30.4ms remaining: 28.1ms
52: learn: 0.5176368 total: 31.3ms remaining: 27.8ms
53: learn: 0.5153116 total: 32ms remaining: 27.3ms
54: learn: 0.5130050 total: 32.2ms remaining: 26.3ms
55: learn: 0.5107425 total: 33.1ms remaining: 26ms
56: learn: 0.5083821 total: 33.6ms remaining: 25.3ms
57: learn: 0.5061264 total: 34.3ms remaining: 24.9ms
58: learn: 0.5037891 total: 35ms remaining: 24.3ms
59: learn: 0.5014108 total: 35.4ms remaining: 23.6ms
60: learn: 0.4991912 total: 35.7ms remaining: 22.8ms
61: learn: 0.4968640 total: 36.3ms remaining: 22.3ms
62: learn: 0.4943204 total: 37ms remaining: 21.8ms
63: learn: 0.4923553 total: 38ms remaining: 21.4ms
64: learn: 0.4903686 total: 38.5ms remaining: 20.7ms
65: learn: 0.4880668 total: 39.3ms remaining: 20.2ms
66: learn: 0.4859492 total: 40ms remaining: 19.7ms
67: learn: 0.4839476 total: 40.1ms remaining: 18.9ms
68: learn: 0.4818279 total: 40.9ms remaining: 18.4ms
69: learn: 0.4799646 total: 41.2ms remaining: 17.7ms
70: learn: 0.4780925 total: 41.8ms remaining: 17.1ms
71: learn: 0.4761372 total: 42.5ms remaining: 16.5ms
72: learn: 0.4741833 total: 43.3ms remaining: 16ms
73: learn: 0.4723118 total: 43.5ms remaining: 15.3ms
74: learn: 0.4703517 total: 43.9ms remaining: 14.6ms
75: learn: 0.4682914 total: 44.5ms remaining: 14ms
76: learn: 0.4662159 total: 45.1ms remaining: 13.5ms
77: learn: 0.4643002 total: 45.6ms remaining: 12.9ms
78: learn: 0.4627185 total: 46.3ms remaining: 12.3ms
79: learn: 0.4607886 total: 46.8ms remaining: 11.7ms
80: learn: 0.4591387 total: 47.5ms remaining: 11.1ms
81: learn: 0.4572590 total: 48.3ms remaining: 10.6ms
82: learn: 0.4552794 total: 49ms remaining: 10ms
83: learn: 0.4535576 total: 49.4ms remaining: 9.41ms
84: learn: 0.4515675 total: 50.1ms remaining: 8.84ms
85: learn: 0.4500313 total: 50.6ms remaining: 8.23ms
86: learn: 0.4484269 total: 51ms remaining: 7.63ms
87: learn: 0.4468399 total: 51.3ms remaining: 6.99ms
88: learn: 0.4452077 total: 52ms remaining: 6.43ms
89: learn: 0.4437499 total: 52.9ms remaining: 5.88ms
90: learn: 0.4421317 total: 53.6ms remaining: 5.3ms
91: learn: 0.4404462 total: 54.4ms remaining: 4.73ms
92: learn: 0.4384857 total: 55ms remaining: 4.14ms
93: learn: 0.4369197 total: 55.7ms remaining: 3.55ms
94: learn: 0.4351167 total: 56.2ms remaining: 2.96ms
95: learn: 0.4335310 total: 56.9ms remaining: 2.37ms
96: learn: 0.4315526 total: 57.7ms remaining: 1.78ms
97: learn: 0.4299308 total: 58.3ms remaining: 1.19ms
98: learn: 0.4283443 total: 59.4ms remaining: 599us
99: learn: 0.4265996 total: 59.9ms remaining: 0us
0: learn: 0.6890516 total: 1.16ms remaining: 115ms
1: learn: 0.6843684 total: 2.2ms remaining: 108ms
2: learn: 0.6798558 total: 3.23ms remaining: 104ms
3: learn: 0.6749312 total: 4.05ms remaining: 97.2ms
4: learn: 0.6708149 total: 4.92ms remaining: 93.6ms
5: learn: 0.6666183 total: 5.81ms remaining: 91ms
6: learn: 0.6614496 total: 6.64ms remaining: 88.2ms
7: learn: 0.6573563 total: 7.5ms remaining: 86.3ms
8: learn: 0.6532401 total: 8.3ms remaining: 83.9ms
9: learn: 0.6500008 total: 9.04ms remaining: 81.3ms
10: learn: 0.6454487 total: 9.93ms remaining: 80.4ms
11: learn: 0.6410700 total: 11ms remaining: 80.3ms
12: learn: 0.6370755 total: 11.9ms remaining: 79.8ms
13: learn: 0.6327961 total: 12.7ms remaining: 78.2ms
14: learn: 0.6294922 total: 13.6ms remaining: 77.3ms
15: learn: 0.6260024 total: 14.3ms remaining: 75.3ms
16: learn: 0.6225606 total: 15.1ms remaining: 73.8ms
17: learn: 0.6194160 total: 15.8ms remaining: 71.9ms
18: learn: 0.6160197 total: 16.7ms remaining: 71.1ms
19: learn: 0.6121442 total: 17.4ms remaining: 69.6ms
20: learn: 0.6080268 total: 18.2ms remaining: 68.6ms
21: learn: 0.6045500 total: 18.9ms remaining: 67ms
22: learn: 0.6005688 total: 19.6ms remaining: 65.8ms
23: learn: 0.5973170 total: 20.5ms remaining: 65ms
24: learn: 0.5939270 total: 21.4ms remaining: 64.3ms
25: learn: 0.5907113 total: 24.2ms remaining: 68.8ms
26: learn: 0.5874044 total: 25.3ms remaining: 68.3ms
27: learn: 0.5838297 total: 26.4ms remaining: 67.9ms
28: learn: 0.5804910 total: 27.4ms remaining: 67ms
29: learn: 0.5767455 total: 28.1ms remaining: 65.6ms
30: learn: 0.5729896 total: 28.9ms remaining: 64.4ms
31: learn: 0.5699794 total: 30ms remaining: 63.7ms
32: learn: 0.5664476 total: 31.1ms remaining: 63.1ms
33: learn: 0.5630630 total: 32.2ms remaining: 62.6ms
34: learn: 0.5606261 total: 33.4ms remaining: 62ms
35: learn: 0.5575230 total: 34.4ms remaining: 61.2ms
36: learn: 0.5543894 total: 35.5ms remaining: 60.5ms
37: learn: 0.5513296 total: 36.5ms remaining: 59.6ms
38: learn: 0.5481648 total: 37.5ms remaining: 58.7ms
39: learn: 0.5451467 total: 38.3ms remaining: 57.5ms
40: learn: 0.5421398 total: 39.4ms remaining: 56.7ms
41: learn: 0.5394561 total: 40.4ms remaining: 55.8ms
42: learn: 0.5365911 total: 41.4ms remaining: 54.9ms
43: learn: 0.5336141 total: 42.3ms remaining: 53.9ms
44: learn: 0.5307000 total: 43.4ms remaining: 53ms
45: learn: 0.5278690 total: 44.3ms remaining: 52ms
46: learn: 0.5250755 total: 45.1ms remaining: 50.8ms
47: learn: 0.5225611 total: 45.7ms remaining: 49.6ms
48: learn: 0.5201245 total: 46.5ms remaining: 48.4ms
49: learn: 0.5169272 total: 47.1ms remaining: 47.1ms
50: learn: 0.5145339 total: 47.4ms remaining: 45.5ms
51: learn: 0.5119153 total: 48.1ms remaining: 44.4ms
52: learn: 0.5093315 total: 48.8ms remaining: 43.3ms
53: learn: 0.5069142 total: 49.6ms remaining: 42.3ms
54: learn: 0.5046413 total: 50ms remaining: 40.9ms
55: learn: 0.5018997 total: 50.7ms remaining: 39.8ms
56: learn: 0.4991637 total: 51.7ms remaining: 39ms
57: learn: 0.4964448 total: 52.4ms remaining: 38ms
58: learn: 0.4940433 total: 53.4ms remaining: 37.1ms
59: learn: 0.4921181 total: 54.3ms remaining: 36.2ms
60: learn: 0.4896671 total: 55.1ms remaining: 35.2ms
61: learn: 0.4878902 total: 56ms remaining: 34.3ms
62: learn: 0.4856388 total: 56.7ms remaining: 33.3ms
63: learn: 0.4829752 total: 57.4ms remaining: 32.3ms
64: learn: 0.4808916 total: 58ms remaining: 31.2ms
65: learn: 0.4790227 total: 58.7ms remaining: 30.2ms
66: learn: 0.4769375 total: 59.3ms remaining: 29.2ms
67: learn: 0.4745963 total: 60ms remaining: 28.3ms
68: learn: 0.4724582 total: 60.9ms remaining: 27.4ms
69: learn: 0.4701525 total: 61.6ms remaining: 26.4ms
70: learn: 0.4681852 total: 62.4ms remaining: 25.5ms
71: learn: 0.4660803 total: 63ms remaining: 24.5ms
72: learn: 0.4639571 total: 63.7ms remaining: 23.5ms
73: learn: 0.4620728 total: 64.1ms remaining: 22.5ms
74: learn: 0.4599836 total: 64.9ms remaining: 21.6ms
75: learn: 0.4576282 total: 65.6ms remaining: 20.7ms
76: learn: 0.4557343 total: 66.2ms remaining: 19.8ms
77: learn: 0.4538526 total: 66.5ms remaining: 18.7ms
78: learn: 0.4522163 total: 67.1ms remaining: 17.8ms
79: learn: 0.4497491 total: 67.9ms remaining: 17ms
80: learn: 0.4477534 total: 68.5ms remaining: 16.1ms
81: learn: 0.4460425 total: 69.4ms remaining: 15.2ms
82: learn: 0.4442584 total: 70.1ms remaining: 14.4ms
83: learn: 0.4423690 total: 70.9ms remaining: 13.5ms
84: learn: 0.4405038 total: 71.6ms remaining: 12.6ms
85: learn: 0.4384172 total: 72.1ms remaining: 11.7ms
86: learn: 0.4362462 total: 72.6ms remaining: 10.9ms
87: learn: 0.4349486 total: 73.4ms remaining: 10ms
88: learn: 0.4333516 total: 74.1ms remaining: 9.16ms
89: learn: 0.4314233 total: 74.9ms remaining: 8.32ms
90: learn: 0.4298179 total: 75.6ms remaining: 7.47ms
91: learn: 0.4279365 total: 76.4ms remaining: 6.64ms
92: learn: 0.4260680 total: 76.9ms remaining: 5.79ms
93: learn: 0.4243747 total: 77.4ms remaining: 4.94ms
94: learn: 0.4224190 total: 78.1ms remaining: 4.11ms
95: learn: 0.4204823 total: 78.7ms remaining: 3.28ms
96: learn: 0.4185932 total: 79.9ms remaining: 2.47ms
97: learn: 0.4171285 total: 80.6ms remaining: 1.65ms
98: learn: 0.4159387 total: 81ms remaining: 818us
99: learn: 0.4141446 total: 81.7ms remaining: 0us
0: learn: 0.6884397 total: 798us remaining: 79.1ms
1: learn: 0.6835109 total: 1.43ms remaining: 70.1ms
2: learn: 0.6795496 total: 2.52ms remaining: 81.6ms
3: learn: 0.6751464 total: 3.49ms remaining: 83.7ms
4: learn: 0.6712628 total: 4.31ms remaining: 81.9ms
5: learn: 0.6671028 total: 5.55ms remaining: 87ms
6: learn: 0.6631048 total: 6.29ms remaining: 83.6ms
7: learn: 0.6589732 total: 7.23ms remaining: 83.1ms
8: learn: 0.6543210 total: 7.99ms remaining: 80.7ms
9: learn: 0.6503692 total: 8.8ms remaining: 79.2ms
10: learn: 0.6460389 total: 9.11ms remaining: 73.7ms
11: learn: 0.6423343 total: 9.88ms remaining: 72.5ms
12: learn: 0.6382826 total: 10.7ms remaining: 71.5ms
13: learn: 0.6341497 total: 11.2ms remaining: 69.1ms
14: learn: 0.6306207 total: 11.9ms remaining: 67.2ms
15: learn: 0.6267048 total: 12.8ms remaining: 67ms
16: learn: 0.6235354 total: 13.4ms remaining: 65.5ms
17: learn: 0.6202240 total: 13.9ms remaining: 63.4ms
18: learn: 0.6164425 total: 14.6ms remaining: 62.3ms
19: learn: 0.6124860 total: 15.1ms remaining: 60.4ms
20: learn: 0.6090163 total: 15.8ms remaining: 59.5ms
21: learn: 0.6057270 total: 16.5ms remaining: 58.6ms
22: learn: 0.6017883 total: 17.3ms remaining: 58ms
23: learn: 0.5982530 total: 17.9ms remaining: 56.8ms
24: learn: 0.5953937 total: 18.9ms remaining: 56.6ms
25: learn: 0.5918257 total: 19.2ms remaining: 54.7ms
26: learn: 0.5884519 total: 19.5ms remaining: 52.7ms
27: learn: 0.5852720 total: 20ms remaining: 51.5ms
28: learn: 0.5822245 total: 20.6ms remaining: 50.4ms
29: learn: 0.5795334 total: 21.2ms remaining: 49.4ms
30: learn: 0.5760984 total: 21.6ms remaining: 48.1ms
31: learn: 0.5726693 total: 22.3ms remaining: 47.3ms
32: learn: 0.5692743 total: 22.8ms remaining: 46.4ms
33: learn: 0.5666976 total: 23.2ms remaining: 45.1ms
34: learn: 0.5634716 total: 23.6ms remaining: 43.9ms
35: learn: 0.5606266 total: 24.1ms remaining: 42.8ms
36: learn: 0.5577354 total: 24.6ms remaining: 41.9ms
37: learn: 0.5545374 total: 25.2ms remaining: 41.1ms
38: learn: 0.5516278 total: 25.7ms remaining: 40.1ms
39: learn: 0.5482702 total: 26.1ms remaining: 39.1ms
40: learn: 0.5451829 total: 26.4ms remaining: 37.9ms
41: learn: 0.5421858 total: 26.9ms remaining: 37.2ms
42: learn: 0.5394945 total: 27.2ms remaining: 36.1ms
43: learn: 0.5367497 total: 27.7ms remaining: 35.2ms
44: learn: 0.5341104 total: 28.1ms remaining: 34.4ms
45: learn: 0.5318895 total: 28.6ms remaining: 33.6ms
46: learn: 0.5292087 total: 29.1ms remaining: 32.8ms
47: learn: 0.5268572 total: 29.7ms remaining: 32.2ms
48: learn: 0.5241924 total: 30.3ms remaining: 31.5ms
49: learn: 0.5215148 total: 30.8ms remaining: 30.8ms
50: learn: 0.5187961 total: 31.3ms remaining: 30ms
51: learn: 0.5163954 total: 31.6ms remaining: 29.2ms
52: learn: 0.5143928 total: 32.6ms remaining: 28.9ms
53: learn: 0.5123380 total: 33.3ms remaining: 28.3ms
54: learn: 0.5104340 total: 33.9ms remaining: 27.8ms
55: learn: 0.5077391 total: 34.5ms remaining: 27.1ms
56: learn: 0.5053542 total: 35.1ms remaining: 26.4ms
57: learn: 0.5027166 total: 35.7ms remaining: 25.9ms
58: learn: 0.5003520 total: 36.1ms remaining: 25.1ms
59: learn: 0.4982657 total: 36.4ms remaining: 24.3ms
60: learn: 0.4955679 total: 36.6ms remaining: 23.4ms
61: learn: 0.4933646 total: 37.3ms remaining: 22.8ms
62: learn: 0.4915750 total: 37.8ms remaining: 22.2ms
63: learn: 0.4894400 total: 38.4ms remaining: 21.6ms
64: learn: 0.4874196 total: 38.8ms remaining: 20.9ms
65: learn: 0.4855573 total: 39.4ms remaining: 20.3ms
66: learn: 0.4832764 total: 40ms remaining: 19.7ms
67: learn: 0.4811310 total: 40.5ms remaining: 19.1ms
68: learn: 0.4789793 total: 41ms remaining: 18.4ms
69: learn: 0.4766580 total: 41.4ms remaining: 17.7ms
70: learn: 0.4744961 total: 42ms remaining: 17.1ms
71: learn: 0.4725618 total: 42.6ms remaining: 16.6ms
72: learn: 0.4704516 total: 43.3ms remaining: 16ms
73: learn: 0.4683780 total: 43.9ms remaining: 15.4ms
74: learn: 0.4660309 total: 44.6ms remaining: 14.9ms
75: learn: 0.4640216 total: 45ms remaining: 14.2ms
76: learn: 0.4621854 total: 45.6ms remaining: 13.6ms
77: learn: 0.4606499 total: 46.2ms remaining: 13ms
78: learn: 0.4588338 total: 46.8ms remaining: 12.4ms
79: learn: 0.4566047 total: 47.3ms remaining: 11.8ms
80: learn: 0.4547340 total: 47.9ms remaining: 11.2ms
81: learn: 0.4526263 total: 48.5ms remaining: 10.6ms
82: learn: 0.4510854 total: 48.9ms remaining: 10ms
83: learn: 0.4493695 total: 49.3ms remaining: 9.39ms
84: learn: 0.4475559 total: 49.9ms remaining: 8.81ms
85: learn: 0.4460767 total: 50.6ms remaining: 8.23ms
86: learn: 0.4441912 total: 51.1ms remaining: 7.63ms
87: learn: 0.4422915 total: 51.8ms remaining: 7.06ms
88: learn: 0.4408521 total: 52.4ms remaining: 6.47ms
89: learn: 0.4390969 total: 53ms remaining: 5.89ms
90: learn: 0.4371253 total: 53.4ms remaining: 5.28ms
91: learn: 0.4354587 total: 54ms remaining: 4.7ms
92: learn: 0.4336762 total: 54.5ms remaining: 4.1ms
93: learn: 0.4316399 total: 55.1ms remaining: 3.52ms
94: learn: 0.4300309 total: 55.7ms remaining: 2.93ms
95: learn: 0.4282519 total: 56.3ms remaining: 2.34ms
96: learn: 0.4270079 total: 56.8ms remaining: 1.76ms
97: learn: 0.4251263 total: 57.5ms remaining: 1.17ms
98: learn: 0.4234855 total: 58.1ms remaining: 587us
99: learn: 0.4217781 total: 58.7ms remaining: 0us
0: learn: 0.6884211 total: 638us remaining: 63.2ms
1: learn: 0.6839877 total: 2.42ms remaining: 119ms
2: learn: 0.6791304 total: 3.69ms remaining: 119ms
3: learn: 0.6745372 total: 5.04ms remaining: 121ms
4: learn: 0.6705564 total: 6.21ms remaining: 118ms
5: learn: 0.6657961 total: 7.39ms remaining: 116ms
6: learn: 0.6618196 total: 8.28ms remaining: 110ms
7: learn: 0.6577615 total: 9.57ms remaining: 110ms
8: learn: 0.6540484 total: 10.8ms remaining: 109ms
9: learn: 0.6503615 total: 11.6ms remaining: 105ms
10: learn: 0.6460645 total: 12ms remaining: 97.2ms
11: learn: 0.6420317 total: 13.3ms remaining: 97.3ms
12: learn: 0.6381292 total: 14.2ms remaining: 95.2ms
13: learn: 0.6339102 total: 14.9ms remaining: 91.7ms
14: learn: 0.6301877 total: 16ms remaining: 90.6ms
15: learn: 0.6264915 total: 17.1ms remaining: 89.9ms
16: learn: 0.6233760 total: 18.1ms remaining: 88.6ms
17: learn: 0.6198275 total: 19.1ms remaining: 87.2ms
18: learn: 0.6163971 total: 20.1ms remaining: 85.9ms
19: learn: 0.6123553 total: 20.9ms remaining: 83.7ms
20: learn: 0.6089223 total: 22ms remaining: 82.9ms
21: learn: 0.6059369 total: 23ms remaining: 81.7ms
22: learn: 0.6028116 total: 24ms remaining: 80.3ms
23: learn: 0.5990138 total: 25ms remaining: 79.3ms
24: learn: 0.5957435 total: 26ms remaining: 77.9ms
25: learn: 0.5925306 total: 26.8ms remaining: 76.2ms
26: learn: 0.5892411 total: 27.6ms remaining: 74.7ms
27: learn: 0.5857035 total: 28.8ms remaining: 74ms
28: learn: 0.5820093 total: 29.4ms remaining: 72ms
29: learn: 0.5790695 total: 30.1ms remaining: 70.1ms
30: learn: 0.5761151 total: 30.8ms remaining: 68.6ms
31: learn: 0.5730415 total: 31.7ms remaining: 67.3ms
32: learn: 0.5701165 total: 32.5ms remaining: 66ms
33: learn: 0.5666366 total: 33.3ms remaining: 64.7ms
34: learn: 0.5632009 total: 33.9ms remaining: 63ms
35: learn: 0.5600635 total: 34.6ms remaining: 61.5ms
36: learn: 0.5569608 total: 34.8ms remaining: 59.2ms
37: learn: 0.5540607 total: 35.3ms remaining: 57.6ms
38: learn: 0.5510633 total: 35.5ms remaining: 55.5ms
39: learn: 0.5483487 total: 36.4ms remaining: 54.5ms
40: learn: 0.5452854 total: 37.6ms remaining: 54.1ms
41: learn: 0.5423161 total: 38.3ms remaining: 52.9ms
42: learn: 0.5392697 total: 38.9ms remaining: 51.6ms
43: learn: 0.5359676 total: 39.5ms remaining: 50.3ms
44: learn: 0.5334258 total: 40.1ms remaining: 49ms
45: learn: 0.5303586 total: 40.6ms remaining: 47.7ms
46: learn: 0.5272792 total: 41.1ms remaining: 46.3ms
47: learn: 0.5248664 total: 41.5ms remaining: 45ms
48: learn: 0.5221360 total: 42.2ms remaining: 43.9ms
49: learn: 0.5195198 total: 42.4ms remaining: 42.4ms
50: learn: 0.5168951 total: 43ms remaining: 41.3ms
51: learn: 0.5146291 total: 43.5ms remaining: 40.2ms
52: learn: 0.5121901 total: 44.1ms remaining: 39.1ms
53: learn: 0.5098933 total: 44.7ms remaining: 38.1ms
54: learn: 0.5080300 total: 45.3ms remaining: 37.1ms
55: learn: 0.5055177 total: 45.8ms remaining: 36ms
56: learn: 0.5030124 total: 46.5ms remaining: 35.1ms
57: learn: 0.5007695 total: 47.2ms remaining: 34.2ms
58: learn: 0.4984228 total: 47.8ms remaining: 33.2ms
59: learn: 0.4960590 total: 48.5ms remaining: 32.3ms
60: learn: 0.4936214 total: 48.9ms remaining: 31.3ms
61: learn: 0.4915400 total: 49.5ms remaining: 30.4ms
62: learn: 0.4893040 total: 50.1ms remaining: 29.4ms
63: learn: 0.4871798 total: 50.8ms remaining: 28.6ms
64: learn: 0.4858563 total: 51.4ms remaining: 27.7ms
65: learn: 0.4834271 total: 51.9ms remaining: 26.8ms
66: learn: 0.4812225 total: 52.1ms remaining: 25.7ms
67: learn: 0.4788882 total: 52.8ms remaining: 24.8ms
68: learn: 0.4768470 total: 53.4ms remaining: 24ms
69: learn: 0.4746989 total: 54ms remaining: 23.1ms
70: learn: 0.4730787 total: 54.5ms remaining: 22.3ms
71: learn: 0.4708940 total: 54.9ms remaining: 21.4ms
72: learn: 0.4687408 total: 55.5ms remaining: 20.5ms
73: learn: 0.4666934 total: 56.1ms remaining: 19.7ms
74: learn: 0.4645537 total: 56.6ms remaining: 18.9ms
75: learn: 0.4627319 total: 57.1ms remaining: 18ms
76: learn: 0.4609235 total: 57.6ms remaining: 17.2ms
77: learn: 0.4592381 total: 58.1ms remaining: 16.4ms
78: learn: 0.4575002 total: 58.4ms remaining: 15.5ms
79: learn: 0.4557680 total: 59.1ms remaining: 14.8ms
80: learn: 0.4537185 total: 59.7ms remaining: 14ms
81: learn: 0.4516379 total: 60.2ms remaining: 13.2ms
82: learn: 0.4495987 total: 60.8ms remaining: 12.4ms
83: learn: 0.4479635 total: 61.4ms remaining: 11.7ms
84: learn: 0.4461101 total: 61.8ms remaining: 10.9ms
85: learn: 0.4440801 total: 62.4ms remaining: 10.2ms
86: learn: 0.4423571 total: 63ms remaining: 9.42ms
87: learn: 0.4402532 total: 63.4ms remaining: 8.64ms
88: learn: 0.4386896 total: 63.9ms remaining: 7.9ms
89: learn: 0.4368157 total: 64.5ms remaining: 7.17ms
90: learn: 0.4352640 total: 64.9ms remaining: 6.42ms
91: learn: 0.4334000 total: 65.4ms remaining: 5.68ms
92: learn: 0.4316505 total: 65.9ms remaining: 4.96ms
93: learn: 0.4299271 total: 66.4ms remaining: 4.24ms
94: learn: 0.4279354 total: 67ms remaining: 3.53ms
95: learn: 0.4261149 total: 67.5ms remaining: 2.81ms
96: learn: 0.4246395 total: 68.1ms remaining: 2.1ms
97: learn: 0.4229854 total: 68.6ms remaining: 1.4ms
98: learn: 0.4214009 total: 69.2ms remaining: 698us
99: learn: 0.4194362 total: 69.7ms remaining: 0us
0: learn: 0.6878325 total: 646us remaining: 64ms
1: learn: 0.6829833 total: 1.47ms remaining: 71.9ms
2: learn: 0.6783131 total: 2.25ms remaining: 72.9ms
3: learn: 0.6737727 total: 3.2ms remaining: 76.9ms
4: learn: 0.6691293 total: 4.49ms remaining: 85.3ms
5: learn: 0.6650105 total: 5.71ms remaining: 89.5ms
6: learn: 0.6608788 total: 6.96ms remaining: 92.4ms
7: learn: 0.6565204 total: 8.41ms remaining: 96.7ms
8: learn: 0.6524720 total: 9.31ms remaining: 94.2ms
9: learn: 0.6474983 total: 10.3ms remaining: 93ms
10: learn: 0.6435726 total: 11.4ms remaining: 92.4ms
11: learn: 0.6388268 total: 12.2ms remaining: 89.7ms
12: learn: 0.6351269 total: 13.1ms remaining: 87.7ms
13: learn: 0.6311666 total: 14.2ms remaining: 87.1ms
14: learn: 0.6277222 total: 15.2ms remaining: 86ms
15: learn: 0.6240896 total: 15.8ms remaining: 83.1ms
16: learn: 0.6204197 total: 16.7ms remaining: 81.6ms
17: learn: 0.6168908 total: 17.9ms remaining: 81.4ms
18: learn: 0.6124770 total: 18.2ms remaining: 77.7ms
19: learn: 0.6086259 total: 19.1ms remaining: 76.5ms
20: learn: 0.6052431 total: 20.1ms remaining: 75.6ms
21: learn: 0.6019313 total: 21ms remaining: 74.6ms
22: learn: 0.5985107 total: 21.9ms remaining: 73.3ms
23: learn: 0.5946564 total: 22.9ms remaining: 72.5ms
24: learn: 0.5909908 total: 23.9ms remaining: 71.6ms
25: learn: 0.5875362 total: 24.4ms remaining: 69.4ms
26: learn: 0.5837100 total: 25.4ms remaining: 68.6ms
27: learn: 0.5800796 total: 26.2ms remaining: 67.3ms
28: learn: 0.5767052 total: 26.4ms remaining: 64.7ms
29: learn: 0.5738579 total: 27.2ms remaining: 63.5ms
30: learn: 0.5707203 total: 28.2ms remaining: 62.8ms
31: learn: 0.5674669 total: 28.4ms remaining: 60.4ms
32: learn: 0.5643868 total: 29.2ms remaining: 59.2ms
33: learn: 0.5607419 total: 29.7ms remaining: 57.6ms
34: learn: 0.5576943 total: 29.9ms remaining: 55.5ms
35: learn: 0.5545097 total: 30.5ms remaining: 54.2ms
36: learn: 0.5512279 total: 31ms remaining: 52.9ms
37: learn: 0.5483413 total: 31.8ms remaining: 51.8ms
38: learn: 0.5457237 total: 32.5ms remaining: 50.8ms
39: learn: 0.5426478 total: 33.1ms remaining: 49.7ms
40: learn: 0.5400138 total: 33.8ms remaining: 48.7ms
41: learn: 0.5371179 total: 34.4ms remaining: 47.5ms
42: learn: 0.5343056 total: 35.1ms remaining: 46.5ms
43: learn: 0.5319724 total: 35.4ms remaining: 45ms
44: learn: 0.5292683 total: 36ms remaining: 43.9ms
45: learn: 0.5263663 total: 36.5ms remaining: 42.9ms
46: learn: 0.5237196 total: 37ms remaining: 41.7ms
47: learn: 0.5209193 total: 37.8ms remaining: 40.9ms
48: learn: 0.5182532 total: 38.5ms remaining: 40ms
49: learn: 0.5157383 total: 39.1ms remaining: 39.1ms
50: learn: 0.5132768 total: 39.8ms remaining: 38.3ms
51: learn: 0.5106957 total: 40.4ms remaining: 37.3ms
52: learn: 0.5082316 total: 41ms remaining: 36.4ms
53: learn: 0.5055883 total: 41.6ms remaining: 35.4ms
54: learn: 0.5030910 total: 42.1ms remaining: 34.4ms
55: learn: 0.5004086 total: 42.8ms remaining: 33.7ms
56: learn: 0.4976497 total: 43.4ms remaining: 32.8ms
57: learn: 0.4958708 total: 44.2ms remaining: 32ms
58: learn: 0.4932535 total: 44.7ms remaining: 31ms
59: learn: 0.4909799 total: 45.2ms remaining: 30.1ms
60: learn: 0.4889763 total: 45.8ms remaining: 29.3ms
61: learn: 0.4866255 total: 46.4ms remaining: 28.4ms
62: learn: 0.4845023 total: 46.8ms remaining: 27.5ms
63: learn: 0.4823934 total: 47.4ms remaining: 26.7ms
64: learn: 0.4807212 total: 48ms remaining: 25.9ms
65: learn: 0.4782510 total: 48.4ms remaining: 24.9ms
66: learn: 0.4765083 total: 48.8ms remaining: 24.1ms
67: learn: 0.4744120 total: 49.5ms remaining: 23.3ms
68: learn: 0.4719467 total: 49.9ms remaining: 22.4ms
69: learn: 0.4698192 total: 50.4ms remaining: 21.6ms
70: learn: 0.4679931 total: 51ms remaining: 20.9ms
71: learn: 0.4661314 total: 51.6ms remaining: 20.1ms
72: learn: 0.4635561 total: 52ms remaining: 19.3ms
73: learn: 0.4608827 total: 52.7ms remaining: 18.5ms
74: learn: 0.4585496 total: 53.3ms remaining: 17.8ms
75: learn: 0.4565222 total: 53.9ms remaining: 17ms
76: learn: 0.4546256 total: 54.5ms remaining: 16.3ms
77: learn: 0.4528901 total: 55ms remaining: 15.5ms
78: learn: 0.4506486 total: 55.5ms remaining: 14.8ms
79: learn: 0.4488937 total: 55.9ms remaining: 14ms
80: learn: 0.4468200 total: 56.5ms remaining: 13.2ms
81: learn: 0.4451943 total: 57.1ms remaining: 12.5ms
82: learn: 0.4431326 total: 57.6ms remaining: 11.8ms
83: learn: 0.4412071 total: 58.2ms remaining: 11.1ms
84: learn: 0.4390748 total: 58.8ms remaining: 10.4ms
85: learn: 0.4377617 total: 59.4ms remaining: 9.66ms
86: learn: 0.4356655 total: 59.9ms remaining: 8.95ms
87: learn: 0.4338199 total: 60.5ms remaining: 8.25ms
88: learn: 0.4319592 total: 61.1ms remaining: 7.55ms
89: learn: 0.4299895 total: 61.7ms remaining: 6.85ms
90: learn: 0.4284256 total: 62.2ms remaining: 6.15ms
91: learn: 0.4269251 total: 62.7ms remaining: 5.45ms
92: learn: 0.4256075 total: 63.2ms remaining: 4.76ms
93: learn: 0.4237744 total: 63.8ms remaining: 4.07ms
94: learn: 0.4221346 total: 64.3ms remaining: 3.39ms
95: learn: 0.4205565 total: 64.5ms remaining: 2.69ms
96: learn: 0.4188508 total: 65.1ms remaining: 2.01ms
97: learn: 0.4173016 total: 65.4ms remaining: 1.33ms
98: learn: 0.4155900 total: 66ms remaining: 666us
99: learn: 0.4137467 total: 66.5ms remaining: 0us
0: learn: 0.6882585 total: 720us remaining: 71.3ms
1: learn: 0.6839777 total: 1.8ms remaining: 88.5ms
2: learn: 0.6792552 total: 3.25ms remaining: 105ms
3: learn: 0.6748659 total: 4.22ms remaining: 101ms
4: learn: 0.6706315 total: 5.1ms remaining: 97ms
5: learn: 0.6666245 total: 6.16ms remaining: 96.5ms
6: learn: 0.6626422 total: 7.18ms remaining: 95.4ms
7: learn: 0.6586781 total: 8.18ms remaining: 94.1ms
8: learn: 0.6552025 total: 9.14ms remaining: 92.4ms
9: learn: 0.6506840 total: 9.91ms remaining: 89.2ms
10: learn: 0.6460304 total: 10.1ms remaining: 81.8ms
11: learn: 0.6419958 total: 10.8ms remaining: 79.1ms
12: learn: 0.6382144 total: 11.5ms remaining: 76.8ms
13: learn: 0.6336491 total: 12ms remaining: 73.5ms
14: learn: 0.6292397 total: 12.7ms remaining: 72.2ms
15: learn: 0.6255244 total: 13.6ms remaining: 71.2ms
16: learn: 0.6219032 total: 14.5ms remaining: 70.6ms
17: learn: 0.6189971 total: 15ms remaining: 68.5ms
18: learn: 0.6153793 total: 15.8ms remaining: 67.4ms
19: learn: 0.6114960 total: 16.5ms remaining: 65.9ms
20: learn: 0.6075407 total: 17.1ms remaining: 64.3ms
21: learn: 0.6039678 total: 17.9ms remaining: 63.3ms
22: learn: 0.6005275 total: 18.6ms remaining: 62.3ms
23: learn: 0.5969838 total: 19.3ms remaining: 61.2ms
24: learn: 0.5940994 total: 19.9ms remaining: 59.7ms
25: learn: 0.5903183 total: 20.2ms remaining: 57.5ms
26: learn: 0.5865176 total: 20.4ms remaining: 55.2ms
27: learn: 0.5827540 total: 21ms remaining: 53.9ms
28: learn: 0.5802146 total: 21.5ms remaining: 52.6ms
29: learn: 0.5768232 total: 22ms remaining: 51.4ms
30: learn: 0.5730392 total: 22.6ms remaining: 50.2ms
31: learn: 0.5706433 total: 23.2ms remaining: 49.4ms
32: learn: 0.5670191 total: 23.9ms remaining: 48.6ms
33: learn: 0.5640741 total: 24.5ms remaining: 47.6ms
34: learn: 0.5603547 total: 25.3ms remaining: 47ms
35: learn: 0.5572766 total: 26ms remaining: 46.2ms
36: learn: 0.5551564 total: 26.9ms remaining: 45.8ms
37: learn: 0.5523044 total: 27.5ms remaining: 44.8ms
38: learn: 0.5491651 total: 28ms remaining: 43.8ms
39: learn: 0.5457140 total: 28.4ms remaining: 42.6ms
40: learn: 0.5429322 total: 29.2ms remaining: 42.1ms
41: learn: 0.5395395 total: 30ms remaining: 41.4ms
42: learn: 0.5373249 total: 30.7ms remaining: 40.7ms
43: learn: 0.5341033 total: 31.3ms remaining: 39.9ms
44: learn: 0.5311522 total: 32ms remaining: 39.1ms
45: learn: 0.5283215 total: 32.6ms remaining: 38.3ms
46: learn: 0.5254838 total: 33.4ms remaining: 37.7ms
47: learn: 0.5232056 total: 34.3ms remaining: 37.2ms
48: learn: 0.5205159 total: 35.1ms remaining: 36.5ms
49: learn: 0.5177927 total: 35.8ms remaining: 35.8ms
50: learn: 0.5151464 total: 36.1ms remaining: 34.7ms
51: learn: 0.5122949 total: 37ms remaining: 34.1ms
52: learn: 0.5106887 total: 37.8ms remaining: 33.5ms
53: learn: 0.5081829 total: 38.6ms remaining: 32.9ms
54: learn: 0.5053364 total: 39.1ms remaining: 32ms
55: learn: 0.5026561 total: 40ms remaining: 31.4ms
56: learn: 0.5004080 total: 40.9ms remaining: 30.9ms
57: learn: 0.4977372 total: 41.9ms remaining: 30.3ms
58: learn: 0.4958501 total: 42.3ms remaining: 29.4ms
59: learn: 0.4935148 total: 43.1ms remaining: 28.7ms
60: learn: 0.4911186 total: 44ms remaining: 28.1ms
61: learn: 0.4887119 total: 44.7ms remaining: 27.4ms
62: learn: 0.4860313 total: 45.4ms remaining: 26.7ms
63: learn: 0.4837995 total: 46.1ms remaining: 25.9ms
64: learn: 0.4817179 total: 47ms remaining: 25.3ms
65: learn: 0.4794803 total: 47.9ms remaining: 24.7ms
66: learn: 0.4770329 total: 48.6ms remaining: 24ms
67: learn: 0.4746189 total: 49.3ms remaining: 23.2ms
68: learn: 0.4721843 total: 50ms remaining: 22.5ms
69: learn: 0.4705194 total: 50.8ms remaining: 21.8ms
70: learn: 0.4681769 total: 51.5ms remaining: 21ms
71: learn: 0.4665577 total: 52.3ms remaining: 20.3ms
72: learn: 0.4644336 total: 53ms remaining: 19.6ms
73: learn: 0.4623078 total: 53.7ms remaining: 18.9ms
74: learn: 0.4597296 total: 54.3ms remaining: 18.1ms
75: learn: 0.4576521 total: 55ms remaining: 17.4ms
76: learn: 0.4558040 total: 55.7ms remaining: 16.7ms
77: learn: 0.4541016 total: 56.4ms remaining: 15.9ms
78: learn: 0.4521654 total: 57.3ms remaining: 15.2ms
79: learn: 0.4498925 total: 58.1ms remaining: 14.5ms
80: learn: 0.4476604 total: 58.9ms remaining: 13.8ms
81: learn: 0.4461599 total: 59.4ms remaining: 13ms
82: learn: 0.4441266 total: 60.1ms remaining: 12.3ms
83: learn: 0.4422789 total: 61.1ms remaining: 11.6ms
84: learn: 0.4402099 total: 61.7ms remaining: 10.9ms
85: learn: 0.4388677 total: 62.7ms remaining: 10.2ms
86: learn: 0.4372308 total: 63.3ms remaining: 9.46ms
87: learn: 0.4353302 total: 64.3ms remaining: 8.76ms
88: learn: 0.4334144 total: 65.1ms remaining: 8.05ms
89: learn: 0.4313791 total: 66ms remaining: 7.33ms
90: learn: 0.4295541 total: 66.7ms remaining: 6.6ms
91: learn: 0.4275814 total: 67.7ms remaining: 5.88ms
92: learn: 0.4258450 total: 68.3ms remaining: 5.14ms
93: learn: 0.4245697 total: 69ms remaining: 4.41ms
94: learn: 0.4229895 total: 69.7ms remaining: 3.67ms
95: learn: 0.4211116 total: 70.5ms remaining: 2.94ms
96: learn: 0.4194295 total: 71.1ms remaining: 2.2ms
97: learn: 0.4179723 total: 71.7ms remaining: 1.46ms
98: learn: 0.4161217 total: 72.4ms remaining: 731us
99: learn: 0.4146871 total: 73.1ms remaining: 0us
0: learn: 0.6881440 total: 745us remaining: 73.8ms
1: learn: 0.6838226 total: 1.46ms remaining: 71.5ms
2: learn: 0.6798522 total: 2.29ms remaining: 74.2ms
3: learn: 0.6753612 total: 2.92ms remaining: 70.1ms
4: learn: 0.6718949 total: 3.67ms remaining: 69.7ms
5: learn: 0.6680771 total: 4.34ms remaining: 68.1ms
6: learn: 0.6634730 total: 4.93ms remaining: 65.6ms
7: learn: 0.6597522 total: 5.75ms remaining: 66.1ms
8: learn: 0.6563034 total: 6.44ms remaining: 65.1ms
9: learn: 0.6521967 total: 6.67ms remaining: 60.1ms
10: learn: 0.6481657 total: 7.4ms remaining: 59.8ms
11: learn: 0.6448143 total: 8.17ms remaining: 59.9ms
12: learn: 0.6420479 total: 8.59ms remaining: 57.5ms
13: learn: 0.6381273 total: 9.26ms remaining: 56.9ms
14: learn: 0.6348032 total: 9.96ms remaining: 56.4ms
15: learn: 0.6309156 total: 10.7ms remaining: 56.3ms
16: learn: 0.6273113 total: 11.5ms remaining: 56.1ms
17: learn: 0.6236722 total: 12.1ms remaining: 55.3ms
18: learn: 0.6193329 total: 12.8ms remaining: 54.6ms
19: learn: 0.6151982 total: 13.5ms remaining: 54ms
20: learn: 0.6114231 total: 14.2ms remaining: 53.3ms
21: learn: 0.6080377 total: 14.8ms remaining: 52.4ms
22: learn: 0.6050294 total: 15.4ms remaining: 51.7ms
23: learn: 0.6013056 total: 16.1ms remaining: 51ms
24: learn: 0.5975738 total: 16.7ms remaining: 50.1ms
25: learn: 0.5942470 total: 17.3ms remaining: 49.3ms
26: learn: 0.5909084 total: 18ms remaining: 48.8ms
27: learn: 0.5876613 total: 18.8ms remaining: 48.3ms
28: learn: 0.5846476 total: 19.6ms remaining: 48ms
29: learn: 0.5813644 total: 20.3ms remaining: 47.3ms
30: learn: 0.5778006 total: 20.9ms remaining: 46.4ms
31: learn: 0.5740806 total: 21.5ms remaining: 45.7ms
32: learn: 0.5706020 total: 22ms remaining: 44.7ms
33: learn: 0.5675473 total: 22.6ms remaining: 43.9ms
34: learn: 0.5644498 total: 23.3ms remaining: 43.3ms
35: learn: 0.5610447 total: 23.9ms remaining: 42.5ms
36: learn: 0.5578353 total: 24.5ms remaining: 41.8ms
37: learn: 0.5551180 total: 25.1ms remaining: 41ms
38: learn: 0.5521754 total: 25.7ms remaining: 40.2ms
39: learn: 0.5491918 total: 26.3ms remaining: 39.5ms
40: learn: 0.5464784 total: 27ms remaining: 38.8ms
41: learn: 0.5434380 total: 27.7ms remaining: 38.2ms
42: learn: 0.5409694 total: 28.4ms remaining: 37.6ms
43: learn: 0.5386926 total: 29ms remaining: 36.9ms
44: learn: 0.5358634 total: 29.6ms remaining: 36.1ms
45: learn: 0.5334981 total: 29.8ms remaining: 34.9ms
46: learn: 0.5305617 total: 30.4ms remaining: 34.2ms
47: learn: 0.5279681 total: 31ms remaining: 33.5ms
48: learn: 0.5252795 total: 31.6ms remaining: 32.9ms
49: learn: 0.5226056 total: 32.2ms remaining: 32.2ms
50: learn: 0.5207011 total: 32.6ms remaining: 31.3ms
51: learn: 0.5180738 total: 33.3ms remaining: 30.7ms
52: learn: 0.5155331 total: 33.9ms remaining: 30ms
53: learn: 0.5129543 total: 34.3ms remaining: 29.2ms
54: learn: 0.5107313 total: 35ms remaining: 28.6ms
55: learn: 0.5080379 total: 35.5ms remaining: 27.9ms
56: learn: 0.5052252 total: 36.1ms remaining: 27.2ms
57: learn: 0.5024864 total: 36.7ms remaining: 26.6ms
58: learn: 0.5003343 total: 37.4ms remaining: 26ms
59: learn: 0.4978079 total: 38ms remaining: 25.3ms
60: learn: 0.4958259 total: 38.3ms remaining: 24.5ms
61: learn: 0.4936826 total: 38.8ms remaining: 23.8ms
62: learn: 0.4918267 total: 39.2ms remaining: 23ms
63: learn: 0.4900561 total: 39.4ms remaining: 22.2ms
64: learn: 0.4878887 total: 40.1ms remaining: 21.6ms
65: learn: 0.4859410 total: 40.8ms remaining: 21ms
66: learn: 0.4837216 total: 41.4ms remaining: 20.4ms
67: learn: 0.4817776 total: 42ms remaining: 19.8ms
68: learn: 0.4794759 total: 42.3ms remaining: 19ms
69: learn: 0.4773828 total: 42.9ms remaining: 18.4ms
70: learn: 0.4755905 total: 43.3ms remaining: 17.7ms
71: learn: 0.4738410 total: 43.8ms remaining: 17ms
72: learn: 0.4721256 total: 44.4ms remaining: 16.4ms
73: learn: 0.4702684 total: 45ms remaining: 15.8ms
74: learn: 0.4681690 total: 45.7ms remaining: 15.2ms
75: learn: 0.4657247 total: 46.4ms remaining: 14.6ms
76: learn: 0.4637225 total: 47ms remaining: 14ms
77: learn: 0.4621415 total: 47.6ms remaining: 13.4ms
78: learn: 0.4608689 total: 48.1ms remaining: 12.8ms
79: learn: 0.4594696 total: 48.4ms remaining: 12.1ms
80: learn: 0.4576645 total: 48.9ms remaining: 11.5ms
81: learn: 0.4557380 total: 49.5ms remaining: 10.9ms
82: learn: 0.4538984 total: 50.1ms remaining: 10.3ms
83: learn: 0.4519214 total: 50.8ms remaining: 9.67ms
84: learn: 0.4498923 total: 51.2ms remaining: 9.03ms
85: learn: 0.4480507 total: 51.8ms remaining: 8.42ms
86: learn: 0.4468536 total: 52.3ms remaining: 7.81ms
87: learn: 0.4454365 total: 52.9ms remaining: 7.21ms
88: learn: 0.4435342 total: 53.4ms remaining: 6.61ms
89: learn: 0.4420734 total: 54ms remaining: 6ms
90: learn: 0.4402998 total: 54.5ms remaining: 5.39ms
91: learn: 0.4386186 total: 55.1ms remaining: 4.79ms
92: learn: 0.4367547 total: 55.9ms remaining: 4.21ms
93: learn: 0.4354804 total: 56.4ms remaining: 3.6ms
94: learn: 0.4337295 total: 56.9ms remaining: 3ms
95: learn: 0.4323046 total: 57.1ms remaining: 2.38ms
96: learn: 0.4305899 total: 57.7ms remaining: 1.78ms
97: learn: 0.4291240 total: 58.2ms remaining: 1.19ms
98: learn: 0.4272748 total: 58.6ms remaining: 591us
99: learn: 0.4256044 total: 59.1ms remaining: 0us
0: learn: 0.6880598 total: 753us remaining: 74.6ms
1: learn: 0.6833246 total: 1.63ms remaining: 79.7ms
2: learn: 0.6789769 total: 2.6ms remaining: 83.9ms
3: learn: 0.6739423 total: 3.31ms remaining: 79.4ms
4: learn: 0.6690233 total: 3.94ms remaining: 74.8ms
5: learn: 0.6655579 total: 4.76ms remaining: 74.7ms
6: learn: 0.6615555 total: 5.56ms remaining: 73.8ms
7: learn: 0.6581583 total: 6.21ms remaining: 71.5ms
8: learn: 0.6542419 total: 6.92ms remaining: 69.9ms
9: learn: 0.6497969 total: 7.5ms remaining: 67.5ms
10: learn: 0.6456478 total: 7.68ms remaining: 62.2ms
11: learn: 0.6415730 total: 7.99ms remaining: 58.6ms
12: learn: 0.6372634 total: 8.66ms remaining: 58ms
13: learn: 0.6326632 total: 9.14ms remaining: 56.1ms
14: learn: 0.6293084 total: 9.72ms remaining: 55.1ms
15: learn: 0.6257282 total: 10.3ms remaining: 54.1ms
16: learn: 0.6216776 total: 10.9ms remaining: 53.3ms
17: learn: 0.6188229 total: 11.3ms remaining: 51.7ms
18: learn: 0.6149747 total: 12.1ms remaining: 51.5ms
19: learn: 0.6112811 total: 12.2ms remaining: 48.9ms
20: learn: 0.6074915 total: 12.8ms remaining: 48.1ms
21: learn: 0.6044135 total: 13.4ms remaining: 47.5ms
22: learn: 0.6009953 total: 14ms remaining: 46.7ms
23: learn: 0.5974793 total: 14.6ms remaining: 46.1ms
24: learn: 0.5937905 total: 15.3ms remaining: 45.8ms
25: learn: 0.5902730 total: 16.2ms remaining: 46ms
26: learn: 0.5875519 total: 17ms remaining: 45.8ms
27: learn: 0.5841550 total: 17.7ms remaining: 45.6ms
28: learn: 0.5808950 total: 18.5ms remaining: 45.3ms
29: learn: 0.5772600 total: 19.4ms remaining: 45.2ms
30: learn: 0.5745535 total: 20ms remaining: 44.5ms
31: learn: 0.5712423 total: 20.4ms remaining: 43.3ms
32: learn: 0.5676332 total: 21ms remaining: 42.6ms
33: learn: 0.5646369 total: 21.5ms remaining: 41.7ms
34: learn: 0.5617155 total: 22.1ms remaining: 41ms
35: learn: 0.5585733 total: 22.6ms remaining: 40.2ms
36: learn: 0.5559330 total: 22.8ms remaining: 38.8ms
37: learn: 0.5529272 total: 23.5ms remaining: 38.4ms
38: learn: 0.5503624 total: 23.7ms remaining: 37.1ms
39: learn: 0.5471022 total: 24.3ms remaining: 36.5ms
40: learn: 0.5445004 total: 25.1ms remaining: 36.1ms
41: learn: 0.5418501 total: 25.7ms remaining: 35.5ms
42: learn: 0.5393487 total: 26.6ms remaining: 35.2ms
43: learn: 0.5365438 total: 27.4ms remaining: 34.9ms
44: learn: 0.5334766 total: 28.2ms remaining: 34.4ms
45: learn: 0.5308641 total: 28.7ms remaining: 33.7ms
46: learn: 0.5284121 total: 29.6ms remaining: 33.3ms
47: learn: 0.5257992 total: 29.9ms remaining: 32.4ms
48: learn: 0.5233899 total: 30.6ms remaining: 31.8ms
49: learn: 0.5203597 total: 31.2ms remaining: 31.2ms
50: learn: 0.5179876 total: 31.5ms remaining: 30.3ms
51: learn: 0.5151740 total: 32ms remaining: 29.5ms
52: learn: 0.5130448 total: 32.8ms remaining: 29.1ms
53: learn: 0.5110259 total: 33.4ms remaining: 28.5ms
54: learn: 0.5090380 total: 33.9ms remaining: 27.8ms
55: learn: 0.5071256 total: 34.7ms remaining: 27.2ms
56: learn: 0.5049034 total: 35ms remaining: 26.4ms
57: learn: 0.5022447 total: 35.6ms remaining: 25.8ms
58: learn: 0.5001021 total: 36.3ms remaining: 25.2ms
59: learn: 0.4979569 total: 37.3ms remaining: 24.9ms
60: learn: 0.4958132 total: 38ms remaining: 24.3ms
61: learn: 0.4932032 total: 38.7ms remaining: 23.7ms
62: learn: 0.4908546 total: 39.5ms remaining: 23.2ms
63: learn: 0.4887150 total: 40ms remaining: 22.5ms
64: learn: 0.4868649 total: 40.7ms remaining: 21.9ms
65: learn: 0.4844882 total: 41.4ms remaining: 21.3ms
66: learn: 0.4823326 total: 42.2ms remaining: 20.8ms
67: learn: 0.4797374 total: 42.7ms remaining: 20.1ms
68: learn: 0.4772965 total: 43.3ms remaining: 19.5ms
69: learn: 0.4751097 total: 43.8ms remaining: 18.8ms
70: learn: 0.4729762 total: 44.4ms remaining: 18.1ms
71: learn: 0.4715059 total: 45ms remaining: 17.5ms
72: learn: 0.4699832 total: 45.5ms remaining: 16.8ms
73: learn: 0.4677990 total: 46.2ms remaining: 16.2ms
74: learn: 0.4661791 total: 46.9ms remaining: 15.6ms
75: learn: 0.4648058 total: 47.8ms remaining: 15.1ms
76: learn: 0.4632243 total: 48.6ms remaining: 14.5ms
77: learn: 0.4615577 total: 49.2ms remaining: 13.9ms
78: learn: 0.4598789 total: 49.6ms remaining: 13.2ms
79: learn: 0.4579248 total: 50.2ms remaining: 12.5ms
80: learn: 0.4558359 total: 50.8ms remaining: 11.9ms
81: learn: 0.4542296 total: 51.1ms remaining: 11.2ms
82: learn: 0.4525182 total: 51.9ms remaining: 10.6ms
83: learn: 0.4506565 total: 52.3ms remaining: 9.95ms
84: learn: 0.4490059 total: 52.6ms remaining: 9.28ms
85: learn: 0.4474648 total: 53.1ms remaining: 8.64ms
86: learn: 0.4458212 total: 53.8ms remaining: 8.04ms
87: learn: 0.4442739 total: 54.5ms remaining: 7.43ms
88: learn: 0.4422842 total: 55.1ms remaining: 6.81ms
89: learn: 0.4407117 total: 56.1ms remaining: 6.23ms
90: learn: 0.4396359 total: 57ms remaining: 5.63ms
91: learn: 0.4382579 total: 57.7ms remaining: 5.01ms
92: learn: 0.4366664 total: 58.3ms remaining: 4.39ms
93: learn: 0.4346557 total: 58.9ms remaining: 3.76ms
94: learn: 0.4327488 total: 59.5ms remaining: 3.13ms
95: learn: 0.4311377 total: 60.2ms remaining: 2.51ms
96: learn: 0.4290571 total: 60.8ms remaining: 1.88ms
97: learn: 0.4274063 total: 61.5ms remaining: 1.25ms
98: learn: 0.4261279 total: 62.2ms remaining: 627us
99: learn: 0.4243579 total: 62.7ms remaining: 0us
0: learn: 0.6882715 total: 622us remaining: 61.6ms
1: learn: 0.6837467 total: 1.23ms remaining: 60.2ms
2: learn: 0.6783988 total: 1.91ms remaining: 61.8ms
3: learn: 0.6736412 total: 2.53ms remaining: 60.7ms
4: learn: 0.6688160 total: 3.2ms remaining: 60.9ms
5: learn: 0.6636849 total: 3.71ms remaining: 58.2ms
6: learn: 0.6595088 total: 4.3ms remaining: 57.2ms
7: learn: 0.6554600 total: 4.94ms remaining: 56.8ms
8: learn: 0.6513102 total: 5.42ms remaining: 54.8ms
9: learn: 0.6474276 total: 5.92ms remaining: 53.2ms
10: learn: 0.6433167 total: 6.45ms remaining: 52.2ms
11: learn: 0.6393555 total: 7.15ms remaining: 52.5ms
12: learn: 0.6353104 total: 7.73ms remaining: 51.7ms
13: learn: 0.6323059 total: 8.34ms remaining: 51.2ms
14: learn: 0.6292336 total: 8.84ms remaining: 50.1ms
15: learn: 0.6254054 total: 9.35ms remaining: 49.1ms
16: learn: 0.6217592 total: 9.99ms remaining: 48.8ms
17: learn: 0.6182798 total: 10.2ms remaining: 46.4ms
18: learn: 0.6145356 total: 10.7ms remaining: 45.7ms
19: learn: 0.6111740 total: 11.3ms remaining: 45.1ms
20: learn: 0.6075515 total: 11.8ms remaining: 44.5ms
21: learn: 0.6036147 total: 12.4ms remaining: 44.1ms
22: learn: 0.5997521 total: 13ms remaining: 43.5ms
23: learn: 0.5954807 total: 13.5ms remaining: 42.8ms
24: learn: 0.5914498 total: 14.1ms remaining: 42.2ms
25: learn: 0.5877992 total: 14.6ms remaining: 41.5ms
26: learn: 0.5840369 total: 15.2ms remaining: 41.1ms
27: learn: 0.5807457 total: 15.8ms remaining: 40.7ms
28: learn: 0.5773241 total: 16.3ms remaining: 39.8ms
29: learn: 0.5752621 total: 16.8ms remaining: 39.1ms
30: learn: 0.5711845 total: 17.4ms remaining: 38.7ms
31: learn: 0.5677277 total: 17.7ms remaining: 37.6ms
32: learn: 0.5642957 total: 18.3ms remaining: 37.2ms
33: learn: 0.5610649 total: 18.8ms remaining: 36.6ms
34: learn: 0.5579598 total: 19.3ms remaining: 35.9ms
35: learn: 0.5543794 total: 19.9ms remaining: 35.3ms
36: learn: 0.5516154 total: 20.5ms remaining: 34.9ms
37: learn: 0.5487977 total: 20.7ms remaining: 33.8ms
38: learn: 0.5459438 total: 21.4ms remaining: 33.4ms
39: learn: 0.5435490 total: 22ms remaining: 33ms
40: learn: 0.5410065 total: 22.5ms remaining: 32.3ms
41: learn: 0.5381863 total: 23.1ms remaining: 31.8ms
42: learn: 0.5346479 total: 23.6ms remaining: 31.3ms
43: learn: 0.5323395 total: 24.4ms remaining: 31ms
44: learn: 0.5297288 total: 25.2ms remaining: 30.8ms
45: learn: 0.5279816 total: 25.8ms remaining: 30.3ms
46: learn: 0.5253830 total: 26.6ms remaining: 30ms
47: learn: 0.5228773 total: 26.9ms remaining: 29.2ms
48: learn: 0.5200328 total: 27.8ms remaining: 29ms
49: learn: 0.5170826 total: 28.4ms remaining: 28.4ms
50: learn: 0.5142298 total: 29.3ms remaining: 28.1ms
51: learn: 0.5117048 total: 30.2ms remaining: 27.9ms
52: learn: 0.5089911 total: 30.9ms remaining: 27.4ms
53: learn: 0.5066490 total: 31.2ms remaining: 26.6ms
54: learn: 0.5040246 total: 32ms remaining: 26.2ms
55: learn: 0.5015074 total: 32.8ms remaining: 25.8ms
56: learn: 0.4983565 total: 33.4ms remaining: 25.2ms
57: learn: 0.4959523 total: 33.6ms remaining: 24.3ms
58: learn: 0.4936506 total: 34.5ms remaining: 24ms
59: learn: 0.4915824 total: 35.4ms remaining: 23.6ms
60: learn: 0.4894619 total: 36.1ms remaining: 23.1ms
61: learn: 0.4870344 total: 37ms remaining: 22.7ms
62: learn: 0.4842965 total: 37.8ms remaining: 22.2ms
63: learn: 0.4817147 total: 38.3ms remaining: 21.5ms
64: learn: 0.4794687 total: 39ms remaining: 21ms
65: learn: 0.4777544 total: 39.5ms remaining: 20.4ms
66: learn: 0.4751102 total: 40.1ms remaining: 19.8ms
67: learn: 0.4732955 total: 40.8ms remaining: 19.2ms
68: learn: 0.4712845 total: 41.5ms remaining: 18.6ms
69: learn: 0.4694769 total: 42.1ms remaining: 18.1ms
70: learn: 0.4680735 total: 42.4ms remaining: 17.3ms
71: learn: 0.4656390 total: 42.9ms remaining: 16.7ms
72: learn: 0.4636848 total: 43.6ms remaining: 16.1ms
73: learn: 0.4617332 total: 44.1ms remaining: 15.5ms
74: learn: 0.4591228 total: 44.8ms remaining: 14.9ms
75: learn: 0.4570065 total: 45.4ms remaining: 14.3ms
76: learn: 0.4554797 total: 45.9ms remaining: 13.7ms
77: learn: 0.4537128 total: 46.7ms remaining: 13.2ms
78: learn: 0.4516644 total: 47.3ms remaining: 12.6ms
79: learn: 0.4494415 total: 48ms remaining: 12ms
80: learn: 0.4472458 total: 48.9ms remaining: 11.5ms
81: learn: 0.4451586 total: 50.1ms remaining: 11ms
82: learn: 0.4432670 total: 50.9ms remaining: 10.4ms
83: learn: 0.4412769 total: 51.6ms remaining: 9.83ms
84: learn: 0.4395748 total: 52.3ms remaining: 9.23ms
85: learn: 0.4377752 total: 52.5ms remaining: 8.55ms
86: learn: 0.4377575 total: 52.7ms remaining: 7.87ms
87: learn: 0.4359400 total: 53.4ms remaining: 7.28ms
88: learn: 0.4339381 total: 54.1ms remaining: 6.69ms
89: learn: 0.4318944 total: 54.8ms remaining: 6.08ms
90: learn: 0.4303627 total: 55.5ms remaining: 5.49ms
91: learn: 0.4287533 total: 56ms remaining: 4.87ms
92: learn: 0.4267085 total: 56.8ms remaining: 4.28ms
93: learn: 0.4250170 total: 57.4ms remaining: 3.67ms
94: learn: 0.4232858 total: 58.2ms remaining: 3.06ms
95: learn: 0.4213435 total: 58.7ms remaining: 2.44ms
96: learn: 0.4196453 total: 59.5ms remaining: 1.84ms
97: learn: 0.4180252 total: 60.3ms remaining: 1.23ms
98: learn: 0.4161187 total: 61.1ms remaining: 616us
99: learn: 0.4144881 total: 61.8ms remaining: 0us
0: learn: 0.6895544 total: 727us remaining: 72.1ms
1: learn: 0.6849260 total: 1.35ms remaining: 66.4ms
2: learn: 0.6810258 total: 1.91ms remaining: 61.9ms
3: learn: 0.6765089 total: 2.35ms remaining: 56.3ms
4: learn: 0.6731004 total: 2.92ms remaining: 55.4ms
5: learn: 0.6686184 total: 3.62ms remaining: 56.7ms
6: learn: 0.6644403 total: 4.29ms remaining: 57ms
7: learn: 0.6598710 total: 4.99ms remaining: 57.4ms
8: learn: 0.6550665 total: 5.64ms remaining: 57ms
9: learn: 0.6515600 total: 6.15ms remaining: 55.4ms
10: learn: 0.6484847 total: 6.82ms remaining: 55.2ms
11: learn: 0.6441061 total: 7.48ms remaining: 54.9ms
12: learn: 0.6395647 total: 8.09ms remaining: 54.1ms
13: learn: 0.6362526 total: 8.77ms remaining: 53.8ms
14: learn: 0.6329626 total: 9.39ms remaining: 53.2ms
15: learn: 0.6290173 total: 9.96ms remaining: 52.3ms
16: learn: 0.6262281 total: 10.4ms remaining: 50.9ms
17: learn: 0.6227024 total: 11ms remaining: 50.2ms
18: learn: 0.6188889 total: 11.2ms remaining: 47.6ms
19: learn: 0.6152467 total: 11.7ms remaining: 47ms
20: learn: 0.6111927 total: 12.4ms remaining: 46.8ms
21: learn: 0.6078590 total: 13.2ms remaining: 46.6ms
22: learn: 0.6043231 total: 14.1ms remaining: 47.1ms
23: learn: 0.6004283 total: 14.9ms remaining: 47ms
24: learn: 0.5967828 total: 15.4ms remaining: 46.3ms
25: learn: 0.5934005 total: 16.2ms remaining: 46ms
26: learn: 0.5897854 total: 16.8ms remaining: 45.5ms
27: learn: 0.5867783 total: 17.4ms remaining: 44.9ms
28: learn: 0.5835423 total: 18.2ms remaining: 44.5ms
29: learn: 0.5799524 total: 18.8ms remaining: 43.9ms
30: learn: 0.5767129 total: 19.6ms remaining: 43.5ms
31: learn: 0.5734765 total: 20.2ms remaining: 42.9ms
32: learn: 0.5697662 total: 20.6ms remaining: 41.9ms
33: learn: 0.5660582 total: 21.2ms remaining: 41.2ms
34: learn: 0.5634641 total: 21.7ms remaining: 40.3ms
35: learn: 0.5603953 total: 21.9ms remaining: 38.9ms
36: learn: 0.5577197 total: 22.6ms remaining: 38.4ms
37: learn: 0.5547286 total: 22.8ms remaining: 37.2ms
38: learn: 0.5513552 total: 23.4ms remaining: 36.6ms
39: learn: 0.5487145 total: 24.1ms remaining: 36.2ms
40: learn: 0.5458332 total: 24.5ms remaining: 35.3ms
41: learn: 0.5430226 total: 24.8ms remaining: 34.3ms
42: learn: 0.5405773 total: 25.5ms remaining: 33.7ms
43: learn: 0.5376341 total: 26ms remaining: 33.1ms
44: learn: 0.5347205 total: 26.3ms remaining: 32.2ms
45: learn: 0.5318596 total: 26.8ms remaining: 31.5ms
46: learn: 0.5296991 total: 27.4ms remaining: 30.9ms
47: learn: 0.5265906 total: 28ms remaining: 30.3ms
48: learn: 0.5243975 total: 28.6ms remaining: 29.8ms
49: learn: 0.5220615 total: 29.2ms remaining: 29.2ms
50: learn: 0.5197911 total: 29.6ms remaining: 28.5ms
51: learn: 0.5174800 total: 30.7ms remaining: 28.3ms
52: learn: 0.5151445 total: 34.9ms remaining: 30.9ms
53: learn: 0.5128888 total: 36.3ms remaining: 31ms
54: learn: 0.5105547 total: 46.1ms remaining: 37.7ms
55: learn: 0.5080203 total: 47.7ms remaining: 37.4ms
56: learn: 0.5050813 total: 48.2ms remaining: 36.4ms
57: learn: 0.5030250 total: 49ms remaining: 35.5ms
58: learn: 0.5007144 total: 50.2ms remaining: 34.9ms
59: learn: 0.4978772 total: 51ms remaining: 34ms
60: learn: 0.4960648 total: 52.4ms remaining: 33.5ms
61: learn: 0.4938557 total: 53.1ms remaining: 32.5ms
62: learn: 0.4912799 total: 53.9ms remaining: 31.7ms
63: learn: 0.4893021 total: 55.3ms remaining: 31.1ms
64: learn: 0.4865816 total: 56.7ms remaining: 30.5ms
65: learn: 0.4843468 total: 57.6ms remaining: 29.7ms
66: learn: 0.4828484 total: 59.4ms remaining: 29.3ms
67: learn: 0.4805846 total: 60.8ms remaining: 28.6ms
68: learn: 0.4784127 total: 61.3ms remaining: 27.5ms
69: learn: 0.4763356 total: 62ms remaining: 26.6ms
70: learn: 0.4742603 total: 63.9ms remaining: 26.1ms
71: learn: 0.4721389 total: 65.4ms remaining: 25.4ms
72: learn: 0.4698023 total: 66.8ms remaining: 24.7ms
73: learn: 0.4679967 total: 67.4ms remaining: 23.7ms
74: learn: 0.4662974 total: 68.7ms remaining: 22.9ms
75: learn: 0.4644232 total: 69.8ms remaining: 22ms
76: learn: 0.4624581 total: 70.5ms remaining: 21.1ms
77: learn: 0.4607757 total: 72.2ms remaining: 20.4ms
78: learn: 0.4585083 total: 73.6ms remaining: 19.6ms
79: learn: 0.4563258 total: 74.7ms remaining: 18.7ms
80: learn: 0.4546436 total: 75.2ms remaining: 17.6ms
81: learn: 0.4530862 total: 76.5ms remaining: 16.8ms
82: learn: 0.4511524 total: 78ms remaining: 16ms
83: learn: 0.4494753 total: 79.3ms remaining: 15.1ms
84: learn: 0.4477776 total: 80.9ms remaining: 14.3ms
85: learn: 0.4456535 total: 81.8ms remaining: 13.3ms
86: learn: 0.4438481 total: 84.2ms remaining: 12.6ms
87: learn: 0.4425067 total: 85.2ms remaining: 11.6ms
88: learn: 0.4404480 total: 86ms remaining: 10.6ms
89: learn: 0.4389404 total: 86.9ms remaining: 9.65ms
90: learn: 0.4374187 total: 88ms remaining: 8.71ms
91: learn: 0.4356672 total: 89.7ms remaining: 7.8ms
92: learn: 0.4343536 total: 91ms remaining: 6.85ms
93: learn: 0.4329625 total: 92.3ms remaining: 5.89ms
94: learn: 0.4312231 total: 93.1ms remaining: 4.9ms
95: learn: 0.4292682 total: 93.9ms remaining: 3.91ms
96: learn: 0.4278009 total: 94.5ms remaining: 2.92ms
97: learn: 0.4256812 total: 94.9ms remaining: 1.94ms
98: learn: 0.4245583 total: 95.8ms remaining: 967us
99: learn: 0.4231246 total: 96.9ms remaining: 0us
0: learn: 0.6899173 total: 1.36ms remaining: 135ms
1: learn: 0.6859439 total: 2.57ms remaining: 126ms
2: learn: 0.6820029 total: 4.32ms remaining: 140ms
3: learn: 0.6775952 total: 5.51ms remaining: 132ms
4: learn: 0.6734936 total: 6.59ms remaining: 125ms
5: learn: 0.6694175 total: 7.89ms remaining: 124ms
6: learn: 0.6650204 total: 8.94ms remaining: 119ms
7: learn: 0.6604569 total: 10.2ms remaining: 117ms
8: learn: 0.6566835 total: 11ms remaining: 112ms
9: learn: 0.6522852 total: 12ms remaining: 108ms
10: learn: 0.6482039 total: 13ms remaining: 105ms
11: learn: 0.6452449 total: 13.8ms remaining: 102ms
12: learn: 0.6413751 total: 14.9ms remaining: 99.4ms
13: learn: 0.6373760 total: 15.6ms remaining: 96.1ms
14: learn: 0.6325962 total: 16.4ms remaining: 93ms
15: learn: 0.6287772 total: 17.3ms remaining: 90.8ms
16: learn: 0.6249406 total: 18ms remaining: 87.8ms
17: learn: 0.6213689 total: 19.2ms remaining: 87.3ms
18: learn: 0.6178202 total: 19.9ms remaining: 84.9ms
19: learn: 0.6138921 total: 20.9ms remaining: 83.7ms
20: learn: 0.6102038 total: 21.9ms remaining: 82.2ms
21: learn: 0.6067466 total: 22.8ms remaining: 80.7ms
22: learn: 0.6034256 total: 23.8ms remaining: 79.6ms
23: learn: 0.5995496 total: 24.6ms remaining: 78ms
24: learn: 0.5958312 total: 25.5ms remaining: 76.6ms
25: learn: 0.5922630 total: 26.6ms remaining: 75.8ms
26: learn: 0.5891180 total: 27.6ms remaining: 74.7ms
27: learn: 0.5855249 total: 28.3ms remaining: 72.7ms
28: learn: 0.5822194 total: 29.6ms remaining: 72.4ms
29: learn: 0.5783025 total: 30.7ms remaining: 71.6ms
30: learn: 0.5749866 total: 31.1ms remaining: 69.3ms
31: learn: 0.5717586 total: 32.2ms remaining: 68.5ms
32: learn: 0.5679180 total: 33.2ms remaining: 67.4ms
33: learn: 0.5647130 total: 34.3ms remaining: 66.6ms
34: learn: 0.5615301 total: 35.1ms remaining: 65.2ms
35: learn: 0.5582356 total: 35.7ms remaining: 63.5ms
36: learn: 0.5552371 total: 36.1ms remaining: 61.5ms
37: learn: 0.5522339 total: 37ms remaining: 60.4ms
38: learn: 0.5490700 total: 38.5ms remaining: 60.2ms
39: learn: 0.5460870 total: 39.6ms remaining: 59.5ms
40: learn: 0.5430329 total: 40.8ms remaining: 58.6ms
41: learn: 0.5394535 total: 41.6ms remaining: 57.4ms
42: learn: 0.5364412 total: 42.4ms remaining: 56.2ms
43: learn: 0.5343403 total: 43.1ms remaining: 54.9ms
44: learn: 0.5311697 total: 44.1ms remaining: 53.9ms
45: learn: 0.5285576 total: 45ms remaining: 52.8ms
46: learn: 0.5254773 total: 45.5ms remaining: 51.3ms
47: learn: 0.5228290 total: 46.6ms remaining: 50.5ms
48: learn: 0.5197728 total: 47.4ms remaining: 49.3ms
49: learn: 0.5170935 total: 48.6ms remaining: 48.6ms
50: learn: 0.5143981 total: 49.6ms remaining: 47.6ms
51: learn: 0.5119282 total: 50.5ms remaining: 46.6ms
52: learn: 0.5089990 total: 51.8ms remaining: 45.9ms
53: learn: 0.5063444 total: 52.5ms remaining: 44.7ms
54: learn: 0.5036184 total: 53.5ms remaining: 43.8ms
55: learn: 0.5008157 total: 54.3ms remaining: 42.7ms
56: learn: 0.4987444 total: 55.2ms remaining: 41.6ms
57: learn: 0.4965341 total: 56ms remaining: 40.6ms
58: learn: 0.4944533 total: 56.5ms remaining: 39.3ms
59: learn: 0.4923812 total: 57.5ms remaining: 38.3ms
60: learn: 0.4898208 total: 58.8ms remaining: 37.6ms
61: learn: 0.4875806 total: 63.1ms remaining: 38.7ms
62: learn: 0.4855243 total: 64.6ms remaining: 37.9ms
63: learn: 0.4829258 total: 65.9ms remaining: 37.1ms
64: learn: 0.4807748 total: 67.5ms remaining: 36.3ms
65: learn: 0.4783937 total: 68.4ms remaining: 35.2ms
66: learn: 0.4765855 total: 69.3ms remaining: 34.1ms
67: learn: 0.4750638 total: 70.2ms remaining: 33.1ms
68: learn: 0.4728485 total: 71ms remaining: 31.9ms
69: learn: 0.4709367 total: 71.8ms remaining: 30.8ms
70: learn: 0.4695238 total: 72.3ms remaining: 29.5ms
71: learn: 0.4671008 total: 73.5ms remaining: 28.6ms
72: learn: 0.4651308 total: 74.3ms remaining: 27.5ms
73: learn: 0.4632167 total: 75.4ms remaining: 26.5ms
74: learn: 0.4613239 total: 76.3ms remaining: 25.4ms
75: learn: 0.4595749 total: 77.3ms remaining: 24.4ms
76: learn: 0.4576102 total: 77.8ms remaining: 23.3ms
77: learn: 0.4555698 total: 78.3ms remaining: 22.1ms
78: learn: 0.4536103 total: 79.3ms remaining: 21.1ms
79: learn: 0.4520065 total: 80.4ms remaining: 20.1ms
80: learn: 0.4503276 total: 81.3ms remaining: 19.1ms
81: learn: 0.4482540 total: 82.3ms remaining: 18.1ms
82: learn: 0.4465758 total: 83.2ms remaining: 17ms
83: learn: 0.4444727 total: 84.3ms remaining: 16.1ms
84: learn: 0.4424653 total: 85.2ms remaining: 15ms
85: learn: 0.4409029 total: 85.5ms remaining: 13.9ms
86: learn: 0.4390786 total: 86.4ms remaining: 12.9ms
87: learn: 0.4372358 total: 87.1ms remaining: 11.9ms
88: learn: 0.4359033 total: 88.1ms remaining: 10.9ms
89: learn: 0.4345957 total: 89.3ms remaining: 9.92ms
90: learn: 0.4333527 total: 89.9ms remaining: 8.89ms
91: learn: 0.4315202 total: 90.7ms remaining: 7.89ms
92: learn: 0.4295818 total: 91.6ms remaining: 6.89ms
93: learn: 0.4275246 total: 92.6ms remaining: 5.91ms
94: learn: 0.4255765 total: 93.1ms remaining: 4.9ms
95: learn: 0.4239125 total: 93.9ms remaining: 3.91ms
96: learn: 0.4222595 total: 94.8ms remaining: 2.93ms
97: learn: 0.4209821 total: 95.7ms remaining: 1.95ms
98: learn: 0.4196025 total: 95.9ms remaining: 968us
99: learn: 0.4180498 total: 97ms remaining: 0us
0: learn: 0.6895196 total: 732us remaining: 72.5ms
1: learn: 0.6848229 total: 1.29ms remaining: 63.5ms
2: learn: 0.6798775 total: 2.07ms remaining: 66.9ms
3: learn: 0.6752787 total: 2.74ms remaining: 65.7ms
4: learn: 0.6702490 total: 3.41ms remaining: 64.9ms
5: learn: 0.6655144 total: 4.13ms remaining: 64.6ms
6: learn: 0.6609475 total: 4.94ms remaining: 65.7ms
7: learn: 0.6563440 total: 5.64ms remaining: 64.8ms
8: learn: 0.6525135 total: 6.43ms remaining: 65ms
9: learn: 0.6486859 total: 7.28ms remaining: 65.5ms
10: learn: 0.6448139 total: 8.25ms remaining: 66.8ms
11: learn: 0.6411210 total: 8.71ms remaining: 63.9ms
12: learn: 0.6382069 total: 9.62ms remaining: 64.4ms
13: learn: 0.6339793 total: 10.2ms remaining: 62.7ms
14: learn: 0.6297542 total: 10.9ms remaining: 61.6ms
15: learn: 0.6262754 total: 11.7ms remaining: 61.4ms
16: learn: 0.6232733 total: 12.4ms remaining: 60.5ms
17: learn: 0.6193642 total: 13ms remaining: 59.1ms
18: learn: 0.6150939 total: 13.6ms remaining: 58ms
19: learn: 0.6115523 total: 13.9ms remaining: 55.4ms
20: learn: 0.6078337 total: 14.6ms remaining: 54.8ms
21: learn: 0.6044395 total: 15.3ms remaining: 54.4ms
22: learn: 0.6008396 total: 16.2ms remaining: 54.4ms
23: learn: 0.5972830 total: 16.8ms remaining: 53.3ms
24: learn: 0.5934103 total: 17.5ms remaining: 52.6ms
25: learn: 0.5901748 total: 18.4ms remaining: 52.3ms
26: learn: 0.5868766 total: 19ms remaining: 51.3ms
27: learn: 0.5835131 total: 19.8ms remaining: 50.8ms
28: learn: 0.5805723 total: 20.6ms remaining: 50.4ms
29: learn: 0.5773479 total: 21.3ms remaining: 49.6ms
30: learn: 0.5736389 total: 21.7ms remaining: 48.3ms
31: learn: 0.5699806 total: 22.3ms remaining: 47.3ms
32: learn: 0.5668918 total: 23.1ms remaining: 46.9ms
33: learn: 0.5633195 total: 23.9ms remaining: 46.5ms
34: learn: 0.5598419 total: 24.6ms remaining: 45.6ms
35: learn: 0.5573127 total: 25.2ms remaining: 44.8ms
36: learn: 0.5542739 total: 25.9ms remaining: 44.1ms
37: learn: 0.5519820 total: 26.8ms remaining: 43.8ms
38: learn: 0.5491087 total: 27.6ms remaining: 43.2ms
39: learn: 0.5465576 total: 28.4ms remaining: 42.6ms
40: learn: 0.5436731 total: 29ms remaining: 41.8ms
41: learn: 0.5409845 total: 29.6ms remaining: 40.9ms
42: learn: 0.5383135 total: 30.2ms remaining: 40.1ms
43: learn: 0.5355018 total: 30.8ms remaining: 39.2ms
44: learn: 0.5333852 total: 31.5ms remaining: 38.6ms
45: learn: 0.5305767 total: 32.2ms remaining: 37.8ms
46: learn: 0.5275610 total: 32.8ms remaining: 37ms
47: learn: 0.5247445 total: 33.4ms remaining: 36.2ms
48: learn: 0.5223355 total: 33.9ms remaining: 35.3ms
49: learn: 0.5195292 total: 34.6ms remaining: 34.6ms
50: learn: 0.5171677 total: 35ms remaining: 33.7ms
51: learn: 0.5149542 total: 35.9ms remaining: 33.2ms
52: learn: 0.5124907 total: 36.7ms remaining: 32.5ms
53: learn: 0.5098953 total: 37.3ms remaining: 31.8ms
54: learn: 0.5072442 total: 38.3ms remaining: 31.4ms
55: learn: 0.5054656 total: 39.2ms remaining: 30.8ms
56: learn: 0.5034813 total: 39.8ms remaining: 30ms
57: learn: 0.5011521 total: 40.5ms remaining: 29.3ms
58: learn: 0.4987473 total: 41.5ms remaining: 28.8ms
59: learn: 0.4965739 total: 42.2ms remaining: 28.1ms
60: learn: 0.4940743 total: 42.7ms remaining: 27.3ms
61: learn: 0.4918511 total: 43.5ms remaining: 26.6ms
62: learn: 0.4903165 total: 44.1ms remaining: 25.9ms
63: learn: 0.4881625 total: 44.9ms remaining: 25.2ms
64: learn: 0.4859965 total: 45.7ms remaining: 24.6ms
65: learn: 0.4841086 total: 46.4ms remaining: 23.9ms
66: learn: 0.4819928 total: 47.4ms remaining: 23.3ms
67: learn: 0.4798261 total: 48ms remaining: 22.6ms
68: learn: 0.4782583 total: 48.5ms remaining: 21.8ms
69: learn: 0.4762403 total: 49.1ms remaining: 21.1ms
70: learn: 0.4737023 total: 49.9ms remaining: 20.4ms
71: learn: 0.4719914 total: 50.4ms remaining: 19.6ms
72: learn: 0.4697693 total: 51.1ms remaining: 18.9ms
73: learn: 0.4677942 total: 51.8ms remaining: 18.2ms
74: learn: 0.4654577 total: 52.4ms remaining: 17.5ms
75: learn: 0.4634901 total: 53.2ms remaining: 16.8ms
76: learn: 0.4613554 total: 53.9ms remaining: 16.1ms
77: learn: 0.4599424 total: 54.5ms remaining: 15.4ms
78: learn: 0.4584380 total: 55.1ms remaining: 14.6ms
79: learn: 0.4566588 total: 56ms remaining: 14ms
80: learn: 0.4544215 total: 56.7ms remaining: 13.3ms
81: learn: 0.4522932 total: 57.3ms remaining: 12.6ms
82: learn: 0.4504139 total: 58.4ms remaining: 12ms
83: learn: 0.4484829 total: 59.3ms remaining: 11.3ms
84: learn: 0.4465528 total: 60.2ms remaining: 10.6ms
85: learn: 0.4447133 total: 61.1ms remaining: 9.94ms
86: learn: 0.4432632 total: 62ms remaining: 9.26ms
87: learn: 0.4414880 total: 62.5ms remaining: 8.52ms
88: learn: 0.4396675 total: 63.2ms remaining: 7.81ms
89: learn: 0.4378557 total: 64ms remaining: 7.11ms
90: learn: 0.4360673 total: 64.7ms remaining: 6.4ms
91: learn: 0.4342336 total: 65.4ms remaining: 5.69ms
92: learn: 0.4325860 total: 66.2ms remaining: 4.98ms
93: learn: 0.4309977 total: 67ms remaining: 4.27ms
94: learn: 0.4295320 total: 67.7ms remaining: 3.56ms
95: learn: 0.4276862 total: 68.5ms remaining: 2.85ms
96: learn: 0.4260535 total: 69.2ms remaining: 2.14ms
97: learn: 0.4244318 total: 70.1ms remaining: 1.43ms
98: learn: 0.4229271 total: 71ms remaining: 717us
99: learn: 0.4214919 total: 72.1ms remaining: 0us
0: learn: 0.6881645 total: 794us remaining: 78.6ms
1: learn: 0.6828074 total: 1.67ms remaining: 81.9ms
2: learn: 0.6783972 total: 2.67ms remaining: 86.5ms
3: learn: 0.6738964 total: 3.5ms remaining: 84ms
4: learn: 0.6689173 total: 4.17ms remaining: 79.2ms
5: learn: 0.6650825 total: 4.96ms remaining: 77.7ms
6: learn: 0.6601666 total: 5.81ms remaining: 77.1ms
7: learn: 0.6554972 total: 7.12ms remaining: 81.9ms
8: learn: 0.6505983 total: 7.93ms remaining: 80.2ms
9: learn: 0.6462642 total: 8.74ms remaining: 78.6ms
10: learn: 0.6414765 total: 9.44ms remaining: 76.4ms
11: learn: 0.6373932 total: 10.3ms remaining: 75.3ms
12: learn: 0.6321228 total: 11.2ms remaining: 74.7ms
13: learn: 0.6283485 total: 11.9ms remaining: 73.4ms
14: learn: 0.6242799 total: 12.5ms remaining: 71ms
15: learn: 0.6194836 total: 13.1ms remaining: 68.8ms
16: learn: 0.6155681 total: 14ms remaining: 68.3ms
17: learn: 0.6117237 total: 14.2ms remaining: 64.6ms
18: learn: 0.6075215 total: 14.8ms remaining: 63.3ms
19: learn: 0.6036330 total: 15.5ms remaining: 62.2ms
20: learn: 0.5997560 total: 16.3ms remaining: 61.3ms
21: learn: 0.5961921 total: 16.9ms remaining: 60ms
22: learn: 0.5921406 total: 17.5ms remaining: 58.6ms
23: learn: 0.5881204 total: 18.2ms remaining: 57.6ms
24: learn: 0.5842689 total: 18.8ms remaining: 56.3ms
25: learn: 0.5804399 total: 19.4ms remaining: 55.1ms
26: learn: 0.5770751 total: 20.1ms remaining: 54.4ms
27: learn: 0.5733741 total: 20.8ms remaining: 53.4ms
28: learn: 0.5696210 total: 21.3ms remaining: 52.1ms
29: learn: 0.5660444 total: 21.8ms remaining: 51ms
30: learn: 0.5626817 total: 22.5ms remaining: 50.1ms
31: learn: 0.5591196 total: 23.1ms remaining: 49ms
32: learn: 0.5557326 total: 23.7ms remaining: 48.2ms
33: learn: 0.5523177 total: 24.4ms remaining: 47.3ms
34: learn: 0.5493842 total: 25.1ms remaining: 46.6ms
35: learn: 0.5460369 total: 25.6ms remaining: 45.5ms
36: learn: 0.5436892 total: 26.1ms remaining: 44.5ms
37: learn: 0.5404256 total: 26.8ms remaining: 43.7ms
38: learn: 0.5370325 total: 27.3ms remaining: 42.8ms
39: learn: 0.5341095 total: 28ms remaining: 42ms
40: learn: 0.5307599 total: 28.7ms remaining: 41.3ms
41: learn: 0.5281249 total: 29.2ms remaining: 40.3ms
42: learn: 0.5247488 total: 29.8ms remaining: 39.5ms
43: learn: 0.5220971 total: 30.5ms remaining: 38.8ms
44: learn: 0.5193386 total: 30.7ms remaining: 37.6ms
45: learn: 0.5168324 total: 31.5ms remaining: 36.9ms
46: learn: 0.5137939 total: 32.2ms remaining: 36.3ms
47: learn: 0.5105831 total: 32.9ms remaining: 35.7ms
48: learn: 0.5081495 total: 33.5ms remaining: 34.9ms
49: learn: 0.5052566 total: 34.2ms remaining: 34.2ms
50: learn: 0.5021816 total: 34.8ms remaining: 33.5ms
51: learn: 0.4991272 total: 35.5ms remaining: 32.8ms
52: learn: 0.4964772 total: 36.2ms remaining: 32.1ms
53: learn: 0.4933487 total: 37.1ms remaining: 31.6ms
54: learn: 0.4906787 total: 37.6ms remaining: 30.8ms
55: learn: 0.4882294 total: 38.3ms remaining: 30.1ms
56: learn: 0.4854180 total: 38.8ms remaining: 29.3ms
57: learn: 0.4831615 total: 39.1ms remaining: 28.3ms
58: learn: 0.4808132 total: 39.3ms remaining: 27.3ms
59: learn: 0.4782683 total: 40.1ms remaining: 26.8ms
60: learn: 0.4757675 total: 40.9ms remaining: 26.1ms
61: learn: 0.4738739 total: 41.5ms remaining: 25.5ms
62: learn: 0.4710584 total: 42.1ms remaining: 24.7ms
63: learn: 0.4687973 total: 42.6ms remaining: 23.9ms
64: learn: 0.4665910 total: 42.7ms remaining: 23ms
65: learn: 0.4640006 total: 43.4ms remaining: 22.4ms
66: learn: 0.4616570 total: 44ms remaining: 21.7ms
67: learn: 0.4592095 total: 44.7ms remaining: 21ms
68: learn: 0.4568761 total: 45.3ms remaining: 20.3ms
69: learn: 0.4543969 total: 45.9ms remaining: 19.7ms
70: learn: 0.4527871 total: 46.7ms remaining: 19.1ms
71: learn: 0.4507332 total: 47.3ms remaining: 18.4ms
72: learn: 0.4482096 total: 47.8ms remaining: 17.7ms
73: learn: 0.4460208 total: 48.5ms remaining: 17ms
74: learn: 0.4435941 total: 49.1ms remaining: 16.4ms
75: learn: 0.4411338 total: 49.8ms remaining: 15.7ms
76: learn: 0.4388022 total: 50.1ms remaining: 15ms
77: learn: 0.4367202 total: 50.7ms remaining: 14.3ms
78: learn: 0.4345595 total: 51.4ms remaining: 13.7ms
79: learn: 0.4324668 total: 52.1ms remaining: 13ms
80: learn: 0.4303298 total: 52.7ms remaining: 12.4ms
81: learn: 0.4282114 total: 53.5ms remaining: 11.8ms
82: learn: 0.4258690 total: 54.2ms remaining: 11.1ms
83: learn: 0.4238294 total: 54.9ms remaining: 10.5ms
84: learn: 0.4217277 total: 55.5ms remaining: 9.8ms
85: learn: 0.4196928 total: 56.2ms remaining: 9.15ms
86: learn: 0.4182239 total: 56.9ms remaining: 8.5ms
87: learn: 0.4162570 total: 57.4ms remaining: 7.83ms
88: learn: 0.4142180 total: 58.1ms remaining: 7.19ms
89: learn: 0.4123849 total: 58.8ms remaining: 6.53ms
90: learn: 0.4102499 total: 59.4ms remaining: 5.88ms
91: learn: 0.4080206 total: 60.1ms remaining: 5.23ms
92: learn: 0.4062764 total: 60.9ms remaining: 4.59ms
93: learn: 0.4045679 total: 61.8ms remaining: 3.94ms
94: learn: 0.4027085 total: 62.2ms remaining: 3.27ms
95: learn: 0.4007954 total: 62.9ms remaining: 2.62ms
96: learn: 0.3990088 total: 63.6ms remaining: 1.97ms
97: learn: 0.3973489 total: 64.2ms remaining: 1.31ms
98: learn: 0.3959789 total: 64.9ms remaining: 655us
99: learn: 0.3947270 total: 65.4ms remaining: 0us
0: learn: 0.6878932 total: 711us remaining: 70.5ms
1: learn: 0.6839973 total: 1.49ms remaining: 73.1ms
2: learn: 0.6793372 total: 2.18ms remaining: 70.6ms
3: learn: 0.6751816 total: 2.78ms remaining: 66.8ms
4: learn: 0.6706232 total: 3.52ms remaining: 67ms
5: learn: 0.6661899 total: 3.84ms remaining: 60.2ms
6: learn: 0.6619799 total: 4.58ms remaining: 60.9ms
7: learn: 0.6578849 total: 5.03ms remaining: 57.9ms
8: learn: 0.6532810 total: 5.71ms remaining: 57.7ms
9: learn: 0.6492916 total: 6.27ms remaining: 56.5ms
10: learn: 0.6452099 total: 7.16ms remaining: 57.9ms
11: learn: 0.6407272 total: 7.44ms remaining: 54.5ms
12: learn: 0.6364104 total: 8.01ms remaining: 53.6ms
13: learn: 0.6328575 total: 8.65ms remaining: 53.2ms
14: learn: 0.6296023 total: 9.25ms remaining: 52.4ms
15: learn: 0.6265584 total: 9.93ms remaining: 52.1ms
16: learn: 0.6233491 total: 10.3ms remaining: 50.4ms
17: learn: 0.6191226 total: 10.9ms remaining: 49.6ms
18: learn: 0.6151361 total: 11.9ms remaining: 50.7ms
19: learn: 0.6117795 total: 12.8ms remaining: 51.4ms
20: learn: 0.6081684 total: 13.4ms remaining: 50.6ms
21: learn: 0.6042276 total: 14.2ms remaining: 50.4ms
22: learn: 0.6006605 total: 15ms remaining: 50.4ms
23: learn: 0.5971764 total: 15.2ms remaining: 48.3ms
24: learn: 0.5942878 total: 15.9ms remaining: 47.8ms
25: learn: 0.5906416 total: 16.8ms remaining: 47.7ms
26: learn: 0.5872862 total: 17.4ms remaining: 47ms
27: learn: 0.5834617 total: 17.9ms remaining: 45.9ms
28: learn: 0.5802959 total: 18.3ms remaining: 44.9ms
29: learn: 0.5769130 total: 19ms remaining: 44.4ms
30: learn: 0.5747204 total: 19.8ms remaining: 44ms
31: learn: 0.5714667 total: 20.5ms remaining: 43.6ms
32: learn: 0.5685505 total: 21.6ms remaining: 43.9ms
33: learn: 0.5654209 total: 22.2ms remaining: 43.1ms
34: learn: 0.5624272 total: 22.9ms remaining: 42.5ms
35: learn: 0.5589781 total: 23.2ms remaining: 41.3ms
36: learn: 0.5554954 total: 23.7ms remaining: 40.4ms
37: learn: 0.5527426 total: 24.4ms remaining: 39.9ms
38: learn: 0.5494957 total: 24.7ms remaining: 38.6ms
39: learn: 0.5462821 total: 25.2ms remaining: 37.9ms
40: learn: 0.5433351 total: 25.7ms remaining: 37ms
41: learn: 0.5402277 total: 26.4ms remaining: 36.4ms
42: learn: 0.5368235 total: 26.9ms remaining: 35.7ms
43: learn: 0.5342293 total: 27.5ms remaining: 35ms
44: learn: 0.5311675 total: 28ms remaining: 34.2ms
45: learn: 0.5286138 total: 28.5ms remaining: 33.5ms
46: learn: 0.5257986 total: 29ms remaining: 32.7ms
47: learn: 0.5230951 total: 29.6ms remaining: 32.1ms
48: learn: 0.5208991 total: 30.2ms remaining: 31.4ms
49: learn: 0.5188314 total: 30.8ms remaining: 30.8ms
50: learn: 0.5161209 total: 31.6ms remaining: 30.3ms
51: learn: 0.5133814 total: 32.3ms remaining: 29.8ms
52: learn: 0.5109164 total: 32.5ms remaining: 28.8ms
53: learn: 0.5086263 total: 33ms remaining: 28.1ms
54: learn: 0.5058213 total: 33.6ms remaining: 27.5ms
55: learn: 0.5037096 total: 34.2ms remaining: 26.9ms
56: learn: 0.5014182 total: 34.7ms remaining: 26.2ms
57: learn: 0.4991026 total: 35.4ms remaining: 25.6ms
58: learn: 0.4961687 total: 36.3ms remaining: 25.2ms
59: learn: 0.4945034 total: 37ms remaining: 24.7ms
60: learn: 0.4922745 total: 37.6ms remaining: 24ms
61: learn: 0.4899098 total: 38.4ms remaining: 23.5ms
62: learn: 0.4876085 total: 39.1ms remaining: 23ms
63: learn: 0.4853648 total: 39.7ms remaining: 22.3ms
64: learn: 0.4830400 total: 40.2ms remaining: 21.6ms
65: learn: 0.4806109 total: 40.4ms remaining: 20.8ms
66: learn: 0.4785673 total: 40.9ms remaining: 20.2ms
67: learn: 0.4761730 total: 41.7ms remaining: 19.6ms
68: learn: 0.4739548 total: 42.4ms remaining: 19.1ms
69: learn: 0.4717292 total: 42.8ms remaining: 18.3ms
70: learn: 0.4697934 total: 43.4ms remaining: 17.7ms
71: learn: 0.4680407 total: 44ms remaining: 17.1ms
72: learn: 0.4658210 total: 44.6ms remaining: 16.5ms
73: learn: 0.4638347 total: 45.1ms remaining: 15.8ms
74: learn: 0.4617776 total: 45.7ms remaining: 15.2ms
75: learn: 0.4597503 total: 46.1ms remaining: 14.6ms
76: learn: 0.4581052 total: 46.7ms remaining: 13.9ms
77: learn: 0.4562682 total: 47.2ms remaining: 13.3ms
78: learn: 0.4545903 total: 47.7ms remaining: 12.7ms
79: learn: 0.4524356 total: 48.4ms remaining: 12.1ms
80: learn: 0.4505757 total: 49ms remaining: 11.5ms
81: learn: 0.4483205 total: 49.5ms remaining: 10.9ms
82: learn: 0.4463437 total: 50.2ms remaining: 10.3ms
83: learn: 0.4440199 total: 51ms remaining: 9.71ms
84: learn: 0.4420932 total: 51.7ms remaining: 9.12ms
85: learn: 0.4403515 total: 52ms remaining: 8.46ms
86: learn: 0.4386037 total: 52.7ms remaining: 7.87ms
87: learn: 0.4368057 total: 53.4ms remaining: 7.28ms
88: learn: 0.4352458 total: 54.1ms remaining: 6.69ms
89: learn: 0.4330688 total: 54.5ms remaining: 6.05ms
90: learn: 0.4311929 total: 54.8ms remaining: 5.42ms
91: learn: 0.4292900 total: 55.4ms remaining: 4.82ms
92: learn: 0.4280034 total: 56.4ms remaining: 4.24ms
93: learn: 0.4265435 total: 57.1ms remaining: 3.65ms
94: learn: 0.4249009 total: 59ms remaining: 3.1ms
95: learn: 0.4234669 total: 59.9ms remaining: 2.5ms
96: learn: 0.4216397 total: 61.1ms remaining: 1.89ms
97: learn: 0.4200636 total: 62ms remaining: 1.26ms
98: learn: 0.4185628 total: 63ms remaining: 636us
99: learn: 0.4172024 total: 63.9ms remaining: 0us
0: learn: 0.6880858 total: 837us remaining: 82.9ms
1: learn: 0.6835325 total: 1.52ms remaining: 74.5ms
2: learn: 0.6790942 total: 2.26ms remaining: 73.1ms
3: learn: 0.6743935 total: 2.81ms remaining: 67.4ms
4: learn: 0.6698191 total: 3.45ms remaining: 65.6ms
5: learn: 0.6646577 total: 4.08ms remaining: 63.9ms
6: learn: 0.6594989 total: 4.69ms remaining: 62.3ms
7: learn: 0.6558044 total: 5.41ms remaining: 62.2ms
8: learn: 0.6515938 total: 6.14ms remaining: 62.1ms
9: learn: 0.6462522 total: 6.49ms remaining: 58.5ms
10: learn: 0.6425140 total: 7.13ms remaining: 57.6ms
11: learn: 0.6381312 total: 7.35ms remaining: 53.9ms
12: learn: 0.6347719 total: 7.9ms remaining: 52.9ms
13: learn: 0.6302025 total: 8.46ms remaining: 52ms
14: learn: 0.6261653 total: 9.1ms remaining: 51.6ms
15: learn: 0.6226721 total: 9.8ms remaining: 51.5ms
16: learn: 0.6187113 total: 10.5ms remaining: 51.2ms
17: learn: 0.6140191 total: 11ms remaining: 50.1ms
18: learn: 0.6096826 total: 11.5ms remaining: 48.9ms
19: learn: 0.6052420 total: 12ms remaining: 47.9ms
20: learn: 0.6017107 total: 12.6ms remaining: 47.4ms
21: learn: 0.5983776 total: 13.3ms remaining: 47.2ms
22: learn: 0.5951994 total: 14ms remaining: 46.7ms
23: learn: 0.5918906 total: 14.6ms remaining: 46.2ms
24: learn: 0.5884141 total: 15.2ms remaining: 45.7ms
25: learn: 0.5841399 total: 15.5ms remaining: 44.1ms
26: learn: 0.5803691 total: 16.1ms remaining: 43.4ms
27: learn: 0.5769200 total: 16.7ms remaining: 42.8ms
28: learn: 0.5736473 total: 17.2ms remaining: 42.1ms
29: learn: 0.5700408 total: 17.8ms remaining: 41.5ms
30: learn: 0.5662298 total: 18.4ms remaining: 40.9ms
31: learn: 0.5626713 total: 19ms remaining: 40.3ms
32: learn: 0.5589932 total: 19.3ms remaining: 39.3ms
33: learn: 0.5555780 total: 19.8ms remaining: 38.5ms
34: learn: 0.5522308 total: 20ms remaining: 37.2ms
35: learn: 0.5489336 total: 20.2ms remaining: 36ms
36: learn: 0.5463816 total: 21ms remaining: 35.7ms
37: learn: 0.5430801 total: 21.8ms remaining: 35.6ms
38: learn: 0.5403064 total: 22.3ms remaining: 34.9ms
39: learn: 0.5369235 total: 23.1ms remaining: 34.7ms
40: learn: 0.5339607 total: 23.4ms remaining: 33.6ms
41: learn: 0.5311299 total: 24.2ms remaining: 33.4ms
42: learn: 0.5281621 total: 25.1ms remaining: 33.3ms
43: learn: 0.5248973 total: 25.9ms remaining: 33ms
44: learn: 0.5222278 total: 26.6ms remaining: 32.5ms
45: learn: 0.5195237 total: 27.4ms remaining: 32.2ms
46: learn: 0.5169016 total: 28.1ms remaining: 31.7ms
47: learn: 0.5141868 total: 28.9ms remaining: 31.3ms
48: learn: 0.5115914 total: 29.6ms remaining: 30.8ms
49: learn: 0.5085887 total: 30.1ms remaining: 30.1ms
50: learn: 0.5062185 total: 31.1ms remaining: 29.9ms
51: learn: 0.5034077 total: 31.8ms remaining: 29.4ms
52: learn: 0.5004999 total: 32.6ms remaining: 28.9ms
53: learn: 0.4984601 total: 33.5ms remaining: 28.5ms
54: learn: 0.4961639 total: 34.2ms remaining: 28ms
55: learn: 0.4931663 total: 35.1ms remaining: 27.6ms
56: learn: 0.4906388 total: 35.6ms remaining: 26.9ms
57: learn: 0.4884438 total: 36.5ms remaining: 26.4ms
58: learn: 0.4861598 total: 37.1ms remaining: 25.8ms
59: learn: 0.4833555 total: 37.8ms remaining: 25.2ms
60: learn: 0.4806488 total: 38.6ms remaining: 24.7ms
61: learn: 0.4783838 total: 39.3ms remaining: 24.1ms
62: learn: 0.4760700 total: 39.9ms remaining: 23.5ms
63: learn: 0.4737210 total: 40.3ms remaining: 22.7ms
64: learn: 0.4708672 total: 41ms remaining: 22.1ms
65: learn: 0.4683877 total: 41.6ms remaining: 21.4ms
66: learn: 0.4662321 total: 42.5ms remaining: 21ms
67: learn: 0.4634618 total: 43.4ms remaining: 20.4ms
68: learn: 0.4613501 total: 44.2ms remaining: 19.9ms
69: learn: 0.4588719 total: 44.9ms remaining: 19.3ms
70: learn: 0.4564823 total: 45.6ms remaining: 18.6ms
71: learn: 0.4544797 total: 45.8ms remaining: 17.8ms
72: learn: 0.4523694 total: 46.5ms remaining: 17.2ms
73: learn: 0.4504473 total: 47ms remaining: 16.5ms
74: learn: 0.4484612 total: 47.8ms remaining: 15.9ms
75: learn: 0.4467771 total: 48.7ms remaining: 15.4ms
76: learn: 0.4443176 total: 49.5ms remaining: 14.8ms
77: learn: 0.4423196 total: 50.4ms remaining: 14.2ms
78: learn: 0.4403209 total: 51.1ms remaining: 13.6ms
79: learn: 0.4384068 total: 52ms remaining: 13ms
80: learn: 0.4363520 total: 52.9ms remaining: 12.4ms
81: learn: 0.4341259 total: 53.7ms remaining: 11.8ms
82: learn: 0.4324370 total: 54.4ms remaining: 11.1ms
83: learn: 0.4305021 total: 55.2ms remaining: 10.5ms
84: learn: 0.4286833 total: 55.9ms remaining: 9.86ms
85: learn: 0.4266801 total: 56.6ms remaining: 9.22ms
86: learn: 0.4247576 total: 57.2ms remaining: 8.54ms
87: learn: 0.4229180 total: 58.4ms remaining: 7.96ms
88: learn: 0.4207467 total: 58.9ms remaining: 7.28ms
89: learn: 0.4188543 total: 59.6ms remaining: 6.63ms
90: learn: 0.4170876 total: 60.7ms remaining: 6ms
91: learn: 0.4154013 total: 61.5ms remaining: 5.35ms
92: learn: 0.4140714 total: 61.9ms remaining: 4.66ms
93: learn: 0.4121182 total: 62.6ms remaining: 4ms
94: learn: 0.4104015 total: 63.3ms remaining: 3.33ms
95: learn: 0.4087088 total: 64.1ms remaining: 2.67ms
96: learn: 0.4069240 total: 65.1ms remaining: 2.01ms
97: learn: 0.4049016 total: 65.8ms remaining: 1.34ms
98: learn: 0.4034390 total: 66.6ms remaining: 672us
99: learn: 0.4018267 total: 67.3ms remaining: 0us
0: learn: 0.6881413 total: 675us remaining: 66.9ms
1: learn: 0.6838407 total: 1.43ms remaining: 70ms
2: learn: 0.6798131 total: 2.15ms remaining: 69.6ms
3: learn: 0.6758858 total: 2.65ms remaining: 63.6ms
4: learn: 0.6712487 total: 3.11ms remaining: 59.2ms
5: learn: 0.6667165 total: 3.58ms remaining: 56ms
6: learn: 0.6624754 total: 3.97ms remaining: 52.8ms
7: learn: 0.6584117 total: 4.68ms remaining: 53.9ms
8: learn: 0.6543664 total: 5.37ms remaining: 54.3ms
9: learn: 0.6500307 total: 6.3ms remaining: 56.7ms
10: learn: 0.6463957 total: 6.76ms remaining: 54.7ms
11: learn: 0.6433796 total: 7.49ms remaining: 55ms
12: learn: 0.6393141 total: 8.41ms remaining: 56.3ms
13: learn: 0.6355468 total: 9.18ms remaining: 56.4ms
14: learn: 0.6316320 total: 10.1ms remaining: 57.4ms
15: learn: 0.6285376 total: 10.9ms remaining: 57.4ms
16: learn: 0.6239545 total: 11.4ms remaining: 55.5ms
17: learn: 0.6199367 total: 12.1ms remaining: 54.9ms
18: learn: 0.6159201 total: 12.7ms remaining: 54.3ms
19: learn: 0.6118337 total: 13.7ms remaining: 54.9ms
20: learn: 0.6088449 total: 14.2ms remaining: 53.4ms
21: learn: 0.6054882 total: 14.8ms remaining: 52.5ms
22: learn: 0.6018578 total: 15.3ms remaining: 51.2ms
23: learn: 0.5985672 total: 15.6ms remaining: 49.5ms
24: learn: 0.5946907 total: 16.3ms remaining: 48.9ms
25: learn: 0.5905861 total: 16.7ms remaining: 47.4ms
26: learn: 0.5873582 total: 17.4ms remaining: 47ms
27: learn: 0.5835815 total: 18.2ms remaining: 46.8ms
28: learn: 0.5802048 total: 19ms remaining: 46.5ms
29: learn: 0.5766559 total: 19.6ms remaining: 45.7ms
30: learn: 0.5741789 total: 20.4ms remaining: 45.5ms
31: learn: 0.5708948 total: 21.1ms remaining: 44.9ms
32: learn: 0.5679576 total: 21.8ms remaining: 44.3ms
33: learn: 0.5653729 total: 22.7ms remaining: 44ms
34: learn: 0.5628168 total: 23.5ms remaining: 43.6ms
35: learn: 0.5597310 total: 24.1ms remaining: 42.8ms
36: learn: 0.5574384 total: 24.6ms remaining: 41.9ms
37: learn: 0.5552292 total: 25ms remaining: 40.8ms
38: learn: 0.5518550 total: 25.5ms remaining: 39.9ms
39: learn: 0.5488987 total: 26.2ms remaining: 39.3ms
40: learn: 0.5458771 total: 27ms remaining: 38.8ms
41: learn: 0.5425878 total: 27.6ms remaining: 38.1ms
42: learn: 0.5398097 total: 28.3ms remaining: 37.6ms
43: learn: 0.5371612 total: 29ms remaining: 36.9ms
44: learn: 0.5340660 total: 29.8ms remaining: 36.5ms
45: learn: 0.5317800 total: 30.4ms remaining: 35.7ms
46: learn: 0.5293149 total: 31.2ms remaining: 35.2ms
47: learn: 0.5266531 total: 32.1ms remaining: 34.8ms
48: learn: 0.5238752 total: 32.6ms remaining: 34ms
49: learn: 0.5207962 total: 33.1ms remaining: 33.1ms
50: learn: 0.5183692 total: 33.3ms remaining: 32ms
51: learn: 0.5154727 total: 34ms remaining: 31.4ms
52: learn: 0.5128088 total: 34.6ms remaining: 30.7ms
53: learn: 0.5106960 total: 35.2ms remaining: 30ms
54: learn: 0.5079387 total: 36ms remaining: 29.4ms
55: learn: 0.5052003 total: 36.7ms remaining: 28.8ms
56: learn: 0.5030968 total: 37ms remaining: 27.9ms
57: learn: 0.5004473 total: 37.7ms remaining: 27.3ms
58: learn: 0.4979869 total: 38.5ms remaining: 26.8ms
59: learn: 0.4954643 total: 39.3ms remaining: 26.2ms
60: learn: 0.4929486 total: 39.9ms remaining: 25.5ms
61: learn: 0.4906020 total: 40.6ms remaining: 24.9ms
62: learn: 0.4885906 total: 41.5ms remaining: 24.4ms
63: learn: 0.4863976 total: 41.9ms remaining: 23.6ms
64: learn: 0.4840766 total: 42.6ms remaining: 22.9ms
65: learn: 0.4818117 total: 43.5ms remaining: 22.4ms
66: learn: 0.4798257 total: 44.1ms remaining: 21.7ms
67: learn: 0.4779516 total: 45.3ms remaining: 21.3ms
68: learn: 0.4760737 total: 46ms remaining: 20.7ms
69: learn: 0.4739915 total: 46.9ms remaining: 20.1ms
70: learn: 0.4720782 total: 47.6ms remaining: 19.5ms
71: learn: 0.4701228 total: 48.5ms remaining: 18.9ms
72: learn: 0.4681061 total: 49.3ms remaining: 18.2ms
73: learn: 0.4663896 total: 49.8ms remaining: 17.5ms
74: learn: 0.4647239 total: 50.8ms remaining: 16.9ms
75: learn: 0.4627239 total: 51.5ms remaining: 16.3ms
76: learn: 0.4608506 total: 52.1ms remaining: 15.6ms
77: learn: 0.4587431 total: 52.8ms remaining: 14.9ms
78: learn: 0.4568555 total: 53.5ms remaining: 14.2ms
79: learn: 0.4548857 total: 54.1ms remaining: 13.5ms
80: learn: 0.4536379 total: 54.4ms remaining: 12.8ms
81: learn: 0.4515869 total: 55.3ms remaining: 12.1ms
82: learn: 0.4493301 total: 56ms remaining: 11.5ms
83: learn: 0.4472792 total: 56.6ms remaining: 10.8ms
84: learn: 0.4454140 total: 57.5ms remaining: 10.1ms
85: learn: 0.4434786 total: 58.2ms remaining: 9.47ms
86: learn: 0.4414590 total: 59ms remaining: 8.81ms
87: learn: 0.4398276 total: 59.7ms remaining: 8.14ms
88: learn: 0.4380012 total: 60.3ms remaining: 7.45ms
89: learn: 0.4362726 total: 60.6ms remaining: 6.73ms
90: learn: 0.4343256 total: 61.2ms remaining: 6.05ms
91: learn: 0.4326818 total: 62ms remaining: 5.39ms
92: learn: 0.4312036 total: 63ms remaining: 4.74ms
93: learn: 0.4293792 total: 63.8ms remaining: 4.07ms
94: learn: 0.4277877 total: 64.6ms remaining: 3.4ms
95: learn: 0.4263225 total: 65.5ms remaining: 2.73ms
96: learn: 0.4247527 total: 66.4ms remaining: 2.05ms
97: learn: 0.4232069 total: 67.3ms remaining: 1.37ms
98: learn: 0.4216454 total: 68ms remaining: 687us
99: learn: 0.4197723 total: 68.7ms remaining: 0us
Evaluating param combo 13/16: {'boosted_cb_model__iterations': 100, 'boosted_cb_model__learning_rate': 0.1, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 8}
Duplicate found for this combo (hash=cfb36c53299c39b3af4068021ff76474). Removing old run: 7b31bff039244611be46df2187b86886
0: learn: 0.6462693 total: 864us remaining: 85.6ms
1: learn: 0.6006126 total: 1.6ms remaining: 78.3ms
2: learn: 0.5631732 total: 2.28ms remaining: 73.7ms
3: learn: 0.5318762 total: 3.23ms remaining: 77.6ms
4: learn: 0.5054558 total: 4.09ms remaining: 77.8ms
5: learn: 0.4799320 total: 5.34ms remaining: 83.7ms
6: learn: 0.4585994 total: 5.99ms remaining: 79.6ms
7: learn: 0.4417764 total: 6.76ms remaining: 77.7ms
8: learn: 0.4225440 total: 8.14ms remaining: 82.3ms
9: learn: 0.4084612 total: 9.87ms remaining: 88.8ms
10: learn: 0.3962466 total: 10.6ms remaining: 85.6ms
11: learn: 0.3845676 total: 11.4ms remaining: 83.6ms
12: learn: 0.3717729 total: 12.6ms remaining: 84.4ms
13: learn: 0.3609883 total: 13.2ms remaining: 81ms
14: learn: 0.3500067 total: 14.4ms remaining: 81.4ms
15: learn: 0.3412218 total: 15.7ms remaining: 82.5ms
16: learn: 0.3315580 total: 17.6ms remaining: 86.1ms
17: learn: 0.3252685 total: 19.4ms remaining: 88.3ms
18: learn: 0.3195212 total: 20.5ms remaining: 87.5ms
19: learn: 0.3143782 total: 21.1ms remaining: 84.5ms
20: learn: 0.3089230 total: 21.9ms remaining: 82.4ms
21: learn: 0.3037625 total: 22.2ms remaining: 78.6ms
22: learn: 0.2990881 total: 22.5ms remaining: 75.3ms
23: learn: 0.2939303 total: 23ms remaining: 72.9ms
24: learn: 0.2891699 total: 24.2ms remaining: 72.5ms
25: learn: 0.2840286 total: 25.8ms remaining: 73.4ms
26: learn: 0.2792264 total: 27.4ms remaining: 74ms
27: learn: 0.2746170 total: 28.6ms remaining: 73.6ms
28: learn: 0.2711557 total: 29.1ms remaining: 71.2ms
29: learn: 0.2693929 total: 30.1ms remaining: 70.2ms
30: learn: 0.2660210 total: 31.2ms remaining: 69.5ms
31: learn: 0.2638487 total: 31.4ms remaining: 66.8ms
32: learn: 0.2615085 total: 31.8ms remaining: 64.6ms
33: learn: 0.2594956 total: 32.1ms remaining: 62.3ms
34: learn: 0.2566762 total: 33.5ms remaining: 62.3ms
35: learn: 0.2542834 total: 33.8ms remaining: 60.2ms
36: learn: 0.2522301 total: 34.1ms remaining: 58ms
37: learn: 0.2495766 total: 34.3ms remaining: 55.9ms
38: learn: 0.2473101 total: 34.7ms remaining: 54.2ms
39: learn: 0.2450895 total: 35.1ms remaining: 52.6ms
40: learn: 0.2426112 total: 35.8ms remaining: 51.5ms
41: learn: 0.2396711 total: 36.6ms remaining: 50.5ms
42: learn: 0.2378629 total: 37.1ms remaining: 49.2ms
43: learn: 0.2352862 total: 37.8ms remaining: 48.1ms
44: learn: 0.2334733 total: 38.5ms remaining: 47ms
45: learn: 0.2314837 total: 39.6ms remaining: 46.5ms
46: learn: 0.2290257 total: 40ms remaining: 45.1ms
47: learn: 0.2272964 total: 40.3ms remaining: 43.6ms
48: learn: 0.2253947 total: 40.8ms remaining: 42.4ms
49: learn: 0.2237654 total: 41ms remaining: 41ms
50: learn: 0.2223406 total: 41.3ms remaining: 39.7ms
51: learn: 0.2210548 total: 41.6ms remaining: 38.4ms
52: learn: 0.2195405 total: 41.8ms remaining: 37.1ms
53: learn: 0.2183775 total: 42ms remaining: 35.8ms
54: learn: 0.2169509 total: 42.2ms remaining: 34.5ms
55: learn: 0.2154372 total: 42.5ms remaining: 33.4ms
56: learn: 0.2137457 total: 42.7ms remaining: 32.2ms
57: learn: 0.2119889 total: 42.9ms remaining: 31.1ms
58: learn: 0.2109968 total: 43.2ms remaining: 30ms
59: learn: 0.2094826 total: 43.4ms remaining: 28.9ms
60: learn: 0.2085079 total: 43.6ms remaining: 27.9ms
61: learn: 0.2071628 total: 44ms remaining: 27ms
62: learn: 0.2062468 total: 44.6ms remaining: 26.2ms
63: learn: 0.2043969 total: 45.2ms remaining: 25.4ms
64: learn: 0.2031529 total: 45.7ms remaining: 24.6ms
65: learn: 0.2014021 total: 46.2ms remaining: 23.8ms
66: learn: 0.2003742 total: 46.7ms remaining: 23ms
67: learn: 0.1991346 total: 47.6ms remaining: 22.4ms
68: learn: 0.1982389 total: 47.9ms remaining: 21.5ms
69: learn: 0.1974659 total: 48.2ms remaining: 20.6ms
70: learn: 0.1961055 total: 48.5ms remaining: 19.8ms
71: learn: 0.1953784 total: 49ms remaining: 19ms
72: learn: 0.1941287 total: 49.4ms remaining: 18.3ms
73: learn: 0.1930324 total: 49.9ms remaining: 17.5ms
74: learn: 0.1924753 total: 50.3ms remaining: 16.8ms
75: learn: 0.1915718 total: 50.9ms remaining: 16.1ms
76: learn: 0.1902028 total: 51.1ms remaining: 15.3ms
77: learn: 0.1892241 total: 51.4ms remaining: 14.5ms
78: learn: 0.1883913 total: 51.7ms remaining: 13.7ms
79: learn: 0.1869245 total: 52ms remaining: 13ms
80: learn: 0.1862597 total: 52.4ms remaining: 12.3ms
81: learn: 0.1850881 total: 52.8ms remaining: 11.6ms
82: learn: 0.1844696 total: 53.1ms remaining: 10.9ms
83: learn: 0.1836129 total: 53.4ms remaining: 10.2ms
84: learn: 0.1831236 total: 53.8ms remaining: 9.5ms
85: learn: 0.1826632 total: 54.2ms remaining: 8.82ms
86: learn: 0.1820750 total: 54.4ms remaining: 8.13ms
87: learn: 0.1812083 total: 54.8ms remaining: 7.47ms
88: learn: 0.1801868 total: 55.1ms remaining: 6.8ms
89: learn: 0.1796862 total: 55.3ms remaining: 6.14ms
90: learn: 0.1789454 total: 55.5ms remaining: 5.49ms
91: learn: 0.1784504 total: 55.8ms remaining: 4.85ms
92: learn: 0.1772510 total: 56.1ms remaining: 4.22ms
93: learn: 0.1768141 total: 56.3ms remaining: 3.59ms
94: learn: 0.1760395 total: 56.6ms remaining: 2.98ms
95: learn: 0.1755551 total: 56.8ms remaining: 2.37ms
96: learn: 0.1747803 total: 57.1ms remaining: 1.76ms
97: learn: 0.1743621 total: 57.4ms remaining: 1.17ms
98: learn: 0.1740438 total: 57.7ms remaining: 582us
99: learn: 0.1732493 total: 58ms remaining: 0us
0: learn: 0.6437160 total: 348us remaining: 34.5ms
1: learn: 0.5949760 total: 1.04ms remaining: 51ms
2: learn: 0.5565073 total: 1.23ms remaining: 39.8ms
3: learn: 0.5265837 total: 1.45ms remaining: 34.9ms
4: learn: 0.5005287 total: 1.68ms remaining: 31.9ms
5: learn: 0.4720428 total: 1.86ms remaining: 29.1ms
6: learn: 0.4484482 total: 2.02ms remaining: 26.8ms
7: learn: 0.4293736 total: 2.22ms remaining: 25.6ms
8: learn: 0.4148507 total: 2.38ms remaining: 24.1ms
9: learn: 0.3977165 total: 2.58ms remaining: 23.2ms
10: learn: 0.3825212 total: 2.76ms remaining: 22.4ms
11: learn: 0.3667856 total: 3.13ms remaining: 22.9ms
12: learn: 0.3547366 total: 3.34ms remaining: 22.3ms
13: learn: 0.3424929 total: 3.73ms remaining: 22.9ms
14: learn: 0.3349308 total: 3.95ms remaining: 22.4ms
15: learn: 0.3224127 total: 4.12ms remaining: 21.7ms
16: learn: 0.3178131 total: 4.34ms remaining: 21.2ms
17: learn: 0.3131939 total: 4.54ms remaining: 20.7ms
18: learn: 0.3045147 total: 4.81ms remaining: 20.5ms
19: learn: 0.2970139 total: 5.09ms remaining: 20.4ms
20: learn: 0.2918113 total: 5.68ms remaining: 21.4ms
21: learn: 0.2854668 total: 6.17ms remaining: 21.9ms
22: learn: 0.2779652 total: 6.65ms remaining: 22.3ms
23: learn: 0.2734233 total: 6.85ms remaining: 21.7ms
24: learn: 0.2685745 total: 7.08ms remaining: 21.3ms
25: learn: 0.2629943 total: 7.29ms remaining: 20.8ms
26: learn: 0.2604321 total: 7.54ms remaining: 20.4ms
27: learn: 0.2566107 total: 7.75ms remaining: 19.9ms
28: learn: 0.2531121 total: 8.95ms remaining: 21.9ms
29: learn: 0.2515028 total: 9.28ms remaining: 21.7ms
30: learn: 0.2482660 total: 9.81ms remaining: 21.8ms
31: learn: 0.2458159 total: 10.4ms remaining: 22.1ms
32: learn: 0.2416483 total: 10.7ms remaining: 21.8ms
33: learn: 0.2386672 total: 10.9ms remaining: 21.2ms
34: learn: 0.2359889 total: 11.3ms remaining: 20.9ms
35: learn: 0.2338575 total: 11.9ms remaining: 21.2ms
36: learn: 0.2298220 total: 12.1ms remaining: 20.7ms
37: learn: 0.2283967 total: 12.5ms remaining: 20.4ms
38: learn: 0.2263530 total: 13.1ms remaining: 20.5ms
39: learn: 0.2246581 total: 13.8ms remaining: 20.7ms
40: learn: 0.2223378 total: 14.2ms remaining: 20.5ms
41: learn: 0.2207038 total: 14.8ms remaining: 20.5ms
42: learn: 0.2197856 total: 15.5ms remaining: 20.6ms
43: learn: 0.2188898 total: 15.7ms remaining: 20ms
44: learn: 0.2176194 total: 16.2ms remaining: 19.7ms
45: learn: 0.2163312 total: 16.5ms remaining: 19.4ms
46: learn: 0.2154455 total: 17.3ms remaining: 19.5ms
47: learn: 0.2139407 total: 17.9ms remaining: 19.4ms
48: learn: 0.2120411 total: 18.8ms remaining: 19.5ms
49: learn: 0.2106386 total: 19.4ms remaining: 19.4ms
50: learn: 0.2091600 total: 19.9ms remaining: 19.1ms
51: learn: 0.2072987 total: 20.4ms remaining: 18.8ms
52: learn: 0.2045983 total: 20.7ms remaining: 18.4ms
53: learn: 0.2034906 total: 21.1ms remaining: 18ms
54: learn: 0.2021212 total: 21.6ms remaining: 17.7ms
55: learn: 0.1991710 total: 22.4ms remaining: 17.6ms
56: learn: 0.1976866 total: 23.3ms remaining: 17.5ms
57: learn: 0.1968438 total: 23.7ms remaining: 17.2ms
58: learn: 0.1956886 total: 24.2ms remaining: 16.8ms
59: learn: 0.1934149 total: 24.8ms remaining: 16.5ms
60: learn: 0.1920875 total: 25.2ms remaining: 16.1ms
61: learn: 0.1911042 total: 25.5ms remaining: 15.6ms
62: learn: 0.1898230 total: 25.7ms remaining: 15.1ms
63: learn: 0.1889307 total: 26ms remaining: 14.6ms
64: learn: 0.1869322 total: 26.3ms remaining: 14.2ms
65: learn: 0.1850626 total: 27.4ms remaining: 14.1ms
66: learn: 0.1842326 total: 27.8ms remaining: 13.7ms
67: learn: 0.1824564 total: 28ms remaining: 13.2ms
68: learn: 0.1816802 total: 28.3ms remaining: 12.7ms
69: learn: 0.1799241 total: 28.7ms remaining: 12.3ms
70: learn: 0.1785063 total: 29.3ms remaining: 12ms
71: learn: 0.1768805 total: 30.3ms remaining: 11.8ms
72: learn: 0.1755565 total: 30.6ms remaining: 11.3ms
73: learn: 0.1741842 total: 31ms remaining: 10.9ms
74: learn: 0.1729470 total: 31.3ms remaining: 10.4ms
75: learn: 0.1717045 total: 31.5ms remaining: 9.96ms
76: learn: 0.1707227 total: 32ms remaining: 9.54ms
77: learn: 0.1697844 total: 32.5ms remaining: 9.18ms
78: learn: 0.1687726 total: 33ms remaining: 8.76ms
79: learn: 0.1677323 total: 33.3ms remaining: 8.32ms
80: learn: 0.1666055 total: 33.5ms remaining: 7.87ms
81: learn: 0.1653523 total: 33.9ms remaining: 7.45ms
82: learn: 0.1644991 total: 34.3ms remaining: 7.02ms
83: learn: 0.1637596 total: 34.6ms remaining: 6.59ms
84: learn: 0.1626667 total: 35ms remaining: 6.17ms
85: learn: 0.1618658 total: 35.5ms remaining: 5.77ms
86: learn: 0.1610996 total: 35.7ms remaining: 5.33ms
87: learn: 0.1604054 total: 35.9ms remaining: 4.89ms
88: learn: 0.1599545 total: 36.1ms remaining: 4.46ms
89: learn: 0.1593649 total: 36.3ms remaining: 4.03ms
90: learn: 0.1588678 total: 36.6ms remaining: 3.62ms
91: learn: 0.1582488 total: 36.7ms remaining: 3.19ms
92: learn: 0.1578052 total: 37ms remaining: 2.79ms
93: learn: 0.1574109 total: 37.3ms remaining: 2.38ms
94: learn: 0.1564849 total: 37.7ms remaining: 1.98ms
95: learn: 0.1554839 total: 38ms remaining: 1.58ms
96: learn: 0.1550581 total: 38.3ms remaining: 1.18ms
97: learn: 0.1541604 total: 39.2ms remaining: 800us
98: learn: 0.1538731 total: 39.4ms remaining: 398us
99: learn: 0.1529953 total: 39.7ms remaining: 0us
0: learn: 0.6454260 total: 322us remaining: 31.9ms
1: learn: 0.6019562 total: 574us remaining: 28.1ms
2: learn: 0.5626070 total: 780us remaining: 25.2ms
3: learn: 0.5307213 total: 1.14ms remaining: 27.4ms
4: learn: 0.5032371 total: 1.39ms remaining: 26.4ms
5: learn: 0.4776693 total: 1.61ms remaining: 25.2ms
6: learn: 0.4566797 total: 1.9ms remaining: 25.2ms
7: learn: 0.4389420 total: 2.13ms remaining: 24.4ms
8: learn: 0.4181396 total: 2.32ms remaining: 23.4ms
9: learn: 0.4027384 total: 2.52ms remaining: 22.7ms
10: learn: 0.3900032 total: 2.68ms remaining: 21.7ms
11: learn: 0.3777932 total: 3.03ms remaining: 22.2ms
12: learn: 0.3636268 total: 3.22ms remaining: 21.6ms
13: learn: 0.3525592 total: 3.46ms remaining: 21.2ms
14: learn: 0.3415734 total: 3.67ms remaining: 20.8ms
15: learn: 0.3322054 total: 3.98ms remaining: 20.9ms
16: learn: 0.3233730 total: 4.34ms remaining: 21.2ms
17: learn: 0.3167153 total: 4.51ms remaining: 20.6ms
18: learn: 0.3106400 total: 4.84ms remaining: 20.6ms
19: learn: 0.3023905 total: 5.43ms remaining: 21.7ms
20: learn: 0.2963086 total: 6.03ms remaining: 22.7ms
21: learn: 0.2904163 total: 6.5ms remaining: 23.1ms
22: learn: 0.2852205 total: 7.08ms remaining: 23.7ms
23: learn: 0.2798857 total: 7.41ms remaining: 23.5ms
24: learn: 0.2744258 total: 7.75ms remaining: 23.2ms
25: learn: 0.2718590 total: 8.03ms remaining: 22.8ms
26: learn: 0.2677934 total: 8.25ms remaining: 22.3ms
27: learn: 0.2637715 total: 8.75ms remaining: 22.5ms
28: learn: 0.2593053 total: 8.97ms remaining: 22ms
29: learn: 0.2574366 total: 9.17ms remaining: 21.4ms
30: learn: 0.2539525 total: 9.48ms remaining: 21.1ms
31: learn: 0.2512715 total: 9.77ms remaining: 20.8ms
32: learn: 0.2481380 total: 10.1ms remaining: 20.5ms
33: learn: 0.2458802 total: 10.3ms remaining: 20.1ms
34: learn: 0.2430230 total: 10.7ms remaining: 19.9ms
35: learn: 0.2402213 total: 11.2ms remaining: 19.8ms
36: learn: 0.2382592 total: 11.9ms remaining: 20.2ms
37: learn: 0.2366223 total: 12.6ms remaining: 20.5ms
38: learn: 0.2346630 total: 13.3ms remaining: 20.8ms
39: learn: 0.2316478 total: 13.6ms remaining: 20.5ms
40: learn: 0.2295176 total: 14.2ms remaining: 20.4ms
41: learn: 0.2275613 total: 14.4ms remaining: 19.9ms
42: learn: 0.2261304 total: 15.3ms remaining: 20.2ms
43: learn: 0.2230182 total: 15.9ms remaining: 20.3ms
44: learn: 0.2213852 total: 16.3ms remaining: 19.9ms
45: learn: 0.2198024 total: 17ms remaining: 20ms
46: learn: 0.2183045 total: 17.5ms remaining: 19.8ms
47: learn: 0.2166827 total: 17.9ms remaining: 19.4ms
48: learn: 0.2136031 total: 18.3ms remaining: 19.1ms
49: learn: 0.2121566 total: 19ms remaining: 19ms
50: learn: 0.2102998 total: 19.7ms remaining: 18.9ms
51: learn: 0.2078778 total: 20.3ms remaining: 18.7ms
52: learn: 0.2060268 total: 20.6ms remaining: 18.2ms
53: learn: 0.2048023 total: 21ms remaining: 17.9ms
54: learn: 0.2033060 total: 21.3ms remaining: 17.4ms
55: learn: 0.2012317 total: 21.6ms remaining: 17ms
56: learn: 0.1991270 total: 21.9ms remaining: 16.5ms
57: learn: 0.1982670 total: 22.3ms remaining: 16.1ms
58: learn: 0.1971084 total: 22.6ms remaining: 15.7ms
59: learn: 0.1955812 total: 23ms remaining: 15.4ms
60: learn: 0.1931865 total: 23.4ms remaining: 14.9ms
61: learn: 0.1918434 total: 23.6ms remaining: 14.5ms
62: learn: 0.1909635 total: 23.9ms remaining: 14ms
63: learn: 0.1886742 total: 24.2ms remaining: 13.6ms
64: learn: 0.1875385 total: 24.6ms remaining: 13.3ms
65: learn: 0.1859142 total: 24.8ms remaining: 12.8ms
66: learn: 0.1842228 total: 25.1ms remaining: 12.4ms
67: learn: 0.1832482 total: 25.4ms remaining: 12ms
68: learn: 0.1817732 total: 25.7ms remaining: 11.5ms
69: learn: 0.1806441 total: 26ms remaining: 11.1ms
70: learn: 0.1797458 total: 26.3ms remaining: 10.7ms
71: learn: 0.1789580 total: 26.5ms remaining: 10.3ms
72: learn: 0.1774616 total: 26.8ms remaining: 9.9ms
73: learn: 0.1765499 total: 27.1ms remaining: 9.51ms
74: learn: 0.1752367 total: 27.4ms remaining: 9.12ms
75: learn: 0.1747060 total: 27.7ms remaining: 8.73ms
76: learn: 0.1740200 total: 28ms remaining: 8.35ms
77: learn: 0.1731577 total: 28.2ms remaining: 7.96ms
78: learn: 0.1725317 total: 28.5ms remaining: 7.56ms
79: learn: 0.1709006 total: 28.8ms remaining: 7.19ms
80: learn: 0.1701828 total: 29ms remaining: 6.81ms
81: learn: 0.1694006 total: 29.3ms remaining: 6.43ms
82: learn: 0.1681916 total: 29.6ms remaining: 6.06ms
83: learn: 0.1674656 total: 29.9ms remaining: 5.69ms
84: learn: 0.1668704 total: 30.2ms remaining: 5.32ms
85: learn: 0.1663118 total: 30.4ms remaining: 4.95ms
86: learn: 0.1658180 total: 30.7ms remaining: 4.59ms
87: learn: 0.1651004 total: 31ms remaining: 4.23ms
88: learn: 0.1644411 total: 31.3ms remaining: 3.87ms
89: learn: 0.1638417 total: 31.6ms remaining: 3.51ms
90: learn: 0.1633229 total: 31.9ms remaining: 3.15ms
91: learn: 0.1629261 total: 32.2ms remaining: 2.8ms
92: learn: 0.1623520 total: 32.4ms remaining: 2.44ms
93: learn: 0.1618410 total: 32.7ms remaining: 2.09ms
94: learn: 0.1611050 total: 33ms remaining: 1.74ms
95: learn: 0.1606811 total: 33.2ms remaining: 1.38ms
96: learn: 0.1592629 total: 33.8ms remaining: 1.04ms
97: learn: 0.1581160 total: 34.3ms remaining: 700us
98: learn: 0.1577026 total: 34.7ms remaining: 350us
99: learn: 0.1573761 total: 35.1ms remaining: 0us
0: learn: 0.6470966 total: 381us remaining: 37.8ms
1: learn: 0.6001838 total: 680us remaining: 33.3ms
2: learn: 0.5604623 total: 899us remaining: 29.1ms
3: learn: 0.5261755 total: 1.22ms remaining: 29.4ms
4: learn: 0.4992383 total: 1.58ms remaining: 30ms
5: learn: 0.4721898 total: 1.88ms remaining: 29.4ms
6: learn: 0.4498006 total: 2.14ms remaining: 28.4ms
7: learn: 0.4314508 total: 2.39ms remaining: 27.5ms
8: learn: 0.4101723 total: 2.62ms remaining: 26.5ms
9: learn: 0.3935443 total: 2.88ms remaining: 25.9ms
10: learn: 0.3798147 total: 3.1ms remaining: 25.1ms
11: learn: 0.3665670 total: 3.43ms remaining: 25.2ms
12: learn: 0.3536267 total: 3.75ms remaining: 25.1ms
13: learn: 0.3420766 total: 4.06ms remaining: 24.9ms
14: learn: 0.3326825 total: 4.3ms remaining: 24.4ms
15: learn: 0.3240086 total: 4.61ms remaining: 24.2ms
16: learn: 0.3134403 total: 4.91ms remaining: 24ms
17: learn: 0.3067898 total: 5.13ms remaining: 23.4ms
18: learn: 0.3003032 total: 5.38ms remaining: 22.9ms
19: learn: 0.2951168 total: 5.66ms remaining: 22.6ms
20: learn: 0.2888849 total: 6ms remaining: 22.6ms
21: learn: 0.2831076 total: 6.21ms remaining: 22ms
22: learn: 0.2763535 total: 6.51ms remaining: 21.8ms
23: learn: 0.2698591 total: 6.82ms remaining: 21.6ms
24: learn: 0.2647141 total: 7.13ms remaining: 21.4ms
25: learn: 0.2590451 total: 7.38ms remaining: 21ms
26: learn: 0.2557403 total: 7.63ms remaining: 20.6ms
27: learn: 0.2526011 total: 7.89ms remaining: 20.3ms
28: learn: 0.2495110 total: 8.02ms remaining: 19.6ms
29: learn: 0.2446900 total: 8.28ms remaining: 19.3ms
30: learn: 0.2404218 total: 8.52ms remaining: 19ms
31: learn: 0.2370704 total: 8.83ms remaining: 18.8ms
32: learn: 0.2333679 total: 9.12ms remaining: 18.5ms
33: learn: 0.2298418 total: 9.42ms remaining: 18.3ms
34: learn: 0.2271491 total: 9.69ms remaining: 18ms
35: learn: 0.2245757 total: 9.97ms remaining: 17.7ms
36: learn: 0.2214767 total: 10.2ms remaining: 17.3ms
37: learn: 0.2191640 total: 10.4ms remaining: 17ms
38: learn: 0.2178533 total: 10.7ms remaining: 16.8ms
39: learn: 0.2163079 total: 11ms remaining: 16.5ms
40: learn: 0.2149998 total: 11.3ms remaining: 16.3ms
41: learn: 0.2135240 total: 11.6ms remaining: 16.1ms
42: learn: 0.2118976 total: 11.9ms remaining: 15.8ms
43: learn: 0.2106812 total: 12.2ms remaining: 15.6ms
44: learn: 0.2087706 total: 12.5ms remaining: 15.3ms
45: learn: 0.2077590 total: 12.8ms remaining: 15ms
46: learn: 0.2062417 total: 13ms remaining: 14.7ms
47: learn: 0.2041230 total: 13.3ms remaining: 14.4ms
48: learn: 0.2029428 total: 13.6ms remaining: 14.1ms
49: learn: 0.2010316 total: 13.8ms remaining: 13.8ms
50: learn: 0.1999470 total: 14.1ms remaining: 13.6ms
51: learn: 0.1986324 total: 14.3ms remaining: 13.2ms
52: learn: 0.1981784 total: 14.5ms remaining: 12.8ms
53: learn: 0.1963600 total: 14.9ms remaining: 12.7ms
54: learn: 0.1951677 total: 15.2ms remaining: 12.5ms
55: learn: 0.1934322 total: 15.6ms remaining: 12.3ms
56: learn: 0.1924811 total: 16ms remaining: 12ms
57: learn: 0.1908243 total: 16.5ms remaining: 11.9ms
58: learn: 0.1892999 total: 16.8ms remaining: 11.7ms
59: learn: 0.1887691 total: 17.1ms remaining: 11.4ms
60: learn: 0.1867416 total: 17.4ms remaining: 11.1ms
61: learn: 0.1851716 total: 17.8ms remaining: 10.9ms
62: learn: 0.1841887 total: 18.2ms remaining: 10.7ms
63: learn: 0.1826161 total: 18.6ms remaining: 10.5ms
64: learn: 0.1807570 total: 19ms remaining: 10.2ms
65: learn: 0.1795807 total: 19.4ms remaining: 9.99ms
66: learn: 0.1786182 total: 19.7ms remaining: 9.73ms
67: learn: 0.1776957 total: 20.1ms remaining: 9.48ms
68: learn: 0.1769061 total: 20.5ms remaining: 9.21ms
69: learn: 0.1754625 total: 20.9ms remaining: 8.95ms
70: learn: 0.1743497 total: 21.3ms remaining: 8.69ms
71: learn: 0.1737500 total: 21.6ms remaining: 8.42ms
72: learn: 0.1730347 total: 22ms remaining: 8.15ms
73: learn: 0.1713745 total: 22.4ms remaining: 7.88ms
74: learn: 0.1697650 total: 22.9ms remaining: 7.62ms
75: learn: 0.1685269 total: 23.3ms remaining: 7.34ms
76: learn: 0.1678087 total: 23.6ms remaining: 7.04ms
77: learn: 0.1667430 total: 23.9ms remaining: 6.75ms
78: learn: 0.1660898 total: 24.3ms remaining: 6.47ms
79: learn: 0.1653770 total: 24.7ms remaining: 6.16ms
80: learn: 0.1645453 total: 25ms remaining: 5.87ms
81: learn: 0.1635048 total: 25.3ms remaining: 5.56ms
82: learn: 0.1628895 total: 25.6ms remaining: 5.25ms
83: learn: 0.1623059 total: 25.9ms remaining: 4.93ms
84: learn: 0.1608705 total: 26.3ms remaining: 4.65ms
85: learn: 0.1593044 total: 26.7ms remaining: 4.35ms
86: learn: 0.1587007 total: 27.1ms remaining: 4.04ms
87: learn: 0.1578239 total: 27.4ms remaining: 3.74ms
88: learn: 0.1567637 total: 27.8ms remaining: 3.43ms
89: learn: 0.1561612 total: 28.1ms remaining: 3.13ms
90: learn: 0.1556924 total: 28.4ms remaining: 2.81ms
91: learn: 0.1550177 total: 28.8ms remaining: 2.5ms
92: learn: 0.1544644 total: 29.1ms remaining: 2.19ms
93: learn: 0.1534878 total: 29.4ms remaining: 1.87ms
94: learn: 0.1530854 total: 29.7ms remaining: 1.56ms
95: learn: 0.1523530 total: 30ms remaining: 1.25ms
96: learn: 0.1518856 total: 30.3ms remaining: 937us
97: learn: 0.1515904 total: 30.7ms remaining: 626us
98: learn: 0.1509596 total: 31ms remaining: 313us
99: learn: 0.1503649 total: 31.2ms remaining: 0us
0: learn: 0.6466390 total: 297us remaining: 29.5ms
1: learn: 0.6043306 total: 552us remaining: 27.1ms
2: learn: 0.5698679 total: 730us remaining: 23.6ms
3: learn: 0.5400060 total: 929us remaining: 22.3ms
4: learn: 0.5139000 total: 1.16ms remaining: 22ms
5: learn: 0.4909190 total: 1.36ms remaining: 21.4ms
6: learn: 0.4663019 total: 1.55ms remaining: 20.6ms
7: learn: 0.4496656 total: 1.75ms remaining: 20.1ms
8: learn: 0.4302446 total: 1.92ms remaining: 19.5ms
9: learn: 0.4155090 total: 2.14ms remaining: 19.3ms
10: learn: 0.4046576 total: 2.31ms remaining: 18.7ms
11: learn: 0.3902395 total: 2.48ms remaining: 18.2ms
12: learn: 0.3781937 total: 2.75ms remaining: 18.4ms
13: learn: 0.3684001 total: 3.12ms remaining: 19.2ms
14: learn: 0.3594224 total: 3.51ms remaining: 19.9ms
15: learn: 0.3506654 total: 3.71ms remaining: 19.5ms
16: learn: 0.3427953 total: 3.9ms remaining: 19ms
17: learn: 0.3375828 total: 4.25ms remaining: 19.4ms
18: learn: 0.3304561 total: 4.65ms remaining: 19.8ms
19: learn: 0.3248426 total: 4.92ms remaining: 19.7ms
20: learn: 0.3187507 total: 5.14ms remaining: 19.3ms
21: learn: 0.3109731 total: 5.43ms remaining: 19.3ms
22: learn: 0.3052623 total: 5.71ms remaining: 19.1ms
23: learn: 0.3002449 total: 6.02ms remaining: 19.1ms
24: learn: 0.2967801 total: 6.44ms remaining: 19.3ms
25: learn: 0.2923089 total: 6.81ms remaining: 19.4ms
26: learn: 0.2868579 total: 7.16ms remaining: 19.3ms
27: learn: 0.2841698 total: 7.74ms remaining: 19.9ms
28: learn: 0.2810342 total: 8.31ms remaining: 20.4ms
29: learn: 0.2780382 total: 8.79ms remaining: 20.5ms
30: learn: 0.2758407 total: 9.15ms remaining: 20.4ms
31: learn: 0.2730709 total: 9.66ms remaining: 20.5ms
32: learn: 0.2704504 total: 10.1ms remaining: 20.5ms
33: learn: 0.2684170 total: 10.4ms remaining: 20.3ms
34: learn: 0.2646933 total: 11ms remaining: 20.4ms
35: learn: 0.2624324 total: 11.4ms remaining: 20.2ms
36: learn: 0.2604280 total: 11.8ms remaining: 20.1ms
37: learn: 0.2579343 total: 12.2ms remaining: 19.9ms
38: learn: 0.2550934 total: 12.6ms remaining: 19.7ms
39: learn: 0.2526229 total: 12.9ms remaining: 19.4ms
40: learn: 0.2500359 total: 13.2ms remaining: 19ms
41: learn: 0.2471391 total: 13.5ms remaining: 18.7ms
42: learn: 0.2454036 total: 13.9ms remaining: 18.5ms
43: learn: 0.2433846 total: 14.2ms remaining: 18.1ms
44: learn: 0.2406459 total: 14.5ms remaining: 17.8ms
45: learn: 0.2380629 total: 14.9ms remaining: 17.5ms
46: learn: 0.2358969 total: 15.2ms remaining: 17.1ms
47: learn: 0.2344105 total: 15.5ms remaining: 16.8ms
48: learn: 0.2324208 total: 15.8ms remaining: 16.4ms
49: learn: 0.2305732 total: 16.1ms remaining: 16.1ms
50: learn: 0.2295668 total: 16.4ms remaining: 15.7ms
51: learn: 0.2277100 total: 16.7ms remaining: 15.4ms
52: learn: 0.2270326 total: 17ms remaining: 15.1ms
53: learn: 0.2254242 total: 17.4ms remaining: 14.9ms
54: learn: 0.2232393 total: 17.7ms remaining: 14.5ms
55: learn: 0.2212543 total: 18.1ms remaining: 14.2ms
56: learn: 0.2193358 total: 18.4ms remaining: 13.9ms
57: learn: 0.2174221 total: 19ms remaining: 13.7ms
58: learn: 0.2156762 total: 19.2ms remaining: 13.3ms
59: learn: 0.2116688 total: 19.5ms remaining: 13ms
60: learn: 0.2099580 total: 19.8ms remaining: 12.7ms
61: learn: 0.2086876 total: 20.1ms remaining: 12.3ms
62: learn: 0.2070665 total: 20.4ms remaining: 12ms
63: learn: 0.2065565 total: 20.6ms remaining: 11.6ms
64: learn: 0.2049650 total: 20.9ms remaining: 11.2ms
65: learn: 0.2031501 total: 21.1ms remaining: 10.9ms
66: learn: 0.2006850 total: 21.4ms remaining: 10.5ms
67: learn: 0.1994539 total: 21.6ms remaining: 10.2ms
68: learn: 0.1982312 total: 21.8ms remaining: 9.82ms
69: learn: 0.1967728 total: 22.1ms remaining: 9.45ms
70: learn: 0.1944350 total: 22.4ms remaining: 9.14ms
71: learn: 0.1926110 total: 22.8ms remaining: 8.87ms
72: learn: 0.1900694 total: 23.2ms remaining: 8.58ms
73: learn: 0.1893151 total: 23.5ms remaining: 8.26ms
74: learn: 0.1883047 total: 23.9ms remaining: 7.96ms
75: learn: 0.1865259 total: 24.2ms remaining: 7.63ms
76: learn: 0.1848260 total: 24.5ms remaining: 7.31ms
77: learn: 0.1833083 total: 24.8ms remaining: 7ms
78: learn: 0.1823737 total: 25.1ms remaining: 6.67ms
79: learn: 0.1803818 total: 25.4ms remaining: 6.34ms
80: learn: 0.1785443 total: 25.7ms remaining: 6.02ms
81: learn: 0.1772803 total: 25.9ms remaining: 5.7ms
82: learn: 0.1763651 total: 26.2ms remaining: 5.37ms
83: learn: 0.1757682 total: 26.5ms remaining: 5.05ms
84: learn: 0.1743110 total: 26.9ms remaining: 4.74ms
85: learn: 0.1725994 total: 27.2ms remaining: 4.43ms
86: learn: 0.1713861 total: 27.5ms remaining: 4.11ms
87: learn: 0.1705274 total: 27.8ms remaining: 3.8ms
88: learn: 0.1692688 total: 28.1ms remaining: 3.48ms
89: learn: 0.1677366 total: 28.9ms remaining: 3.21ms
90: learn: 0.1669654 total: 29.2ms remaining: 2.89ms
91: learn: 0.1662445 total: 29.6ms remaining: 2.57ms
92: learn: 0.1646721 total: 30.2ms remaining: 2.27ms
93: learn: 0.1637357 total: 30.7ms remaining: 1.96ms
94: learn: 0.1622838 total: 31ms remaining: 1.63ms
95: learn: 0.1612547 total: 31.3ms remaining: 1.3ms
96: learn: 0.1604476 total: 31.7ms remaining: 978us
97: learn: 0.1595644 total: 32ms remaining: 654us
98: learn: 0.1585538 total: 32.3ms remaining: 326us
99: learn: 0.1579433 total: 32.6ms remaining: 0us
0: learn: 0.6501714 total: 324us remaining: 32.1ms
1: learn: 0.6066480 total: 566us remaining: 27.8ms
2: learn: 0.5657020 total: 796us remaining: 25.8ms
3: learn: 0.5314888 total: 1.46ms remaining: 34.9ms
4: learn: 0.5033352 total: 2.07ms remaining: 39.3ms
5: learn: 0.4746589 total: 2.65ms remaining: 41.5ms
6: learn: 0.4545285 total: 3.05ms remaining: 40.5ms
7: learn: 0.4381047 total: 3.53ms remaining: 40.6ms
8: learn: 0.4177237 total: 3.77ms remaining: 38.1ms
9: learn: 0.4023522 total: 4.11ms remaining: 37ms
10: learn: 0.3914068 total: 4.45ms remaining: 36ms
11: learn: 0.3787263 total: 5.13ms remaining: 37.6ms
12: learn: 0.3650055 total: 5.62ms remaining: 37.6ms
13: learn: 0.3522909 total: 6.26ms remaining: 38.4ms
14: learn: 0.3408388 total: 6.51ms remaining: 36.9ms
15: learn: 0.3301578 total: 7.14ms remaining: 37.5ms
16: learn: 0.3207327 total: 7.57ms remaining: 37ms
17: learn: 0.3122013 total: 8.03ms remaining: 36.6ms
18: learn: 0.3065739 total: 8.32ms remaining: 35.5ms
19: learn: 0.2986789 total: 9.07ms remaining: 36.3ms
20: learn: 0.2920784 total: 9.66ms remaining: 36.3ms
21: learn: 0.2861408 total: 10.3ms remaining: 36.4ms
22: learn: 0.2806232 total: 10.8ms remaining: 36.3ms
23: learn: 0.2757454 total: 11.1ms remaining: 35.2ms
24: learn: 0.2715914 total: 11.5ms remaining: 34.5ms
25: learn: 0.2662000 total: 11.7ms remaining: 33.3ms
26: learn: 0.2620222 total: 12ms remaining: 32.5ms
27: learn: 0.2586619 total: 12.6ms remaining: 32.4ms
28: learn: 0.2559411 total: 13.4ms remaining: 32.8ms
29: learn: 0.2521874 total: 13.6ms remaining: 31.7ms
30: learn: 0.2487332 total: 13.9ms remaining: 31ms
31: learn: 0.2455035 total: 14.8ms remaining: 31.5ms
32: learn: 0.2428433 total: 15.5ms remaining: 31.5ms
33: learn: 0.2410762 total: 15.9ms remaining: 30.8ms
34: learn: 0.2382084 total: 16.7ms remaining: 30.9ms
35: learn: 0.2366788 total: 16.9ms remaining: 30.1ms
36: learn: 0.2347484 total: 17.7ms remaining: 30.2ms
37: learn: 0.2324811 total: 18.3ms remaining: 29.9ms
38: learn: 0.2302696 total: 18.9ms remaining: 29.6ms
39: learn: 0.2282705 total: 19.6ms remaining: 29.5ms
40: learn: 0.2261110 total: 19.9ms remaining: 28.6ms
41: learn: 0.2238719 total: 20.1ms remaining: 27.7ms
42: learn: 0.2215777 total: 20.4ms remaining: 27ms
43: learn: 0.2196766 total: 21.2ms remaining: 26.9ms
44: learn: 0.2185631 total: 22.1ms remaining: 27ms
45: learn: 0.2166483 total: 22.5ms remaining: 26.4ms
46: learn: 0.2139387 total: 22.8ms remaining: 25.7ms
47: learn: 0.2129512 total: 23ms remaining: 24.9ms
48: learn: 0.2119949 total: 23.5ms remaining: 24.5ms
49: learn: 0.2109265 total: 23.8ms remaining: 23.8ms
50: learn: 0.2093739 total: 24.7ms remaining: 23.7ms
51: learn: 0.2079054 total: 25ms remaining: 23.1ms
52: learn: 0.2064375 total: 25.3ms remaining: 22.5ms
53: learn: 0.2055025 total: 25.9ms remaining: 22.1ms
54: learn: 0.2040856 total: 26.6ms remaining: 21.7ms
55: learn: 0.2028301 total: 27ms remaining: 21.2ms
56: learn: 0.2014687 total: 27.5ms remaining: 20.8ms
57: learn: 0.1993156 total: 28.1ms remaining: 20.3ms
58: learn: 0.1972837 total: 28.9ms remaining: 20.1ms
59: learn: 0.1959902 total: 29.6ms remaining: 19.8ms
60: learn: 0.1952245 total: 30.3ms remaining: 19.4ms
61: learn: 0.1934942 total: 30.5ms remaining: 18.7ms
62: learn: 0.1919841 total: 30.7ms remaining: 18ms
63: learn: 0.1905876 total: 31.1ms remaining: 17.5ms
64: learn: 0.1894580 total: 31.5ms remaining: 17ms
65: learn: 0.1874826 total: 32.1ms remaining: 16.5ms
66: learn: 0.1863395 total: 32.7ms remaining: 16.1ms
67: learn: 0.1849459 total: 32.9ms remaining: 15.5ms
68: learn: 0.1838012 total: 33.2ms remaining: 14.9ms
69: learn: 0.1827711 total: 33.6ms remaining: 14.4ms
70: learn: 0.1814848 total: 33.9ms remaining: 13.8ms
71: learn: 0.1800869 total: 34.4ms remaining: 13.4ms
72: learn: 0.1783025 total: 35.2ms remaining: 13ms
73: learn: 0.1772895 total: 35.8ms remaining: 12.6ms
74: learn: 0.1763496 total: 36.9ms remaining: 12.3ms
75: learn: 0.1754821 total: 37.5ms remaining: 11.8ms
76: learn: 0.1746590 total: 38.2ms remaining: 11.4ms
77: learn: 0.1740616 total: 38.9ms remaining: 11ms
78: learn: 0.1731370 total: 39.9ms remaining: 10.6ms
79: learn: 0.1724127 total: 40.8ms remaining: 10.2ms
80: learn: 0.1714950 total: 41.5ms remaining: 9.74ms
81: learn: 0.1706543 total: 41.9ms remaining: 9.2ms
82: learn: 0.1689667 total: 42.2ms remaining: 8.64ms
83: learn: 0.1679040 total: 42.5ms remaining: 8.09ms
84: learn: 0.1671496 total: 43.2ms remaining: 7.63ms
85: learn: 0.1661126 total: 43.8ms remaining: 7.13ms
86: learn: 0.1647861 total: 44.4ms remaining: 6.64ms
87: learn: 0.1638029 total: 44.7ms remaining: 6.1ms
88: learn: 0.1630324 total: 44.9ms remaining: 5.55ms
89: learn: 0.1622552 total: 45.3ms remaining: 5.03ms
90: learn: 0.1614442 total: 45.6ms remaining: 4.51ms
91: learn: 0.1607039 total: 46ms remaining: 4ms
92: learn: 0.1600161 total: 46.6ms remaining: 3.51ms
93: learn: 0.1588467 total: 46.9ms remaining: 2.99ms
94: learn: 0.1581478 total: 47.1ms remaining: 2.48ms
95: learn: 0.1577160 total: 47.3ms remaining: 1.97ms
96: learn: 0.1572810 total: 47.6ms remaining: 1.47ms
97: learn: 0.1568207 total: 47.9ms remaining: 978us
98: learn: 0.1559880 total: 48.2ms remaining: 487us
99: learn: 0.1552295 total: 48.7ms remaining: 0us
0: learn: 0.6481110 total: 563us remaining: 55.8ms
1: learn: 0.6015437 total: 1.01ms remaining: 49.7ms
2: learn: 0.5637141 total: 1.21ms remaining: 39ms
3: learn: 0.5285975 total: 1.6ms remaining: 38.5ms
4: learn: 0.5021036 total: 1.92ms remaining: 36.5ms
5: learn: 0.4762132 total: 2.4ms remaining: 37.7ms
6: learn: 0.4571360 total: 2.84ms remaining: 37.7ms
7: learn: 0.4392327 total: 3.5ms remaining: 40.3ms
8: learn: 0.4235303 total: 3.7ms remaining: 37.5ms
9: learn: 0.4067796 total: 4.4ms remaining: 39.6ms
10: learn: 0.3956489 total: 4.61ms remaining: 37.3ms
11: learn: 0.3834309 total: 4.96ms remaining: 36.4ms
12: learn: 0.3706993 total: 5.62ms remaining: 37.6ms
13: learn: 0.3593553 total: 6.19ms remaining: 38ms
14: learn: 0.3495868 total: 6.64ms remaining: 37.7ms
15: learn: 0.3380214 total: 7.18ms remaining: 37.7ms
16: learn: 0.3303325 total: 7.7ms remaining: 37.6ms
17: learn: 0.3239962 total: 8.24ms remaining: 37.5ms
18: learn: 0.3173096 total: 8.77ms remaining: 37.4ms
19: learn: 0.3130535 total: 9.14ms remaining: 36.6ms
20: learn: 0.3053856 total: 9.45ms remaining: 35.5ms
21: learn: 0.2967920 total: 9.67ms remaining: 34.3ms
22: learn: 0.2911773 total: 9.87ms remaining: 33.1ms
23: learn: 0.2846182 total: 10.2ms remaining: 32.4ms
24: learn: 0.2808455 total: 10.5ms remaining: 31.6ms
25: learn: 0.2780712 total: 10.9ms remaining: 31ms
26: learn: 0.2744681 total: 11.5ms remaining: 31.1ms
27: learn: 0.2696076 total: 11.8ms remaining: 30.3ms
28: learn: 0.2644929 total: 12.4ms remaining: 30.3ms
29: learn: 0.2616771 total: 12.7ms remaining: 29.5ms
30: learn: 0.2582177 total: 12.9ms remaining: 28.8ms
31: learn: 0.2542861 total: 13.4ms remaining: 28.4ms
32: learn: 0.2512674 total: 14ms remaining: 28.5ms
33: learn: 0.2476332 total: 14.5ms remaining: 28.1ms
34: learn: 0.2439946 total: 15ms remaining: 27.8ms
35: learn: 0.2408321 total: 15.6ms remaining: 27.8ms
36: learn: 0.2375090 total: 16ms remaining: 27.3ms
37: learn: 0.2354509 total: 16.2ms remaining: 26.5ms
38: learn: 0.2341470 total: 16.5ms remaining: 25.8ms
39: learn: 0.2321929 total: 16.7ms remaining: 25ms
40: learn: 0.2300608 total: 17ms remaining: 24.4ms
41: learn: 0.2276641 total: 17.4ms remaining: 24.1ms
42: learn: 0.2262292 total: 18ms remaining: 23.8ms
43: learn: 0.2238001 total: 18.1ms remaining: 23.1ms
44: learn: 0.2225238 total: 18.4ms remaining: 22.5ms
45: learn: 0.2207839 total: 18.6ms remaining: 21.8ms
46: learn: 0.2192435 total: 18.9ms remaining: 21.3ms
47: learn: 0.2170573 total: 19.1ms remaining: 20.7ms
48: learn: 0.2151633 total: 19.8ms remaining: 20.6ms
49: learn: 0.2135362 total: 20.5ms remaining: 20.5ms
50: learn: 0.2118303 total: 21.4ms remaining: 20.5ms
51: learn: 0.2096032 total: 22.3ms remaining: 20.5ms
52: learn: 0.2073696 total: 22.8ms remaining: 20.2ms
53: learn: 0.2065992 total: 23ms remaining: 19.6ms
54: learn: 0.2047594 total: 23.7ms remaining: 19.4ms
55: learn: 0.2027697 total: 24.3ms remaining: 19.1ms
56: learn: 0.2012123 total: 24.8ms remaining: 18.7ms
57: learn: 0.1988549 total: 25.1ms remaining: 18.2ms
58: learn: 0.1968074 total: 25.4ms remaining: 17.6ms
59: learn: 0.1947502 total: 25.6ms remaining: 17.1ms
60: learn: 0.1923815 total: 25.9ms remaining: 16.6ms
61: learn: 0.1912902 total: 26.3ms remaining: 16.1ms
62: learn: 0.1899180 total: 26.6ms remaining: 15.6ms
63: learn: 0.1874070 total: 27.5ms remaining: 15.5ms
64: learn: 0.1862103 total: 28.1ms remaining: 15.1ms
65: learn: 0.1850574 total: 29.3ms remaining: 15.1ms
66: learn: 0.1831118 total: 29.7ms remaining: 14.6ms
67: learn: 0.1815357 total: 30.5ms remaining: 14.3ms
68: learn: 0.1797526 total: 31ms remaining: 13.9ms
69: learn: 0.1786087 total: 31.6ms remaining: 13.6ms
70: learn: 0.1774149 total: 32.3ms remaining: 13.2ms
71: learn: 0.1767038 total: 33ms remaining: 12.8ms
72: learn: 0.1753482 total: 33.5ms remaining: 12.4ms
73: learn: 0.1738405 total: 33.8ms remaining: 11.9ms
74: learn: 0.1724542 total: 34.6ms remaining: 11.5ms
75: learn: 0.1714198 total: 35.1ms remaining: 11.1ms
76: learn: 0.1700085 total: 35.4ms remaining: 10.6ms
77: learn: 0.1694224 total: 35.6ms remaining: 10ms
78: learn: 0.1681729 total: 35.9ms remaining: 9.54ms
79: learn: 0.1669886 total: 36.1ms remaining: 9.03ms
80: learn: 0.1654271 total: 36.3ms remaining: 8.52ms
81: learn: 0.1642109 total: 36.6ms remaining: 8.03ms
82: learn: 0.1626449 total: 36.8ms remaining: 7.54ms
83: learn: 0.1614115 total: 37.3ms remaining: 7.1ms
84: learn: 0.1596543 total: 37.6ms remaining: 6.63ms
85: learn: 0.1590043 total: 37.8ms remaining: 6.16ms
86: learn: 0.1582783 total: 38.1ms remaining: 5.7ms
87: learn: 0.1568860 total: 38.3ms remaining: 5.23ms
88: learn: 0.1559547 total: 38.6ms remaining: 4.78ms
89: learn: 0.1552091 total: 39.1ms remaining: 4.34ms
90: learn: 0.1544308 total: 39.4ms remaining: 3.89ms
91: learn: 0.1535623 total: 39.7ms remaining: 3.46ms
92: learn: 0.1530276 total: 40ms remaining: 3.01ms
93: learn: 0.1522294 total: 40.3ms remaining: 2.57ms
94: learn: 0.1508219 total: 40.5ms remaining: 2.13ms
95: learn: 0.1501568 total: 40.9ms remaining: 1.7ms
96: learn: 0.1495230 total: 41.3ms remaining: 1.28ms
97: learn: 0.1485514 total: 42.1ms remaining: 859us
98: learn: 0.1474101 total: 43ms remaining: 434us
99: learn: 0.1464165 total: 43.8ms remaining: 0us
0: learn: 0.6515506 total: 389us remaining: 38.5ms
1: learn: 0.6073513 total: 748us remaining: 36.7ms
2: learn: 0.5704869 total: 1.11ms remaining: 35.8ms
3: learn: 0.5398446 total: 1.39ms remaining: 33.3ms
4: learn: 0.5111154 total: 1.61ms remaining: 30.6ms
5: learn: 0.4857031 total: 2.08ms remaining: 32.5ms
6: learn: 0.4649189 total: 2.44ms remaining: 32.5ms
7: learn: 0.4473799 total: 2.67ms remaining: 30.7ms
8: learn: 0.4283679 total: 2.95ms remaining: 29.8ms
9: learn: 0.4129013 total: 3.19ms remaining: 28.7ms
10: learn: 0.3981913 total: 3.39ms remaining: 27.4ms
11: learn: 0.3860850 total: 3.69ms remaining: 27ms
12: learn: 0.3747197 total: 3.94ms remaining: 26.4ms
13: learn: 0.3643286 total: 4.27ms remaining: 26.2ms
14: learn: 0.3560632 total: 4.41ms remaining: 25ms
15: learn: 0.3493369 total: 4.69ms remaining: 24.6ms
16: learn: 0.3405992 total: 4.9ms remaining: 23.9ms
17: learn: 0.3364019 total: 5.1ms remaining: 23.2ms
18: learn: 0.3284740 total: 5.3ms remaining: 22.6ms
19: learn: 0.3207455 total: 5.97ms remaining: 23.9ms
20: learn: 0.3136963 total: 6.42ms remaining: 24.2ms
21: learn: 0.3068343 total: 6.75ms remaining: 23.9ms
22: learn: 0.3015201 total: 7.24ms remaining: 24.2ms
23: learn: 0.2973018 total: 7.6ms remaining: 24.1ms
24: learn: 0.2925741 total: 7.9ms remaining: 23.7ms
25: learn: 0.2863699 total: 8.16ms remaining: 23.2ms
26: learn: 0.2828654 total: 9.03ms remaining: 24.4ms
27: learn: 0.2796819 total: 9.58ms remaining: 24.6ms
28: learn: 0.2773885 total: 9.82ms remaining: 24ms
29: learn: 0.2764725 total: 10.1ms remaining: 23.7ms
30: learn: 0.2727544 total: 10.6ms remaining: 23.7ms
31: learn: 0.2691427 total: 11.4ms remaining: 24.2ms
32: learn: 0.2657733 total: 11.8ms remaining: 24ms
33: learn: 0.2627910 total: 12.8ms remaining: 24.8ms
34: learn: 0.2593242 total: 13.5ms remaining: 25.1ms
35: learn: 0.2568494 total: 14ms remaining: 25ms
36: learn: 0.2522878 total: 15.1ms remaining: 25.8ms
37: learn: 0.2495150 total: 15.7ms remaining: 25.7ms
38: learn: 0.2464842 total: 16.1ms remaining: 25.2ms
39: learn: 0.2445408 total: 16.5ms remaining: 24.7ms
40: learn: 0.2418177 total: 16.7ms remaining: 24.1ms
41: learn: 0.2396222 total: 17ms remaining: 23.5ms
42: learn: 0.2366848 total: 17.4ms remaining: 23.1ms
43: learn: 0.2330100 total: 18ms remaining: 22.9ms
44: learn: 0.2310290 total: 18.3ms remaining: 22.4ms
45: learn: 0.2291962 total: 18.5ms remaining: 21.8ms
46: learn: 0.2279771 total: 18.7ms remaining: 21.1ms
47: learn: 0.2257751 total: 19ms remaining: 20.5ms
48: learn: 0.2228804 total: 19.2ms remaining: 20ms
49: learn: 0.2201211 total: 19.5ms remaining: 19.5ms
50: learn: 0.2173346 total: 19.9ms remaining: 19.1ms
51: learn: 0.2157845 total: 20.2ms remaining: 18.7ms
52: learn: 0.2144211 total: 20.4ms remaining: 18.1ms
53: learn: 0.2125434 total: 20.7ms remaining: 17.6ms
54: learn: 0.2108065 total: 20.9ms remaining: 17.1ms
55: learn: 0.2092356 total: 21.1ms remaining: 16.6ms
56: learn: 0.2081446 total: 21.5ms remaining: 16.2ms
57: learn: 0.2066968 total: 21.8ms remaining: 15.8ms
58: learn: 0.2044598 total: 22.2ms remaining: 15.5ms
59: learn: 0.2029064 total: 22.8ms remaining: 15.2ms
60: learn: 0.2009449 total: 23.6ms remaining: 15.1ms
61: learn: 0.1997688 total: 24.5ms remaining: 15ms
62: learn: 0.1985798 total: 24.7ms remaining: 14.5ms
63: learn: 0.1964098 total: 25.2ms remaining: 14.2ms
64: learn: 0.1953435 total: 25.7ms remaining: 13.9ms
65: learn: 0.1935257 total: 26ms remaining: 13.4ms
66: learn: 0.1919715 total: 26.4ms remaining: 13ms
67: learn: 0.1910014 total: 26.6ms remaining: 12.5ms
68: learn: 0.1894297 total: 27.1ms remaining: 12.2ms
69: learn: 0.1884763 total: 27.8ms remaining: 11.9ms
70: learn: 0.1866306 total: 28.1ms remaining: 11.5ms
71: learn: 0.1849277 total: 28.6ms remaining: 11.1ms
72: learn: 0.1837207 total: 29.3ms remaining: 10.8ms
73: learn: 0.1823749 total: 29.9ms remaining: 10.5ms
74: learn: 0.1811123 total: 30.7ms remaining: 10.2ms
75: learn: 0.1802044 total: 31.2ms remaining: 9.87ms
76: learn: 0.1787988 total: 31.8ms remaining: 9.49ms
77: learn: 0.1780456 total: 32.4ms remaining: 9.15ms
78: learn: 0.1769628 total: 33.2ms remaining: 8.83ms
79: learn: 0.1761383 total: 33.8ms remaining: 8.44ms
80: learn: 0.1746551 total: 34.1ms remaining: 7.99ms
81: learn: 0.1732442 total: 34.3ms remaining: 7.53ms
82: learn: 0.1724555 total: 34.5ms remaining: 7.07ms
83: learn: 0.1714454 total: 34.8ms remaining: 6.63ms
84: learn: 0.1707684 total: 35ms remaining: 6.17ms
85: learn: 0.1697459 total: 35.3ms remaining: 5.74ms
86: learn: 0.1687502 total: 35.8ms remaining: 5.35ms
87: learn: 0.1681338 total: 36.6ms remaining: 5ms
88: learn: 0.1670008 total: 37.3ms remaining: 4.61ms
89: learn: 0.1662372 total: 38.1ms remaining: 4.23ms
90: learn: 0.1653360 total: 38.7ms remaining: 3.82ms
91: learn: 0.1640089 total: 39.2ms remaining: 3.41ms
92: learn: 0.1633292 total: 39.9ms remaining: 3ms
93: learn: 0.1627398 total: 40.2ms remaining: 2.57ms
94: learn: 0.1621147 total: 40.5ms remaining: 2.13ms
95: learn: 0.1616381 total: 40.8ms remaining: 1.7ms
96: learn: 0.1609506 total: 41.3ms remaining: 1.28ms
97: learn: 0.1599289 total: 41.9ms remaining: 854us
98: learn: 0.1591254 total: 42.3ms remaining: 427us
99: learn: 0.1584201 total: 43ms remaining: 0us
0: learn: 0.6455145 total: 321us remaining: 31.8ms
1: learn: 0.5988760 total: 709us remaining: 34.8ms
2: learn: 0.5594107 total: 887us remaining: 28.7ms
3: learn: 0.5244881 total: 1.27ms remaining: 30.6ms
4: learn: 0.4973720 total: 1.59ms remaining: 30.1ms
5: learn: 0.4741560 total: 2.18ms remaining: 34.2ms
6: learn: 0.4507672 total: 2.98ms remaining: 39.6ms
7: learn: 0.4334544 total: 3.99ms remaining: 45.9ms
8: learn: 0.4127818 total: 4.69ms remaining: 47.4ms
9: learn: 0.3964144 total: 5.26ms remaining: 47.4ms
10: learn: 0.3815367 total: 6.07ms remaining: 49.1ms
11: learn: 0.3706824 total: 6.93ms remaining: 50.8ms
12: learn: 0.3603382 total: 7.56ms remaining: 50.6ms
13: learn: 0.3494307 total: 8.09ms remaining: 49.7ms
14: learn: 0.3417150 total: 8.57ms remaining: 48.5ms
15: learn: 0.3334102 total: 9.49ms remaining: 49.8ms
16: learn: 0.3233884 total: 9.82ms remaining: 48ms
17: learn: 0.3176727 total: 10.2ms remaining: 46.6ms
18: learn: 0.3103917 total: 10.8ms remaining: 46.2ms
19: learn: 0.3034642 total: 11.5ms remaining: 46ms
20: learn: 0.2980677 total: 12.3ms remaining: 46.3ms
21: learn: 0.2918482 total: 12.8ms remaining: 45.3ms
22: learn: 0.2854194 total: 13.4ms remaining: 44.8ms
23: learn: 0.2799811 total: 14.1ms remaining: 44.7ms
24: learn: 0.2749249 total: 14.7ms remaining: 44ms
25: learn: 0.2704717 total: 15.4ms remaining: 43.7ms
26: learn: 0.2662482 total: 15.9ms remaining: 42.9ms
27: learn: 0.2633140 total: 16.5ms remaining: 42.3ms
28: learn: 0.2597887 total: 17.3ms remaining: 42.3ms
29: learn: 0.2568451 total: 17.7ms remaining: 41.4ms
30: learn: 0.2544384 total: 18.2ms remaining: 40.5ms
31: learn: 0.2516443 total: 18.8ms remaining: 39.9ms
32: learn: 0.2467225 total: 19.5ms remaining: 39.6ms
33: learn: 0.2444546 total: 20ms remaining: 38.9ms
34: learn: 0.2425635 total: 20.3ms remaining: 37.7ms
35: learn: 0.2394869 total: 20.6ms remaining: 36.6ms
36: learn: 0.2373269 total: 21.1ms remaining: 36ms
37: learn: 0.2343314 total: 21.6ms remaining: 35.2ms
38: learn: 0.2317488 total: 21.8ms remaining: 34.1ms
39: learn: 0.2306135 total: 22.3ms remaining: 33.5ms
40: learn: 0.2289222 total: 23ms remaining: 33.1ms
41: learn: 0.2267190 total: 23.2ms remaining: 32.1ms
42: learn: 0.2254531 total: 23.5ms remaining: 31.2ms
43: learn: 0.2235827 total: 24ms remaining: 30.6ms
44: learn: 0.2210251 total: 25.2ms remaining: 30.8ms
45: learn: 0.2194663 total: 25.9ms remaining: 30.4ms
46: learn: 0.2180275 total: 26.5ms remaining: 29.9ms
47: learn: 0.2169634 total: 27.4ms remaining: 29.6ms
48: learn: 0.2141994 total: 27.8ms remaining: 28.9ms
49: learn: 0.2126256 total: 28.1ms remaining: 28.1ms
50: learn: 0.2115400 total: 28.8ms remaining: 27.7ms
51: learn: 0.2099402 total: 29.4ms remaining: 27.1ms
52: learn: 0.2078684 total: 29.9ms remaining: 26.5ms
53: learn: 0.2068950 total: 30.3ms remaining: 25.8ms
54: learn: 0.2058179 total: 30.5ms remaining: 25ms
55: learn: 0.2040213 total: 30.8ms remaining: 24.2ms
56: learn: 0.2023684 total: 31.3ms remaining: 23.6ms
57: learn: 0.2005247 total: 31.5ms remaining: 22.8ms
58: learn: 0.1995970 total: 32ms remaining: 22.2ms
59: learn: 0.1984756 total: 32.6ms remaining: 21.7ms
60: learn: 0.1970708 total: 33.1ms remaining: 21.1ms
61: learn: 0.1957965 total: 33.8ms remaining: 20.7ms
62: learn: 0.1944455 total: 34.3ms remaining: 20.2ms
63: learn: 0.1929219 total: 34.6ms remaining: 19.4ms
64: learn: 0.1913913 total: 34.9ms remaining: 18.8ms
65: learn: 0.1902021 total: 35.1ms remaining: 18.1ms
66: learn: 0.1889744 total: 35.3ms remaining: 17.4ms
67: learn: 0.1871736 total: 35.6ms remaining: 16.8ms
68: learn: 0.1859543 total: 36.1ms remaining: 16.2ms
69: learn: 0.1845997 total: 36.6ms remaining: 15.7ms
70: learn: 0.1835376 total: 37ms remaining: 15.1ms
71: learn: 0.1825610 total: 37.4ms remaining: 14.5ms
72: learn: 0.1807745 total: 38ms remaining: 14.1ms
73: learn: 0.1800400 total: 38.5ms remaining: 13.5ms
74: learn: 0.1793376 total: 39.3ms remaining: 13.1ms
75: learn: 0.1784109 total: 40.4ms remaining: 12.7ms
76: learn: 0.1773741 total: 40.6ms remaining: 12.1ms
77: learn: 0.1761102 total: 40.9ms remaining: 11.5ms
78: learn: 0.1746922 total: 41.3ms remaining: 11ms
79: learn: 0.1738470 total: 42ms remaining: 10.5ms
80: learn: 0.1730243 total: 42.4ms remaining: 9.95ms
81: learn: 0.1721958 total: 43.1ms remaining: 9.45ms
82: learn: 0.1713148 total: 43.6ms remaining: 8.93ms
83: learn: 0.1707098 total: 44ms remaining: 8.39ms
84: learn: 0.1699495 total: 44.7ms remaining: 7.89ms
85: learn: 0.1691816 total: 45ms remaining: 7.33ms
86: learn: 0.1682771 total: 45.4ms remaining: 6.78ms
87: learn: 0.1674974 total: 45.9ms remaining: 6.26ms
88: learn: 0.1669940 total: 46.4ms remaining: 5.73ms
89: learn: 0.1660191 total: 46.7ms remaining: 5.18ms
90: learn: 0.1654985 total: 47.3ms remaining: 4.68ms
91: learn: 0.1649944 total: 47.8ms remaining: 4.15ms
92: learn: 0.1641069 total: 48.3ms remaining: 3.64ms
93: learn: 0.1630292 total: 48.6ms remaining: 3.1ms
94: learn: 0.1623293 total: 49.2ms remaining: 2.59ms
95: learn: 0.1617627 total: 50.1ms remaining: 2.09ms
96: learn: 0.1609515 total: 50.5ms remaining: 1.56ms
97: learn: 0.1597019 total: 50.9ms remaining: 1.04ms
98: learn: 0.1591249 total: 51.2ms remaining: 517us
99: learn: 0.1585563 total: 51.8ms remaining: 0us
0: learn: 0.6503257 total: 308us remaining: 30.5ms
1: learn: 0.6091405 total: 713us remaining: 35ms
2: learn: 0.5749262 total: 884us remaining: 28.6ms
3: learn: 0.5438978 total: 1.12ms remaining: 26.8ms
4: learn: 0.5155700 total: 1.32ms remaining: 25.2ms
5: learn: 0.4927320 total: 1.55ms remaining: 24.2ms
6: learn: 0.4727667 total: 1.71ms remaining: 22.7ms
7: learn: 0.4557302 total: 1.9ms remaining: 21.9ms
8: learn: 0.4388609 total: 2.12ms remaining: 21.4ms
9: learn: 0.4247644 total: 2.27ms remaining: 20.4ms
10: learn: 0.4144140 total: 2.41ms remaining: 19.5ms
11: learn: 0.4019024 total: 2.66ms remaining: 19.5ms
12: learn: 0.3916899 total: 2.99ms remaining: 20ms
13: learn: 0.3815831 total: 3.37ms remaining: 20.7ms
14: learn: 0.3717996 total: 3.71ms remaining: 21ms
15: learn: 0.3647230 total: 4.05ms remaining: 21.3ms
16: learn: 0.3545912 total: 4.36ms remaining: 21.3ms
17: learn: 0.3502944 total: 4.61ms remaining: 21ms
18: learn: 0.3428250 total: 4.91ms remaining: 20.9ms
19: learn: 0.3374503 total: 5.2ms remaining: 20.8ms
20: learn: 0.3317534 total: 5.56ms remaining: 20.9ms
21: learn: 0.3248470 total: 6ms remaining: 21.3ms
22: learn: 0.3197077 total: 6.29ms remaining: 21.1ms
23: learn: 0.3149752 total: 6.6ms remaining: 20.9ms
24: learn: 0.3115804 total: 7.03ms remaining: 21.1ms
25: learn: 0.3073465 total: 7.44ms remaining: 21.2ms
26: learn: 0.3014933 total: 7.84ms remaining: 21.2ms
27: learn: 0.2982788 total: 8.25ms remaining: 21.2ms
28: learn: 0.2954439 total: 8.71ms remaining: 21.3ms
29: learn: 0.2917875 total: 9.29ms remaining: 21.7ms
30: learn: 0.2900107 total: 9.6ms remaining: 21.4ms
31: learn: 0.2864544 total: 9.92ms remaining: 21.1ms
32: learn: 0.2810382 total: 10.5ms remaining: 21.3ms
33: learn: 0.2787524 total: 10.8ms remaining: 21ms
34: learn: 0.2764095 total: 11.1ms remaining: 20.7ms
35: learn: 0.2748109 total: 11.5ms remaining: 20.5ms
36: learn: 0.2728280 total: 11.9ms remaining: 20.3ms
37: learn: 0.2703365 total: 12.4ms remaining: 20.2ms
38: learn: 0.2683979 total: 12.7ms remaining: 19.9ms
39: learn: 0.2667767 total: 13.1ms remaining: 19.6ms
40: learn: 0.2652805 total: 13.5ms remaining: 19.4ms
41: learn: 0.2621843 total: 13.9ms remaining: 19.1ms
42: learn: 0.2596001 total: 14.4ms remaining: 19ms
43: learn: 0.2571810 total: 14.7ms remaining: 18.8ms
44: learn: 0.2554186 total: 15.2ms remaining: 18.5ms
45: learn: 0.2533093 total: 15.6ms remaining: 18.3ms
46: learn: 0.2511507 total: 16ms remaining: 18ms
47: learn: 0.2495800 total: 16.2ms remaining: 17.6ms
48: learn: 0.2474001 total: 16.6ms remaining: 17.2ms
49: learn: 0.2461070 total: 17ms remaining: 17ms
50: learn: 0.2445824 total: 17.4ms remaining: 16.7ms
51: learn: 0.2411191 total: 17.7ms remaining: 16.4ms
52: learn: 0.2386823 total: 18.1ms remaining: 16.1ms
53: learn: 0.2375267 total: 18.4ms remaining: 15.7ms
54: learn: 0.2359654 total: 18.7ms remaining: 15.3ms
55: learn: 0.2335567 total: 19.1ms remaining: 15ms
56: learn: 0.2323313 total: 19.4ms remaining: 14.6ms
57: learn: 0.2301428 total: 19.7ms remaining: 14.3ms
58: learn: 0.2273682 total: 20ms remaining: 13.9ms
59: learn: 0.2245088 total: 20.4ms remaining: 13.6ms
60: learn: 0.2229013 total: 20.9ms remaining: 13.3ms
61: learn: 0.2218228 total: 21.2ms remaining: 13ms
62: learn: 0.2203296 total: 21.6ms remaining: 12.7ms
63: learn: 0.2190087 total: 22ms remaining: 12.4ms
64: learn: 0.2178656 total: 22.4ms remaining: 12ms
65: learn: 0.2156503 total: 22.8ms remaining: 11.7ms
66: learn: 0.2148563 total: 23ms remaining: 11.3ms
67: learn: 0.2126004 total: 23.4ms remaining: 11ms
68: learn: 0.2111062 total: 23.7ms remaining: 10.6ms
69: learn: 0.2096250 total: 24ms remaining: 10.3ms
70: learn: 0.2083270 total: 24.3ms remaining: 9.93ms
71: learn: 0.2070989 total: 24.8ms remaining: 9.63ms
72: learn: 0.2061435 total: 25.2ms remaining: 9.31ms
73: learn: 0.2045066 total: 25.5ms remaining: 8.95ms
74: learn: 0.2031592 total: 25.8ms remaining: 8.61ms
75: learn: 0.2015893 total: 26.2ms remaining: 8.29ms
76: learn: 0.2006151 total: 26.7ms remaining: 7.98ms
77: learn: 0.1988155 total: 27.1ms remaining: 7.64ms
78: learn: 0.1977362 total: 27.5ms remaining: 7.31ms
79: learn: 0.1967043 total: 27.8ms remaining: 6.96ms
80: learn: 0.1956287 total: 28.3ms remaining: 6.63ms
81: learn: 0.1942310 total: 28.8ms remaining: 6.32ms
82: learn: 0.1928918 total: 29.4ms remaining: 6.02ms
83: learn: 0.1919528 total: 30.1ms remaining: 5.73ms
84: learn: 0.1906315 total: 30.5ms remaining: 5.38ms
85: learn: 0.1895164 total: 30.9ms remaining: 5.03ms
86: learn: 0.1887474 total: 31.3ms remaining: 4.67ms
87: learn: 0.1877110 total: 31.7ms remaining: 4.32ms
88: learn: 0.1866921 total: 32.2ms remaining: 3.98ms
89: learn: 0.1856475 total: 32.7ms remaining: 3.63ms
90: learn: 0.1849670 total: 33ms remaining: 3.26ms
91: learn: 0.1839782 total: 33.4ms remaining: 2.91ms
92: learn: 0.1831375 total: 33.8ms remaining: 2.54ms
93: learn: 0.1818797 total: 34.2ms remaining: 2.18ms
94: learn: 0.1810629 total: 34.6ms remaining: 1.82ms
95: learn: 0.1801821 total: 35ms remaining: 1.46ms
96: learn: 0.1794811 total: 35.3ms remaining: 1.09ms
97: learn: 0.1788221 total: 35.7ms remaining: 728us
98: learn: 0.1770942 total: 36.1ms remaining: 364us
99: learn: 0.1766127 total: 36.5ms remaining: 0us
0: learn: 0.6515750 total: 396us remaining: 39.2ms
1: learn: 0.6134885 total: 661us remaining: 32.4ms
2: learn: 0.5837400 total: 1.28ms remaining: 41.3ms
3: learn: 0.5489154 total: 1.77ms remaining: 42.4ms
4: learn: 0.5169299 total: 2.21ms remaining: 41.9ms
5: learn: 0.4907425 total: 2.46ms remaining: 38.5ms
6: learn: 0.4713171 total: 3.03ms remaining: 40.3ms
7: learn: 0.4534210 total: 3.17ms remaining: 36.5ms
8: learn: 0.4367045 total: 3.66ms remaining: 37ms
9: learn: 0.4205350 total: 3.95ms remaining: 35.6ms
10: learn: 0.4056829 total: 4.23ms remaining: 34.2ms
11: learn: 0.3903177 total: 4.59ms remaining: 33.6ms
12: learn: 0.3766978 total: 4.82ms remaining: 32.3ms
13: learn: 0.3669745 total: 5.12ms remaining: 31.4ms
14: learn: 0.3548570 total: 5.34ms remaining: 30.3ms
15: learn: 0.3456593 total: 5.62ms remaining: 29.5ms
16: learn: 0.3378650 total: 5.98ms remaining: 29.2ms
17: learn: 0.3310731 total: 6.29ms remaining: 28.6ms
18: learn: 0.3233461 total: 6.62ms remaining: 28.2ms
19: learn: 0.3165295 total: 7ms remaining: 28ms
20: learn: 0.3094843 total: 7.32ms remaining: 27.5ms
21: learn: 0.3020178 total: 7.62ms remaining: 27ms
22: learn: 0.2965073 total: 8ms remaining: 26.8ms
23: learn: 0.2896890 total: 8.41ms remaining: 26.6ms
24: learn: 0.2844204 total: 8.79ms remaining: 26.4ms
25: learn: 0.2797083 total: 9.04ms remaining: 25.7ms
26: learn: 0.2751533 total: 9.4ms remaining: 25.4ms
27: learn: 0.2715553 total: 9.78ms remaining: 25.2ms
28: learn: 0.2691510 total: 9.95ms remaining: 24.4ms
29: learn: 0.2671634 total: 10.3ms remaining: 24.1ms
30: learn: 0.2650620 total: 10.5ms remaining: 23.4ms
31: learn: 0.2613732 total: 11ms remaining: 23.3ms
32: learn: 0.2592097 total: 11.3ms remaining: 23ms
33: learn: 0.2555304 total: 11.8ms remaining: 22.9ms
34: learn: 0.2532133 total: 12ms remaining: 22.4ms
35: learn: 0.2505760 total: 12.4ms remaining: 22.1ms
36: learn: 0.2488293 total: 12.8ms remaining: 21.8ms
37: learn: 0.2453197 total: 13ms remaining: 21.2ms
38: learn: 0.2419295 total: 13.4ms remaining: 20.9ms
39: learn: 0.2391228 total: 13.7ms remaining: 20.6ms
40: learn: 0.2363322 total: 13.9ms remaining: 20ms
41: learn: 0.2343441 total: 14.2ms remaining: 19.6ms
42: learn: 0.2325132 total: 14.5ms remaining: 19.2ms
43: learn: 0.2296161 total: 14.8ms remaining: 18.8ms
44: learn: 0.2287773 total: 15ms remaining: 18.3ms
45: learn: 0.2272871 total: 15.3ms remaining: 17.9ms
46: learn: 0.2260889 total: 15.5ms remaining: 17.5ms
47: learn: 0.2244358 total: 16ms remaining: 17.4ms
48: learn: 0.2231578 total: 16.4ms remaining: 17.1ms
49: learn: 0.2215341 total: 16.9ms remaining: 16.9ms
50: learn: 0.2182713 total: 17.2ms remaining: 16.5ms
51: learn: 0.2166147 total: 17.5ms remaining: 16.2ms
52: learn: 0.2155089 total: 17.9ms remaining: 15.9ms
53: learn: 0.2140156 total: 18.3ms remaining: 15.6ms
54: learn: 0.2128961 total: 18.6ms remaining: 15.2ms
55: learn: 0.2114523 total: 18.9ms remaining: 14.8ms
56: learn: 0.2098777 total: 19.2ms remaining: 14.5ms
57: learn: 0.2080078 total: 19.5ms remaining: 14.1ms
58: learn: 0.2066818 total: 19.8ms remaining: 13.8ms
59: learn: 0.2052481 total: 20.1ms remaining: 13.4ms
60: learn: 0.2035334 total: 20.4ms remaining: 13ms
61: learn: 0.2021471 total: 20.7ms remaining: 12.7ms
62: learn: 0.2011057 total: 21ms remaining: 12.3ms
63: learn: 0.1996851 total: 21.3ms remaining: 12ms
64: learn: 0.1979320 total: 21.6ms remaining: 11.6ms
65: learn: 0.1969688 total: 22ms remaining: 11.3ms
66: learn: 0.1956253 total: 22.4ms remaining: 11ms
67: learn: 0.1944589 total: 22.7ms remaining: 10.7ms
68: learn: 0.1929730 total: 23ms remaining: 10.3ms
69: learn: 0.1916246 total: 23.5ms remaining: 10.1ms
70: learn: 0.1904793 total: 23.8ms remaining: 9.74ms
71: learn: 0.1896955 total: 24.2ms remaining: 9.4ms
72: learn: 0.1875003 total: 24.5ms remaining: 9.05ms
73: learn: 0.1860530 total: 24.7ms remaining: 8.69ms
74: learn: 0.1847521 total: 25ms remaining: 8.32ms
75: learn: 0.1836835 total: 25.3ms remaining: 7.99ms
76: learn: 0.1830485 total: 25.8ms remaining: 7.7ms
77: learn: 0.1814416 total: 26.1ms remaining: 7.37ms
78: learn: 0.1804926 total: 26.4ms remaining: 7.01ms
79: learn: 0.1796085 total: 26.6ms remaining: 6.65ms
80: learn: 0.1788568 total: 27ms remaining: 6.32ms
81: learn: 0.1779278 total: 27.5ms remaining: 6.03ms
82: learn: 0.1769908 total: 27.7ms remaining: 5.67ms
83: learn: 0.1763817 total: 27.9ms remaining: 5.32ms
84: learn: 0.1754758 total: 28.1ms remaining: 4.97ms
85: learn: 0.1743761 total: 28.4ms remaining: 4.62ms
86: learn: 0.1730073 total: 28.8ms remaining: 4.31ms
87: learn: 0.1723445 total: 29.1ms remaining: 3.96ms
88: learn: 0.1712899 total: 29.6ms remaining: 3.66ms
89: learn: 0.1708731 total: 30.2ms remaining: 3.35ms
90: learn: 0.1695320 total: 30.7ms remaining: 3.03ms
91: learn: 0.1688723 total: 31.1ms remaining: 2.7ms
92: learn: 0.1683870 total: 31.6ms remaining: 2.38ms
93: learn: 0.1673684 total: 32.1ms remaining: 2.05ms
94: learn: 0.1666907 total: 32.4ms remaining: 1.7ms
95: learn: 0.1659053 total: 32.6ms remaining: 1.36ms
96: learn: 0.1652727 total: 33.2ms remaining: 1.02ms
97: learn: 0.1645047 total: 33.4ms remaining: 682us
98: learn: 0.1640110 total: 33.7ms remaining: 340us
99: learn: 0.1634089 total: 34ms remaining: 0us
0: learn: 0.6495190 total: 280us remaining: 27.7ms
1: learn: 0.6095526 total: 580us remaining: 28.5ms
2: learn: 0.5719992 total: 767us remaining: 24.8ms
3: learn: 0.5425656 total: 955us remaining: 22.9ms
4: learn: 0.5159659 total: 1.14ms remaining: 21.6ms
5: learn: 0.4917545 total: 1.33ms remaining: 20.9ms
6: learn: 0.4701324 total: 1.5ms remaining: 19.9ms
7: learn: 0.4524320 total: 1.66ms remaining: 19.1ms
8: learn: 0.4325252 total: 1.84ms remaining: 18.7ms
9: learn: 0.4165980 total: 2.05ms remaining: 18.5ms
10: learn: 0.4035100 total: 2.23ms remaining: 18ms
11: learn: 0.3907931 total: 2.42ms remaining: 17.7ms
12: learn: 0.3789276 total: 2.61ms remaining: 17.5ms
13: learn: 0.3688594 total: 2.86ms remaining: 17.6ms
14: learn: 0.3595050 total: 3.03ms remaining: 17.2ms
15: learn: 0.3510985 total: 3.24ms remaining: 17ms
16: learn: 0.3435509 total: 3.46ms remaining: 16.9ms
17: learn: 0.3372297 total: 3.61ms remaining: 16.5ms
18: learn: 0.3310716 total: 3.8ms remaining: 16.2ms
19: learn: 0.3258688 total: 4.08ms remaining: 16.3ms
20: learn: 0.3195430 total: 4.33ms remaining: 16.3ms
21: learn: 0.3125519 total: 4.55ms remaining: 16.1ms
22: learn: 0.3074617 total: 4.75ms remaining: 15.9ms
23: learn: 0.3031872 total: 4.87ms remaining: 15.4ms
24: learn: 0.2996652 total: 5.08ms remaining: 15.3ms
25: learn: 0.2955970 total: 5.3ms remaining: 15.1ms
26: learn: 0.2919958 total: 5.52ms remaining: 14.9ms
27: learn: 0.2880784 total: 5.7ms remaining: 14.7ms
28: learn: 0.2850178 total: 5.88ms remaining: 14.4ms
29: learn: 0.2831896 total: 6.03ms remaining: 14.1ms
30: learn: 0.2807488 total: 6.25ms remaining: 13.9ms
31: learn: 0.2781251 total: 6.38ms remaining: 13.6ms
32: learn: 0.2755400 total: 6.79ms remaining: 13.8ms
33: learn: 0.2735858 total: 7.09ms remaining: 13.8ms
34: learn: 0.2709428 total: 7.37ms remaining: 13.7ms
35: learn: 0.2680968 total: 7.59ms remaining: 13.5ms
36: learn: 0.2653391 total: 7.92ms remaining: 13.5ms
37: learn: 0.2622118 total: 8.33ms remaining: 13.6ms
38: learn: 0.2589166 total: 8.68ms remaining: 13.6ms
39: learn: 0.2560415 total: 9.37ms remaining: 14.1ms
40: learn: 0.2545774 total: 9.75ms remaining: 14ms
41: learn: 0.2521408 total: 10.1ms remaining: 14ms
42: learn: 0.2508771 total: 10.4ms remaining: 13.8ms
43: learn: 0.2500696 total: 10.6ms remaining: 13.5ms
44: learn: 0.2485904 total: 11ms remaining: 13.5ms
45: learn: 0.2465004 total: 11.4ms remaining: 13.4ms
46: learn: 0.2446957 total: 11.7ms remaining: 13.2ms
47: learn: 0.2427096 total: 12.1ms remaining: 13.1ms
48: learn: 0.2415083 total: 12.5ms remaining: 13ms
49: learn: 0.2387823 total: 12.9ms remaining: 12.9ms
50: learn: 0.2376992 total: 13.2ms remaining: 12.7ms
51: learn: 0.2345011 total: 13.6ms remaining: 12.6ms
52: learn: 0.2324914 total: 13.9ms remaining: 12.3ms
53: learn: 0.2311739 total: 14.2ms remaining: 12.1ms
54: learn: 0.2288382 total: 14.6ms remaining: 12ms
55: learn: 0.2271510 total: 15ms remaining: 11.8ms
56: learn: 0.2258501 total: 15.3ms remaining: 11.6ms
57: learn: 0.2239054 total: 15.7ms remaining: 11.3ms
58: learn: 0.2216254 total: 16ms remaining: 11.1ms
59: learn: 0.2189058 total: 16.4ms remaining: 10.9ms
60: learn: 0.2167674 total: 16.7ms remaining: 10.7ms
61: learn: 0.2157021 total: 17ms remaining: 10.4ms
62: learn: 0.2141794 total: 17.5ms remaining: 10.3ms
63: learn: 0.2122613 total: 17.9ms remaining: 10.1ms
64: learn: 0.2108818 total: 18.3ms remaining: 9.83ms
65: learn: 0.2095270 total: 18.9ms remaining: 9.71ms
66: learn: 0.2087435 total: 19.2ms remaining: 9.46ms
67: learn: 0.2074101 total: 19.5ms remaining: 9.16ms
68: learn: 0.2057417 total: 19.8ms remaining: 8.88ms
69: learn: 0.2038966 total: 20.1ms remaining: 8.61ms
70: learn: 0.2025303 total: 20.6ms remaining: 8.4ms
71: learn: 0.2015057 total: 20.9ms remaining: 8.12ms
72: learn: 0.2004784 total: 21.3ms remaining: 7.86ms
73: learn: 0.1986380 total: 21.5ms remaining: 7.55ms
74: learn: 0.1967574 total: 21.8ms remaining: 7.28ms
75: learn: 0.1958965 total: 22.2ms remaining: 7.01ms
76: learn: 0.1950152 total: 22.5ms remaining: 6.72ms
77: learn: 0.1942672 total: 22.7ms remaining: 6.42ms
78: learn: 0.1922748 total: 23.1ms remaining: 6.13ms
79: learn: 0.1915469 total: 23.3ms remaining: 5.84ms
80: learn: 0.1905149 total: 23.6ms remaining: 5.54ms
81: learn: 0.1889936 total: 24ms remaining: 5.28ms
82: learn: 0.1884547 total: 24.3ms remaining: 4.98ms
83: learn: 0.1879366 total: 24.6ms remaining: 4.68ms
84: learn: 0.1861980 total: 24.9ms remaining: 4.39ms
85: learn: 0.1854796 total: 25.1ms remaining: 4.09ms
86: learn: 0.1836693 total: 25.4ms remaining: 3.79ms
87: learn: 0.1829421 total: 25.6ms remaining: 3.48ms
88: learn: 0.1814350 total: 25.9ms remaining: 3.2ms
89: learn: 0.1810181 total: 26.2ms remaining: 2.91ms
90: learn: 0.1806212 total: 26.7ms remaining: 2.64ms
91: learn: 0.1795526 total: 26.9ms remaining: 2.34ms
92: learn: 0.1791160 total: 27.2ms remaining: 2.05ms
93: learn: 0.1787314 total: 27.5ms remaining: 1.75ms
94: learn: 0.1782327 total: 27.8ms remaining: 1.46ms
95: learn: 0.1778748 total: 28ms remaining: 1.17ms
96: learn: 0.1769739 total: 28.2ms remaining: 871us
97: learn: 0.1765155 total: 28.4ms remaining: 579us
98: learn: 0.1761686 total: 28.8ms remaining: 291us
99: learn: 0.1757469 total: 29ms remaining: 0us
0: learn: 0.6487726 total: 270us remaining: 26.8ms
1: learn: 0.6054752 total: 465us remaining: 22.8ms
2: learn: 0.5690516 total: 637us remaining: 20.6ms
3: learn: 0.5376892 total: 832us remaining: 20ms
4: learn: 0.5107400 total: 1.01ms remaining: 19.2ms
5: learn: 0.4860125 total: 1.26ms remaining: 19.8ms
6: learn: 0.4650487 total: 1.49ms remaining: 19.8ms
7: learn: 0.4474529 total: 1.73ms remaining: 19.9ms
8: learn: 0.4286769 total: 1.91ms remaining: 19.3ms
9: learn: 0.4136039 total: 2.12ms remaining: 19.1ms
10: learn: 0.4019389 total: 2.27ms remaining: 18.4ms
11: learn: 0.3894000 total: 2.49ms remaining: 18.2ms
12: learn: 0.3778865 total: 2.73ms remaining: 18.3ms
13: learn: 0.3672801 total: 2.94ms remaining: 18ms
14: learn: 0.3576247 total: 3.11ms remaining: 17.6ms
15: learn: 0.3495637 total: 3.36ms remaining: 17.7ms
16: learn: 0.3412530 total: 3.63ms remaining: 17.7ms
17: learn: 0.3350939 total: 3.83ms remaining: 17.4ms
18: learn: 0.3290651 total: 4.01ms remaining: 17.1ms
19: learn: 0.3248385 total: 4.25ms remaining: 17ms
20: learn: 0.3197584 total: 4.49ms remaining: 16.9ms
21: learn: 0.3123746 total: 4.76ms remaining: 16.9ms
22: learn: 0.3067161 total: 5.01ms remaining: 16.8ms
23: learn: 0.3018604 total: 5.27ms remaining: 16.7ms
24: learn: 0.2972128 total: 5.52ms remaining: 16.6ms
25: learn: 0.2947712 total: 5.87ms remaining: 16.7ms
26: learn: 0.2912988 total: 6.16ms remaining: 16.7ms
27: learn: 0.2870765 total: 6.54ms remaining: 16.8ms
28: learn: 0.2831592 total: 6.88ms remaining: 16.8ms
29: learn: 0.2813999 total: 7.12ms remaining: 16.6ms
30: learn: 0.2782774 total: 7.42ms remaining: 16.5ms
31: learn: 0.2757420 total: 7.59ms remaining: 16.1ms
32: learn: 0.2733458 total: 7.87ms remaining: 16ms
33: learn: 0.2714703 total: 8.03ms remaining: 15.6ms
34: learn: 0.2692580 total: 8.18ms remaining: 15.2ms
35: learn: 0.2669193 total: 8.65ms remaining: 15.4ms
36: learn: 0.2654010 total: 8.99ms remaining: 15.3ms
37: learn: 0.2626552 total: 9.32ms remaining: 15.2ms
38: learn: 0.2591544 total: 9.65ms remaining: 15.1ms
39: learn: 0.2568921 total: 10ms remaining: 15ms
40: learn: 0.2539529 total: 10.3ms remaining: 14.8ms
41: learn: 0.2519482 total: 10.5ms remaining: 14.5ms
42: learn: 0.2496161 total: 10.8ms remaining: 14.3ms
43: learn: 0.2481991 total: 11.1ms remaining: 14.2ms
44: learn: 0.2469000 total: 11.5ms remaining: 14ms
45: learn: 0.2455710 total: 11.8ms remaining: 13.8ms
46: learn: 0.2415597 total: 12.1ms remaining: 13.7ms
47: learn: 0.2399522 total: 12.4ms remaining: 13.5ms
48: learn: 0.2386716 total: 12.7ms remaining: 13.2ms
49: learn: 0.2371674 total: 12.9ms remaining: 12.9ms
50: learn: 0.2356024 total: 13.2ms remaining: 12.7ms
51: learn: 0.2339625 total: 13.5ms remaining: 12.4ms
52: learn: 0.2329458 total: 13.7ms remaining: 12.2ms
53: learn: 0.2315437 total: 14.1ms remaining: 12ms
54: learn: 0.2301420 total: 14.3ms remaining: 11.7ms
55: learn: 0.2274817 total: 14.5ms remaining: 11.4ms
56: learn: 0.2258392 total: 14.8ms remaining: 11.2ms
57: learn: 0.2239846 total: 15.1ms remaining: 10.9ms
58: learn: 0.2215748 total: 15.3ms remaining: 10.6ms
59: learn: 0.2188303 total: 15.7ms remaining: 10.5ms
60: learn: 0.2169148 total: 16ms remaining: 10.2ms
61: learn: 0.2145196 total: 16.4ms remaining: 10ms
62: learn: 0.2131793 total: 16.8ms remaining: 9.85ms
63: learn: 0.2115461 total: 17.2ms remaining: 9.69ms
64: learn: 0.2092351 total: 17.6ms remaining: 9.48ms
65: learn: 0.2070521 total: 18ms remaining: 9.26ms
66: learn: 0.2059674 total: 18.2ms remaining: 8.98ms
67: learn: 0.2041380 total: 18.5ms remaining: 8.69ms
68: learn: 0.2031281 total: 18.8ms remaining: 8.44ms
69: learn: 0.2018882 total: 19.2ms remaining: 8.21ms
70: learn: 0.1998817 total: 19.4ms remaining: 7.91ms
71: learn: 0.1988290 total: 19.8ms remaining: 7.71ms
72: learn: 0.1978987 total: 20.1ms remaining: 7.44ms
73: learn: 0.1969798 total: 20.5ms remaining: 7.2ms
74: learn: 0.1951727 total: 20.9ms remaining: 6.96ms
75: learn: 0.1942894 total: 21.1ms remaining: 6.67ms
76: learn: 0.1931971 total: 21.4ms remaining: 6.4ms
77: learn: 0.1918851 total: 21.8ms remaining: 6.14ms
78: learn: 0.1910988 total: 22.1ms remaining: 5.89ms
79: learn: 0.1901434 total: 22.4ms remaining: 5.61ms
80: learn: 0.1893449 total: 22.9ms remaining: 5.37ms
81: learn: 0.1884621 total: 23.4ms remaining: 5.13ms
82: learn: 0.1876637 total: 23.7ms remaining: 4.86ms
83: learn: 0.1863474 total: 24.1ms remaining: 4.58ms
84: learn: 0.1854177 total: 24.3ms remaining: 4.28ms
85: learn: 0.1848509 total: 24.7ms remaining: 4.01ms
86: learn: 0.1830131 total: 25ms remaining: 3.73ms
87: learn: 0.1814085 total: 25.3ms remaining: 3.45ms
88: learn: 0.1803102 total: 25.8ms remaining: 3.19ms
89: learn: 0.1791514 total: 26.2ms remaining: 2.92ms
90: learn: 0.1778142 total: 26.5ms remaining: 2.62ms
91: learn: 0.1772303 total: 26.9ms remaining: 2.34ms
92: learn: 0.1762585 total: 27.3ms remaining: 2.06ms
93: learn: 0.1758778 total: 27.6ms remaining: 1.76ms
94: learn: 0.1750961 total: 28ms remaining: 1.47ms
95: learn: 0.1743590 total: 28.3ms remaining: 1.18ms
96: learn: 0.1735165 total: 28.6ms remaining: 884us
97: learn: 0.1729827 total: 29.1ms remaining: 593us
98: learn: 0.1722555 total: 29.3ms remaining: 295us
99: learn: 0.1718057 total: 29.5ms remaining: 0us
0: learn: 0.6470146 total: 346us remaining: 34.3ms
1: learn: 0.6064299 total: 660us remaining: 32.4ms
2: learn: 0.5691283 total: 848us remaining: 27.4ms
3: learn: 0.5366569 total: 1.07ms remaining: 25.8ms
4: learn: 0.5098000 total: 1.27ms remaining: 24.2ms
5: learn: 0.4832216 total: 1.46ms remaining: 22.9ms
6: learn: 0.4596274 total: 1.65ms remaining: 21.9ms
7: learn: 0.4437963 total: 1.83ms remaining: 21.1ms
8: learn: 0.4236547 total: 2ms remaining: 20.2ms
9: learn: 0.4091465 total: 2.19ms remaining: 19.7ms
10: learn: 0.3961872 total: 2.35ms remaining: 19ms
11: learn: 0.3834046 total: 2.61ms remaining: 19.1ms
12: learn: 0.3710681 total: 2.93ms remaining: 19.6ms
13: learn: 0.3595058 total: 3.27ms remaining: 20.1ms
14: learn: 0.3509086 total: 3.46ms remaining: 19.6ms
15: learn: 0.3450153 total: 3.66ms remaining: 19.2ms
16: learn: 0.3371444 total: 3.86ms remaining: 18.8ms
17: learn: 0.3298510 total: 4.09ms remaining: 18.6ms
18: learn: 0.3232019 total: 4.24ms remaining: 18.1ms
19: learn: 0.3162856 total: 4.49ms remaining: 18ms
20: learn: 0.3111943 total: 4.84ms remaining: 18.2ms
21: learn: 0.3057793 total: 5.07ms remaining: 18ms
22: learn: 0.3007321 total: 5.32ms remaining: 17.8ms
23: learn: 0.2962390 total: 5.49ms remaining: 17.4ms
24: learn: 0.2927857 total: 5.66ms remaining: 17ms
25: learn: 0.2900541 total: 5.94ms remaining: 16.9ms
26: learn: 0.2865934 total: 6.14ms remaining: 16.6ms
27: learn: 0.2819302 total: 6.38ms remaining: 16.4ms
28: learn: 0.2778663 total: 6.54ms remaining: 16ms
29: learn: 0.2737055 total: 6.76ms remaining: 15.8ms
30: learn: 0.2722615 total: 6.91ms remaining: 15.4ms
31: learn: 0.2689709 total: 7.21ms remaining: 15.3ms
32: learn: 0.2664546 total: 7.41ms remaining: 15ms
33: learn: 0.2629107 total: 7.7ms remaining: 14.9ms
34: learn: 0.2603216 total: 7.93ms remaining: 14.7ms
35: learn: 0.2574981 total: 8.19ms remaining: 14.6ms
36: learn: 0.2557634 total: 8.34ms remaining: 14.2ms
37: learn: 0.2525677 total: 8.55ms remaining: 13.9ms
38: learn: 0.2487591 total: 8.86ms remaining: 13.8ms
39: learn: 0.2454105 total: 9.05ms remaining: 13.6ms
40: learn: 0.2435551 total: 9.65ms remaining: 13.9ms
41: learn: 0.2401939 total: 10.1ms remaining: 13.9ms
42: learn: 0.2387428 total: 10.2ms remaining: 13.6ms
43: learn: 0.2376608 total: 10.5ms remaining: 13.4ms
44: learn: 0.2358510 total: 10.8ms remaining: 13.2ms
45: learn: 0.2332892 total: 11ms remaining: 13ms
46: learn: 0.2316049 total: 11.2ms remaining: 12.6ms
47: learn: 0.2293167 total: 11.5ms remaining: 12.4ms
48: learn: 0.2264321 total: 11.7ms remaining: 12.2ms
49: learn: 0.2240784 total: 12.2ms remaining: 12.2ms
50: learn: 0.2226746 total: 13ms remaining: 12.4ms
51: learn: 0.2192451 total: 13.7ms remaining: 12.7ms
52: learn: 0.2177215 total: 14.3ms remaining: 12.6ms
53: learn: 0.2143575 total: 14.9ms remaining: 12.7ms
54: learn: 0.2118161 total: 16.1ms remaining: 13.2ms
55: learn: 0.2094860 total: 17.1ms remaining: 13.4ms
56: learn: 0.2079612 total: 17.9ms remaining: 13.5ms
57: learn: 0.2048245 total: 18.9ms remaining: 13.7ms
58: learn: 0.2022788 total: 19.8ms remaining: 13.8ms
59: learn: 0.2009432 total: 20.5ms remaining: 13.7ms
60: learn: 0.1997459 total: 21.1ms remaining: 13.5ms
61: learn: 0.1973434 total: 22ms remaining: 13.5ms
62: learn: 0.1940305 total: 22.7ms remaining: 13.3ms
63: learn: 0.1931598 total: 23.4ms remaining: 13.1ms
64: learn: 0.1905972 total: 24.2ms remaining: 13ms
65: learn: 0.1892452 total: 25.1ms remaining: 12.9ms
66: learn: 0.1876009 total: 25.9ms remaining: 12.7ms
67: learn: 0.1854832 total: 26.8ms remaining: 12.6ms
68: learn: 0.1834037 total: 27.5ms remaining: 12.4ms
69: learn: 0.1822009 total: 28ms remaining: 12ms
70: learn: 0.1805010 total: 28.6ms remaining: 11.7ms
71: learn: 0.1791419 total: 29.2ms remaining: 11.4ms
72: learn: 0.1780161 total: 29.7ms remaining: 11ms
73: learn: 0.1769993 total: 30.3ms remaining: 10.6ms
74: learn: 0.1761321 total: 31.1ms remaining: 10.4ms
75: learn: 0.1741401 total: 31.8ms remaining: 10ms
76: learn: 0.1727571 total: 32.3ms remaining: 9.66ms
77: learn: 0.1715988 total: 32.9ms remaining: 9.27ms
78: learn: 0.1704649 total: 33.3ms remaining: 8.85ms
79: learn: 0.1697879 total: 33.9ms remaining: 8.47ms
80: learn: 0.1689765 total: 34.6ms remaining: 8.11ms
81: learn: 0.1684358 total: 35.1ms remaining: 7.69ms
82: learn: 0.1668083 total: 35.6ms remaining: 7.29ms
83: learn: 0.1651321 total: 36ms remaining: 6.85ms
84: learn: 0.1639347 total: 36.5ms remaining: 6.43ms
85: learn: 0.1623101 total: 36.8ms remaining: 5.99ms
86: learn: 0.1606518 total: 37.2ms remaining: 5.55ms
87: learn: 0.1598402 total: 37.6ms remaining: 5.12ms
88: learn: 0.1591425 total: 37.8ms remaining: 4.67ms
89: learn: 0.1579675 total: 38ms remaining: 4.22ms
90: learn: 0.1574750 total: 38.3ms remaining: 3.79ms
91: learn: 0.1561653 total: 38.5ms remaining: 3.35ms
92: learn: 0.1556096 total: 38.8ms remaining: 2.92ms
93: learn: 0.1549666 total: 39.5ms remaining: 2.52ms
94: learn: 0.1541801 total: 40.1ms remaining: 2.11ms
95: learn: 0.1532460 total: 40.6ms remaining: 1.69ms
96: learn: 0.1518631 total: 41ms remaining: 1.27ms
97: learn: 0.1506036 total: 41.3ms remaining: 843us
98: learn: 0.1491278 total: 41.8ms remaining: 422us
99: learn: 0.1482352 total: 42.1ms remaining: 0us
0: learn: 0.6531507 total: 248us remaining: 24.6ms
1: learn: 0.6065162 total: 482us remaining: 23.6ms
2: learn: 0.5670336 total: 636us remaining: 20.6ms
3: learn: 0.5358549 total: 946us remaining: 22.7ms
4: learn: 0.5074412 total: 1.14ms remaining: 21.6ms
5: learn: 0.4804679 total: 1.36ms remaining: 21.3ms
6: learn: 0.4657784 total: 1.61ms remaining: 21.5ms
7: learn: 0.4441830 total: 1.93ms remaining: 22.2ms
8: learn: 0.4246293 total: 2.13ms remaining: 21.6ms
9: learn: 0.4082806 total: 2.41ms remaining: 21.7ms
10: learn: 0.3971826 total: 2.71ms remaining: 22ms
11: learn: 0.3859011 total: 3.03ms remaining: 22.3ms
12: learn: 0.3732273 total: 3.31ms remaining: 22.2ms
13: learn: 0.3599254 total: 3.6ms remaining: 22.1ms
14: learn: 0.3520452 total: 3.77ms remaining: 21.3ms
15: learn: 0.3434055 total: 4.05ms remaining: 21.2ms
16: learn: 0.3346383 total: 4.36ms remaining: 21.3ms
17: learn: 0.3283062 total: 4.69ms remaining: 21.4ms
18: learn: 0.3212423 total: 4.93ms remaining: 21ms
19: learn: 0.3155269 total: 5.13ms remaining: 20.5ms
20: learn: 0.3091024 total: 5.48ms remaining: 20.6ms
21: learn: 0.3038824 total: 5.76ms remaining: 20.4ms
22: learn: 0.2960116 total: 6.05ms remaining: 20.3ms
23: learn: 0.2915019 total: 6.4ms remaining: 20.3ms
24: learn: 0.2851576 total: 6.64ms remaining: 19.9ms
25: learn: 0.2824845 total: 6.88ms remaining: 19.6ms
26: learn: 0.2779683 total: 7.27ms remaining: 19.7ms
27: learn: 0.2739306 total: 7.58ms remaining: 19.5ms
28: learn: 0.2711215 total: 7.9ms remaining: 19.3ms
29: learn: 0.2672378 total: 8.25ms remaining: 19.3ms
30: learn: 0.2642976 total: 8.58ms remaining: 19.1ms
31: learn: 0.2610812 total: 8.89ms remaining: 18.9ms
32: learn: 0.2570838 total: 9.21ms remaining: 18.7ms
33: learn: 0.2537967 total: 9.6ms remaining: 18.6ms
34: learn: 0.2509818 total: 9.89ms remaining: 18.4ms
35: learn: 0.2490697 total: 10.3ms remaining: 18.3ms
36: learn: 0.2463164 total: 10.5ms remaining: 17.9ms
37: learn: 0.2441294 total: 10.9ms remaining: 17.7ms
38: learn: 0.2423599 total: 11.2ms remaining: 17.6ms
39: learn: 0.2392679 total: 11.5ms remaining: 17.3ms
40: learn: 0.2373584 total: 11.9ms remaining: 17.2ms
41: learn: 0.2351607 total: 12.2ms remaining: 16.9ms
42: learn: 0.2329397 total: 12.5ms remaining: 16.6ms
43: learn: 0.2311672 total: 12.9ms remaining: 16.4ms
44: learn: 0.2291315 total: 13.3ms remaining: 16.2ms
45: learn: 0.2273520 total: 13.6ms remaining: 16ms
46: learn: 0.2248574 total: 13.9ms remaining: 15.7ms
47: learn: 0.2237799 total: 14.2ms remaining: 15.4ms
48: learn: 0.2229512 total: 14.5ms remaining: 15.1ms
49: learn: 0.2207943 total: 14.8ms remaining: 14.8ms
50: learn: 0.2191466 total: 14.9ms remaining: 14.3ms
51: learn: 0.2167604 total: 15.2ms remaining: 14.1ms
52: learn: 0.2150212 total: 15.6ms remaining: 13.8ms
53: learn: 0.2143156 total: 15.8ms remaining: 13.4ms
54: learn: 0.2129308 total: 16.1ms remaining: 13.2ms
55: learn: 0.2115729 total: 16.4ms remaining: 12.9ms
56: learn: 0.2096498 total: 16.8ms remaining: 12.7ms
57: learn: 0.2069896 total: 17.1ms remaining: 12.4ms
58: learn: 0.2057671 total: 17.4ms remaining: 12.1ms
59: learn: 0.2044193 total: 17.8ms remaining: 11.9ms
60: learn: 0.2030844 total: 18.2ms remaining: 11.6ms
61: learn: 0.2013383 total: 18.6ms remaining: 11.4ms
62: learn: 0.1998751 total: 19ms remaining: 11.1ms
63: learn: 0.1980976 total: 19.3ms remaining: 10.8ms
64: learn: 0.1971064 total: 19.6ms remaining: 10.6ms
65: learn: 0.1953909 total: 19.9ms remaining: 10.3ms
66: learn: 0.1939258 total: 20.2ms remaining: 9.96ms
67: learn: 0.1927908 total: 20.5ms remaining: 9.65ms
68: learn: 0.1908997 total: 20.8ms remaining: 9.35ms
69: learn: 0.1897857 total: 21.2ms remaining: 9.07ms
70: learn: 0.1885897 total: 21.5ms remaining: 8.78ms
71: learn: 0.1875013 total: 22ms remaining: 8.54ms
72: learn: 0.1861197 total: 22.4ms remaining: 8.27ms
73: learn: 0.1851799 total: 22.7ms remaining: 7.98ms
74: learn: 0.1844384 total: 23ms remaining: 7.66ms
75: learn: 0.1833483 total: 23.4ms remaining: 7.38ms
76: learn: 0.1823807 total: 23.7ms remaining: 7.09ms
77: learn: 0.1813433 total: 24.1ms remaining: 6.8ms
78: learn: 0.1799584 total: 24.5ms remaining: 6.52ms
79: learn: 0.1790604 total: 24.9ms remaining: 6.22ms
80: learn: 0.1773258 total: 25.3ms remaining: 5.93ms
81: learn: 0.1764698 total: 25.7ms remaining: 5.64ms
82: learn: 0.1751124 total: 26ms remaining: 5.33ms
83: learn: 0.1740194 total: 26.3ms remaining: 5.01ms
84: learn: 0.1732525 total: 26.7ms remaining: 4.71ms
85: learn: 0.1724760 total: 27ms remaining: 4.4ms
86: learn: 0.1713105 total: 27.3ms remaining: 4.08ms
87: learn: 0.1706636 total: 27.5ms remaining: 3.75ms
88: learn: 0.1698617 total: 27.8ms remaining: 3.44ms
89: learn: 0.1686562 total: 28.2ms remaining: 3.14ms
90: learn: 0.1678511 total: 28.7ms remaining: 2.83ms
91: learn: 0.1671549 total: 29.1ms remaining: 2.53ms
92: learn: 0.1663800 total: 30ms remaining: 2.26ms
93: learn: 0.1648383 total: 30.6ms remaining: 1.96ms
94: learn: 0.1639513 total: 30.9ms remaining: 1.63ms
95: learn: 0.1628442 total: 31.3ms remaining: 1.3ms
96: learn: 0.1620735 total: 31.7ms remaining: 978us
97: learn: 0.1613418 total: 32ms remaining: 653us
98: learn: 0.1606256 total: 32.4ms remaining: 327us
99: learn: 0.1599078 total: 32.9ms remaining: 0us
0: learn: 0.6532610 total: 504us remaining: 49.9ms
1: learn: 0.6102409 total: 1.09ms remaining: 53.2ms
2: learn: 0.5726766 total: 1.56ms remaining: 50.6ms
3: learn: 0.5399936 total: 2.47ms remaining: 59.3ms
4: learn: 0.5149233 total: 2.73ms remaining: 51.9ms
5: learn: 0.4883109 total: 3.26ms remaining: 51ms
6: learn: 0.4641478 total: 3.59ms remaining: 47.7ms
7: learn: 0.4460774 total: 4.09ms remaining: 47ms
8: learn: 0.4323097 total: 4.23ms remaining: 42.8ms
9: learn: 0.4157583 total: 4.67ms remaining: 42ms
10: learn: 0.4022919 total: 5.27ms remaining: 42.6ms
11: learn: 0.3898014 total: 6.01ms remaining: 44.1ms
12: learn: 0.3788049 total: 6.95ms remaining: 46.5ms
13: learn: 0.3683505 total: 7.38ms remaining: 45.4ms
14: learn: 0.3577059 total: 7.74ms remaining: 43.8ms
15: learn: 0.3493490 total: 8.43ms remaining: 44.3ms
16: learn: 0.3384982 total: 9.86ms remaining: 48.1ms
17: learn: 0.3329396 total: 10.2ms remaining: 46.3ms
18: learn: 0.3265991 total: 10.5ms remaining: 44.6ms
19: learn: 0.3205432 total: 10.8ms remaining: 43.3ms
20: learn: 0.3157322 total: 11.4ms remaining: 42.9ms
21: learn: 0.3109822 total: 11.8ms remaining: 41.8ms
22: learn: 0.3063239 total: 12.2ms remaining: 40.7ms
23: learn: 0.3018177 total: 12.4ms remaining: 39.3ms
24: learn: 0.2976308 total: 12.7ms remaining: 38.2ms
25: learn: 0.2931462 total: 13.1ms remaining: 37.2ms
26: learn: 0.2895543 total: 13.6ms remaining: 36.9ms
27: learn: 0.2858063 total: 14.6ms remaining: 37.5ms
28: learn: 0.2817349 total: 14.9ms remaining: 36.4ms
29: learn: 0.2783638 total: 15.2ms remaining: 35.5ms
30: learn: 0.2755228 total: 15.5ms remaining: 34.4ms
31: learn: 0.2726879 total: 15.8ms remaining: 33.6ms
32: learn: 0.2707994 total: 16.1ms remaining: 32.6ms
33: learn: 0.2671753 total: 16.5ms remaining: 32.1ms
34: learn: 0.2642467 total: 16.8ms remaining: 31.2ms
35: learn: 0.2618695 total: 17.1ms remaining: 30.4ms
36: learn: 0.2601981 total: 17.5ms remaining: 29.7ms
37: learn: 0.2580945 total: 17.8ms remaining: 29ms
38: learn: 0.2563573 total: 18.4ms remaining: 28.7ms
39: learn: 0.2534017 total: 19.3ms remaining: 29ms
40: learn: 0.2503710 total: 19.5ms remaining: 28.1ms
41: learn: 0.2467603 total: 19.8ms remaining: 27.3ms
42: learn: 0.2445181 total: 20.1ms remaining: 26.6ms
43: learn: 0.2436733 total: 20.3ms remaining: 25.8ms
44: learn: 0.2424590 total: 20.6ms remaining: 25.2ms
45: learn: 0.2413170 total: 20.9ms remaining: 24.5ms
46: learn: 0.2395461 total: 21.3ms remaining: 24ms
47: learn: 0.2376051 total: 21.7ms remaining: 23.5ms
48: learn: 0.2348047 total: 22.3ms remaining: 23.2ms
49: learn: 0.2325867 total: 22.9ms remaining: 22.9ms
50: learn: 0.2312478 total: 23.1ms remaining: 22.2ms
51: learn: 0.2292347 total: 23.9ms remaining: 22.1ms
52: learn: 0.2264443 total: 25ms remaining: 22.1ms
53: learn: 0.2240809 total: 25.9ms remaining: 22ms
54: learn: 0.2224813 total: 26.3ms remaining: 21.5ms
55: learn: 0.2197070 total: 26.6ms remaining: 20.9ms
56: learn: 0.2181432 total: 27.1ms remaining: 20.4ms
57: learn: 0.2173565 total: 27.6ms remaining: 20ms
58: learn: 0.2157150 total: 27.9ms remaining: 19.4ms
59: learn: 0.2140966 total: 28.6ms remaining: 19ms
60: learn: 0.2115956 total: 28.9ms remaining: 18.5ms
61: learn: 0.2100308 total: 30.4ms remaining: 18.7ms
62: learn: 0.2074310 total: 31.1ms remaining: 18.3ms
63: learn: 0.2057503 total: 32.1ms remaining: 18.1ms
64: learn: 0.2047251 total: 32.8ms remaining: 17.6ms
65: learn: 0.2034574 total: 33.4ms remaining: 17.2ms
66: learn: 0.2023031 total: 34ms remaining: 16.7ms
67: learn: 0.2013246 total: 35.1ms remaining: 16.5ms
68: learn: 0.1994649 total: 35.6ms remaining: 16ms
69: learn: 0.1984366 total: 36.1ms remaining: 15.5ms
70: learn: 0.1971174 total: 36.7ms remaining: 15ms
71: learn: 0.1957417 total: 37.7ms remaining: 14.7ms
72: learn: 0.1949303 total: 38ms remaining: 14ms
73: learn: 0.1929753 total: 38.3ms remaining: 13.4ms
74: learn: 0.1922421 total: 38.8ms remaining: 12.9ms
75: learn: 0.1910603 total: 39.4ms remaining: 12.4ms
76: learn: 0.1900257 total: 40ms remaining: 11.9ms
77: learn: 0.1890963 total: 40.5ms remaining: 11.4ms
78: learn: 0.1880508 total: 41.2ms remaining: 11ms
79: learn: 0.1872142 total: 42.3ms remaining: 10.6ms
80: learn: 0.1863501 total: 42.6ms remaining: 10ms
81: learn: 0.1855408 total: 43.1ms remaining: 9.45ms
82: learn: 0.1840863 total: 43.6ms remaining: 8.93ms
83: learn: 0.1833803 total: 44ms remaining: 8.38ms
84: learn: 0.1822225 total: 44.6ms remaining: 7.87ms
85: learn: 0.1807301 total: 45.6ms remaining: 7.42ms
86: learn: 0.1798001 total: 46.2ms remaining: 6.9ms
87: learn: 0.1790189 total: 46.8ms remaining: 6.38ms
88: learn: 0.1781547 total: 47.4ms remaining: 5.86ms
89: learn: 0.1770911 total: 48.1ms remaining: 5.35ms
90: learn: 0.1762956 total: 48.6ms remaining: 4.8ms
91: learn: 0.1754951 total: 48.9ms remaining: 4.25ms
92: learn: 0.1744672 total: 49.3ms remaining: 3.71ms
93: learn: 0.1735861 total: 49.9ms remaining: 3.18ms
94: learn: 0.1726698 total: 50.4ms remaining: 2.65ms
95: learn: 0.1721135 total: 50.9ms remaining: 2.12ms
96: learn: 0.1715908 total: 51.3ms remaining: 1.59ms
97: learn: 0.1710334 total: 51.9ms remaining: 1.06ms
98: learn: 0.1706777 total: 52.2ms remaining: 527us
99: learn: 0.1703117 total: 52.5ms remaining: 0us
0: learn: 0.6478370 total: 445us remaining: 44.1ms
1: learn: 0.6050464 total: 681us remaining: 33.4ms
2: learn: 0.5715635 total: 839us remaining: 27.1ms
3: learn: 0.5388645 total: 1.13ms remaining: 27.1ms
4: learn: 0.5127410 total: 1.24ms remaining: 23.6ms
5: learn: 0.4894968 total: 1.49ms remaining: 23.3ms
6: learn: 0.4690824 total: 1.68ms remaining: 22.4ms
7: learn: 0.4527488 total: 1.89ms remaining: 21.8ms
8: learn: 0.4367211 total: 2ms remaining: 20.3ms
9: learn: 0.4197791 total: 2.27ms remaining: 20.4ms
10: learn: 0.4076903 total: 2.54ms remaining: 20.6ms
11: learn: 0.3953699 total: 2.85ms remaining: 20.9ms
12: learn: 0.3835805 total: 3.13ms remaining: 20.9ms
13: learn: 0.3706580 total: 3.32ms remaining: 20.4ms
14: learn: 0.3632837 total: 3.54ms remaining: 20.1ms
15: learn: 0.3557629 total: 3.75ms remaining: 19.7ms
16: learn: 0.3465254 total: 3.96ms remaining: 19.3ms
17: learn: 0.3424635 total: 4.16ms remaining: 19ms
18: learn: 0.3340110 total: 4.36ms remaining: 18.6ms
19: learn: 0.3290275 total: 4.61ms remaining: 18.4ms
20: learn: 0.3221192 total: 4.9ms remaining: 18.4ms
21: learn: 0.3160905 total: 5.3ms remaining: 18.8ms
22: learn: 0.3106294 total: 5.47ms remaining: 18.3ms
23: learn: 0.3056046 total: 5.72ms remaining: 18.1ms
24: learn: 0.3024425 total: 5.97ms remaining: 17.9ms
25: learn: 0.2989832 total: 6.27ms remaining: 17.9ms
26: learn: 0.2944404 total: 6.66ms remaining: 18ms
27: learn: 0.2902619 total: 6.88ms remaining: 17.7ms
28: learn: 0.2872351 total: 7.1ms remaining: 17.4ms
29: learn: 0.2832049 total: 7.38ms remaining: 17.2ms
30: learn: 0.2811090 total: 7.71ms remaining: 17.2ms
31: learn: 0.2773095 total: 8.03ms remaining: 17.1ms
32: learn: 0.2748773 total: 8.3ms remaining: 16.8ms
33: learn: 0.2728913 total: 8.61ms remaining: 16.7ms
34: learn: 0.2704194 total: 9.27ms remaining: 17.2ms
35: learn: 0.2677007 total: 9.85ms remaining: 17.5ms
36: learn: 0.2654716 total: 10.4ms remaining: 17.8ms
37: learn: 0.2625555 total: 10.7ms remaining: 17.5ms
38: learn: 0.2588682 total: 11ms remaining: 17.2ms
39: learn: 0.2557431 total: 11.2ms remaining: 16.8ms
40: learn: 0.2541972 total: 11.4ms remaining: 16.4ms
41: learn: 0.2533046 total: 12ms remaining: 16.6ms
42: learn: 0.2508947 total: 12.5ms remaining: 16.5ms
43: learn: 0.2488413 total: 13.2ms remaining: 16.8ms
44: learn: 0.2460522 total: 13.8ms remaining: 16.8ms
45: learn: 0.2437101 total: 14ms remaining: 16.4ms
46: learn: 0.2420143 total: 14.5ms remaining: 16.3ms
47: learn: 0.2394717 total: 15.3ms remaining: 16.6ms
48: learn: 0.2377849 total: 15.6ms remaining: 16.3ms
49: learn: 0.2356327 total: 15.9ms remaining: 15.9ms
50: learn: 0.2333147 total: 16.5ms remaining: 15.8ms
51: learn: 0.2314324 total: 17ms remaining: 15.7ms
52: learn: 0.2295085 total: 17.3ms remaining: 15.3ms
53: learn: 0.2276318 total: 17.5ms remaining: 14.9ms
54: learn: 0.2268899 total: 17.7ms remaining: 14.5ms
55: learn: 0.2237974 total: 18ms remaining: 14.1ms
56: learn: 0.2224561 total: 18.3ms remaining: 13.8ms
57: learn: 0.2192132 total: 18.6ms remaining: 13.5ms
58: learn: 0.2173932 total: 19ms remaining: 13.2ms
59: learn: 0.2150435 total: 19.6ms remaining: 13.1ms
60: learn: 0.2132824 total: 19.9ms remaining: 12.7ms
61: learn: 0.2119521 total: 20.2ms remaining: 12.4ms
62: learn: 0.2102608 total: 20.5ms remaining: 12ms
63: learn: 0.2068338 total: 21.1ms remaining: 11.9ms
64: learn: 0.2054391 total: 22.3ms remaining: 12ms
65: learn: 0.2037807 total: 22.6ms remaining: 11.6ms
66: learn: 0.2022487 total: 22.9ms remaining: 11.3ms
67: learn: 0.1999168 total: 23.6ms remaining: 11.1ms
68: learn: 0.1984479 total: 24ms remaining: 10.8ms
69: learn: 0.1970045 total: 24.6ms remaining: 10.5ms
70: learn: 0.1956535 total: 25.1ms remaining: 10.2ms
71: learn: 0.1942315 total: 25.5ms remaining: 9.9ms
72: learn: 0.1932206 total: 26.2ms remaining: 9.68ms
73: learn: 0.1917156 total: 26.5ms remaining: 9.32ms
74: learn: 0.1897797 total: 27.5ms remaining: 9.16ms
75: learn: 0.1888298 total: 27.9ms remaining: 8.81ms
76: learn: 0.1875736 total: 28.5ms remaining: 8.51ms
77: learn: 0.1861071 total: 29.2ms remaining: 8.23ms
78: learn: 0.1849552 total: 30ms remaining: 7.96ms
79: learn: 0.1830319 total: 30.6ms remaining: 7.64ms
80: learn: 0.1811466 total: 31ms remaining: 7.27ms
81: learn: 0.1805334 total: 31.3ms remaining: 6.87ms
82: learn: 0.1784587 total: 31.5ms remaining: 6.46ms
83: learn: 0.1777659 total: 31.8ms remaining: 6.05ms
84: learn: 0.1770975 total: 32ms remaining: 5.64ms
85: learn: 0.1754173 total: 32.4ms remaining: 5.28ms
86: learn: 0.1737798 total: 33.2ms remaining: 4.96ms
87: learn: 0.1726413 total: 33.7ms remaining: 4.6ms
88: learn: 0.1712179 total: 34.1ms remaining: 4.21ms
89: learn: 0.1706006 total: 34.4ms remaining: 3.83ms
90: learn: 0.1688359 total: 34.7ms remaining: 3.43ms
91: learn: 0.1683922 total: 35.3ms remaining: 3.06ms
92: learn: 0.1679554 total: 35.7ms remaining: 2.69ms
93: learn: 0.1668017 total: 36.2ms remaining: 2.31ms
94: learn: 0.1659074 total: 37ms remaining: 1.95ms
95: learn: 0.1646166 total: 37.7ms remaining: 1.57ms
96: learn: 0.1642003 total: 39.1ms remaining: 1.21ms
97: learn: 0.1631870 total: 39.5ms remaining: 805us
98: learn: 0.1620912 total: 39.8ms remaining: 401us
99: learn: 0.1612658 total: 40.1ms remaining: 0us
0: learn: 0.6489461 total: 390us remaining: 38.7ms
1: learn: 0.6064240 total: 1.03ms remaining: 50.6ms
2: learn: 0.5700715 total: 1.41ms remaining: 45.7ms
3: learn: 0.5400758 total: 1.85ms remaining: 44.4ms
4: learn: 0.5145103 total: 2.14ms remaining: 40.6ms
5: learn: 0.4866673 total: 2.48ms remaining: 38.9ms
6: learn: 0.4673804 total: 3.32ms remaining: 44.1ms
7: learn: 0.4481874 total: 3.9ms remaining: 44.9ms
8: learn: 0.4300384 total: 4.47ms remaining: 45.2ms
9: learn: 0.4134195 total: 4.76ms remaining: 42.9ms
10: learn: 0.3962502 total: 5.13ms remaining: 41.5ms
11: learn: 0.3811386 total: 5.63ms remaining: 41.3ms
12: learn: 0.3693335 total: 5.92ms remaining: 39.6ms
13: learn: 0.3598914 total: 6.26ms remaining: 38.5ms
14: learn: 0.3516374 total: 6.98ms remaining: 39.5ms
15: learn: 0.3406200 total: 7.87ms remaining: 41.3ms
16: learn: 0.3323188 total: 8.2ms remaining: 40ms
17: learn: 0.3250314 total: 8.34ms remaining: 38ms
18: learn: 0.3192270 total: 8.76ms remaining: 37.3ms
19: learn: 0.3128178 total: 9.35ms remaining: 37.4ms
20: learn: 0.3059293 total: 9.96ms remaining: 37.5ms
21: learn: 0.2987314 total: 10.5ms remaining: 37.2ms
22: learn: 0.2940187 total: 11ms remaining: 37ms
23: learn: 0.2900778 total: 11.8ms remaining: 37.3ms
24: learn: 0.2872265 total: 12.4ms remaining: 37.3ms
25: learn: 0.2842832 total: 13ms remaining: 36.9ms
26: learn: 0.2804576 total: 13.5ms remaining: 36.4ms
27: learn: 0.2770150 total: 14.3ms remaining: 36.8ms
28: learn: 0.2748851 total: 14.6ms remaining: 35.7ms
29: learn: 0.2718988 total: 15.8ms remaining: 37ms
30: learn: 0.2689076 total: 16.1ms remaining: 35.7ms
31: learn: 0.2655543 total: 16.4ms remaining: 34.9ms
32: learn: 0.2633803 total: 17.2ms remaining: 34.9ms
33: learn: 0.2607521 total: 17.4ms remaining: 33.8ms
34: learn: 0.2586816 total: 17.8ms remaining: 33ms
35: learn: 0.2561481 total: 18.1ms remaining: 32.3ms
36: learn: 0.2535555 total: 18.5ms remaining: 31.6ms
37: learn: 0.2498320 total: 19ms remaining: 31ms
38: learn: 0.2465524 total: 19.7ms remaining: 30.8ms
39: learn: 0.2453466 total: 20.3ms remaining: 30.4ms
40: learn: 0.2437222 total: 20.9ms remaining: 30ms
41: learn: 0.2416669 total: 21.4ms remaining: 29.6ms
42: learn: 0.2391285 total: 22.4ms remaining: 29.7ms
43: learn: 0.2365899 total: 22.7ms remaining: 28.9ms
44: learn: 0.2348016 total: 23ms remaining: 28.1ms
45: learn: 0.2321103 total: 23.4ms remaining: 27.4ms
46: learn: 0.2307285 total: 23.7ms remaining: 26.8ms
47: learn: 0.2292365 total: 24.3ms remaining: 26.4ms
48: learn: 0.2281501 total: 24.5ms remaining: 25.5ms
49: learn: 0.2266474 total: 25.1ms remaining: 25.1ms
50: learn: 0.2254592 total: 25.3ms remaining: 24.3ms
51: learn: 0.2225412 total: 25.5ms remaining: 23.6ms
52: learn: 0.2209814 total: 25.7ms remaining: 22.8ms
53: learn: 0.2188452 total: 25.9ms remaining: 22.1ms
54: learn: 0.2176260 total: 26.4ms remaining: 21.6ms
55: learn: 0.2160541 total: 27.1ms remaining: 21.3ms
56: learn: 0.2145296 total: 27.8ms remaining: 21ms
57: learn: 0.2131576 total: 28.4ms remaining: 20.5ms
58: learn: 0.2112919 total: 29ms remaining: 20.1ms
59: learn: 0.2099507 total: 29.2ms remaining: 19.5ms
60: learn: 0.2078440 total: 29.8ms remaining: 19ms
61: learn: 0.2063141 total: 30.4ms remaining: 18.6ms
62: learn: 0.2047629 total: 30.8ms remaining: 18.1ms
63: learn: 0.2038695 total: 31.6ms remaining: 17.8ms
64: learn: 0.2023354 total: 32.1ms remaining: 17.3ms
65: learn: 0.1996688 total: 32.7ms remaining: 16.9ms
66: learn: 0.1973198 total: 34ms remaining: 16.8ms
67: learn: 0.1959162 total: 34.4ms remaining: 16.2ms
68: learn: 0.1951752 total: 34.7ms remaining: 15.6ms
69: learn: 0.1940283 total: 35.8ms remaining: 15.4ms
70: learn: 0.1930343 total: 36.5ms remaining: 14.9ms
71: learn: 0.1907450 total: 37ms remaining: 14.4ms
72: learn: 0.1887311 total: 37.5ms remaining: 13.9ms
73: learn: 0.1874915 total: 38ms remaining: 13.4ms
74: learn: 0.1860438 total: 38.8ms remaining: 12.9ms
75: learn: 0.1851532 total: 39.4ms remaining: 12.4ms
76: learn: 0.1843621 total: 39.9ms remaining: 11.9ms
77: learn: 0.1828291 total: 40.3ms remaining: 11.4ms
78: learn: 0.1819787 total: 40.7ms remaining: 10.8ms
79: learn: 0.1800943 total: 41.5ms remaining: 10.4ms
80: learn: 0.1789557 total: 41.9ms remaining: 9.82ms
81: learn: 0.1783137 total: 42.2ms remaining: 9.27ms
82: learn: 0.1776746 total: 42.5ms remaining: 8.71ms
83: learn: 0.1766376 total: 43ms remaining: 8.19ms
84: learn: 0.1754587 total: 43.5ms remaining: 7.68ms
85: learn: 0.1747746 total: 44.1ms remaining: 7.18ms
86: learn: 0.1734613 total: 44.4ms remaining: 6.63ms
87: learn: 0.1723720 total: 44.7ms remaining: 6.1ms
88: learn: 0.1716154 total: 45.1ms remaining: 5.57ms
89: learn: 0.1705611 total: 45.7ms remaining: 5.08ms
90: learn: 0.1699904 total: 46ms remaining: 4.55ms
91: learn: 0.1692174 total: 46.3ms remaining: 4.03ms
92: learn: 0.1685566 total: 46.6ms remaining: 3.51ms
93: learn: 0.1679728 total: 47ms remaining: 3ms
94: learn: 0.1674829 total: 47.3ms remaining: 2.49ms
95: learn: 0.1669377 total: 47.9ms remaining: 2ms
96: learn: 0.1663326 total: 48.1ms remaining: 1.49ms
97: learn: 0.1654778 total: 48.9ms remaining: 997us
98: learn: 0.1643119 total: 49.6ms remaining: 501us
99: learn: 0.1631584 total: 50ms remaining: 0us
0: learn: 0.6590402 total: 411us remaining: 40.8ms
1: learn: 0.6114009 total: 709us remaining: 34.8ms
2: learn: 0.5810295 total: 976us remaining: 31.6ms
3: learn: 0.5494286 total: 1.25ms remaining: 30.1ms
4: learn: 0.5218481 total: 1.48ms remaining: 28.2ms
5: learn: 0.4961409 total: 1.67ms remaining: 26.2ms
6: learn: 0.4825933 total: 1.93ms remaining: 25.7ms
7: learn: 0.4612439 total: 2.13ms remaining: 24.5ms
8: learn: 0.4405597 total: 2.42ms remaining: 24.4ms
9: learn: 0.4236230 total: 2.58ms remaining: 23.3ms
10: learn: 0.4086284 total: 2.8ms remaining: 22.6ms
11: learn: 0.3966478 total: 3.02ms remaining: 22.1ms
12: learn: 0.3845862 total: 3.29ms remaining: 22ms
13: learn: 0.3734868 total: 3.55ms remaining: 21.8ms
14: learn: 0.3612373 total: 3.77ms remaining: 21.3ms
15: learn: 0.3563945 total: 3.93ms remaining: 20.6ms
16: learn: 0.3520736 total: 4.13ms remaining: 20.2ms
17: learn: 0.3437252 total: 4.33ms remaining: 19.7ms
18: learn: 0.3366232 total: 4.56ms remaining: 19.4ms
19: learn: 0.3305014 total: 4.81ms remaining: 19.3ms
20: learn: 0.3237168 total: 5.09ms remaining: 19.1ms
21: learn: 0.3165086 total: 5.27ms remaining: 18.7ms
22: learn: 0.3115073 total: 5.52ms remaining: 18.5ms
23: learn: 0.3070963 total: 5.9ms remaining: 18.7ms
24: learn: 0.3018950 total: 6.18ms remaining: 18.6ms
25: learn: 0.2978404 total: 6.47ms remaining: 18.4ms
26: learn: 0.2944708 total: 6.73ms remaining: 18.2ms
27: learn: 0.2896176 total: 7.03ms remaining: 18.1ms
28: learn: 0.2862532 total: 7.38ms remaining: 18.1ms
29: learn: 0.2832470 total: 7.61ms remaining: 17.8ms
30: learn: 0.2798789 total: 7.91ms remaining: 17.6ms
31: learn: 0.2760300 total: 8.16ms remaining: 17.3ms
32: learn: 0.2725581 total: 8.35ms remaining: 17ms
33: learn: 0.2704228 total: 8.55ms remaining: 16.6ms
34: learn: 0.2677841 total: 8.81ms remaining: 16.4ms
35: learn: 0.2653151 total: 9.09ms remaining: 16.2ms
36: learn: 0.2633276 total: 9.54ms remaining: 16.2ms
37: learn: 0.2602114 total: 9.91ms remaining: 16.2ms
38: learn: 0.2575564 total: 10.2ms remaining: 15.9ms
39: learn: 0.2550264 total: 10.5ms remaining: 15.7ms
40: learn: 0.2531548 total: 10.8ms remaining: 15.6ms
41: learn: 0.2507302 total: 11.2ms remaining: 15.4ms
42: learn: 0.2494902 total: 11.6ms remaining: 15.4ms
43: learn: 0.2463801 total: 12ms remaining: 15.2ms
44: learn: 0.2447248 total: 12.2ms remaining: 14.9ms
45: learn: 0.2424518 total: 12.5ms remaining: 14.7ms
46: learn: 0.2411762 total: 12.7ms remaining: 14.3ms
47: learn: 0.2387374 total: 13ms remaining: 14.1ms
48: learn: 0.2362627 total: 13.3ms remaining: 13.8ms
49: learn: 0.2354837 total: 13.4ms remaining: 13.4ms
50: learn: 0.2334826 total: 13.6ms remaining: 13.1ms
51: learn: 0.2320311 total: 13.9ms remaining: 12.8ms
52: learn: 0.2304605 total: 14.3ms remaining: 12.6ms
53: learn: 0.2283454 total: 14.8ms remaining: 12.6ms
54: learn: 0.2259308 total: 15.2ms remaining: 12.5ms
55: learn: 0.2243436 total: 15.7ms remaining: 12.3ms
56: learn: 0.2234152 total: 16.2ms remaining: 12.2ms
57: learn: 0.2222624 total: 16.8ms remaining: 12.2ms
58: learn: 0.2198506 total: 17.3ms remaining: 12.1ms
59: learn: 0.2180131 total: 17.8ms remaining: 11.9ms
60: learn: 0.2153596 total: 18.2ms remaining: 11.6ms
61: learn: 0.2135086 total: 18.8ms remaining: 11.5ms
62: learn: 0.2112580 total: 19.6ms remaining: 11.5ms
63: learn: 0.2097674 total: 19.9ms remaining: 11.2ms
64: learn: 0.2083025 total: 20.4ms remaining: 11ms
65: learn: 0.2067273 total: 21ms remaining: 10.8ms
66: learn: 0.2052166 total: 21.4ms remaining: 10.5ms
67: learn: 0.2045802 total: 22.1ms remaining: 10.4ms
68: learn: 0.2034975 total: 22.5ms remaining: 10.1ms
69: learn: 0.2027905 total: 22.8ms remaining: 9.78ms
70: learn: 0.2018563 total: 23.4ms remaining: 9.58ms
71: learn: 0.2000458 total: 23.8ms remaining: 9.26ms
72: learn: 0.1986833 total: 24.1ms remaining: 8.9ms
73: learn: 0.1981076 total: 24.4ms remaining: 8.56ms
74: learn: 0.1975341 total: 24.6ms remaining: 8.22ms
75: learn: 0.1966905 total: 25.3ms remaining: 8ms
76: learn: 0.1956072 total: 25.7ms remaining: 7.66ms
77: learn: 0.1944255 total: 25.9ms remaining: 7.32ms
78: learn: 0.1935610 total: 26.3ms remaining: 7ms
79: learn: 0.1927345 total: 26.7ms remaining: 6.68ms
80: learn: 0.1919683 total: 27.1ms remaining: 6.36ms
81: learn: 0.1910788 total: 27.4ms remaining: 6.02ms
82: learn: 0.1899076 total: 28.1ms remaining: 5.75ms
83: learn: 0.1886907 total: 28.7ms remaining: 5.47ms
84: learn: 0.1880866 total: 29ms remaining: 5.12ms
85: learn: 0.1877364 total: 29.8ms remaining: 4.85ms
86: learn: 0.1869338 total: 30.2ms remaining: 4.51ms
87: learn: 0.1860941 total: 30.7ms remaining: 4.19ms
88: learn: 0.1855313 total: 31.6ms remaining: 3.91ms
89: learn: 0.1846593 total: 32.1ms remaining: 3.57ms
90: learn: 0.1840153 total: 32.8ms remaining: 3.25ms
91: learn: 0.1833519 total: 33.1ms remaining: 2.88ms
92: learn: 0.1817814 total: 33.4ms remaining: 2.51ms
93: learn: 0.1806517 total: 33.6ms remaining: 2.15ms
94: learn: 0.1801285 total: 34ms remaining: 1.79ms
95: learn: 0.1795813 total: 34.6ms remaining: 1.44ms
96: learn: 0.1790231 total: 34.9ms remaining: 1.08ms
97: learn: 0.1784280 total: 35.4ms remaining: 722us
98: learn: 0.1779345 total: 35.6ms remaining: 360us
99: learn: 0.1763333 total: 36.2ms remaining: 0us
0: learn: 0.6577557 total: 345us remaining: 34.2ms
1: learn: 0.6115590 total: 635us remaining: 31.1ms
2: learn: 0.5782661 total: 848us remaining: 27.4ms
3: learn: 0.5504365 total: 1.11ms remaining: 26.6ms
4: learn: 0.5239138 total: 1.39ms remaining: 26.4ms
5: learn: 0.4960141 total: 1.57ms remaining: 24.7ms
6: learn: 0.4751299 total: 1.79ms remaining: 23.8ms
7: learn: 0.4522825 total: 1.96ms remaining: 22.6ms
8: learn: 0.4350004 total: 2.2ms remaining: 22.3ms
9: learn: 0.4172532 total: 2.36ms remaining: 21.2ms
10: learn: 0.4031272 total: 2.61ms remaining: 21.2ms
11: learn: 0.3894778 total: 2.81ms remaining: 20.6ms
12: learn: 0.3759268 total: 3.01ms remaining: 20.1ms
13: learn: 0.3646396 total: 3.23ms remaining: 19.9ms
14: learn: 0.3550212 total: 3.5ms remaining: 19.8ms
15: learn: 0.3459478 total: 3.75ms remaining: 19.7ms
16: learn: 0.3387418 total: 3.95ms remaining: 19.3ms
17: learn: 0.3317651 total: 4.13ms remaining: 18.8ms
18: learn: 0.3245328 total: 4.33ms remaining: 18.5ms
19: learn: 0.3186379 total: 4.54ms remaining: 18.2ms
20: learn: 0.3123720 total: 4.8ms remaining: 18ms
21: learn: 0.3066610 total: 4.97ms remaining: 17.6ms
22: learn: 0.3021609 total: 5.1ms remaining: 17.1ms
23: learn: 0.2981775 total: 5.41ms remaining: 17.1ms
24: learn: 0.2947302 total: 5.6ms remaining: 16.8ms
25: learn: 0.2894158 total: 5.91ms remaining: 16.8ms
26: learn: 0.2849985 total: 6.15ms remaining: 16.6ms
27: learn: 0.2816687 total: 6.38ms remaining: 16.4ms
28: learn: 0.2784744 total: 6.64ms remaining: 16.3ms
29: learn: 0.2753041 total: 6.94ms remaining: 16.2ms
30: learn: 0.2704793 total: 7.3ms remaining: 16.3ms
31: learn: 0.2655361 total: 7.61ms remaining: 16.2ms
32: learn: 0.2598502 total: 7.84ms remaining: 15.9ms
33: learn: 0.2568552 total: 8.11ms remaining: 15.7ms
34: learn: 0.2540340 total: 8.43ms remaining: 15.7ms
35: learn: 0.2503535 total: 8.82ms remaining: 15.7ms
36: learn: 0.2479424 total: 9.13ms remaining: 15.5ms
37: learn: 0.2453326 total: 9.49ms remaining: 15.5ms
38: learn: 0.2425282 total: 9.74ms remaining: 15.2ms
39: learn: 0.2398649 total: 9.99ms remaining: 15ms
40: learn: 0.2385556 total: 10.3ms remaining: 14.8ms
41: learn: 0.2365283 total: 10.5ms remaining: 14.6ms
42: learn: 0.2331381 total: 10.8ms remaining: 14.3ms
43: learn: 0.2300152 total: 11.2ms remaining: 14.2ms
44: learn: 0.2279245 total: 11.5ms remaining: 14ms
45: learn: 0.2263903 total: 11.6ms remaining: 13.7ms
46: learn: 0.2238257 total: 11.9ms remaining: 13.5ms
47: learn: 0.2216545 total: 12.2ms remaining: 13.3ms
48: learn: 0.2196501 total: 12.6ms remaining: 13.1ms
49: learn: 0.2168023 total: 12.8ms remaining: 12.8ms
50: learn: 0.2145388 total: 13ms remaining: 12.5ms
51: learn: 0.2132267 total: 13.2ms remaining: 12.2ms
52: learn: 0.2103353 total: 13.5ms remaining: 12ms
53: learn: 0.2089741 total: 13.8ms remaining: 11.7ms
54: learn: 0.2074504 total: 14.1ms remaining: 11.6ms
55: learn: 0.2053822 total: 14.4ms remaining: 11.4ms
56: learn: 0.2031189 total: 14.7ms remaining: 11.1ms
57: learn: 0.2017266 total: 15ms remaining: 10.9ms
58: learn: 0.1996794 total: 15.4ms remaining: 10.7ms
59: learn: 0.1972371 total: 15.7ms remaining: 10.4ms
60: learn: 0.1949181 total: 16ms remaining: 10.2ms
61: learn: 0.1912835 total: 16.3ms remaining: 9.99ms
62: learn: 0.1902439 total: 16.6ms remaining: 9.73ms
63: learn: 0.1884128 total: 17ms remaining: 9.54ms
64: learn: 0.1868456 total: 17.4ms remaining: 9.36ms
65: learn: 0.1856003 total: 17.8ms remaining: 9.14ms
66: learn: 0.1835379 total: 18.2ms remaining: 8.96ms
67: learn: 0.1825284 total: 18.5ms remaining: 8.69ms
68: learn: 0.1804093 total: 18.8ms remaining: 8.44ms
69: learn: 0.1791994 total: 19.1ms remaining: 8.2ms
70: learn: 0.1783252 total: 19.4ms remaining: 7.93ms
71: learn: 0.1765206 total: 19.7ms remaining: 7.67ms
72: learn: 0.1748911 total: 20.2ms remaining: 7.49ms
73: learn: 0.1736684 total: 20.5ms remaining: 7.2ms
74: learn: 0.1727610 total: 20.8ms remaining: 6.94ms
75: learn: 0.1704809 total: 21.1ms remaining: 6.67ms
76: learn: 0.1688713 total: 21.5ms remaining: 6.41ms
77: learn: 0.1679488 total: 21.9ms remaining: 6.17ms
78: learn: 0.1667283 total: 22.2ms remaining: 5.91ms
79: learn: 0.1657982 total: 22.7ms remaining: 5.67ms
80: learn: 0.1642468 total: 23.1ms remaining: 5.43ms
81: learn: 0.1631564 total: 23.5ms remaining: 5.17ms
82: learn: 0.1624591 total: 23.8ms remaining: 4.87ms
83: learn: 0.1611993 total: 24.5ms remaining: 4.67ms
84: learn: 0.1597753 total: 24.8ms remaining: 4.38ms
85: learn: 0.1589474 total: 25.3ms remaining: 4.11ms
86: learn: 0.1580195 total: 25.8ms remaining: 3.85ms
87: learn: 0.1575410 total: 26.3ms remaining: 3.58ms
88: learn: 0.1563160 total: 26.8ms remaining: 3.31ms
89: learn: 0.1557749 total: 27.4ms remaining: 3.05ms
90: learn: 0.1549278 total: 27.7ms remaining: 2.74ms
91: learn: 0.1534022 total: 28ms remaining: 2.43ms
92: learn: 0.1514222 total: 28.4ms remaining: 2.13ms
93: learn: 0.1508935 total: 28.9ms remaining: 1.85ms
94: learn: 0.1492736 total: 29.3ms remaining: 1.54ms
95: learn: 0.1484335 total: 29.8ms remaining: 1.24ms
96: learn: 0.1476766 total: 30ms remaining: 928us
97: learn: 0.1466290 total: 30.6ms remaining: 625us
98: learn: 0.1455973 total: 30.9ms remaining: 312us
99: learn: 0.1440446 total: 31.3ms remaining: 0us
0: learn: 0.6583442 total: 364us remaining: 36.1ms
1: learn: 0.6160693 total: 903us remaining: 44.2ms
2: learn: 0.5795190 total: 1.24ms remaining: 40.2ms
3: learn: 0.5466177 total: 1.54ms remaining: 37.1ms
4: learn: 0.5207080 total: 1.75ms remaining: 33.3ms
5: learn: 0.4950930 total: 2.19ms remaining: 34.4ms
6: learn: 0.4717440 total: 2.38ms remaining: 31.7ms
7: learn: 0.4536730 total: 2.61ms remaining: 30ms
8: learn: 0.4349721 total: 2.79ms remaining: 28.2ms
9: learn: 0.4202796 total: 3.24ms remaining: 29.2ms
10: learn: 0.4077376 total: 3.76ms remaining: 30.4ms
11: learn: 0.3960184 total: 4.28ms remaining: 31.4ms
12: learn: 0.3826414 total: 4.81ms remaining: 32.2ms
13: learn: 0.3722208 total: 5.05ms remaining: 31ms
14: learn: 0.3627836 total: 5.41ms remaining: 30.6ms
15: learn: 0.3526643 total: 5.62ms remaining: 29.5ms
16: learn: 0.3433313 total: 5.95ms remaining: 29.1ms
17: learn: 0.3337858 total: 6.27ms remaining: 28.6ms
18: learn: 0.3281039 total: 6.55ms remaining: 27.9ms
19: learn: 0.3204933 total: 6.98ms remaining: 27.9ms
20: learn: 0.3172523 total: 7.49ms remaining: 28.2ms
21: learn: 0.3112378 total: 8.19ms remaining: 29ms
22: learn: 0.3055845 total: 8.47ms remaining: 28.4ms
23: learn: 0.3017372 total: 8.78ms remaining: 27.8ms
24: learn: 0.2956202 total: 9.58ms remaining: 28.8ms
25: learn: 0.2904468 total: 10.1ms remaining: 28.8ms
26: learn: 0.2863467 total: 10.6ms remaining: 28.6ms
27: learn: 0.2831806 total: 10.9ms remaining: 27.9ms
28: learn: 0.2799353 total: 11.7ms remaining: 28.7ms
29: learn: 0.2769338 total: 12.2ms remaining: 28.4ms
30: learn: 0.2725319 total: 12.6ms remaining: 28ms
31: learn: 0.2690325 total: 12.8ms remaining: 27.2ms
32: learn: 0.2653722 total: 13.1ms remaining: 26.7ms
33: learn: 0.2626298 total: 13.8ms remaining: 26.8ms
34: learn: 0.2613544 total: 14.6ms remaining: 27.1ms
35: learn: 0.2590320 total: 14.8ms remaining: 26.3ms
36: learn: 0.2566611 total: 15.1ms remaining: 25.8ms
37: learn: 0.2535962 total: 15.6ms remaining: 25.5ms
38: learn: 0.2497372 total: 16.3ms remaining: 25.4ms
39: learn: 0.2470181 total: 17ms remaining: 25.4ms
40: learn: 0.2445742 total: 17.4ms remaining: 25ms
41: learn: 0.2430848 total: 18ms remaining: 24.9ms
42: learn: 0.2422891 total: 18.5ms remaining: 24.6ms
43: learn: 0.2397990 total: 19.6ms remaining: 25ms
44: learn: 0.2371706 total: 20.7ms remaining: 25.2ms
45: learn: 0.2352512 total: 21.2ms remaining: 24.9ms
46: learn: 0.2327897 total: 22.2ms remaining: 25ms
47: learn: 0.2308399 total: 22.5ms remaining: 24.4ms
48: learn: 0.2296719 total: 22.7ms remaining: 23.6ms
49: learn: 0.2277718 total: 23ms remaining: 23ms
50: learn: 0.2263527 total: 23.4ms remaining: 22.5ms
51: learn: 0.2248207 total: 23.6ms remaining: 21.8ms
52: learn: 0.2232927 total: 24.3ms remaining: 21.5ms
53: learn: 0.2217390 total: 25.1ms remaining: 21.3ms
54: learn: 0.2188630 total: 25.5ms remaining: 20.8ms
55: learn: 0.2174118 total: 25.8ms remaining: 20.3ms
56: learn: 0.2154217 total: 26.5ms remaining: 20ms
57: learn: 0.2132996 total: 27.1ms remaining: 19.6ms
58: learn: 0.2119245 total: 27.4ms remaining: 19.1ms
59: learn: 0.2092403 total: 27.8ms remaining: 18.5ms
60: learn: 0.2081820 total: 28.2ms remaining: 18.1ms
61: learn: 0.2069303 total: 28.5ms remaining: 17.5ms
62: learn: 0.2057730 total: 29.1ms remaining: 17.1ms
63: learn: 0.2043888 total: 29.9ms remaining: 16.8ms
64: learn: 0.2033712 total: 30.4ms remaining: 16.4ms
65: learn: 0.2023271 total: 30.8ms remaining: 15.8ms
66: learn: 0.2007830 total: 31.2ms remaining: 15.4ms
67: learn: 0.1984698 total: 31.7ms remaining: 14.9ms
68: learn: 0.1971153 total: 32.2ms remaining: 14.5ms
69: learn: 0.1960784 total: 32.9ms remaining: 14.1ms
70: learn: 0.1949722 total: 33.3ms remaining: 13.6ms
71: learn: 0.1941931 total: 33.5ms remaining: 13ms
72: learn: 0.1930148 total: 33.8ms remaining: 12.5ms
73: learn: 0.1922700 total: 34ms remaining: 11.9ms
74: learn: 0.1912845 total: 34.4ms remaining: 11.5ms
75: learn: 0.1902818 total: 34.8ms remaining: 11ms
76: learn: 0.1879536 total: 35.1ms remaining: 10.5ms
77: learn: 0.1870270 total: 35.4ms remaining: 9.98ms
78: learn: 0.1848320 total: 35.7ms remaining: 9.49ms
79: learn: 0.1838008 total: 36ms remaining: 9ms
80: learn: 0.1828718 total: 36.3ms remaining: 8.51ms
81: learn: 0.1812732 total: 36.6ms remaining: 8.03ms
82: learn: 0.1803925 total: 36.9ms remaining: 7.56ms
83: learn: 0.1797710 total: 37.2ms remaining: 7.08ms
84: learn: 0.1789050 total: 37.5ms remaining: 6.61ms
85: learn: 0.1783196 total: 37.8ms remaining: 6.16ms
86: learn: 0.1777546 total: 38.3ms remaining: 5.72ms
87: learn: 0.1765896 total: 38.6ms remaining: 5.27ms
88: learn: 0.1757566 total: 38.9ms remaining: 4.8ms
89: learn: 0.1753380 total: 39.5ms remaining: 4.38ms
90: learn: 0.1738739 total: 40ms remaining: 3.96ms
91: learn: 0.1732464 total: 40.4ms remaining: 3.51ms
92: learn: 0.1719087 total: 40.7ms remaining: 3.06ms
93: learn: 0.1714328 total: 41.1ms remaining: 2.62ms
94: learn: 0.1701988 total: 41.4ms remaining: 2.18ms
95: learn: 0.1696867 total: 41.7ms remaining: 1.74ms
96: learn: 0.1691051 total: 42.1ms remaining: 1.3ms
97: learn: 0.1682508 total: 42.8ms remaining: 873us
98: learn: 0.1678090 total: 43.8ms remaining: 442us
99: learn: 0.1672186 total: 44.4ms remaining: 0us
0: learn: 0.6475054 total: 354us remaining: 35.1ms
1: learn: 0.6071370 total: 566us remaining: 27.7ms
2: learn: 0.5688928 total: 825us remaining: 26.7ms
3: learn: 0.5371729 total: 1.2ms remaining: 28.8ms
4: learn: 0.5086493 total: 1.73ms remaining: 32.9ms
5: learn: 0.4795564 total: 2.08ms remaining: 32.5ms
6: learn: 0.4579098 total: 2.44ms remaining: 32.5ms
7: learn: 0.4308290 total: 2.75ms remaining: 31.7ms
8: learn: 0.4093250 total: 3.09ms remaining: 31.2ms
9: learn: 0.3941826 total: 3.35ms remaining: 30.1ms
10: learn: 0.3800904 total: 3.56ms remaining: 28.8ms
11: learn: 0.3639103 total: 4.05ms remaining: 29.7ms
12: learn: 0.3496290 total: 4.64ms remaining: 31ms
13: learn: 0.3407169 total: 5.05ms remaining: 31.1ms
14: learn: 0.3297829 total: 6.13ms remaining: 34.7ms
15: learn: 0.3189842 total: 6.52ms remaining: 34.2ms
16: learn: 0.3114926 total: 6.91ms remaining: 33.7ms
17: learn: 0.3031443 total: 7.18ms remaining: 32.7ms
18: learn: 0.2955707 total: 7.52ms remaining: 32ms
19: learn: 0.2880411 total: 7.88ms remaining: 31.5ms
20: learn: 0.2805200 total: 8.32ms remaining: 31.3ms
21: learn: 0.2726320 total: 8.68ms remaining: 30.8ms
22: learn: 0.2675664 total: 8.85ms remaining: 29.6ms
23: learn: 0.2634753 total: 9.22ms remaining: 29.2ms
24: learn: 0.2596825 total: 9.62ms remaining: 28.9ms
25: learn: 0.2559484 total: 10ms remaining: 28.5ms
26: learn: 0.2515160 total: 10.5ms remaining: 28.4ms
27: learn: 0.2483297 total: 10.9ms remaining: 28.2ms
28: learn: 0.2462752 total: 11.3ms remaining: 27.7ms
29: learn: 0.2429713 total: 11.8ms remaining: 27.4ms
30: learn: 0.2395684 total: 12.1ms remaining: 26.9ms
31: learn: 0.2364600 total: 12.4ms remaining: 26.4ms
32: learn: 0.2339638 total: 12.8ms remaining: 26.1ms
33: learn: 0.2316042 total: 13.3ms remaining: 25.9ms
34: learn: 0.2292298 total: 13.7ms remaining: 25.5ms
35: learn: 0.2264514 total: 14.2ms remaining: 25.2ms
36: learn: 0.2223391 total: 14.7ms remaining: 25.1ms
37: learn: 0.2194596 total: 15.3ms remaining: 25ms
38: learn: 0.2165141 total: 15.5ms remaining: 24.3ms
39: learn: 0.2141335 total: 15.9ms remaining: 23.8ms
40: learn: 0.2121124 total: 16.4ms remaining: 23.6ms
41: learn: 0.2102779 total: 16.8ms remaining: 23.1ms
42: learn: 0.2082953 total: 17.2ms remaining: 22.8ms
43: learn: 0.2072916 total: 17.5ms remaining: 22.3ms
44: learn: 0.2049074 total: 17.9ms remaining: 21.9ms
45: learn: 0.2036420 total: 18.3ms remaining: 21.5ms
46: learn: 0.2014209 total: 19ms remaining: 21.4ms
47: learn: 0.1999690 total: 19.3ms remaining: 20.9ms
48: learn: 0.1990182 total: 19.8ms remaining: 20.6ms
49: learn: 0.1973890 total: 20.1ms remaining: 20.1ms
50: learn: 0.1938346 total: 20.6ms remaining: 19.8ms
51: learn: 0.1927559 total: 21.1ms remaining: 19.5ms
52: learn: 0.1916926 total: 21.7ms remaining: 19.2ms
53: learn: 0.1904656 total: 22ms remaining: 18.8ms
54: learn: 0.1892644 total: 22.5ms remaining: 18.4ms
55: learn: 0.1881377 total: 22.8ms remaining: 17.9ms
56: learn: 0.1865936 total: 23.2ms remaining: 17.5ms
57: learn: 0.1847520 total: 24ms remaining: 17.4ms
58: learn: 0.1834779 total: 24.6ms remaining: 17.1ms
59: learn: 0.1821843 total: 25.2ms remaining: 16.8ms
60: learn: 0.1803011 total: 26.1ms remaining: 16.7ms
61: learn: 0.1777742 total: 26.6ms remaining: 16.3ms
62: learn: 0.1772984 total: 27ms remaining: 15.9ms
63: learn: 0.1764391 total: 27.4ms remaining: 15.4ms
64: learn: 0.1757728 total: 27.7ms remaining: 14.9ms
65: learn: 0.1743333 total: 28ms remaining: 14.4ms
66: learn: 0.1731950 total: 28.3ms remaining: 14ms
67: learn: 0.1703715 total: 28.6ms remaining: 13.5ms
68: learn: 0.1688254 total: 29ms remaining: 13ms
69: learn: 0.1671513 total: 29.9ms remaining: 12.8ms
70: learn: 0.1655474 total: 30.5ms remaining: 12.4ms
71: learn: 0.1646720 total: 31.1ms remaining: 12.1ms
72: learn: 0.1632495 total: 31.6ms remaining: 11.7ms
73: learn: 0.1618558 total: 32.3ms remaining: 11.4ms
74: learn: 0.1613448 total: 32.9ms remaining: 11ms
75: learn: 0.1605755 total: 33.5ms remaining: 10.6ms
76: learn: 0.1590612 total: 34.2ms remaining: 10.2ms
77: learn: 0.1576873 total: 34.4ms remaining: 9.69ms
78: learn: 0.1568384 total: 34.7ms remaining: 9.21ms
79: learn: 0.1560417 total: 35ms remaining: 8.76ms
80: learn: 0.1553088 total: 35.6ms remaining: 8.35ms
81: learn: 0.1543515 total: 35.9ms remaining: 7.88ms
82: learn: 0.1536344 total: 36.7ms remaining: 7.51ms
83: learn: 0.1528763 total: 37.4ms remaining: 7.12ms
84: learn: 0.1522848 total: 38ms remaining: 6.7ms
85: learn: 0.1503108 total: 38.4ms remaining: 6.26ms
86: learn: 0.1489568 total: 38.7ms remaining: 5.79ms
87: learn: 0.1480627 total: 39.1ms remaining: 5.33ms
88: learn: 0.1472803 total: 39.8ms remaining: 4.91ms
89: learn: 0.1463411 total: 40ms remaining: 4.44ms
90: learn: 0.1457018 total: 40.2ms remaining: 3.98ms
91: learn: 0.1451322 total: 40.4ms remaining: 3.52ms
92: learn: 0.1439193 total: 40.6ms remaining: 3.06ms
93: learn: 0.1426374 total: 40.8ms remaining: 2.6ms
94: learn: 0.1411973 total: 41.2ms remaining: 2.17ms
95: learn: 0.1407247 total: 41.7ms remaining: 1.74ms
96: learn: 0.1402650 total: 42.1ms remaining: 1.3ms
97: learn: 0.1398238 total: 42.5ms remaining: 868us
98: learn: 0.1394731 total: 43.1ms remaining: 434us
99: learn: 0.1391102 total: 43.3ms remaining: 0us
0: learn: 0.6426234 total: 542us remaining: 53.7ms
1: learn: 0.5966344 total: 936us remaining: 45.9ms
2: learn: 0.5610290 total: 1.16ms remaining: 37.4ms
3: learn: 0.5301488 total: 1.65ms remaining: 39.5ms
4: learn: 0.5029961 total: 2.09ms remaining: 39.7ms
5: learn: 0.4793889 total: 2.5ms remaining: 39.2ms
6: learn: 0.4547454 total: 2.72ms remaining: 36.1ms
7: learn: 0.4372305 total: 3.19ms remaining: 36.6ms
8: learn: 0.4185281 total: 3.48ms remaining: 35.2ms
9: learn: 0.4020008 total: 4.04ms remaining: 36.4ms
10: learn: 0.3875013 total: 4.24ms remaining: 34.3ms
11: learn: 0.3742145 total: 4.51ms remaining: 33.1ms
12: learn: 0.3622586 total: 4.97ms remaining: 33.2ms
13: learn: 0.3518018 total: 5.24ms remaining: 32.2ms
14: learn: 0.3435048 total: 5.6ms remaining: 31.7ms
15: learn: 0.3326371 total: 5.82ms remaining: 30.6ms
16: learn: 0.3257278 total: 6.05ms remaining: 29.6ms
17: learn: 0.3167057 total: 6.32ms remaining: 28.8ms
18: learn: 0.3097649 total: 6.49ms remaining: 27.7ms
19: learn: 0.3042157 total: 6.8ms remaining: 27.2ms
20: learn: 0.2982697 total: 7.14ms remaining: 26.9ms
21: learn: 0.2921152 total: 7.42ms remaining: 26.3ms
22: learn: 0.2875303 total: 7.86ms remaining: 26.3ms
23: learn: 0.2834689 total: 8.46ms remaining: 26.8ms
24: learn: 0.2785533 total: 8.88ms remaining: 26.6ms
25: learn: 0.2762674 total: 9.18ms remaining: 26.1ms
26: learn: 0.2729498 total: 9.42ms remaining: 25.5ms
27: learn: 0.2685974 total: 9.67ms remaining: 24.9ms
28: learn: 0.2648926 total: 9.92ms remaining: 24.3ms
29: learn: 0.2629856 total: 10.2ms remaining: 23.7ms
30: learn: 0.2605379 total: 10.7ms remaining: 23.7ms
31: learn: 0.2583238 total: 11.1ms remaining: 23.5ms
32: learn: 0.2549441 total: 11.9ms remaining: 24.1ms
33: learn: 0.2528828 total: 12.1ms remaining: 23.6ms
34: learn: 0.2500136 total: 12.4ms remaining: 23ms
35: learn: 0.2472779 total: 12.7ms remaining: 22.5ms
36: learn: 0.2443514 total: 13.2ms remaining: 22.6ms
37: learn: 0.2422131 total: 13.6ms remaining: 22.1ms
38: learn: 0.2400465 total: 13.8ms remaining: 21.5ms
39: learn: 0.2378067 total: 14ms remaining: 21ms
40: learn: 0.2362807 total: 14.2ms remaining: 20.5ms
41: learn: 0.2342833 total: 14.5ms remaining: 20ms
42: learn: 0.2325844 total: 14.7ms remaining: 19.5ms
43: learn: 0.2309996 total: 14.9ms remaining: 19ms
44: learn: 0.2297499 total: 15.1ms remaining: 18.5ms
45: learn: 0.2277609 total: 15.4ms remaining: 18.1ms
46: learn: 0.2248966 total: 16.2ms remaining: 18.3ms
47: learn: 0.2233626 total: 16.5ms remaining: 17.9ms
48: learn: 0.2210270 total: 16.7ms remaining: 17.4ms
49: learn: 0.2200753 total: 17.1ms remaining: 17.1ms
50: learn: 0.2187332 total: 17.3ms remaining: 16.6ms
51: learn: 0.2163816 total: 17.6ms remaining: 16.3ms
52: learn: 0.2149486 total: 18.1ms remaining: 16.1ms
53: learn: 0.2138791 total: 18.7ms remaining: 15.9ms
54: learn: 0.2127221 total: 19ms remaining: 15.6ms
55: learn: 0.2106814 total: 19.5ms remaining: 15.3ms
56: learn: 0.2094962 total: 20ms remaining: 15.1ms
57: learn: 0.2069710 total: 20.3ms remaining: 14.7ms
58: learn: 0.2056220 total: 20.7ms remaining: 14.4ms
59: learn: 0.2035830 total: 21.1ms remaining: 14.1ms
60: learn: 0.2030326 total: 21.3ms remaining: 13.6ms
61: learn: 0.2013024 total: 22ms remaining: 13.5ms
62: learn: 0.1998871 total: 22.3ms remaining: 13.1ms
63: learn: 0.1983454 total: 22.7ms remaining: 12.7ms
64: learn: 0.1963874 total: 23ms remaining: 12.4ms
65: learn: 0.1944519 total: 23.3ms remaining: 12ms
66: learn: 0.1928225 total: 23.8ms remaining: 11.7ms
67: learn: 0.1914112 total: 24.3ms remaining: 11.4ms
68: learn: 0.1897957 total: 24.7ms remaining: 11.1ms
69: learn: 0.1890089 total: 25.3ms remaining: 10.8ms
70: learn: 0.1879103 total: 25.9ms remaining: 10.6ms
71: learn: 0.1863096 total: 26.4ms remaining: 10.3ms
72: learn: 0.1852996 total: 27ms remaining: 9.97ms
73: learn: 0.1844780 total: 27.6ms remaining: 9.7ms
74: learn: 0.1826719 total: 27.8ms remaining: 9.27ms
75: learn: 0.1818801 total: 28ms remaining: 8.85ms
76: learn: 0.1810469 total: 28.3ms remaining: 8.44ms
77: learn: 0.1801707 total: 28.5ms remaining: 8.04ms
78: learn: 0.1788266 total: 28.7ms remaining: 7.64ms
79: learn: 0.1774748 total: 30ms remaining: 7.51ms
80: learn: 0.1769838 total: 30.4ms remaining: 7.12ms
81: learn: 0.1755796 total: 30.7ms remaining: 6.74ms
82: learn: 0.1747006 total: 31ms remaining: 6.35ms
83: learn: 0.1733876 total: 31.3ms remaining: 5.96ms
84: learn: 0.1723565 total: 31.5ms remaining: 5.55ms
85: learn: 0.1717806 total: 31.9ms remaining: 5.19ms
86: learn: 0.1710007 total: 32.2ms remaining: 4.81ms
87: learn: 0.1701884 total: 32.5ms remaining: 4.42ms
88: learn: 0.1691883 total: 32.8ms remaining: 4.05ms
89: learn: 0.1687620 total: 33.1ms remaining: 3.68ms
90: learn: 0.1679230 total: 33.5ms remaining: 3.31ms
91: learn: 0.1669217 total: 33.7ms remaining: 2.93ms
92: learn: 0.1661735 total: 34ms remaining: 2.56ms
93: learn: 0.1652063 total: 34.3ms remaining: 2.19ms
94: learn: 0.1641294 total: 35.1ms remaining: 1.84ms
95: learn: 0.1635903 total: 35.4ms remaining: 1.47ms
96: learn: 0.1629956 total: 35.7ms remaining: 1.1ms
97: learn: 0.1625569 total: 36.3ms remaining: 739us
98: learn: 0.1621201 total: 36.6ms remaining: 369us
99: learn: 0.1617013 total: 36.9ms remaining: 0us
0: learn: 0.6457342 total: 299us remaining: 29.7ms
1: learn: 0.6010041 total: 695us remaining: 34.1ms
2: learn: 0.5607770 total: 1.31ms remaining: 42.4ms
3: learn: 0.5254774 total: 1.81ms remaining: 43.4ms
4: learn: 0.4970300 total: 2.38ms remaining: 45.1ms
5: learn: 0.4712240 total: 2.77ms remaining: 43.4ms
6: learn: 0.4472541 total: 3.03ms remaining: 40.2ms
7: learn: 0.4302047 total: 3.24ms remaining: 37.2ms
8: learn: 0.4096284 total: 3.84ms remaining: 38.9ms
9: learn: 0.3943730 total: 4.25ms remaining: 38.3ms
10: learn: 0.3825996 total: 4.44ms remaining: 35.9ms
11: learn: 0.3682381 total: 4.77ms remaining: 35ms
12: learn: 0.3542027 total: 5.09ms remaining: 34.1ms
13: learn: 0.3416179 total: 5.39ms remaining: 33.1ms
14: learn: 0.3315787 total: 5.76ms remaining: 32.7ms
15: learn: 0.3215391 total: 6.16ms remaining: 32.3ms
16: learn: 0.3128192 total: 6.62ms remaining: 32.3ms
17: learn: 0.3060670 total: 6.96ms remaining: 31.7ms
18: learn: 0.2996001 total: 7.21ms remaining: 30.7ms
19: learn: 0.2933228 total: 7.59ms remaining: 30.3ms
20: learn: 0.2865127 total: 8.25ms remaining: 31ms
21: learn: 0.2823357 total: 8.76ms remaining: 31ms
22: learn: 0.2769019 total: 9.38ms remaining: 31.4ms
23: learn: 0.2713227 total: 9.79ms remaining: 31ms
24: learn: 0.2655899 total: 10ms remaining: 30.1ms
25: learn: 0.2602218 total: 10.7ms remaining: 30.5ms
26: learn: 0.2562777 total: 11.3ms remaining: 30.6ms
27: learn: 0.2537223 total: 11.7ms remaining: 30.2ms
28: learn: 0.2511636 total: 12ms remaining: 29.5ms
29: learn: 0.2464586 total: 12.3ms remaining: 28.6ms
30: learn: 0.2430363 total: 12.5ms remaining: 27.9ms
31: learn: 0.2402076 total: 12.8ms remaining: 27.2ms
32: learn: 0.2372463 total: 13ms remaining: 26.4ms
33: learn: 0.2343819 total: 13.3ms remaining: 25.8ms
34: learn: 0.2322473 total: 13.5ms remaining: 25.1ms
35: learn: 0.2294492 total: 13.8ms remaining: 24.5ms
36: learn: 0.2274605 total: 14.1ms remaining: 24.1ms
37: learn: 0.2247957 total: 14.4ms remaining: 23.5ms
38: learn: 0.2220142 total: 15ms remaining: 23.4ms
39: learn: 0.2205209 total: 15.7ms remaining: 23.6ms
40: learn: 0.2186829 total: 16.4ms remaining: 23.6ms
41: learn: 0.2163716 total: 16.7ms remaining: 23ms
42: learn: 0.2141476 total: 17ms remaining: 22.5ms
43: learn: 0.2117608 total: 17.4ms remaining: 22.1ms
44: learn: 0.2093530 total: 17.9ms remaining: 21.8ms
45: learn: 0.2075321 total: 18.2ms remaining: 21.3ms
46: learn: 0.2060894 total: 18.6ms remaining: 21ms
47: learn: 0.2051089 total: 18.8ms remaining: 20.3ms
48: learn: 0.2037288 total: 19ms remaining: 19.8ms
49: learn: 0.2020560 total: 19.3ms remaining: 19.3ms
50: learn: 0.2012479 total: 19.7ms remaining: 18.9ms
51: learn: 0.2000097 total: 20.1ms remaining: 18.5ms
52: learn: 0.1985358 total: 20.3ms remaining: 18ms
53: learn: 0.1966868 total: 20.9ms remaining: 17.8ms
54: learn: 0.1953039 total: 21.5ms remaining: 17.6ms
55: learn: 0.1942397 total: 22.2ms remaining: 17.5ms
56: learn: 0.1930919 total: 22.8ms remaining: 17.2ms
57: learn: 0.1918476 total: 23.8ms remaining: 17.2ms
58: learn: 0.1905491 total: 24.1ms remaining: 16.8ms
59: learn: 0.1895913 total: 24.5ms remaining: 16.3ms
60: learn: 0.1876756 total: 24.8ms remaining: 15.9ms
61: learn: 0.1864385 total: 25.5ms remaining: 15.6ms
62: learn: 0.1857486 total: 26.2ms remaining: 15.4ms
63: learn: 0.1843478 total: 27.2ms remaining: 15.3ms
64: learn: 0.1832449 total: 27.4ms remaining: 14.8ms
65: learn: 0.1824715 total: 27.7ms remaining: 14.3ms
66: learn: 0.1813436 total: 28ms remaining: 13.8ms
67: learn: 0.1802899 total: 28.3ms remaining: 13.3ms
68: learn: 0.1795508 total: 28.9ms remaining: 13ms
69: learn: 0.1785407 total: 29.4ms remaining: 12.6ms
70: learn: 0.1773668 total: 29.8ms remaining: 12.2ms
71: learn: 0.1753779 total: 30.2ms remaining: 11.7ms
72: learn: 0.1744473 total: 30.4ms remaining: 11.2ms
73: learn: 0.1731742 total: 30.7ms remaining: 10.8ms
74: learn: 0.1719759 total: 31.9ms remaining: 10.6ms
75: learn: 0.1708276 total: 32.2ms remaining: 10.2ms
76: learn: 0.1700271 total: 32.9ms remaining: 9.83ms
77: learn: 0.1691305 total: 33.4ms remaining: 9.43ms
78: learn: 0.1681001 total: 34.1ms remaining: 9.06ms
79: learn: 0.1671839 total: 34.8ms remaining: 8.71ms
80: learn: 0.1660925 total: 35.6ms remaining: 8.35ms
81: learn: 0.1651765 total: 36.3ms remaining: 7.96ms
82: learn: 0.1640415 total: 36.7ms remaining: 7.51ms
83: learn: 0.1632295 total: 37.1ms remaining: 7.07ms
84: learn: 0.1629375 total: 37.6ms remaining: 6.63ms
85: learn: 0.1618696 total: 37.8ms remaining: 6.16ms
86: learn: 0.1616127 total: 38.1ms remaining: 5.7ms
87: learn: 0.1608334 total: 38.6ms remaining: 5.27ms
88: learn: 0.1599306 total: 39.1ms remaining: 4.83ms
89: learn: 0.1594392 total: 39.6ms remaining: 4.39ms
90: learn: 0.1584703 total: 40.3ms remaining: 3.98ms
91: learn: 0.1577935 total: 40.9ms remaining: 3.55ms
92: learn: 0.1573067 total: 41.8ms remaining: 3.15ms
93: learn: 0.1565912 total: 42.8ms remaining: 2.73ms
94: learn: 0.1559672 total: 43.5ms remaining: 2.29ms
95: learn: 0.1553522 total: 43.9ms remaining: 1.83ms
96: learn: 0.1543906 total: 44.3ms remaining: 1.37ms
97: learn: 0.1535468 total: 44.7ms remaining: 912us
98: learn: 0.1528294 total: 45.1ms remaining: 455us
99: learn: 0.1522443 total: 45.7ms remaining: 0us
0: learn: 0.6495311 total: 299us remaining: 29.6ms
1: learn: 0.6040737 total: 500us remaining: 24.5ms
2: learn: 0.5672379 total: 737us remaining: 23.8ms
3: learn: 0.5406197 total: 973us remaining: 23.4ms
4: learn: 0.5151644 total: 1.23ms remaining: 23.4ms
5: learn: 0.4914903 total: 1.37ms remaining: 21.5ms
6: learn: 0.4692605 total: 1.55ms remaining: 20.6ms
7: learn: 0.4519017 total: 1.78ms remaining: 20.5ms
8: learn: 0.4380025 total: 2.5ms remaining: 25.3ms
9: learn: 0.4235189 total: 2.65ms remaining: 23.8ms
10: learn: 0.4113278 total: 2.96ms remaining: 24ms
11: learn: 0.3991034 total: 3.2ms remaining: 23.4ms
12: learn: 0.3858848 total: 3.48ms remaining: 23.3ms
13: learn: 0.3728550 total: 3.82ms remaining: 23.4ms
14: learn: 0.3636348 total: 4.05ms remaining: 22.9ms
15: learn: 0.3534055 total: 4.3ms remaining: 22.6ms
16: learn: 0.3436837 total: 4.6ms remaining: 22.5ms
17: learn: 0.3372644 total: 5.09ms remaining: 23.2ms
18: learn: 0.3313884 total: 5.68ms remaining: 24.2ms
19: learn: 0.3262682 total: 6.32ms remaining: 25.3ms
20: learn: 0.3172674 total: 6.69ms remaining: 25.2ms
21: learn: 0.3110035 total: 6.99ms remaining: 24.8ms
22: learn: 0.3032744 total: 7.33ms remaining: 24.5ms
23: learn: 0.2978424 total: 7.79ms remaining: 24.7ms
24: learn: 0.2923408 total: 8.44ms remaining: 25.3ms
25: learn: 0.2896119 total: 8.69ms remaining: 24.7ms
26: learn: 0.2859146 total: 8.88ms remaining: 24ms
27: learn: 0.2821476 total: 9.1ms remaining: 23.4ms
28: learn: 0.2781989 total: 9.32ms remaining: 22.8ms
29: learn: 0.2739802 total: 9.57ms remaining: 22.3ms
30: learn: 0.2710510 total: 9.89ms remaining: 22ms
31: learn: 0.2675356 total: 10.1ms remaining: 21.5ms
32: learn: 0.2641957 total: 10.3ms remaining: 20.9ms
33: learn: 0.2593319 total: 10.5ms remaining: 20.4ms
34: learn: 0.2557795 total: 10.7ms remaining: 19.8ms
35: learn: 0.2535475 total: 10.8ms remaining: 19.3ms
36: learn: 0.2505609 total: 11.1ms remaining: 18.9ms
37: learn: 0.2480422 total: 11.3ms remaining: 18.5ms
38: learn: 0.2455669 total: 11.5ms remaining: 18ms
39: learn: 0.2439872 total: 12ms remaining: 18ms
40: learn: 0.2422787 total: 12.1ms remaining: 17.5ms
41: learn: 0.2409530 total: 12.6ms remaining: 17.3ms
42: learn: 0.2383699 total: 12.9ms remaining: 17.1ms
43: learn: 0.2357869 total: 13.5ms remaining: 17.2ms
44: learn: 0.2337662 total: 13.8ms remaining: 16.9ms
45: learn: 0.2326745 total: 14.1ms remaining: 16.5ms
46: learn: 0.2309611 total: 14.3ms remaining: 16.2ms
47: learn: 0.2293255 total: 14.6ms remaining: 15.8ms
48: learn: 0.2276355 total: 14.9ms remaining: 15.5ms
49: learn: 0.2256338 total: 15.2ms remaining: 15.2ms
50: learn: 0.2230134 total: 15.4ms remaining: 14.8ms
51: learn: 0.2215946 total: 15.6ms remaining: 14.4ms
52: learn: 0.2190297 total: 15.8ms remaining: 14ms
53: learn: 0.2169878 total: 16ms remaining: 13.6ms
54: learn: 0.2153076 total: 16.2ms remaining: 13.2ms
55: learn: 0.2139220 total: 16.4ms remaining: 12.9ms
56: learn: 0.2124221 total: 16.6ms remaining: 12.5ms
57: learn: 0.2108960 total: 16.8ms remaining: 12.2ms
58: learn: 0.2088332 total: 17.1ms remaining: 11.9ms
59: learn: 0.2074714 total: 17.3ms remaining: 11.5ms
60: learn: 0.2060640 total: 17.5ms remaining: 11.2ms
61: learn: 0.2055411 total: 17.8ms remaining: 10.9ms
62: learn: 0.2045437 total: 18.1ms remaining: 10.6ms
63: learn: 0.2027296 total: 18.3ms remaining: 10.3ms
64: learn: 0.2002059 total: 18.5ms remaining: 9.99ms
65: learn: 0.1987645 total: 18.8ms remaining: 9.66ms
66: learn: 0.1965463 total: 19ms remaining: 9.34ms
67: learn: 0.1950846 total: 19.4ms remaining: 9.13ms
68: learn: 0.1938663 total: 19.7ms remaining: 8.83ms
69: learn: 0.1921205 total: 19.9ms remaining: 8.52ms
70: learn: 0.1898144 total: 20.1ms remaining: 8.2ms
71: learn: 0.1890448 total: 20.3ms remaining: 7.91ms
72: learn: 0.1877640 total: 20.6ms remaining: 7.62ms
73: learn: 0.1861304 total: 20.9ms remaining: 7.33ms
74: learn: 0.1838183 total: 21.1ms remaining: 7.03ms
75: learn: 0.1817840 total: 21.3ms remaining: 6.74ms
76: learn: 0.1811012 total: 21.6ms remaining: 6.44ms
77: learn: 0.1801302 total: 21.8ms remaining: 6.14ms
78: learn: 0.1788358 total: 22ms remaining: 5.86ms
79: learn: 0.1771961 total: 22.7ms remaining: 5.67ms
80: learn: 0.1765127 total: 22.9ms remaining: 5.38ms
81: learn: 0.1752301 total: 23.2ms remaining: 5.09ms
82: learn: 0.1746056 total: 23.5ms remaining: 4.82ms
83: learn: 0.1732466 total: 23.9ms remaining: 4.54ms
84: learn: 0.1724781 total: 24.1ms remaining: 4.25ms
85: learn: 0.1711030 total: 24.3ms remaining: 3.95ms
86: learn: 0.1702846 total: 24.5ms remaining: 3.66ms
87: learn: 0.1697629 total: 24.8ms remaining: 3.38ms
88: learn: 0.1691562 total: 25.2ms remaining: 3.12ms
89: learn: 0.1683050 total: 25.5ms remaining: 2.83ms
90: learn: 0.1675551 total: 25.7ms remaining: 2.54ms
91: learn: 0.1668608 total: 25.9ms remaining: 2.25ms
92: learn: 0.1659986 total: 26.2ms remaining: 1.97ms
93: learn: 0.1652068 total: 26.3ms remaining: 1.68ms
94: learn: 0.1642219 total: 26.5ms remaining: 1.4ms
95: learn: 0.1635650 total: 26.7ms remaining: 1.11ms
96: learn: 0.1630106 total: 27ms remaining: 835us
97: learn: 0.1618195 total: 27.2ms remaining: 556us
98: learn: 0.1612227 total: 27.6ms remaining: 278us
99: learn: 0.1607275 total: 28.1ms remaining: 0us
Evaluating param combo 14/16: {'boosted_cb_model__iterations': 100, 'boosted_cb_model__learning_rate': 0.1, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 16}
Duplicate found for this combo (hash=a3460f9ae7f64a9daef2957b0cabf029). Removing old run: 3225fd9f8c16448aaae28f30edb5a0bf
0: learn: 0.6462693 total: 477us remaining: 47.2ms
1: learn: 0.6006126 total: 805us remaining: 39.5ms
2: learn: 0.5631732 total: 1.2ms remaining: 38.9ms
3: learn: 0.5318762 total: 1.81ms remaining: 43.6ms
4: learn: 0.5054558 total: 2.39ms remaining: 45.4ms
5: learn: 0.4799320 total: 2.71ms remaining: 42.5ms
6: learn: 0.4585994 total: 2.99ms remaining: 39.7ms
7: learn: 0.4417764 total: 3.22ms remaining: 37.1ms
8: learn: 0.4225440 total: 3.39ms remaining: 34.3ms
9: learn: 0.4084612 total: 3.69ms remaining: 33.3ms
10: learn: 0.3962466 total: 4.04ms remaining: 32.6ms
11: learn: 0.3845676 total: 4.28ms remaining: 31.4ms
12: learn: 0.3717729 total: 5.05ms remaining: 33.8ms
13: learn: 0.3609883 total: 6.07ms remaining: 37.3ms
14: learn: 0.3500067 total: 6.59ms remaining: 37.3ms
15: learn: 0.3412218 total: 7.65ms remaining: 40.1ms
16: learn: 0.3315580 total: 8.84ms remaining: 43.2ms
17: learn: 0.3252685 total: 9.34ms remaining: 42.6ms
18: learn: 0.3195212 total: 9.85ms remaining: 42ms
19: learn: 0.3143782 total: 10.8ms remaining: 43.3ms
20: learn: 0.3089230 total: 11.5ms remaining: 43.3ms
21: learn: 0.3037625 total: 12.1ms remaining: 42.9ms
22: learn: 0.2990881 total: 12.6ms remaining: 42.2ms
23: learn: 0.2939303 total: 14.1ms remaining: 44.8ms
24: learn: 0.2891699 total: 14.7ms remaining: 44.2ms
25: learn: 0.2840286 total: 15.4ms remaining: 43.7ms
26: learn: 0.2792264 total: 16ms remaining: 43.4ms
27: learn: 0.2746170 total: 17.3ms remaining: 44.4ms
28: learn: 0.2711557 total: 17.7ms remaining: 43.4ms
29: learn: 0.2693929 total: 18.4ms remaining: 42.9ms
30: learn: 0.2660210 total: 18.9ms remaining: 42.1ms
31: learn: 0.2638487 total: 19.3ms remaining: 41.1ms
32: learn: 0.2615085 total: 20.1ms remaining: 40.8ms
33: learn: 0.2594956 total: 21.2ms remaining: 41.1ms
34: learn: 0.2566762 total: 22ms remaining: 40.8ms
35: learn: 0.2542834 total: 22.7ms remaining: 40.4ms
36: learn: 0.2522301 total: 23.2ms remaining: 39.6ms
37: learn: 0.2495766 total: 23.6ms remaining: 38.5ms
38: learn: 0.2473101 total: 24.1ms remaining: 37.7ms
39: learn: 0.2450895 total: 24.3ms remaining: 36.4ms
40: learn: 0.2426112 total: 24.6ms remaining: 35.4ms
41: learn: 0.2396711 total: 25.2ms remaining: 34.9ms
42: learn: 0.2378629 total: 25.9ms remaining: 34.3ms
43: learn: 0.2352862 total: 26.4ms remaining: 33.6ms
44: learn: 0.2334733 total: 26.8ms remaining: 32.8ms
45: learn: 0.2314837 total: 27.3ms remaining: 32ms
46: learn: 0.2290257 total: 27.7ms remaining: 31.2ms
47: learn: 0.2272964 total: 28.2ms remaining: 30.5ms
48: learn: 0.2253947 total: 28.5ms remaining: 29.7ms
49: learn: 0.2237654 total: 29.2ms remaining: 29.2ms
50: learn: 0.2223406 total: 29.6ms remaining: 28.5ms
51: learn: 0.2210548 total: 30ms remaining: 27.7ms
52: learn: 0.2195405 total: 30.2ms remaining: 26.8ms
53: learn: 0.2183775 total: 30.5ms remaining: 26ms
54: learn: 0.2169509 total: 30.7ms remaining: 25.1ms
55: learn: 0.2154372 total: 31ms remaining: 24.3ms
56: learn: 0.2137457 total: 31.5ms remaining: 23.8ms
57: learn: 0.2119889 total: 31.7ms remaining: 23ms
58: learn: 0.2109968 total: 32ms remaining: 22.2ms
59: learn: 0.2094826 total: 32.2ms remaining: 21.4ms
60: learn: 0.2085079 total: 32.5ms remaining: 20.8ms
61: learn: 0.2071628 total: 32.7ms remaining: 20.1ms
62: learn: 0.2062468 total: 33.3ms remaining: 19.6ms
63: learn: 0.2043969 total: 33.5ms remaining: 18.9ms
64: learn: 0.2031529 total: 33.8ms remaining: 18.2ms
65: learn: 0.2014021 total: 34ms remaining: 17.5ms
66: learn: 0.2003742 total: 34.2ms remaining: 16.8ms
67: learn: 0.1991346 total: 34.4ms remaining: 16.2ms
68: learn: 0.1982389 total: 34.7ms remaining: 15.6ms
69: learn: 0.1974659 total: 35.5ms remaining: 15.2ms
70: learn: 0.1961055 total: 35.8ms remaining: 14.6ms
71: learn: 0.1953784 total: 36.1ms remaining: 14ms
72: learn: 0.1941287 total: 36.3ms remaining: 13.4ms
73: learn: 0.1930324 total: 37ms remaining: 13ms
74: learn: 0.1924753 total: 37.7ms remaining: 12.6ms
75: learn: 0.1915718 total: 38.1ms remaining: 12ms
76: learn: 0.1902028 total: 38.4ms remaining: 11.5ms
77: learn: 0.1892241 total: 38.6ms remaining: 10.9ms
78: learn: 0.1883913 total: 39ms remaining: 10.4ms
79: learn: 0.1869245 total: 39.2ms remaining: 9.8ms
80: learn: 0.1862597 total: 39.4ms remaining: 9.25ms
81: learn: 0.1850881 total: 39.6ms remaining: 8.7ms
82: learn: 0.1844696 total: 39.9ms remaining: 8.18ms
83: learn: 0.1836129 total: 40.6ms remaining: 7.73ms
84: learn: 0.1831236 total: 41.2ms remaining: 7.28ms
85: learn: 0.1826632 total: 41.5ms remaining: 6.75ms
86: learn: 0.1820750 total: 42.2ms remaining: 6.31ms
87: learn: 0.1812083 total: 42.6ms remaining: 5.81ms
88: learn: 0.1801868 total: 42.9ms remaining: 5.3ms
89: learn: 0.1796862 total: 43.1ms remaining: 4.78ms
90: learn: 0.1789454 total: 43.3ms remaining: 4.28ms
91: learn: 0.1784504 total: 43.5ms remaining: 3.79ms
92: learn: 0.1772510 total: 43.8ms remaining: 3.29ms
93: learn: 0.1768141 total: 44.1ms remaining: 2.81ms
94: learn: 0.1760395 total: 44.3ms remaining: 2.33ms
95: learn: 0.1755551 total: 44.5ms remaining: 1.85ms
96: learn: 0.1747803 total: 44.7ms remaining: 1.38ms
97: learn: 0.1743621 total: 45ms remaining: 917us
98: learn: 0.1740438 total: 45.3ms remaining: 457us
99: learn: 0.1732493 total: 45.6ms remaining: 0us
0: learn: 0.6437160 total: 265us remaining: 26.3ms
1: learn: 0.5949760 total: 488us remaining: 23.9ms
2: learn: 0.5565073 total: 671us remaining: 21.7ms
3: learn: 0.5265837 total: 906us remaining: 21.8ms
4: learn: 0.5005287 total: 1.29ms remaining: 24.6ms
5: learn: 0.4720428 total: 1.65ms remaining: 25.8ms
6: learn: 0.4484482 total: 1.91ms remaining: 25.4ms
7: learn: 0.4293736 total: 2.22ms remaining: 25.5ms
8: learn: 0.4148507 total: 2.42ms remaining: 24.5ms
9: learn: 0.3977165 total: 2.69ms remaining: 24.2ms
10: learn: 0.3825212 total: 3.57ms remaining: 28.9ms
11: learn: 0.3667856 total: 3.77ms remaining: 27.7ms
12: learn: 0.3547366 total: 4ms remaining: 26.8ms
13: learn: 0.3424929 total: 4.24ms remaining: 26ms
14: learn: 0.3349308 total: 4.46ms remaining: 25.3ms
15: learn: 0.3224127 total: 4.66ms remaining: 24.4ms
16: learn: 0.3178131 total: 4.81ms remaining: 23.5ms
17: learn: 0.3131939 total: 5.1ms remaining: 23.2ms
18: learn: 0.3045147 total: 5.31ms remaining: 22.6ms
19: learn: 0.2970139 total: 5.5ms remaining: 22ms
20: learn: 0.2918113 total: 5.74ms remaining: 21.6ms
21: learn: 0.2854668 total: 5.92ms remaining: 21ms
22: learn: 0.2779652 total: 6.07ms remaining: 20.3ms
23: learn: 0.2734233 total: 6.33ms remaining: 20ms
24: learn: 0.2685745 total: 6.76ms remaining: 20.3ms
25: learn: 0.2629943 total: 7.15ms remaining: 20.4ms
26: learn: 0.2604321 total: 7.46ms remaining: 20.2ms
27: learn: 0.2566107 total: 7.75ms remaining: 19.9ms
28: learn: 0.2531121 total: 8.17ms remaining: 20ms
29: learn: 0.2515028 total: 8.46ms remaining: 19.8ms
30: learn: 0.2482660 total: 8.84ms remaining: 19.7ms
31: learn: 0.2458159 total: 9.25ms remaining: 19.7ms
32: learn: 0.2416483 total: 9.63ms remaining: 19.6ms
33: learn: 0.2386672 total: 10.1ms remaining: 19.7ms
34: learn: 0.2359889 total: 10.5ms remaining: 19.5ms
35: learn: 0.2338575 total: 10.8ms remaining: 19.2ms
36: learn: 0.2298220 total: 11.1ms remaining: 19ms
37: learn: 0.2283967 total: 11.5ms remaining: 18.8ms
38: learn: 0.2263530 total: 11.9ms remaining: 18.6ms
39: learn: 0.2246581 total: 12.3ms remaining: 18.4ms
40: learn: 0.2223378 total: 12.7ms remaining: 18.3ms
41: learn: 0.2207038 total: 13ms remaining: 18ms
42: learn: 0.2197856 total: 13.4ms remaining: 17.8ms
43: learn: 0.2188898 total: 13.7ms remaining: 17.5ms
44: learn: 0.2176194 total: 13.9ms remaining: 17ms
45: learn: 0.2163312 total: 14.3ms remaining: 16.8ms
46: learn: 0.2154455 total: 14.7ms remaining: 16.6ms
47: learn: 0.2139407 total: 15ms remaining: 16.2ms
48: learn: 0.2120411 total: 15.4ms remaining: 16ms
49: learn: 0.2106386 total: 15.8ms remaining: 15.8ms
50: learn: 0.2091600 total: 16.2ms remaining: 15.6ms
51: learn: 0.2072987 total: 16.5ms remaining: 15.2ms
52: learn: 0.2045983 total: 16.9ms remaining: 15ms
53: learn: 0.2034906 total: 17.3ms remaining: 14.7ms
54: learn: 0.2021212 total: 17.7ms remaining: 14.5ms
55: learn: 0.1991710 total: 18.2ms remaining: 14.3ms
56: learn: 0.1976866 total: 18.5ms remaining: 13.9ms
57: learn: 0.1968438 total: 18.8ms remaining: 13.6ms
58: learn: 0.1956886 total: 19.1ms remaining: 13.3ms
59: learn: 0.1934149 total: 19.6ms remaining: 13.1ms
60: learn: 0.1920875 total: 20.1ms remaining: 12.8ms
61: learn: 0.1911042 total: 20.5ms remaining: 12.5ms
62: learn: 0.1898230 total: 20.8ms remaining: 12.2ms
63: learn: 0.1889307 total: 21.1ms remaining: 11.9ms
64: learn: 0.1869322 total: 21.5ms remaining: 11.6ms
65: learn: 0.1850626 total: 21.9ms remaining: 11.3ms
66: learn: 0.1842326 total: 22.4ms remaining: 11ms
67: learn: 0.1824564 total: 22.7ms remaining: 10.7ms
68: learn: 0.1816802 total: 23.1ms remaining: 10.4ms
69: learn: 0.1799241 total: 23.5ms remaining: 10.1ms
70: learn: 0.1785063 total: 23.8ms remaining: 9.71ms
71: learn: 0.1768805 total: 24.1ms remaining: 9.38ms
72: learn: 0.1755565 total: 24.6ms remaining: 9.09ms
73: learn: 0.1741842 total: 24.9ms remaining: 8.76ms
74: learn: 0.1729470 total: 25.4ms remaining: 8.46ms
75: learn: 0.1717045 total: 25.8ms remaining: 8.15ms
76: learn: 0.1707227 total: 26.2ms remaining: 7.82ms
77: learn: 0.1697844 total: 26.4ms remaining: 7.45ms
78: learn: 0.1687726 total: 26.8ms remaining: 7.13ms
79: learn: 0.1677323 total: 27.3ms remaining: 6.82ms
80: learn: 0.1666055 total: 27.7ms remaining: 6.49ms
81: learn: 0.1653523 total: 28ms remaining: 6.14ms
82: learn: 0.1644991 total: 28.4ms remaining: 5.81ms
83: learn: 0.1637596 total: 28.8ms remaining: 5.49ms
84: learn: 0.1626667 total: 29.3ms remaining: 5.17ms
85: learn: 0.1618658 total: 29.8ms remaining: 4.85ms
86: learn: 0.1610996 total: 30.2ms remaining: 4.51ms
87: learn: 0.1604054 total: 30.5ms remaining: 4.16ms
88: learn: 0.1599545 total: 30.8ms remaining: 3.81ms
89: learn: 0.1593649 total: 31.2ms remaining: 3.46ms
90: learn: 0.1588678 total: 31.6ms remaining: 3.13ms
91: learn: 0.1582488 total: 32.1ms remaining: 2.79ms
92: learn: 0.1578052 total: 32.4ms remaining: 2.44ms
93: learn: 0.1574109 total: 32.7ms remaining: 2.08ms
94: learn: 0.1564849 total: 33ms remaining: 1.74ms
95: learn: 0.1554839 total: 33.4ms remaining: 1.39ms
96: learn: 0.1550581 total: 33.9ms remaining: 1.05ms
97: learn: 0.1541604 total: 34.2ms remaining: 697us
98: learn: 0.1538731 total: 34.4ms remaining: 347us
99: learn: 0.1529953 total: 34.7ms remaining: 0us
0: learn: 0.6454260 total: 301us remaining: 29.8ms
1: learn: 0.6019562 total: 517us remaining: 25.4ms
2: learn: 0.5626070 total: 707us remaining: 22.9ms
3: learn: 0.5307213 total: 932us remaining: 22.4ms
4: learn: 0.5032371 total: 1.14ms remaining: 21.6ms
5: learn: 0.4776693 total: 1.34ms remaining: 21ms
6: learn: 0.4566797 total: 1.54ms remaining: 20.4ms
7: learn: 0.4389420 total: 1.75ms remaining: 20.1ms
8: learn: 0.4181396 total: 1.92ms remaining: 19.4ms
9: learn: 0.4027384 total: 2.12ms remaining: 19.1ms
10: learn: 0.3900032 total: 2.29ms remaining: 18.5ms
11: learn: 0.3777932 total: 2.54ms remaining: 18.6ms
12: learn: 0.3636268 total: 2.74ms remaining: 18.3ms
13: learn: 0.3525592 total: 2.99ms remaining: 18.4ms
14: learn: 0.3415734 total: 3.17ms remaining: 18ms
15: learn: 0.3322054 total: 3.64ms remaining: 19.1ms
16: learn: 0.3233730 total: 4.05ms remaining: 19.8ms
17: learn: 0.3167153 total: 4.54ms remaining: 20.7ms
18: learn: 0.3106400 total: 4.96ms remaining: 21.1ms
19: learn: 0.3023905 total: 5.36ms remaining: 21.4ms
20: learn: 0.2963086 total: 5.7ms remaining: 21.5ms
21: learn: 0.2904163 total: 6.03ms remaining: 21.4ms
22: learn: 0.2852205 total: 6.44ms remaining: 21.6ms
23: learn: 0.2798857 total: 6.88ms remaining: 21.8ms
24: learn: 0.2744258 total: 7.13ms remaining: 21.4ms
25: learn: 0.2718590 total: 7.44ms remaining: 21.2ms
26: learn: 0.2677934 total: 7.74ms remaining: 20.9ms
27: learn: 0.2637715 total: 8.03ms remaining: 20.6ms
28: learn: 0.2593053 total: 8.37ms remaining: 20.5ms
29: learn: 0.2574366 total: 8.69ms remaining: 20.3ms
30: learn: 0.2539525 total: 9.04ms remaining: 20.1ms
31: learn: 0.2512715 total: 9.21ms remaining: 19.6ms
32: learn: 0.2481380 total: 9.49ms remaining: 19.3ms
33: learn: 0.2458802 total: 9.74ms remaining: 18.9ms
34: learn: 0.2430230 total: 10.1ms remaining: 18.8ms
35: learn: 0.2402213 total: 10.3ms remaining: 18.3ms
36: learn: 0.2382592 total: 10.6ms remaining: 18ms
37: learn: 0.2366223 total: 10.8ms remaining: 17.7ms
38: learn: 0.2346630 total: 11.1ms remaining: 17.4ms
39: learn: 0.2316478 total: 11.8ms remaining: 17.7ms
40: learn: 0.2295176 total: 12.2ms remaining: 17.6ms
41: learn: 0.2275613 total: 12.5ms remaining: 17.2ms
42: learn: 0.2261304 total: 12.8ms remaining: 16.9ms
43: learn: 0.2230182 total: 13.2ms remaining: 16.8ms
44: learn: 0.2213852 total: 13.7ms remaining: 16.7ms
45: learn: 0.2198024 total: 14.3ms remaining: 16.7ms
46: learn: 0.2183045 total: 14.7ms remaining: 16.6ms
47: learn: 0.2166827 total: 15ms remaining: 16.2ms
48: learn: 0.2136031 total: 15.8ms remaining: 16.5ms
49: learn: 0.2121566 total: 16.5ms remaining: 16.5ms
50: learn: 0.2102998 total: 17.4ms remaining: 16.7ms
51: learn: 0.2078778 total: 18.2ms remaining: 16.8ms
52: learn: 0.2060268 total: 18.8ms remaining: 16.7ms
53: learn: 0.2048023 total: 19.6ms remaining: 16.7ms
54: learn: 0.2033060 total: 20ms remaining: 16.4ms
55: learn: 0.2012317 total: 20.4ms remaining: 16ms
56: learn: 0.1991270 total: 21ms remaining: 15.9ms
57: learn: 0.1982670 total: 21.9ms remaining: 15.9ms
58: learn: 0.1971084 total: 22.5ms remaining: 15.7ms
59: learn: 0.1955812 total: 23.2ms remaining: 15.5ms
60: learn: 0.1931865 total: 23.9ms remaining: 15.3ms
61: learn: 0.1918434 total: 24.6ms remaining: 15.1ms
62: learn: 0.1909635 total: 25.2ms remaining: 14.8ms
63: learn: 0.1886742 total: 25.8ms remaining: 14.5ms
64: learn: 0.1875385 total: 26.2ms remaining: 14.1ms
65: learn: 0.1859142 total: 26.7ms remaining: 13.7ms
66: learn: 0.1842228 total: 27.1ms remaining: 13.3ms
67: learn: 0.1832482 total: 27.7ms remaining: 13ms
68: learn: 0.1817732 total: 28.3ms remaining: 12.7ms
69: learn: 0.1806441 total: 29.1ms remaining: 12.5ms
70: learn: 0.1797458 total: 29.9ms remaining: 12.2ms
71: learn: 0.1789580 total: 30.4ms remaining: 11.8ms
72: learn: 0.1774616 total: 30.8ms remaining: 11.4ms
73: learn: 0.1765499 total: 31.3ms remaining: 11ms
74: learn: 0.1752367 total: 31.6ms remaining: 10.5ms
75: learn: 0.1747060 total: 32.2ms remaining: 10.2ms
76: learn: 0.1740200 total: 32.6ms remaining: 9.74ms
77: learn: 0.1731577 total: 33.1ms remaining: 9.34ms
78: learn: 0.1725317 total: 33.6ms remaining: 8.93ms
79: learn: 0.1709006 total: 34ms remaining: 8.51ms
80: learn: 0.1701828 total: 34.6ms remaining: 8.12ms
81: learn: 0.1694006 total: 35.3ms remaining: 7.75ms
82: learn: 0.1681916 total: 35.9ms remaining: 7.34ms
83: learn: 0.1674656 total: 36.2ms remaining: 6.9ms
84: learn: 0.1668704 total: 36.5ms remaining: 6.44ms
85: learn: 0.1663118 total: 37ms remaining: 6.02ms
86: learn: 0.1658180 total: 37.5ms remaining: 5.61ms
87: learn: 0.1651004 total: 38.1ms remaining: 5.2ms
88: learn: 0.1644411 total: 38.6ms remaining: 4.77ms
89: learn: 0.1638417 total: 39.1ms remaining: 4.34ms
90: learn: 0.1633229 total: 39.5ms remaining: 3.9ms
91: learn: 0.1629261 total: 39.8ms remaining: 3.46ms
92: learn: 0.1623520 total: 40.5ms remaining: 3.04ms
93: learn: 0.1618410 total: 41ms remaining: 2.61ms
94: learn: 0.1611050 total: 41.6ms remaining: 2.19ms
95: learn: 0.1606811 total: 42ms remaining: 1.75ms
96: learn: 0.1592629 total: 42.2ms remaining: 1.3ms
97: learn: 0.1581160 total: 42.5ms remaining: 867us
98: learn: 0.1577026 total: 42.9ms remaining: 433us
99: learn: 0.1573761 total: 43.4ms remaining: 0us
0: learn: 0.6470966 total: 296us remaining: 29.3ms
1: learn: 0.6001838 total: 588us remaining: 28.8ms
2: learn: 0.5604623 total: 805us remaining: 26ms
3: learn: 0.5261755 total: 1.07ms remaining: 25.8ms
4: learn: 0.4992383 total: 1.24ms remaining: 23.6ms
5: learn: 0.4721898 total: 1.47ms remaining: 23ms
6: learn: 0.4498006 total: 1.81ms remaining: 24ms
7: learn: 0.4314508 total: 2.07ms remaining: 23.9ms
8: learn: 0.4101723 total: 2.26ms remaining: 22.9ms
9: learn: 0.3935443 total: 2.46ms remaining: 22.2ms
10: learn: 0.3798147 total: 2.63ms remaining: 21.2ms
11: learn: 0.3665670 total: 2.85ms remaining: 20.9ms
12: learn: 0.3536267 total: 3.09ms remaining: 20.7ms
13: learn: 0.3420766 total: 3.33ms remaining: 20.4ms
14: learn: 0.3326825 total: 3.56ms remaining: 20.2ms
15: learn: 0.3240086 total: 3.8ms remaining: 19.9ms
16: learn: 0.3134403 total: 4.09ms remaining: 20ms
17: learn: 0.3067898 total: 4.27ms remaining: 19.4ms
18: learn: 0.3003032 total: 4.42ms remaining: 18.8ms
19: learn: 0.2951168 total: 4.72ms remaining: 18.9ms
20: learn: 0.2888849 total: 4.99ms remaining: 18.8ms
21: learn: 0.2831076 total: 5.17ms remaining: 18.3ms
22: learn: 0.2763535 total: 5.36ms remaining: 17.9ms
23: learn: 0.2698591 total: 5.55ms remaining: 17.6ms
24: learn: 0.2647141 total: 5.74ms remaining: 17.2ms
25: learn: 0.2590451 total: 6.08ms remaining: 17.3ms
26: learn: 0.2557403 total: 6.42ms remaining: 17.4ms
27: learn: 0.2526011 total: 8.53ms remaining: 21.9ms
28: learn: 0.2495110 total: 8.91ms remaining: 21.8ms
29: learn: 0.2446900 total: 11.5ms remaining: 26.9ms
30: learn: 0.2404218 total: 12.4ms remaining: 27.6ms
31: learn: 0.2370704 total: 13.1ms remaining: 27.9ms
32: learn: 0.2333679 total: 13.4ms remaining: 27.3ms
33: learn: 0.2298418 total: 13.8ms remaining: 26.8ms
34: learn: 0.2271491 total: 14.3ms remaining: 26.5ms
35: learn: 0.2245757 total: 14.5ms remaining: 25.8ms
36: learn: 0.2214767 total: 14.7ms remaining: 25.1ms
37: learn: 0.2191640 total: 15.1ms remaining: 24.6ms
38: learn: 0.2178533 total: 15.8ms remaining: 24.7ms
39: learn: 0.2163079 total: 16.1ms remaining: 24.1ms
40: learn: 0.2149998 total: 16.4ms remaining: 23.6ms
41: learn: 0.2135240 total: 16.7ms remaining: 23ms
42: learn: 0.2118976 total: 17ms remaining: 22.6ms
43: learn: 0.2106812 total: 17.4ms remaining: 22.1ms
44: learn: 0.2087706 total: 17.7ms remaining: 21.6ms
45: learn: 0.2077590 total: 18ms remaining: 21.1ms
46: learn: 0.2062417 total: 18.2ms remaining: 20.5ms
47: learn: 0.2041230 total: 18.5ms remaining: 20ms
48: learn: 0.2029428 total: 18.7ms remaining: 19.5ms
49: learn: 0.2010316 total: 19ms remaining: 19ms
50: learn: 0.1999470 total: 19.4ms remaining: 18.6ms
51: learn: 0.1986324 total: 19.6ms remaining: 18.1ms
52: learn: 0.1981784 total: 19.7ms remaining: 17.5ms
53: learn: 0.1963600 total: 20.1ms remaining: 17.1ms
54: learn: 0.1951677 total: 20.6ms remaining: 16.8ms
55: learn: 0.1934322 total: 20.9ms remaining: 16.4ms
56: learn: 0.1924811 total: 21.1ms remaining: 15.9ms
57: learn: 0.1908243 total: 21.5ms remaining: 15.6ms
58: learn: 0.1892999 total: 21.9ms remaining: 15.2ms
59: learn: 0.1887691 total: 22.5ms remaining: 15ms
60: learn: 0.1867416 total: 22.9ms remaining: 14.6ms
61: learn: 0.1851716 total: 23.3ms remaining: 14.3ms
62: learn: 0.1841887 total: 23.6ms remaining: 13.9ms
63: learn: 0.1826161 total: 24.1ms remaining: 13.6ms
64: learn: 0.1807570 total: 24.7ms remaining: 13.3ms
65: learn: 0.1795807 total: 25.2ms remaining: 13ms
66: learn: 0.1786182 total: 25.6ms remaining: 12.6ms
67: learn: 0.1776957 total: 26ms remaining: 12.2ms
68: learn: 0.1769061 total: 26.4ms remaining: 11.9ms
69: learn: 0.1754625 total: 26.8ms remaining: 11.5ms
70: learn: 0.1743497 total: 27.1ms remaining: 11.1ms
71: learn: 0.1737500 total: 27.6ms remaining: 10.7ms
72: learn: 0.1730347 total: 28ms remaining: 10.4ms
73: learn: 0.1713745 total: 28.5ms remaining: 10ms
74: learn: 0.1697650 total: 29ms remaining: 9.66ms
75: learn: 0.1685269 total: 29.4ms remaining: 9.28ms
76: learn: 0.1678087 total: 29.8ms remaining: 8.9ms
77: learn: 0.1667430 total: 30.2ms remaining: 8.51ms
78: learn: 0.1660898 total: 30.6ms remaining: 8.14ms
79: learn: 0.1653770 total: 31ms remaining: 7.74ms
80: learn: 0.1645453 total: 31.5ms remaining: 7.39ms
81: learn: 0.1635048 total: 31.9ms remaining: 7.01ms
82: learn: 0.1628895 total: 32.4ms remaining: 6.63ms
83: learn: 0.1623059 total: 32.8ms remaining: 6.25ms
84: learn: 0.1608705 total: 33.2ms remaining: 5.86ms
85: learn: 0.1593044 total: 33.9ms remaining: 5.52ms
86: learn: 0.1587007 total: 34.3ms remaining: 5.13ms
87: learn: 0.1578239 total: 34.7ms remaining: 4.73ms
88: learn: 0.1567637 total: 35.1ms remaining: 4.34ms
89: learn: 0.1561612 total: 35.6ms remaining: 3.96ms
90: learn: 0.1556924 total: 36.2ms remaining: 3.58ms
91: learn: 0.1550177 total: 36.6ms remaining: 3.18ms
92: learn: 0.1544644 total: 37ms remaining: 2.79ms
93: learn: 0.1534878 total: 37.7ms remaining: 2.4ms
94: learn: 0.1530854 total: 38ms remaining: 2ms
95: learn: 0.1523530 total: 38.5ms remaining: 1.6ms
96: learn: 0.1518856 total: 39.1ms remaining: 1.21ms
97: learn: 0.1515904 total: 39.6ms remaining: 807us
98: learn: 0.1509596 total: 40.3ms remaining: 406us
99: learn: 0.1503649 total: 40.7ms remaining: 0us
0: learn: 0.6466390 total: 399us remaining: 39.6ms
1: learn: 0.6043306 total: 728us remaining: 35.7ms
2: learn: 0.5698679 total: 995us remaining: 32.2ms
3: learn: 0.5400060 total: 1.44ms remaining: 34.7ms
4: learn: 0.5139000 total: 1.77ms remaining: 33.7ms
5: learn: 0.4909190 total: 2.3ms remaining: 36ms
6: learn: 0.4663019 total: 2.83ms remaining: 37.6ms
7: learn: 0.4496656 total: 3.34ms remaining: 38.4ms
8: learn: 0.4302446 total: 3.95ms remaining: 40ms
9: learn: 0.4155090 total: 4.73ms remaining: 42.6ms
10: learn: 0.4046576 total: 5.28ms remaining: 42.7ms
11: learn: 0.3902395 total: 5.7ms remaining: 41.8ms
12: learn: 0.3781937 total: 6.22ms remaining: 41.6ms
13: learn: 0.3684001 total: 6.64ms remaining: 40.8ms
14: learn: 0.3594224 total: 7.08ms remaining: 40.1ms
15: learn: 0.3506654 total: 7.43ms remaining: 39ms
16: learn: 0.3427953 total: 8.02ms remaining: 39.1ms
17: learn: 0.3375828 total: 8.45ms remaining: 38.5ms
18: learn: 0.3304561 total: 8.87ms remaining: 37.8ms
19: learn: 0.3248426 total: 9.29ms remaining: 37.2ms
20: learn: 0.3187507 total: 9.78ms remaining: 36.8ms
21: learn: 0.3109731 total: 10.3ms remaining: 36.5ms
22: learn: 0.3052623 total: 10.7ms remaining: 35.8ms
23: learn: 0.3002449 total: 11.1ms remaining: 35.1ms
24: learn: 0.2967801 total: 11.6ms remaining: 34.7ms
25: learn: 0.2923089 total: 12ms remaining: 34.3ms
26: learn: 0.2868579 total: 12.5ms remaining: 33.8ms
27: learn: 0.2841698 total: 13.3ms remaining: 34.3ms
28: learn: 0.2810342 total: 13.7ms remaining: 33.6ms
29: learn: 0.2780382 total: 14.1ms remaining: 33ms
30: learn: 0.2758407 total: 14.5ms remaining: 32.2ms
31: learn: 0.2730709 total: 14.9ms remaining: 31.6ms
32: learn: 0.2704504 total: 15.3ms remaining: 31.1ms
33: learn: 0.2684170 total: 15.7ms remaining: 30.5ms
34: learn: 0.2646933 total: 16.3ms remaining: 30.2ms
35: learn: 0.2624324 total: 16.7ms remaining: 29.8ms
36: learn: 0.2604280 total: 17.2ms remaining: 29.3ms
37: learn: 0.2579343 total: 17.6ms remaining: 28.7ms
38: learn: 0.2550934 total: 18.2ms remaining: 28.4ms
39: learn: 0.2526229 total: 18.6ms remaining: 27.8ms
40: learn: 0.2500359 total: 18.9ms remaining: 27.2ms
41: learn: 0.2471391 total: 19.2ms remaining: 26.5ms
42: learn: 0.2454036 total: 19.6ms remaining: 25.9ms
43: learn: 0.2433846 total: 19.9ms remaining: 25.4ms
44: learn: 0.2406459 total: 20.3ms remaining: 24.8ms
45: learn: 0.2380629 total: 21.3ms remaining: 25ms
46: learn: 0.2358969 total: 21.9ms remaining: 24.7ms
47: learn: 0.2344105 total: 22.3ms remaining: 24.2ms
48: learn: 0.2324208 total: 22.8ms remaining: 23.7ms
49: learn: 0.2305732 total: 23.3ms remaining: 23.3ms
50: learn: 0.2295668 total: 24.3ms remaining: 23.4ms
51: learn: 0.2277100 total: 24.8ms remaining: 22.9ms
52: learn: 0.2270326 total: 25.2ms remaining: 22.4ms
53: learn: 0.2254242 total: 25.6ms remaining: 21.8ms
54: learn: 0.2232393 total: 26ms remaining: 21.3ms
55: learn: 0.2212543 total: 26.5ms remaining: 20.8ms
56: learn: 0.2193358 total: 27.3ms remaining: 20.6ms
57: learn: 0.2174221 total: 28ms remaining: 20.3ms
58: learn: 0.2156762 total: 28.6ms remaining: 19.9ms
59: learn: 0.2116688 total: 29.2ms remaining: 19.4ms
60: learn: 0.2099580 total: 29.7ms remaining: 19ms
61: learn: 0.2086876 total: 30.2ms remaining: 18.5ms
62: learn: 0.2070665 total: 30.8ms remaining: 18.1ms
63: learn: 0.2065565 total: 31.7ms remaining: 17.8ms
64: learn: 0.2049650 total: 32.3ms remaining: 17.4ms
65: learn: 0.2031501 total: 32.7ms remaining: 16.8ms
66: learn: 0.2006850 total: 33.1ms remaining: 16.3ms
67: learn: 0.1994539 total: 33.7ms remaining: 15.8ms
68: learn: 0.1982312 total: 34.2ms remaining: 15.3ms
69: learn: 0.1967728 total: 34.7ms remaining: 14.9ms
70: learn: 0.1944350 total: 35.2ms remaining: 14.4ms
71: learn: 0.1926110 total: 35.7ms remaining: 13.9ms
72: learn: 0.1900694 total: 36.2ms remaining: 13.4ms
73: learn: 0.1893151 total: 36.6ms remaining: 12.9ms
74: learn: 0.1883047 total: 37ms remaining: 12.3ms
75: learn: 0.1865259 total: 37.5ms remaining: 11.8ms
76: learn: 0.1848260 total: 37.8ms remaining: 11.3ms
77: learn: 0.1833083 total: 38.2ms remaining: 10.8ms
78: learn: 0.1823737 total: 38.5ms remaining: 10.2ms
79: learn: 0.1803818 total: 38.9ms remaining: 9.73ms
80: learn: 0.1785443 total: 39.3ms remaining: 9.21ms
81: learn: 0.1772803 total: 39.6ms remaining: 8.7ms
82: learn: 0.1763651 total: 40ms remaining: 8.2ms
83: learn: 0.1757682 total: 40.4ms remaining: 7.7ms
84: learn: 0.1743110 total: 40.8ms remaining: 7.19ms
85: learn: 0.1725994 total: 41.6ms remaining: 6.77ms
86: learn: 0.1713861 total: 42.1ms remaining: 6.29ms
87: learn: 0.1705274 total: 42.7ms remaining: 5.82ms
88: learn: 0.1692688 total: 43.1ms remaining: 5.33ms
89: learn: 0.1677366 total: 43.7ms remaining: 4.86ms
90: learn: 0.1669654 total: 44.2ms remaining: 4.37ms
91: learn: 0.1662445 total: 44.7ms remaining: 3.89ms
92: learn: 0.1646721 total: 45.2ms remaining: 3.4ms
93: learn: 0.1637357 total: 46ms remaining: 2.93ms
94: learn: 0.1622838 total: 46.5ms remaining: 2.45ms
95: learn: 0.1612547 total: 46.9ms remaining: 1.95ms
96: learn: 0.1604476 total: 47.3ms remaining: 1.46ms
97: learn: 0.1595644 total: 47.8ms remaining: 975us
98: learn: 0.1585538 total: 48.5ms remaining: 489us
99: learn: 0.1579433 total: 49ms remaining: 0us
0: learn: 0.6501714 total: 647us remaining: 64.1ms
1: learn: 0.6066480 total: 1.55ms remaining: 76.1ms
2: learn: 0.5657020 total: 1.91ms remaining: 61.7ms
3: learn: 0.5314888 total: 2.5ms remaining: 59.9ms
4: learn: 0.5033352 total: 2.98ms remaining: 56.6ms
5: learn: 0.4746589 total: 3.5ms remaining: 54.9ms
6: learn: 0.4545285 total: 4.15ms remaining: 55.1ms
7: learn: 0.4381047 total: 4.83ms remaining: 55.6ms
8: learn: 0.4177237 total: 5.31ms remaining: 53.7ms
9: learn: 0.4023522 total: 5.79ms remaining: 52.1ms
10: learn: 0.3914068 total: 6.18ms remaining: 50ms
11: learn: 0.3787263 total: 6.82ms remaining: 50ms
12: learn: 0.3650055 total: 7.71ms remaining: 51.6ms
13: learn: 0.3522909 total: 8.61ms remaining: 52.9ms
14: learn: 0.3408388 total: 9.45ms remaining: 53.5ms
15: learn: 0.3301578 total: 10.1ms remaining: 53ms
16: learn: 0.3207327 total: 10.6ms remaining: 51.7ms
17: learn: 0.3122013 total: 11.3ms remaining: 51.6ms
18: learn: 0.3065739 total: 12.2ms remaining: 52.1ms
19: learn: 0.2986789 total: 13ms remaining: 51.8ms
20: learn: 0.2920784 total: 13.4ms remaining: 50.3ms
21: learn: 0.2861408 total: 14ms remaining: 49.6ms
22: learn: 0.2806232 total: 14.5ms remaining: 48.4ms
23: learn: 0.2757454 total: 15ms remaining: 47.5ms
24: learn: 0.2715914 total: 15.4ms remaining: 46.1ms
25: learn: 0.2662000 total: 16ms remaining: 45.6ms
26: learn: 0.2620222 total: 16.5ms remaining: 44.6ms
27: learn: 0.2586619 total: 16.9ms remaining: 43.5ms
28: learn: 0.2559411 total: 17.3ms remaining: 42.4ms
29: learn: 0.2521874 total: 18ms remaining: 42ms
30: learn: 0.2487332 total: 18.7ms remaining: 41.6ms
31: learn: 0.2455035 total: 20.5ms remaining: 43.6ms
32: learn: 0.2428433 total: 21.1ms remaining: 42.9ms
33: learn: 0.2410762 total: 21.5ms remaining: 41.7ms
34: learn: 0.2382084 total: 22.1ms remaining: 41ms
35: learn: 0.2366788 total: 22.5ms remaining: 40ms
36: learn: 0.2347484 total: 23ms remaining: 39.1ms
37: learn: 0.2324811 total: 23.3ms remaining: 38.1ms
38: learn: 0.2302696 total: 23.7ms remaining: 37.1ms
39: learn: 0.2282705 total: 24.1ms remaining: 36.2ms
40: learn: 0.2261110 total: 24.6ms remaining: 35.4ms
41: learn: 0.2238719 total: 24.9ms remaining: 34.4ms
42: learn: 0.2215777 total: 25.3ms remaining: 33.6ms
43: learn: 0.2196766 total: 25.9ms remaining: 33ms
44: learn: 0.2185631 total: 26.3ms remaining: 32.2ms
45: learn: 0.2166483 total: 26.8ms remaining: 31.4ms
46: learn: 0.2139387 total: 27.2ms remaining: 30.6ms
47: learn: 0.2129512 total: 27.6ms remaining: 29.8ms
48: learn: 0.2119949 total: 28ms remaining: 29.1ms
49: learn: 0.2109265 total: 28.3ms remaining: 28.3ms
50: learn: 0.2093739 total: 28.8ms remaining: 27.7ms
51: learn: 0.2079054 total: 29.2ms remaining: 27ms
52: learn: 0.2064375 total: 29.6ms remaining: 26.3ms
53: learn: 0.2055025 total: 30ms remaining: 25.6ms
54: learn: 0.2040856 total: 30.4ms remaining: 24.9ms
55: learn: 0.2028301 total: 30.8ms remaining: 24.2ms
56: learn: 0.2014687 total: 31.2ms remaining: 23.6ms
57: learn: 0.1993156 total: 31.7ms remaining: 22.9ms
58: learn: 0.1972837 total: 32.1ms remaining: 22.3ms
59: learn: 0.1959902 total: 32.4ms remaining: 21.6ms
60: learn: 0.1952245 total: 32.8ms remaining: 20.9ms
61: learn: 0.1934942 total: 33.1ms remaining: 20.3ms
62: learn: 0.1919841 total: 33.6ms remaining: 19.7ms
63: learn: 0.1905876 total: 34ms remaining: 19.1ms
64: learn: 0.1894580 total: 34.5ms remaining: 18.6ms
65: learn: 0.1874826 total: 34.9ms remaining: 18ms
66: learn: 0.1863395 total: 35.3ms remaining: 17.4ms
67: learn: 0.1849459 total: 35.9ms remaining: 16.9ms
68: learn: 0.1838012 total: 36.3ms remaining: 16.3ms
69: learn: 0.1827711 total: 36.7ms remaining: 15.7ms
70: learn: 0.1814848 total: 37.1ms remaining: 15.1ms
71: learn: 0.1800869 total: 37.5ms remaining: 14.6ms
72: learn: 0.1783025 total: 37.9ms remaining: 14ms
73: learn: 0.1772895 total: 38.3ms remaining: 13.4ms
74: learn: 0.1763496 total: 38.7ms remaining: 12.9ms
75: learn: 0.1754821 total: 39.1ms remaining: 12.3ms
76: learn: 0.1746590 total: 39.5ms remaining: 11.8ms
77: learn: 0.1740616 total: 39.8ms remaining: 11.2ms
78: learn: 0.1731370 total: 40.2ms remaining: 10.7ms
79: learn: 0.1724127 total: 40.7ms remaining: 10.2ms
80: learn: 0.1714950 total: 41.1ms remaining: 9.63ms
81: learn: 0.1706543 total: 41.4ms remaining: 9.1ms
82: learn: 0.1689667 total: 41.8ms remaining: 8.56ms
83: learn: 0.1679040 total: 42.1ms remaining: 8.03ms
84: learn: 0.1671496 total: 42.5ms remaining: 7.5ms
85: learn: 0.1661126 total: 43.3ms remaining: 7.06ms
86: learn: 0.1647861 total: 43.8ms remaining: 6.54ms
87: learn: 0.1638029 total: 44.2ms remaining: 6.02ms
88: learn: 0.1630324 total: 44.5ms remaining: 5.5ms
89: learn: 0.1622552 total: 45ms remaining: 5ms
90: learn: 0.1614442 total: 45.3ms remaining: 4.48ms
91: learn: 0.1607039 total: 45.6ms remaining: 3.97ms
92: learn: 0.1600161 total: 45.9ms remaining: 3.46ms
93: learn: 0.1588467 total: 46.2ms remaining: 2.95ms
94: learn: 0.1581478 total: 46.7ms remaining: 2.46ms
95: learn: 0.1577160 total: 47.1ms remaining: 1.96ms
96: learn: 0.1572810 total: 47.6ms remaining: 1.47ms
97: learn: 0.1568207 total: 47.9ms remaining: 977us
98: learn: 0.1559880 total: 48.2ms remaining: 487us
99: learn: 0.1552295 total: 48.5ms remaining: 0us
0: learn: 0.6481110 total: 296us remaining: 29.4ms
1: learn: 0.6015437 total: 560us remaining: 27.5ms
2: learn: 0.5637141 total: 749us remaining: 24.2ms
3: learn: 0.5285975 total: 1.01ms remaining: 24.2ms
4: learn: 0.5021036 total: 1.21ms remaining: 23ms
5: learn: 0.4762132 total: 1.44ms remaining: 22.6ms
6: learn: 0.4571360 total: 1.64ms remaining: 21.8ms
7: learn: 0.4392327 total: 1.88ms remaining: 21.7ms
8: learn: 0.4235303 total: 2.02ms remaining: 20.4ms
9: learn: 0.4067796 total: 2.25ms remaining: 20.3ms
10: learn: 0.3956489 total: 2.42ms remaining: 19.6ms
11: learn: 0.3834309 total: 2.66ms remaining: 19.5ms
12: learn: 0.3706993 total: 2.87ms remaining: 19.2ms
13: learn: 0.3593553 total: 3.16ms remaining: 19.4ms
14: learn: 0.3495868 total: 3.48ms remaining: 19.7ms
15: learn: 0.3380214 total: 3.87ms remaining: 20.3ms
16: learn: 0.3303325 total: 4.2ms remaining: 20.5ms
17: learn: 0.3239962 total: 4.45ms remaining: 20.3ms
18: learn: 0.3173096 total: 4.78ms remaining: 20.4ms
19: learn: 0.3130535 total: 5.1ms remaining: 20.4ms
20: learn: 0.3053856 total: 5.38ms remaining: 20.2ms
21: learn: 0.2967920 total: 5.79ms remaining: 20.5ms
22: learn: 0.2911773 total: 6.17ms remaining: 20.7ms
23: learn: 0.2846182 total: 6.47ms remaining: 20.5ms
24: learn: 0.2808455 total: 6.82ms remaining: 20.5ms
25: learn: 0.2780712 total: 7.16ms remaining: 20.4ms
26: learn: 0.2744681 total: 7.52ms remaining: 20.3ms
27: learn: 0.2696076 total: 7.88ms remaining: 20.3ms
28: learn: 0.2644929 total: 8.21ms remaining: 20.1ms
29: learn: 0.2616771 total: 8.53ms remaining: 19.9ms
30: learn: 0.2582177 total: 8.89ms remaining: 19.8ms
31: learn: 0.2542861 total: 9.22ms remaining: 19.6ms
32: learn: 0.2512674 total: 9.56ms remaining: 19.4ms
33: learn: 0.2476332 total: 9.87ms remaining: 19.2ms
34: learn: 0.2439946 total: 10.2ms remaining: 19ms
35: learn: 0.2408321 total: 10.5ms remaining: 18.7ms
36: learn: 0.2375090 total: 10.8ms remaining: 18.5ms
37: learn: 0.2354509 total: 11.1ms remaining: 18.1ms
38: learn: 0.2341470 total: 11.5ms remaining: 17.9ms
39: learn: 0.2321929 total: 12ms remaining: 18ms
40: learn: 0.2300608 total: 12.7ms remaining: 18.3ms
41: learn: 0.2276641 total: 13.1ms remaining: 18.1ms
42: learn: 0.2262292 total: 13.4ms remaining: 17.7ms
43: learn: 0.2238001 total: 13.7ms remaining: 17.4ms
44: learn: 0.2225238 total: 13.9ms remaining: 17ms
45: learn: 0.2207839 total: 14.3ms remaining: 16.8ms
46: learn: 0.2192435 total: 14.7ms remaining: 16.6ms
47: learn: 0.2170573 total: 15ms remaining: 16.3ms
48: learn: 0.2151633 total: 15.3ms remaining: 15.9ms
49: learn: 0.2135362 total: 15.6ms remaining: 15.6ms
50: learn: 0.2118303 total: 15.9ms remaining: 15.3ms
51: learn: 0.2096032 total: 16.2ms remaining: 15ms
52: learn: 0.2073696 total: 16.5ms remaining: 14.7ms
53: learn: 0.2065992 total: 16.8ms remaining: 14.3ms
54: learn: 0.2047594 total: 17.1ms remaining: 14ms
55: learn: 0.2027697 total: 17.6ms remaining: 13.9ms
56: learn: 0.2012123 total: 17.9ms remaining: 13.5ms
57: learn: 0.1988549 total: 18.3ms remaining: 13.3ms
58: learn: 0.1968074 total: 18.7ms remaining: 13ms
59: learn: 0.1947502 total: 18.9ms remaining: 12.6ms
60: learn: 0.1923815 total: 19.2ms remaining: 12.3ms
61: learn: 0.1912902 total: 19.5ms remaining: 11.9ms
62: learn: 0.1899180 total: 19.8ms remaining: 11.6ms
63: learn: 0.1874070 total: 20.1ms remaining: 11.3ms
64: learn: 0.1862103 total: 20.5ms remaining: 11ms
65: learn: 0.1850574 total: 20.9ms remaining: 10.7ms
66: learn: 0.1831118 total: 21.2ms remaining: 10.5ms
67: learn: 0.1815357 total: 21.7ms remaining: 10.2ms
68: learn: 0.1797526 total: 22.1ms remaining: 9.91ms
69: learn: 0.1786087 total: 22.4ms remaining: 9.61ms
70: learn: 0.1774149 total: 22.7ms remaining: 9.26ms
71: learn: 0.1767038 total: 22.9ms remaining: 8.9ms
72: learn: 0.1753482 total: 23.3ms remaining: 8.6ms
73: learn: 0.1738405 total: 23.7ms remaining: 8.31ms
74: learn: 0.1724542 total: 24ms remaining: 8.02ms
75: learn: 0.1714198 total: 24.4ms remaining: 7.71ms
76: learn: 0.1700085 total: 24.8ms remaining: 7.4ms
77: learn: 0.1694224 total: 25.1ms remaining: 7.09ms
78: learn: 0.1681729 total: 25.4ms remaining: 6.75ms
79: learn: 0.1669886 total: 25.7ms remaining: 6.42ms
80: learn: 0.1654271 total: 26ms remaining: 6.1ms
81: learn: 0.1642109 total: 26.3ms remaining: 5.78ms
82: learn: 0.1626449 total: 26.7ms remaining: 5.47ms
83: learn: 0.1614115 total: 27.2ms remaining: 5.17ms
84: learn: 0.1596543 total: 27.5ms remaining: 4.86ms
85: learn: 0.1590043 total: 27.8ms remaining: 4.53ms
86: learn: 0.1582783 total: 28ms remaining: 4.19ms
87: learn: 0.1568860 total: 28.3ms remaining: 3.85ms
88: learn: 0.1559547 total: 28.5ms remaining: 3.52ms
89: learn: 0.1552091 total: 28.7ms remaining: 3.19ms
90: learn: 0.1544308 total: 29.1ms remaining: 2.88ms
91: learn: 0.1535623 total: 29.3ms remaining: 2.55ms
92: learn: 0.1530276 total: 29.7ms remaining: 2.23ms
93: learn: 0.1522294 total: 29.9ms remaining: 1.91ms
94: learn: 0.1508219 total: 30.3ms remaining: 1.59ms
95: learn: 0.1501568 total: 30.6ms remaining: 1.27ms
96: learn: 0.1495230 total: 30.9ms remaining: 956us
97: learn: 0.1485514 total: 31.2ms remaining: 636us
98: learn: 0.1474101 total: 31.6ms remaining: 318us
99: learn: 0.1464165 total: 31.9ms remaining: 0us
0: learn: 0.6515506 total: 805us remaining: 79.8ms
1: learn: 0.6073513 total: 1.35ms remaining: 66.4ms
2: learn: 0.5704869 total: 1.99ms remaining: 64.4ms
3: learn: 0.5398446 total: 2.44ms remaining: 58.5ms
4: learn: 0.5111154 total: 3.01ms remaining: 57.2ms
5: learn: 0.4857031 total: 3.31ms remaining: 51.9ms
6: learn: 0.4649189 total: 3.71ms remaining: 49.3ms
7: learn: 0.4473799 total: 4.07ms remaining: 46.8ms
8: learn: 0.4283679 total: 4.56ms remaining: 46.1ms
9: learn: 0.4129013 total: 4.98ms remaining: 44.8ms
10: learn: 0.3981913 total: 5.14ms remaining: 41.6ms
11: learn: 0.3860850 total: 5.58ms remaining: 41ms
12: learn: 0.3747197 total: 6.03ms remaining: 40.3ms
13: learn: 0.3643286 total: 7.07ms remaining: 43.4ms
14: learn: 0.3560632 total: 7.24ms remaining: 41ms
15: learn: 0.3493369 total: 7.67ms remaining: 40.2ms
16: learn: 0.3405992 total: 8.17ms remaining: 39.9ms
17: learn: 0.3364019 total: 8.48ms remaining: 38.6ms
18: learn: 0.3284740 total: 8.89ms remaining: 37.9ms
19: learn: 0.3207455 total: 9.45ms remaining: 37.8ms
20: learn: 0.3136963 total: 10.2ms remaining: 38.2ms
21: learn: 0.3068343 total: 10.6ms remaining: 37.7ms
22: learn: 0.3015201 total: 10.8ms remaining: 36.2ms
23: learn: 0.2973018 total: 11.2ms remaining: 35.6ms
24: learn: 0.2925741 total: 11.7ms remaining: 35.2ms
25: learn: 0.2863699 total: 12.2ms remaining: 34.6ms
26: learn: 0.2828654 total: 12.5ms remaining: 33.9ms
27: learn: 0.2796819 total: 12.9ms remaining: 33.2ms
28: learn: 0.2773885 total: 13.1ms remaining: 32.2ms
29: learn: 0.2764725 total: 13.3ms remaining: 31.1ms
30: learn: 0.2727544 total: 13.5ms remaining: 30.1ms
31: learn: 0.2691427 total: 14ms remaining: 29.7ms
32: learn: 0.2657733 total: 14.5ms remaining: 29.4ms
33: learn: 0.2627910 total: 15.1ms remaining: 29.3ms
34: learn: 0.2593242 total: 15.3ms remaining: 28.5ms
35: learn: 0.2568494 total: 16ms remaining: 28.5ms
36: learn: 0.2522878 total: 16.5ms remaining: 28.1ms
37: learn: 0.2495150 total: 16.9ms remaining: 27.5ms
38: learn: 0.2464842 total: 17.2ms remaining: 27ms
39: learn: 0.2445408 total: 17.6ms remaining: 26.3ms
40: learn: 0.2418177 total: 18ms remaining: 25.9ms
41: learn: 0.2396222 total: 18.4ms remaining: 25.4ms
42: learn: 0.2366848 total: 18.7ms remaining: 24.8ms
43: learn: 0.2330100 total: 19.2ms remaining: 24.4ms
44: learn: 0.2310290 total: 19.5ms remaining: 23.8ms
45: learn: 0.2291962 total: 19.8ms remaining: 23.2ms
46: learn: 0.2279771 total: 20.2ms remaining: 22.8ms
47: learn: 0.2257751 total: 20.5ms remaining: 22.2ms
48: learn: 0.2228804 total: 20.7ms remaining: 21.6ms
49: learn: 0.2201211 total: 21ms remaining: 21ms
50: learn: 0.2173346 total: 21.3ms remaining: 20.5ms
51: learn: 0.2157845 total: 21.7ms remaining: 20ms
52: learn: 0.2144211 total: 21.9ms remaining: 19.4ms
53: learn: 0.2125434 total: 22.3ms remaining: 19ms
54: learn: 0.2108065 total: 22.6ms remaining: 18.5ms
55: learn: 0.2092356 total: 22.9ms remaining: 18ms
56: learn: 0.2081446 total: 23.2ms remaining: 17.5ms
57: learn: 0.2066968 total: 23.4ms remaining: 16.9ms
58: learn: 0.2044598 total: 23.6ms remaining: 16.4ms
59: learn: 0.2029064 total: 24ms remaining: 16ms
60: learn: 0.2009449 total: 24.3ms remaining: 15.5ms
61: learn: 0.1997688 total: 24.5ms remaining: 15ms
62: learn: 0.1985798 total: 24.9ms remaining: 14.6ms
63: learn: 0.1964098 total: 25.3ms remaining: 14.3ms
64: learn: 0.1953435 total: 25.6ms remaining: 13.8ms
65: learn: 0.1935257 total: 25.8ms remaining: 13.3ms
66: learn: 0.1919715 total: 26ms remaining: 12.8ms
67: learn: 0.1910014 total: 26.3ms remaining: 12.4ms
68: learn: 0.1894297 total: 26.7ms remaining: 12ms
69: learn: 0.1884763 total: 26.9ms remaining: 11.5ms
70: learn: 0.1866306 total: 27.4ms remaining: 11.2ms
71: learn: 0.1849277 total: 27.8ms remaining: 10.8ms
72: learn: 0.1837207 total: 28.1ms remaining: 10.4ms
73: learn: 0.1823749 total: 28.3ms remaining: 9.95ms
74: learn: 0.1811123 total: 28.6ms remaining: 9.54ms
75: learn: 0.1802044 total: 28.9ms remaining: 9.13ms
76: learn: 0.1787988 total: 29.2ms remaining: 8.71ms
77: learn: 0.1780456 total: 29.4ms remaining: 8.29ms
78: learn: 0.1769628 total: 29.8ms remaining: 7.91ms
79: learn: 0.1761383 total: 30.2ms remaining: 7.55ms
80: learn: 0.1746551 total: 30.6ms remaining: 7.17ms
81: learn: 0.1732442 total: 30.9ms remaining: 6.78ms
82: learn: 0.1724555 total: 31.2ms remaining: 6.4ms
83: learn: 0.1714454 total: 31.5ms remaining: 6ms
84: learn: 0.1707684 total: 31.7ms remaining: 5.59ms
85: learn: 0.1697459 total: 31.9ms remaining: 5.2ms
86: learn: 0.1687502 total: 32.2ms remaining: 4.81ms
87: learn: 0.1681338 total: 32.6ms remaining: 4.45ms
88: learn: 0.1670008 total: 33ms remaining: 4.08ms
89: learn: 0.1662372 total: 33.3ms remaining: 3.7ms
90: learn: 0.1653360 total: 33.5ms remaining: 3.31ms
91: learn: 0.1640089 total: 33.8ms remaining: 2.94ms
92: learn: 0.1633292 total: 34.2ms remaining: 2.58ms
93: learn: 0.1627398 total: 34.6ms remaining: 2.21ms
94: learn: 0.1621147 total: 34.8ms remaining: 1.83ms
95: learn: 0.1616381 total: 35.2ms remaining: 1.47ms
96: learn: 0.1609506 total: 35.8ms remaining: 1.11ms
97: learn: 0.1599289 total: 36.1ms remaining: 737us
98: learn: 0.1591254 total: 36.4ms remaining: 367us
99: learn: 0.1584201 total: 36.6ms remaining: 0us
0: learn: 0.6455145 total: 300us remaining: 29.7ms
1: learn: 0.5988760 total: 549us remaining: 26.9ms
2: learn: 0.5594107 total: 711us remaining: 23ms
3: learn: 0.5244881 total: 911us remaining: 21.9ms
4: learn: 0.4973720 total: 1.08ms remaining: 20.5ms
5: learn: 0.4741560 total: 1.27ms remaining: 19.9ms
6: learn: 0.4507672 total: 1.51ms remaining: 20.1ms
7: learn: 0.4334544 total: 1.7ms remaining: 19.5ms
8: learn: 0.4127818 total: 1.88ms remaining: 19ms
9: learn: 0.3964144 total: 2.06ms remaining: 18.5ms
10: learn: 0.3815367 total: 2.21ms remaining: 17.8ms
11: learn: 0.3706824 total: 2.45ms remaining: 18ms
12: learn: 0.3603382 total: 2.67ms remaining: 17.9ms
13: learn: 0.3494307 total: 2.9ms remaining: 17.8ms
14: learn: 0.3417150 total: 3.12ms remaining: 17.7ms
15: learn: 0.3334102 total: 3.33ms remaining: 17.5ms
16: learn: 0.3233884 total: 3.52ms remaining: 17.2ms
17: learn: 0.3176727 total: 3.68ms remaining: 16.8ms
18: learn: 0.3103917 total: 3.85ms remaining: 16.4ms
19: learn: 0.3034642 total: 4.05ms remaining: 16.2ms
20: learn: 0.2980677 total: 4.26ms remaining: 16ms
21: learn: 0.2918482 total: 4.44ms remaining: 15.8ms
22: learn: 0.2854194 total: 4.72ms remaining: 15.8ms
23: learn: 0.2799811 total: 4.98ms remaining: 15.8ms
24: learn: 0.2749249 total: 5.19ms remaining: 15.6ms
25: learn: 0.2704717 total: 5.41ms remaining: 15.4ms
26: learn: 0.2662482 total: 5.63ms remaining: 15.2ms
27: learn: 0.2633140 total: 5.83ms remaining: 15ms
28: learn: 0.2597887 total: 6.18ms remaining: 15.1ms
29: learn: 0.2568451 total: 6.4ms remaining: 14.9ms
30: learn: 0.2544384 total: 6.56ms remaining: 14.6ms
31: learn: 0.2516443 total: 6.77ms remaining: 14.4ms
32: learn: 0.2467225 total: 6.97ms remaining: 14.2ms
33: learn: 0.2444546 total: 7.32ms remaining: 14.2ms
34: learn: 0.2425635 total: 7.57ms remaining: 14.1ms
35: learn: 0.2394869 total: 7.9ms remaining: 14ms
36: learn: 0.2373269 total: 8.22ms remaining: 14ms
37: learn: 0.2343314 total: 8.43ms remaining: 13.8ms
38: learn: 0.2317488 total: 8.63ms remaining: 13.5ms
39: learn: 0.2306135 total: 8.8ms remaining: 13.2ms
40: learn: 0.2289222 total: 9.03ms remaining: 13ms
41: learn: 0.2267190 total: 9.33ms remaining: 12.9ms
42: learn: 0.2254531 total: 9.55ms remaining: 12.7ms
43: learn: 0.2235827 total: 9.76ms remaining: 12.4ms
44: learn: 0.2210251 total: 9.96ms remaining: 12.2ms
45: learn: 0.2194663 total: 10.2ms remaining: 11.9ms
46: learn: 0.2180275 total: 10.3ms remaining: 11.7ms
47: learn: 0.2169634 total: 10.7ms remaining: 11.6ms
48: learn: 0.2141994 total: 10.9ms remaining: 11.3ms
49: learn: 0.2126256 total: 11.1ms remaining: 11.1ms
50: learn: 0.2115400 total: 11.3ms remaining: 10.8ms
51: learn: 0.2099402 total: 11.6ms remaining: 10.7ms
52: learn: 0.2078684 total: 11.8ms remaining: 10.5ms
53: learn: 0.2068950 total: 12.1ms remaining: 10.3ms
54: learn: 0.2058179 total: 12.3ms remaining: 10.1ms
55: learn: 0.2040213 total: 12.5ms remaining: 9.86ms
56: learn: 0.2023684 total: 12.8ms remaining: 9.66ms
57: learn: 0.2005247 total: 13.1ms remaining: 9.45ms
58: learn: 0.1995970 total: 13.3ms remaining: 9.21ms
59: learn: 0.1984756 total: 13.5ms remaining: 8.97ms
60: learn: 0.1970708 total: 13.7ms remaining: 8.74ms
61: learn: 0.1957965 total: 13.9ms remaining: 8.53ms
62: learn: 0.1944455 total: 14.2ms remaining: 8.31ms
63: learn: 0.1929219 total: 14.4ms remaining: 8.1ms
64: learn: 0.1913913 total: 14.6ms remaining: 7.87ms
65: learn: 0.1902021 total: 14.8ms remaining: 7.64ms
66: learn: 0.1889744 total: 15ms remaining: 7.41ms
67: learn: 0.1871736 total: 15.4ms remaining: 7.26ms
68: learn: 0.1859543 total: 15.8ms remaining: 7.08ms
69: learn: 0.1845997 total: 16ms remaining: 6.85ms
70: learn: 0.1835376 total: 16.3ms remaining: 6.66ms
71: learn: 0.1825610 total: 16.7ms remaining: 6.48ms
72: learn: 0.1807745 total: 16.9ms remaining: 6.27ms
73: learn: 0.1800400 total: 17.3ms remaining: 6.08ms
74: learn: 0.1793376 total: 17.6ms remaining: 5.86ms
75: learn: 0.1784109 total: 17.9ms remaining: 5.64ms
76: learn: 0.1773741 total: 18.2ms remaining: 5.45ms
77: learn: 0.1761102 total: 18.5ms remaining: 5.21ms
78: learn: 0.1746922 total: 18.7ms remaining: 4.97ms
79: learn: 0.1738470 total: 19.1ms remaining: 4.76ms
80: learn: 0.1730243 total: 19.4ms remaining: 4.55ms
81: learn: 0.1721958 total: 19.8ms remaining: 4.34ms
82: learn: 0.1713148 total: 20ms remaining: 4.09ms
83: learn: 0.1707098 total: 20.2ms remaining: 3.84ms
84: learn: 0.1699495 total: 20.4ms remaining: 3.61ms
85: learn: 0.1691816 total: 20.7ms remaining: 3.36ms
86: learn: 0.1682771 total: 20.9ms remaining: 3.12ms
87: learn: 0.1674974 total: 21.3ms remaining: 2.9ms
88: learn: 0.1669940 total: 21.5ms remaining: 2.65ms
89: learn: 0.1660191 total: 21.7ms remaining: 2.41ms
90: learn: 0.1654985 total: 21.9ms remaining: 2.17ms
91: learn: 0.1649944 total: 22.3ms remaining: 1.94ms
92: learn: 0.1641069 total: 22.5ms remaining: 1.69ms
93: learn: 0.1630292 total: 23.1ms remaining: 1.47ms
94: learn: 0.1623293 total: 23.4ms remaining: 1.23ms
95: learn: 0.1617627 total: 23.6ms remaining: 982us
96: learn: 0.1609515 total: 23.9ms remaining: 739us
97: learn: 0.1597019 total: 24.1ms remaining: 492us
98: learn: 0.1591249 total: 24.3ms remaining: 245us
99: learn: 0.1585563 total: 24.5ms remaining: 0us
0: learn: 0.6503257 total: 247us remaining: 24.5ms
1: learn: 0.6091405 total: 657us remaining: 32.2ms
2: learn: 0.5749262 total: 812us remaining: 26.3ms
3: learn: 0.5438978 total: 1.01ms remaining: 24.4ms
4: learn: 0.5155700 total: 1.18ms remaining: 22.5ms
5: learn: 0.4927320 total: 1.36ms remaining: 21.3ms
6: learn: 0.4727667 total: 1.52ms remaining: 20.2ms
7: learn: 0.4557302 total: 1.7ms remaining: 19.5ms
8: learn: 0.4388609 total: 1.88ms remaining: 19ms
9: learn: 0.4247644 total: 2.01ms remaining: 18.1ms
10: learn: 0.4144140 total: 2.18ms remaining: 17.6ms
11: learn: 0.4019024 total: 2.35ms remaining: 17.2ms
12: learn: 0.3916899 total: 2.6ms remaining: 17.4ms
13: learn: 0.3815831 total: 3.25ms remaining: 20ms
14: learn: 0.3717996 total: 3.56ms remaining: 20.1ms
15: learn: 0.3647230 total: 3.89ms remaining: 20.4ms
16: learn: 0.3545912 total: 4.26ms remaining: 20.8ms
17: learn: 0.3502944 total: 4.46ms remaining: 20.3ms
18: learn: 0.3428250 total: 4.67ms remaining: 19.9ms
19: learn: 0.3374503 total: 4.93ms remaining: 19.7ms
20: learn: 0.3317534 total: 5.35ms remaining: 20.1ms
21: learn: 0.3248470 total: 5.6ms remaining: 19.9ms
22: learn: 0.3197077 total: 5.76ms remaining: 19.3ms
23: learn: 0.3149752 total: 5.97ms remaining: 18.9ms
24: learn: 0.3115804 total: 6.17ms remaining: 18.5ms
25: learn: 0.3073465 total: 6.36ms remaining: 18.1ms
26: learn: 0.3014933 total: 6.56ms remaining: 17.7ms
27: learn: 0.2982788 total: 6.74ms remaining: 17.3ms
28: learn: 0.2954439 total: 6.91ms remaining: 16.9ms
29: learn: 0.2917875 total: 7.29ms remaining: 17ms
30: learn: 0.2900107 total: 7.46ms remaining: 16.6ms
31: learn: 0.2864544 total: 7.66ms remaining: 16.3ms
32: learn: 0.2810382 total: 7.89ms remaining: 16ms
33: learn: 0.2787524 total: 8.11ms remaining: 15.7ms
34: learn: 0.2764095 total: 8.33ms remaining: 15.5ms
35: learn: 0.2748109 total: 8.54ms remaining: 15.2ms
36: learn: 0.2728280 total: 8.72ms remaining: 14.8ms
37: learn: 0.2703365 total: 8.9ms remaining: 14.5ms
38: learn: 0.2683979 total: 9.09ms remaining: 14.2ms
39: learn: 0.2667767 total: 9.28ms remaining: 13.9ms
40: learn: 0.2652805 total: 9.56ms remaining: 13.8ms
41: learn: 0.2621843 total: 9.77ms remaining: 13.5ms
42: learn: 0.2596001 total: 9.94ms remaining: 13.2ms
43: learn: 0.2571810 total: 10.1ms remaining: 12.9ms
44: learn: 0.2554186 total: 10.3ms remaining: 12.6ms
45: learn: 0.2533093 total: 10.5ms remaining: 12.4ms
46: learn: 0.2511507 total: 10.8ms remaining: 12.2ms
47: learn: 0.2495800 total: 11.1ms remaining: 12ms
48: learn: 0.2474001 total: 11.4ms remaining: 11.8ms
49: learn: 0.2461070 total: 11.6ms remaining: 11.6ms
50: learn: 0.2445824 total: 11.9ms remaining: 11.4ms
51: learn: 0.2411191 total: 12ms remaining: 11.1ms
52: learn: 0.2386823 total: 12.2ms remaining: 10.9ms
53: learn: 0.2375267 total: 12.4ms remaining: 10.6ms
54: learn: 0.2359654 total: 12.7ms remaining: 10.4ms
55: learn: 0.2335567 total: 13ms remaining: 10.2ms
56: learn: 0.2323313 total: 13.1ms remaining: 9.9ms
57: learn: 0.2301428 total: 13.3ms remaining: 9.66ms
58: learn: 0.2273682 total: 13.6ms remaining: 9.45ms
59: learn: 0.2245088 total: 13.9ms remaining: 9.29ms
60: learn: 0.2229013 total: 14.2ms remaining: 9.06ms
61: learn: 0.2218228 total: 14.4ms remaining: 8.83ms
62: learn: 0.2203296 total: 14.6ms remaining: 8.59ms
63: learn: 0.2190087 total: 14.9ms remaining: 8.36ms
64: learn: 0.2178656 total: 15.1ms remaining: 8.15ms
65: learn: 0.2156503 total: 15.4ms remaining: 7.92ms
66: learn: 0.2148563 total: 15.6ms remaining: 7.68ms
67: learn: 0.2126004 total: 15.9ms remaining: 7.5ms
68: learn: 0.2111062 total: 16.4ms remaining: 7.35ms
69: learn: 0.2096250 total: 16.9ms remaining: 7.25ms
70: learn: 0.2083270 total: 17.3ms remaining: 7.07ms
71: learn: 0.2070989 total: 17.5ms remaining: 6.82ms
72: learn: 0.2061435 total: 17.9ms remaining: 6.61ms
73: learn: 0.2045066 total: 18.1ms remaining: 6.37ms
74: learn: 0.2031592 total: 18.4ms remaining: 6.14ms
75: learn: 0.2015893 total: 18.7ms remaining: 5.91ms
76: learn: 0.2006151 total: 18.9ms remaining: 5.66ms
77: learn: 0.1988155 total: 19.1ms remaining: 5.4ms
78: learn: 0.1977362 total: 19.4ms remaining: 5.16ms
79: learn: 0.1967043 total: 19.7ms remaining: 4.91ms
80: learn: 0.1956287 total: 20ms remaining: 4.68ms
81: learn: 0.1942310 total: 20.2ms remaining: 4.44ms
82: learn: 0.1928918 total: 20.6ms remaining: 4.22ms
83: learn: 0.1919528 total: 21ms remaining: 4.01ms
84: learn: 0.1906315 total: 21.4ms remaining: 3.78ms
85: learn: 0.1895164 total: 21.8ms remaining: 3.55ms
86: learn: 0.1887474 total: 22.1ms remaining: 3.31ms
87: learn: 0.1877110 total: 22.4ms remaining: 3.06ms
88: learn: 0.1866921 total: 22.7ms remaining: 2.8ms
89: learn: 0.1856475 total: 23ms remaining: 2.55ms
90: learn: 0.1849670 total: 23.3ms remaining: 2.31ms
91: learn: 0.1839782 total: 23.7ms remaining: 2.06ms
92: learn: 0.1831375 total: 23.9ms remaining: 1.8ms
93: learn: 0.1818797 total: 24.1ms remaining: 1.54ms
94: learn: 0.1810629 total: 24.4ms remaining: 1.28ms
95: learn: 0.1801821 total: 24.7ms remaining: 1.03ms
96: learn: 0.1794811 total: 24.9ms remaining: 771us
97: learn: 0.1788221 total: 25.2ms remaining: 514us
98: learn: 0.1770942 total: 25.5ms remaining: 257us
99: learn: 0.1766127 total: 25.8ms remaining: 0us
0: learn: 0.6515750 total: 353us remaining: 35ms
1: learn: 0.6134885 total: 586us remaining: 28.7ms
2: learn: 0.5837400 total: 788us remaining: 25.5ms
3: learn: 0.5489154 total: 1.01ms remaining: 24.3ms
4: learn: 0.5169299 total: 1.23ms remaining: 23.4ms
5: learn: 0.4907425 total: 1.42ms remaining: 22.3ms
6: learn: 0.4713171 total: 1.66ms remaining: 22.1ms
7: learn: 0.4534210 total: 1.81ms remaining: 20.8ms
8: learn: 0.4367045 total: 1.98ms remaining: 20.1ms
9: learn: 0.4205350 total: 2.15ms remaining: 19.3ms
10: learn: 0.4056829 total: 2.36ms remaining: 19.1ms
11: learn: 0.3903177 total: 2.69ms remaining: 19.7ms
12: learn: 0.3766978 total: 2.97ms remaining: 19.9ms
13: learn: 0.3669745 total: 3.14ms remaining: 19.3ms
14: learn: 0.3548570 total: 3.34ms remaining: 18.9ms
15: learn: 0.3456593 total: 3.54ms remaining: 18.6ms
16: learn: 0.3378650 total: 3.69ms remaining: 18ms
17: learn: 0.3310731 total: 3.83ms remaining: 17.4ms
18: learn: 0.3233461 total: 4.11ms remaining: 17.5ms
19: learn: 0.3165295 total: 4.31ms remaining: 17.2ms
20: learn: 0.3094843 total: 4.56ms remaining: 17.2ms
21: learn: 0.3020178 total: 4.75ms remaining: 16.9ms
22: learn: 0.2965073 total: 4.98ms remaining: 16.7ms
23: learn: 0.2896890 total: 5.17ms remaining: 16.4ms
24: learn: 0.2844204 total: 5.4ms remaining: 16.2ms
25: learn: 0.2797083 total: 5.63ms remaining: 16ms
26: learn: 0.2751533 total: 5.82ms remaining: 15.7ms
27: learn: 0.2715553 total: 6.02ms remaining: 15.5ms
28: learn: 0.2691510 total: 6.26ms remaining: 15.3ms
29: learn: 0.2671634 total: 6.67ms remaining: 15.6ms
30: learn: 0.2650620 total: 6.83ms remaining: 15.2ms
31: learn: 0.2613732 total: 7.06ms remaining: 15ms
32: learn: 0.2592097 total: 7.32ms remaining: 14.9ms
33: learn: 0.2555304 total: 7.58ms remaining: 14.7ms
34: learn: 0.2532133 total: 7.78ms remaining: 14.4ms
35: learn: 0.2505760 total: 8.1ms remaining: 14.4ms
36: learn: 0.2488293 total: 8.36ms remaining: 14.2ms
37: learn: 0.2453197 total: 8.53ms remaining: 13.9ms
38: learn: 0.2419295 total: 8.82ms remaining: 13.8ms
39: learn: 0.2391228 total: 9.02ms remaining: 13.5ms
40: learn: 0.2363322 total: 9.18ms remaining: 13.2ms
41: learn: 0.2343441 total: 9.46ms remaining: 13.1ms
42: learn: 0.2325132 total: 9.81ms remaining: 13ms
43: learn: 0.2296161 total: 10.1ms remaining: 12.8ms
44: learn: 0.2287773 total: 10.3ms remaining: 12.6ms
45: learn: 0.2272871 total: 10.5ms remaining: 12.3ms
46: learn: 0.2260889 total: 10.7ms remaining: 12.1ms
47: learn: 0.2244358 total: 11ms remaining: 11.9ms
48: learn: 0.2231578 total: 11.3ms remaining: 11.7ms
49: learn: 0.2215341 total: 11.5ms remaining: 11.5ms
50: learn: 0.2182713 total: 11.8ms remaining: 11.3ms
51: learn: 0.2166147 total: 12.2ms remaining: 11.3ms
52: learn: 0.2155089 total: 12.6ms remaining: 11.2ms
53: learn: 0.2140156 total: 13ms remaining: 11.1ms
54: learn: 0.2128961 total: 13.4ms remaining: 10.9ms
55: learn: 0.2114523 total: 13.5ms remaining: 10.6ms
56: learn: 0.2098777 total: 13.8ms remaining: 10.4ms
57: learn: 0.2080078 total: 14.3ms remaining: 10.4ms
58: learn: 0.2066818 total: 14.6ms remaining: 10.2ms
59: learn: 0.2052481 total: 14.9ms remaining: 9.93ms
60: learn: 0.2035334 total: 15.4ms remaining: 9.85ms
61: learn: 0.2021471 total: 15.9ms remaining: 9.72ms
62: learn: 0.2011057 total: 16.1ms remaining: 9.47ms
63: learn: 0.1996851 total: 16.7ms remaining: 9.37ms
64: learn: 0.1979320 total: 17.2ms remaining: 9.25ms
65: learn: 0.1969688 total: 17.8ms remaining: 9.19ms
66: learn: 0.1956253 total: 18.1ms remaining: 8.93ms
67: learn: 0.1944589 total: 18.4ms remaining: 8.68ms
68: learn: 0.1929730 total: 18.8ms remaining: 8.45ms
69: learn: 0.1916246 total: 19.4ms remaining: 8.33ms
70: learn: 0.1904793 total: 19.9ms remaining: 8.14ms
71: learn: 0.1896955 total: 20.6ms remaining: 8ms
72: learn: 0.1875003 total: 21.2ms remaining: 7.84ms
73: learn: 0.1860530 total: 21.5ms remaining: 7.55ms
74: learn: 0.1847521 total: 22.3ms remaining: 7.43ms
75: learn: 0.1836835 total: 22.7ms remaining: 7.17ms
76: learn: 0.1830485 total: 23ms remaining: 6.87ms
77: learn: 0.1814416 total: 23.4ms remaining: 6.59ms
78: learn: 0.1804926 total: 23.8ms remaining: 6.33ms
79: learn: 0.1796085 total: 24.5ms remaining: 6.13ms
80: learn: 0.1788568 total: 25.2ms remaining: 5.91ms
81: learn: 0.1779278 total: 25.4ms remaining: 5.58ms
82: learn: 0.1769908 total: 25.8ms remaining: 5.29ms
83: learn: 0.1763817 total: 26.2ms remaining: 4.99ms
84: learn: 0.1754758 total: 26.8ms remaining: 4.72ms
85: learn: 0.1743761 total: 27.1ms remaining: 4.42ms
86: learn: 0.1730073 total: 27.7ms remaining: 4.13ms
87: learn: 0.1723445 total: 28.2ms remaining: 3.85ms
88: learn: 0.1712899 total: 28.5ms remaining: 3.52ms
89: learn: 0.1708731 total: 28.9ms remaining: 3.21ms
90: learn: 0.1695320 total: 29.2ms remaining: 2.89ms
91: learn: 0.1688723 total: 29.6ms remaining: 2.58ms
92: learn: 0.1683870 total: 30.2ms remaining: 2.27ms
93: learn: 0.1673684 total: 30.5ms remaining: 1.95ms
94: learn: 0.1666907 total: 30.9ms remaining: 1.63ms
95: learn: 0.1659053 total: 31.2ms remaining: 1.3ms
96: learn: 0.1652727 total: 31.5ms remaining: 972us
97: learn: 0.1645047 total: 31.8ms remaining: 647us
98: learn: 0.1640110 total: 32ms remaining: 323us
99: learn: 0.1634089 total: 32.2ms remaining: 0us
0: learn: 0.6495190 total: 305us remaining: 30.3ms
1: learn: 0.6095526 total: 925us remaining: 45.3ms
2: learn: 0.5719992 total: 1.17ms remaining: 38ms
3: learn: 0.5425656 total: 1.54ms remaining: 36.9ms
4: learn: 0.5159659 total: 1.76ms remaining: 33.4ms
5: learn: 0.4917545 total: 1.95ms remaining: 30.6ms
6: learn: 0.4701324 total: 2.1ms remaining: 27.9ms
7: learn: 0.4524320 total: 2.27ms remaining: 26.1ms
8: learn: 0.4325252 total: 2.42ms remaining: 24.4ms
9: learn: 0.4165980 total: 2.66ms remaining: 24ms
10: learn: 0.4035100 total: 2.84ms remaining: 23ms
11: learn: 0.3907931 total: 3.07ms remaining: 22.5ms
12: learn: 0.3789276 total: 3.25ms remaining: 21.7ms
13: learn: 0.3688594 total: 3.49ms remaining: 21.4ms
14: learn: 0.3595050 total: 3.67ms remaining: 20.8ms
15: learn: 0.3510985 total: 3.88ms remaining: 20.4ms
16: learn: 0.3435509 total: 4.1ms remaining: 20ms
17: learn: 0.3372297 total: 4.26ms remaining: 19.4ms
18: learn: 0.3310716 total: 4.41ms remaining: 18.8ms
19: learn: 0.3258688 total: 4.61ms remaining: 18.4ms
20: learn: 0.3195430 total: 4.91ms remaining: 18.5ms
21: learn: 0.3125519 total: 5.14ms remaining: 18.2ms
22: learn: 0.3074617 total: 5.33ms remaining: 17.9ms
23: learn: 0.3031872 total: 5.46ms remaining: 17.3ms
24: learn: 0.2996652 total: 5.65ms remaining: 17ms
25: learn: 0.2955970 total: 5.83ms remaining: 16.6ms
26: learn: 0.2919958 total: 6.18ms remaining: 16.7ms
27: learn: 0.2880784 total: 6.36ms remaining: 16.4ms
28: learn: 0.2850178 total: 6.6ms remaining: 16.2ms
29: learn: 0.2831896 total: 6.78ms remaining: 15.8ms
30: learn: 0.2807488 total: 7.16ms remaining: 15.9ms
31: learn: 0.2781251 total: 7.32ms remaining: 15.6ms
32: learn: 0.2755400 total: 7.63ms remaining: 15.5ms
33: learn: 0.2735858 total: 7.79ms remaining: 15.1ms
34: learn: 0.2709428 total: 8.03ms remaining: 14.9ms
35: learn: 0.2680968 total: 8.31ms remaining: 14.8ms
36: learn: 0.2653391 total: 8.61ms remaining: 14.7ms
37: learn: 0.2622118 total: 8.89ms remaining: 14.5ms
38: learn: 0.2589166 total: 9.15ms remaining: 14.3ms
39: learn: 0.2560415 total: 9.52ms remaining: 14.3ms
40: learn: 0.2545774 total: 9.73ms remaining: 14ms
41: learn: 0.2521408 total: 9.98ms remaining: 13.8ms
42: learn: 0.2508771 total: 10.2ms remaining: 13.5ms
43: learn: 0.2500696 total: 10.4ms remaining: 13.2ms
44: learn: 0.2485904 total: 10.7ms remaining: 13ms
45: learn: 0.2465004 total: 11.1ms remaining: 13ms
46: learn: 0.2446957 total: 11.3ms remaining: 12.7ms
47: learn: 0.2427096 total: 11.6ms remaining: 12.6ms
48: learn: 0.2415083 total: 12.1ms remaining: 12.6ms
49: learn: 0.2387823 total: 12.4ms remaining: 12.4ms
50: learn: 0.2376992 total: 12.8ms remaining: 12.3ms
51: learn: 0.2345011 total: 13.3ms remaining: 12.3ms
52: learn: 0.2324914 total: 13.6ms remaining: 12.1ms
53: learn: 0.2311739 total: 13.9ms remaining: 11.8ms
54: learn: 0.2288382 total: 14.3ms remaining: 11.7ms
55: learn: 0.2271510 total: 14.6ms remaining: 11.5ms
56: learn: 0.2258501 total: 15ms remaining: 11.3ms
57: learn: 0.2239054 total: 15.3ms remaining: 11.1ms
58: learn: 0.2216254 total: 15.7ms remaining: 10.9ms
59: learn: 0.2189058 total: 15.9ms remaining: 10.6ms
60: learn: 0.2167674 total: 16.3ms remaining: 10.4ms
61: learn: 0.2157021 total: 16.6ms remaining: 10.2ms
62: learn: 0.2141794 total: 17.1ms remaining: 10ms
63: learn: 0.2122613 total: 17.5ms remaining: 9.82ms
64: learn: 0.2108818 total: 17.8ms remaining: 9.59ms
65: learn: 0.2095270 total: 18.1ms remaining: 9.34ms
66: learn: 0.2087435 total: 18.4ms remaining: 9.04ms
67: learn: 0.2074101 total: 18.6ms remaining: 8.75ms
68: learn: 0.2057417 total: 18.8ms remaining: 8.46ms
69: learn: 0.2038966 total: 19.1ms remaining: 8.19ms
70: learn: 0.2025303 total: 19.5ms remaining: 7.94ms
71: learn: 0.2015057 total: 19.7ms remaining: 7.67ms
72: learn: 0.2004784 total: 20ms remaining: 7.4ms
73: learn: 0.1986380 total: 20.2ms remaining: 7.11ms
74: learn: 0.1967574 total: 20.7ms remaining: 6.88ms
75: learn: 0.1958965 total: 21.1ms remaining: 6.67ms
76: learn: 0.1950152 total: 21.5ms remaining: 6.42ms
77: learn: 0.1942672 total: 21.9ms remaining: 6.17ms
78: learn: 0.1922748 total: 22.3ms remaining: 5.93ms
79: learn: 0.1915469 total: 22.6ms remaining: 5.65ms
80: learn: 0.1905149 total: 22.9ms remaining: 5.38ms
81: learn: 0.1889936 total: 23.4ms remaining: 5.13ms
82: learn: 0.1884547 total: 23.7ms remaining: 4.85ms
83: learn: 0.1879366 total: 24ms remaining: 4.57ms
84: learn: 0.1861980 total: 24.4ms remaining: 4.3ms
85: learn: 0.1854796 total: 24.8ms remaining: 4.03ms
86: learn: 0.1836693 total: 25.1ms remaining: 3.74ms
87: learn: 0.1829421 total: 25.3ms remaining: 3.45ms
88: learn: 0.1814350 total: 25.6ms remaining: 3.16ms
89: learn: 0.1810181 total: 25.8ms remaining: 2.86ms
90: learn: 0.1806212 total: 26ms remaining: 2.57ms
91: learn: 0.1795526 total: 26.2ms remaining: 2.28ms
92: learn: 0.1791160 total: 26.5ms remaining: 2ms
93: learn: 0.1787314 total: 26.7ms remaining: 1.71ms
94: learn: 0.1782327 total: 27ms remaining: 1.42ms
95: learn: 0.1778748 total: 27.3ms remaining: 1.14ms
96: learn: 0.1769739 total: 27.6ms remaining: 852us
97: learn: 0.1765155 total: 27.8ms remaining: 566us
98: learn: 0.1761686 total: 28ms remaining: 283us
99: learn: 0.1757469 total: 28.2ms remaining: 0us
0: learn: 0.6487726 total: 303us remaining: 30ms
1: learn: 0.6054752 total: 498us remaining: 24.4ms
2: learn: 0.5690516 total: 664us remaining: 21.5ms
3: learn: 0.5376892 total: 915us remaining: 22ms
4: learn: 0.5107400 total: 1.08ms remaining: 20.6ms
5: learn: 0.4860125 total: 1.3ms remaining: 20.4ms
6: learn: 0.4650487 total: 1.5ms remaining: 20ms
7: learn: 0.4474529 total: 1.67ms remaining: 19.2ms
8: learn: 0.4286769 total: 1.85ms remaining: 18.7ms
9: learn: 0.4136039 total: 2.04ms remaining: 18.3ms
10: learn: 0.4019389 total: 2.19ms remaining: 17.8ms
11: learn: 0.3894000 total: 2.4ms remaining: 17.6ms
12: learn: 0.3778865 total: 2.64ms remaining: 17.7ms
13: learn: 0.3672801 total: 2.85ms remaining: 17.5ms
14: learn: 0.3576247 total: 3.09ms remaining: 17.5ms
15: learn: 0.3495637 total: 3.32ms remaining: 17.4ms
16: learn: 0.3412530 total: 3.56ms remaining: 17.4ms
17: learn: 0.3350939 total: 3.73ms remaining: 17ms
18: learn: 0.3290651 total: 3.89ms remaining: 16.6ms
19: learn: 0.3248385 total: 4.09ms remaining: 16.3ms
20: learn: 0.3197584 total: 4.3ms remaining: 16.2ms
21: learn: 0.3123746 total: 4.58ms remaining: 16.3ms
22: learn: 0.3067161 total: 4.89ms remaining: 16.4ms
23: learn: 0.3018604 total: 5.23ms remaining: 16.6ms
24: learn: 0.2972128 total: 5.57ms remaining: 16.7ms
25: learn: 0.2947712 total: 5.87ms remaining: 16.7ms
26: learn: 0.2912988 total: 6.23ms remaining: 16.8ms
27: learn: 0.2870765 total: 6.53ms remaining: 16.8ms
28: learn: 0.2831592 total: 6.8ms remaining: 16.7ms
29: learn: 0.2813999 total: 7.08ms remaining: 16.5ms
30: learn: 0.2782774 total: 7.45ms remaining: 16.6ms
31: learn: 0.2757420 total: 7.56ms remaining: 16.1ms
32: learn: 0.2733458 total: 7.9ms remaining: 16ms
33: learn: 0.2714703 total: 8.15ms remaining: 15.8ms
34: learn: 0.2692580 total: 8.35ms remaining: 15.5ms
35: learn: 0.2669193 total: 8.75ms remaining: 15.6ms
36: learn: 0.2654010 total: 9.14ms remaining: 15.6ms
37: learn: 0.2626552 total: 9.6ms remaining: 15.7ms
38: learn: 0.2591544 total: 9.92ms remaining: 15.5ms
39: learn: 0.2568921 total: 10.3ms remaining: 15.4ms
40: learn: 0.2539529 total: 10.7ms remaining: 15.4ms
41: learn: 0.2519482 total: 11.1ms remaining: 15.3ms
42: learn: 0.2496161 total: 11.5ms remaining: 15.3ms
43: learn: 0.2481991 total: 12ms remaining: 15.2ms
44: learn: 0.2469000 total: 12.3ms remaining: 15.1ms
45: learn: 0.2455710 total: 12.7ms remaining: 14.9ms
46: learn: 0.2415597 total: 13ms remaining: 14.7ms
47: learn: 0.2399522 total: 13.4ms remaining: 14.6ms
48: learn: 0.2386716 total: 13.8ms remaining: 14.4ms
49: learn: 0.2371674 total: 14ms remaining: 14ms
50: learn: 0.2356024 total: 14.3ms remaining: 13.7ms
51: learn: 0.2339625 total: 14.6ms remaining: 13.4ms
52: learn: 0.2329458 total: 14.9ms remaining: 13.3ms
53: learn: 0.2315437 total: 15.3ms remaining: 13ms
54: learn: 0.2301420 total: 15.6ms remaining: 12.8ms
55: learn: 0.2274817 total: 15.9ms remaining: 12.5ms
56: learn: 0.2258392 total: 16.3ms remaining: 12.3ms
57: learn: 0.2239846 total: 16.7ms remaining: 12.1ms
58: learn: 0.2215748 total: 17ms remaining: 11.8ms
59: learn: 0.2188303 total: 17.4ms remaining: 11.6ms
60: learn: 0.2169148 total: 17.7ms remaining: 11.3ms
61: learn: 0.2145196 total: 18.2ms remaining: 11.1ms
62: learn: 0.2131793 total: 18.4ms remaining: 10.8ms
63: learn: 0.2115461 total: 18.9ms remaining: 10.6ms
64: learn: 0.2092351 total: 19.3ms remaining: 10.4ms
65: learn: 0.2070521 total: 19.6ms remaining: 10.1ms
66: learn: 0.2059674 total: 20ms remaining: 9.85ms
67: learn: 0.2041380 total: 20.2ms remaining: 9.5ms
68: learn: 0.2031281 total: 20.4ms remaining: 9.17ms
69: learn: 0.2018882 total: 20.6ms remaining: 8.82ms
70: learn: 0.1998817 total: 20.9ms remaining: 8.54ms
71: learn: 0.1988290 total: 21.2ms remaining: 8.25ms
72: learn: 0.1978987 total: 21.8ms remaining: 8.06ms
73: learn: 0.1969798 total: 22.2ms remaining: 7.8ms
74: learn: 0.1951727 total: 22.5ms remaining: 7.5ms
75: learn: 0.1942894 total: 23.1ms remaining: 7.3ms
76: learn: 0.1931971 total: 23.5ms remaining: 7.02ms
77: learn: 0.1918851 total: 24ms remaining: 6.77ms
78: learn: 0.1910988 total: 24.9ms remaining: 6.63ms
79: learn: 0.1901434 total: 25.1ms remaining: 6.29ms
80: learn: 0.1893449 total: 25.4ms remaining: 5.95ms
81: learn: 0.1884621 total: 25.7ms remaining: 5.64ms
82: learn: 0.1876637 total: 26ms remaining: 5.32ms
83: learn: 0.1863474 total: 26.2ms remaining: 4.99ms
84: learn: 0.1854177 total: 26.4ms remaining: 4.66ms
85: learn: 0.1848509 total: 26.8ms remaining: 4.36ms
86: learn: 0.1830131 total: 27.4ms remaining: 4.09ms
87: learn: 0.1814085 total: 27.6ms remaining: 3.76ms
88: learn: 0.1803102 total: 27.8ms remaining: 3.44ms
89: learn: 0.1791514 total: 28ms remaining: 3.11ms
90: learn: 0.1778142 total: 28.2ms remaining: 2.79ms
91: learn: 0.1772303 total: 28.4ms remaining: 2.47ms
92: learn: 0.1762585 total: 28.6ms remaining: 2.15ms
93: learn: 0.1758778 total: 28.8ms remaining: 1.83ms
94: learn: 0.1750961 total: 29ms remaining: 1.53ms
95: learn: 0.1743590 total: 29.7ms remaining: 1.24ms
96: learn: 0.1735165 total: 30.3ms remaining: 936us
97: learn: 0.1729827 total: 30.8ms remaining: 628us
98: learn: 0.1722555 total: 31.2ms remaining: 315us
99: learn: 0.1718057 total: 31.8ms remaining: 0us
0: learn: 0.6470146 total: 325us remaining: 32.2ms
1: learn: 0.6064299 total: 830us remaining: 40.7ms
2: learn: 0.5691283 total: 1.08ms remaining: 35.1ms
3: learn: 0.5366569 total: 1.31ms remaining: 31.4ms
4: learn: 0.5098000 total: 1.48ms remaining: 28.1ms
5: learn: 0.4832216 total: 1.83ms remaining: 28.7ms
6: learn: 0.4596274 total: 2.05ms remaining: 27.3ms
7: learn: 0.4437963 total: 2.35ms remaining: 27.1ms
8: learn: 0.4236547 total: 2.61ms remaining: 26.4ms
9: learn: 0.4091465 total: 2.97ms remaining: 26.8ms
10: learn: 0.3961872 total: 3.23ms remaining: 26.2ms
11: learn: 0.3834046 total: 3.66ms remaining: 26.8ms
12: learn: 0.3710681 total: 3.93ms remaining: 26.3ms
13: learn: 0.3595058 total: 4.36ms remaining: 26.8ms
14: learn: 0.3509086 total: 4.73ms remaining: 26.8ms
15: learn: 0.3450153 total: 5.24ms remaining: 27.5ms
16: learn: 0.3371444 total: 5.66ms remaining: 27.6ms
17: learn: 0.3298510 total: 6.11ms remaining: 27.8ms
18: learn: 0.3232019 total: 6.29ms remaining: 26.8ms
19: learn: 0.3162856 total: 6.71ms remaining: 26.8ms
20: learn: 0.3111943 total: 6.99ms remaining: 26.3ms
21: learn: 0.3057793 total: 7.29ms remaining: 25.8ms
22: learn: 0.3007321 total: 7.81ms remaining: 26.1ms
23: learn: 0.2962390 total: 8.16ms remaining: 25.9ms
24: learn: 0.2927857 total: 8.43ms remaining: 25.3ms
25: learn: 0.2900541 total: 8.7ms remaining: 24.8ms
26: learn: 0.2865934 total: 8.9ms remaining: 24.1ms
27: learn: 0.2819302 total: 9.41ms remaining: 24.2ms
28: learn: 0.2778663 total: 9.79ms remaining: 24ms
29: learn: 0.2737055 total: 10.3ms remaining: 24ms
30: learn: 0.2722615 total: 10.6ms remaining: 23.6ms
31: learn: 0.2689709 total: 11.2ms remaining: 23.9ms
32: learn: 0.2664546 total: 11.6ms remaining: 23.6ms
33: learn: 0.2629107 total: 12.4ms remaining: 24.1ms
34: learn: 0.2603216 total: 12.7ms remaining: 23.7ms
35: learn: 0.2574981 total: 13.1ms remaining: 23.3ms
36: learn: 0.2557634 total: 13.4ms remaining: 22.8ms
37: learn: 0.2525677 total: 13.8ms remaining: 22.5ms
38: learn: 0.2487591 total: 14.1ms remaining: 22ms
39: learn: 0.2454105 total: 14.3ms remaining: 21.5ms
40: learn: 0.2435551 total: 14.6ms remaining: 21ms
41: learn: 0.2401939 total: 15ms remaining: 20.7ms
42: learn: 0.2387428 total: 15.2ms remaining: 20.1ms
43: learn: 0.2376608 total: 15.5ms remaining: 19.7ms
44: learn: 0.2358510 total: 15.9ms remaining: 19.4ms
45: learn: 0.2332892 total: 16.2ms remaining: 19ms
46: learn: 0.2316049 total: 16.6ms remaining: 18.7ms
47: learn: 0.2293167 total: 17ms remaining: 18.4ms
48: learn: 0.2264321 total: 17.2ms remaining: 17.9ms
49: learn: 0.2240784 total: 17.6ms remaining: 17.6ms
50: learn: 0.2226746 total: 18.1ms remaining: 17.4ms
51: learn: 0.2192451 total: 18.6ms remaining: 17.2ms
52: learn: 0.2177215 total: 19.1ms remaining: 16.9ms
53: learn: 0.2143575 total: 19.4ms remaining: 16.5ms
54: learn: 0.2118161 total: 19.8ms remaining: 16.2ms
55: learn: 0.2094860 total: 20ms remaining: 15.7ms
56: learn: 0.2079612 total: 20.3ms remaining: 15.3ms
57: learn: 0.2048245 total: 20.5ms remaining: 14.8ms
58: learn: 0.2022788 total: 20.8ms remaining: 14.5ms
59: learn: 0.2009432 total: 21.2ms remaining: 14.1ms
60: learn: 0.1997459 total: 21.6ms remaining: 13.8ms
61: learn: 0.1973434 total: 22ms remaining: 13.5ms
62: learn: 0.1940305 total: 22.5ms remaining: 13.2ms
63: learn: 0.1931598 total: 23.2ms remaining: 13ms
64: learn: 0.1905972 total: 23.7ms remaining: 12.8ms
65: learn: 0.1892452 total: 24.4ms remaining: 12.6ms
66: learn: 0.1876009 total: 24.8ms remaining: 12.2ms
67: learn: 0.1854832 total: 25.2ms remaining: 11.9ms
68: learn: 0.1834037 total: 25.4ms remaining: 11.4ms
69: learn: 0.1822009 total: 26ms remaining: 11.2ms
70: learn: 0.1805010 total: 26.5ms remaining: 10.8ms
71: learn: 0.1791419 total: 27ms remaining: 10.5ms
72: learn: 0.1780161 total: 27.4ms remaining: 10.1ms
73: learn: 0.1769993 total: 27.9ms remaining: 9.81ms
74: learn: 0.1761321 total: 28.1ms remaining: 9.38ms
75: learn: 0.1741401 total: 28.4ms remaining: 8.97ms
76: learn: 0.1727571 total: 28.8ms remaining: 8.6ms
77: learn: 0.1715988 total: 29.2ms remaining: 8.23ms
78: learn: 0.1704649 total: 29.8ms remaining: 7.93ms
79: learn: 0.1697879 total: 30.2ms remaining: 7.54ms
80: learn: 0.1689765 total: 30.4ms remaining: 7.13ms
81: learn: 0.1684358 total: 30.8ms remaining: 6.76ms
82: learn: 0.1668083 total: 31.2ms remaining: 6.39ms
83: learn: 0.1651321 total: 31.6ms remaining: 6.02ms
84: learn: 0.1639347 total: 32.3ms remaining: 5.7ms
85: learn: 0.1623101 total: 32.9ms remaining: 5.35ms
86: learn: 0.1606518 total: 33.2ms remaining: 4.96ms
87: learn: 0.1598402 total: 33.7ms remaining: 4.6ms
88: learn: 0.1591425 total: 33.9ms remaining: 4.19ms
89: learn: 0.1579675 total: 34.2ms remaining: 3.8ms
90: learn: 0.1574750 total: 34.4ms remaining: 3.4ms
91: learn: 0.1561653 total: 34.7ms remaining: 3.02ms
92: learn: 0.1556096 total: 35ms remaining: 2.63ms
93: learn: 0.1549666 total: 35.4ms remaining: 2.26ms
94: learn: 0.1541801 total: 35.7ms remaining: 1.88ms
95: learn: 0.1532460 total: 36ms remaining: 1.5ms
96: learn: 0.1518631 total: 36.3ms remaining: 1.12ms
97: learn: 0.1506036 total: 36.5ms remaining: 745us
98: learn: 0.1491278 total: 36.9ms remaining: 373us
99: learn: 0.1482352 total: 37.7ms remaining: 0us
0: learn: 0.6531507 total: 363us remaining: 36ms
1: learn: 0.6065162 total: 567us remaining: 27.8ms
2: learn: 0.5670336 total: 717us remaining: 23.2ms
3: learn: 0.5358549 total: 934us remaining: 22.4ms
4: learn: 0.5074412 total: 1.09ms remaining: 20.8ms
5: learn: 0.4804679 total: 1.29ms remaining: 20.2ms
6: learn: 0.4657784 total: 1.44ms remaining: 19.1ms
7: learn: 0.4441830 total: 1.61ms remaining: 18.5ms
8: learn: 0.4246293 total: 1.78ms remaining: 18ms
9: learn: 0.4082806 total: 1.95ms remaining: 17.6ms
10: learn: 0.3971826 total: 2.11ms remaining: 17ms
11: learn: 0.3859011 total: 2.36ms remaining: 17.3ms
12: learn: 0.3732273 total: 2.54ms remaining: 17ms
13: learn: 0.3599254 total: 2.76ms remaining: 17ms
14: learn: 0.3520452 total: 2.92ms remaining: 16.6ms
15: learn: 0.3434055 total: 3.13ms remaining: 16.4ms
16: learn: 0.3346383 total: 3.34ms remaining: 16.3ms
17: learn: 0.3283062 total: 3.58ms remaining: 16.3ms
18: learn: 0.3212423 total: 3.76ms remaining: 16ms
19: learn: 0.3155269 total: 4.14ms remaining: 16.6ms
20: learn: 0.3091024 total: 4.5ms remaining: 16.9ms
21: learn: 0.3038824 total: 4.69ms remaining: 16.6ms
22: learn: 0.2960116 total: 4.91ms remaining: 16.4ms
23: learn: 0.2915019 total: 5.14ms remaining: 16.3ms
24: learn: 0.2851576 total: 5.28ms remaining: 15.9ms
25: learn: 0.2824845 total: 5.49ms remaining: 15.6ms
26: learn: 0.2779683 total: 5.67ms remaining: 15.3ms
27: learn: 0.2739306 total: 5.99ms remaining: 15.4ms
28: learn: 0.2711215 total: 6.31ms remaining: 15.4ms
29: learn: 0.2672378 total: 6.67ms remaining: 15.6ms
30: learn: 0.2642976 total: 6.91ms remaining: 15.4ms
31: learn: 0.2610812 total: 7.24ms remaining: 15.4ms
32: learn: 0.2570838 total: 7.6ms remaining: 15.4ms
33: learn: 0.2537967 total: 7.96ms remaining: 15.5ms
34: learn: 0.2509818 total: 8.35ms remaining: 15.5ms
35: learn: 0.2490697 total: 8.73ms remaining: 15.5ms
36: learn: 0.2463164 total: 9.01ms remaining: 15.3ms
37: learn: 0.2441294 total: 9.46ms remaining: 15.4ms
38: learn: 0.2423599 total: 9.89ms remaining: 15.5ms
39: learn: 0.2392679 total: 10.1ms remaining: 15.2ms
40: learn: 0.2373584 total: 10.7ms remaining: 15.4ms
41: learn: 0.2351607 total: 11.2ms remaining: 15.4ms
42: learn: 0.2329397 total: 11.7ms remaining: 15.6ms
43: learn: 0.2311672 total: 12.3ms remaining: 15.7ms
44: learn: 0.2291315 total: 13ms remaining: 15.8ms
45: learn: 0.2273520 total: 13.5ms remaining: 15.8ms
46: learn: 0.2248574 total: 14ms remaining: 15.7ms
47: learn: 0.2237799 total: 14.6ms remaining: 15.8ms
48: learn: 0.2229512 total: 15.2ms remaining: 15.9ms
49: learn: 0.2207943 total: 15.8ms remaining: 15.8ms
50: learn: 0.2191466 total: 16.1ms remaining: 15.5ms
51: learn: 0.2167604 total: 16.3ms remaining: 15.1ms
52: learn: 0.2150212 total: 16.5ms remaining: 14.7ms
53: learn: 0.2143156 total: 16.7ms remaining: 14.2ms
54: learn: 0.2129308 total: 16.9ms remaining: 13.8ms
55: learn: 0.2115729 total: 17.1ms remaining: 13.4ms
56: learn: 0.2096498 total: 17.5ms remaining: 13.2ms
57: learn: 0.2069896 total: 17.7ms remaining: 12.8ms
58: learn: 0.2057671 total: 17.9ms remaining: 12.5ms
59: learn: 0.2044193 total: 18.2ms remaining: 12.2ms
60: learn: 0.2030844 total: 18.5ms remaining: 11.8ms
61: learn: 0.2013383 total: 18.7ms remaining: 11.5ms
62: learn: 0.1998751 total: 19.2ms remaining: 11.3ms
63: learn: 0.1980976 total: 19.6ms remaining: 11ms
64: learn: 0.1971064 total: 20.1ms remaining: 10.8ms
65: learn: 0.1953909 total: 20.5ms remaining: 10.6ms
66: learn: 0.1939258 total: 20.8ms remaining: 10.3ms
67: learn: 0.1927908 total: 21ms remaining: 9.89ms
68: learn: 0.1908997 total: 21.8ms remaining: 9.78ms
69: learn: 0.1897857 total: 22.3ms remaining: 9.55ms
70: learn: 0.1885897 total: 22.9ms remaining: 9.35ms
71: learn: 0.1875013 total: 23.4ms remaining: 9.12ms
72: learn: 0.1861197 total: 24.1ms remaining: 8.92ms
73: learn: 0.1851799 total: 24.7ms remaining: 8.68ms
74: learn: 0.1844384 total: 25.2ms remaining: 8.39ms
75: learn: 0.1833483 total: 25.5ms remaining: 8.05ms
76: learn: 0.1823807 total: 26.2ms remaining: 7.81ms
77: learn: 0.1813433 total: 26.4ms remaining: 7.45ms
78: learn: 0.1799584 total: 26.7ms remaining: 7.1ms
79: learn: 0.1790604 total: 27ms remaining: 6.75ms
80: learn: 0.1773258 total: 27.2ms remaining: 6.38ms
81: learn: 0.1764698 total: 27.4ms remaining: 6.02ms
82: learn: 0.1751124 total: 27.6ms remaining: 5.66ms
83: learn: 0.1740194 total: 28.1ms remaining: 5.35ms
84: learn: 0.1732525 total: 29.3ms remaining: 5.17ms
85: learn: 0.1724760 total: 30.2ms remaining: 4.92ms
86: learn: 0.1713105 total: 30.9ms remaining: 4.61ms
87: learn: 0.1706636 total: 31.5ms remaining: 4.3ms
88: learn: 0.1698617 total: 32.1ms remaining: 3.97ms
89: learn: 0.1686562 total: 32.7ms remaining: 3.64ms
90: learn: 0.1678511 total: 33.3ms remaining: 3.29ms
91: learn: 0.1671549 total: 33.5ms remaining: 2.91ms
92: learn: 0.1663800 total: 34.2ms remaining: 2.57ms
93: learn: 0.1648383 total: 34.5ms remaining: 2.2ms
94: learn: 0.1639513 total: 34.8ms remaining: 1.83ms
95: learn: 0.1628442 total: 35ms remaining: 1.46ms
96: learn: 0.1620735 total: 35.3ms remaining: 1.09ms
97: learn: 0.1613418 total: 35.5ms remaining: 723us
98: learn: 0.1606256 total: 35.7ms remaining: 360us
99: learn: 0.1599078 total: 36ms remaining: 0us
0: learn: 0.6532610 total: 309us remaining: 30.7ms
1: learn: 0.6102409 total: 537us remaining: 26.3ms
2: learn: 0.5726766 total: 704us remaining: 22.8ms
3: learn: 0.5399936 total: 1.02ms remaining: 24.6ms
4: learn: 0.5149233 total: 1.22ms remaining: 23.2ms
5: learn: 0.4883109 total: 1.44ms remaining: 22.6ms
6: learn: 0.4641478 total: 1.66ms remaining: 22.1ms
7: learn: 0.4460774 total: 1.96ms remaining: 22.5ms
8: learn: 0.4323097 total: 2.08ms remaining: 21ms
9: learn: 0.4157583 total: 2.51ms remaining: 22.6ms
10: learn: 0.4022919 total: 2.7ms remaining: 21.9ms
11: learn: 0.3898014 total: 2.93ms remaining: 21.5ms
12: learn: 0.3788049 total: 3.14ms remaining: 21ms
13: learn: 0.3683505 total: 3.34ms remaining: 20.5ms
14: learn: 0.3577059 total: 3.63ms remaining: 20.6ms
15: learn: 0.3493490 total: 3.89ms remaining: 20.4ms
16: learn: 0.3384982 total: 4.22ms remaining: 20.6ms
17: learn: 0.3329396 total: 4.57ms remaining: 20.8ms
18: learn: 0.3265991 total: 4.78ms remaining: 20.4ms
19: learn: 0.3205432 total: 5.03ms remaining: 20.1ms
20: learn: 0.3157322 total: 5.26ms remaining: 19.8ms
21: learn: 0.3109822 total: 5.59ms remaining: 19.8ms
22: learn: 0.3063239 total: 5.79ms remaining: 19.4ms
23: learn: 0.3018177 total: 5.99ms remaining: 19ms
24: learn: 0.2976308 total: 6.15ms remaining: 18.4ms
25: learn: 0.2931462 total: 6.48ms remaining: 18.4ms
26: learn: 0.2895543 total: 6.81ms remaining: 18.4ms
27: learn: 0.2858063 total: 7.18ms remaining: 18.5ms
28: learn: 0.2817349 total: 7.37ms remaining: 18ms
29: learn: 0.2783638 total: 7.76ms remaining: 18.1ms
30: learn: 0.2755228 total: 8.04ms remaining: 17.9ms
31: learn: 0.2726879 total: 8.25ms remaining: 17.5ms
32: learn: 0.2707994 total: 8.42ms remaining: 17.1ms
33: learn: 0.2671753 total: 8.64ms remaining: 16.8ms
34: learn: 0.2642467 total: 8.97ms remaining: 16.7ms
35: learn: 0.2618695 total: 9.35ms remaining: 16.6ms
36: learn: 0.2601981 total: 9.54ms remaining: 16.2ms
37: learn: 0.2580945 total: 9.75ms remaining: 15.9ms
38: learn: 0.2563573 total: 10.1ms remaining: 15.8ms
39: learn: 0.2534017 total: 10.4ms remaining: 15.5ms
40: learn: 0.2503710 total: 10.6ms remaining: 15.3ms
41: learn: 0.2467603 total: 10.9ms remaining: 15ms
42: learn: 0.2445181 total: 11.2ms remaining: 14.8ms
43: learn: 0.2436733 total: 11.5ms remaining: 14.6ms
44: learn: 0.2424590 total: 11.9ms remaining: 14.5ms
45: learn: 0.2413170 total: 12.2ms remaining: 14.3ms
46: learn: 0.2395461 total: 12.5ms remaining: 14.1ms
47: learn: 0.2376051 total: 12.7ms remaining: 13.8ms
48: learn: 0.2348047 total: 12.9ms remaining: 13.5ms
49: learn: 0.2325867 total: 13.2ms remaining: 13.2ms
50: learn: 0.2312478 total: 13.5ms remaining: 12.9ms
51: learn: 0.2292347 total: 13.7ms remaining: 12.6ms
52: learn: 0.2264443 total: 14.2ms remaining: 12.6ms
53: learn: 0.2240809 total: 14.6ms remaining: 12.5ms
54: learn: 0.2224813 total: 15ms remaining: 12.3ms
55: learn: 0.2197070 total: 15.4ms remaining: 12.1ms
56: learn: 0.2181432 total: 15.7ms remaining: 11.8ms
57: learn: 0.2173565 total: 16ms remaining: 11.6ms
58: learn: 0.2157150 total: 16.3ms remaining: 11.3ms
59: learn: 0.2140966 total: 16.8ms remaining: 11.2ms
60: learn: 0.2115956 total: 17.3ms remaining: 11.1ms
61: learn: 0.2100308 total: 17.7ms remaining: 10.8ms
62: learn: 0.2074310 total: 18ms remaining: 10.6ms
63: learn: 0.2057503 total: 18.5ms remaining: 10.4ms
64: learn: 0.2047251 total: 18.8ms remaining: 10.1ms
65: learn: 0.2034574 total: 19ms remaining: 9.79ms
66: learn: 0.2023031 total: 19.4ms remaining: 9.56ms
67: learn: 0.2013246 total: 19.9ms remaining: 9.37ms
68: learn: 0.1994649 total: 20.3ms remaining: 9.1ms
69: learn: 0.1984366 total: 20.7ms remaining: 8.88ms
70: learn: 0.1971174 total: 21ms remaining: 8.57ms
71: learn: 0.1957417 total: 21.5ms remaining: 8.35ms
72: learn: 0.1949303 total: 21.8ms remaining: 8.06ms
73: learn: 0.1929753 total: 22.4ms remaining: 7.87ms
74: learn: 0.1922421 total: 22.9ms remaining: 7.62ms
75: learn: 0.1910603 total: 23.4ms remaining: 7.4ms
76: learn: 0.1900257 total: 24.1ms remaining: 7.19ms
77: learn: 0.1890963 total: 24.3ms remaining: 6.85ms
78: learn: 0.1880508 total: 24.6ms remaining: 6.55ms
79: learn: 0.1872142 total: 25.5ms remaining: 6.37ms
80: learn: 0.1863501 total: 26.3ms remaining: 6.18ms
81: learn: 0.1855408 total: 26.9ms remaining: 5.91ms
82: learn: 0.1840863 total: 27.6ms remaining: 5.66ms
83: learn: 0.1833803 total: 28.1ms remaining: 5.35ms
84: learn: 0.1822225 total: 28.4ms remaining: 5.02ms
85: learn: 0.1807301 total: 28.9ms remaining: 4.7ms
86: learn: 0.1798001 total: 29.3ms remaining: 4.38ms
87: learn: 0.1790189 total: 29.6ms remaining: 4.04ms
88: learn: 0.1781547 total: 29.9ms remaining: 3.69ms
89: learn: 0.1770911 total: 30.2ms remaining: 3.35ms
90: learn: 0.1762956 total: 30.9ms remaining: 3.06ms
91: learn: 0.1754951 total: 31.5ms remaining: 2.74ms
92: learn: 0.1744672 total: 32.1ms remaining: 2.41ms
93: learn: 0.1735861 total: 32.4ms remaining: 2.07ms
94: learn: 0.1726698 total: 32.7ms remaining: 1.72ms
95: learn: 0.1721135 total: 33.6ms remaining: 1.4ms
96: learn: 0.1715908 total: 33.8ms remaining: 1.05ms
97: learn: 0.1710334 total: 34.1ms remaining: 695us
98: learn: 0.1706777 total: 34.3ms remaining: 346us
99: learn: 0.1703117 total: 35ms remaining: 0us
0: learn: 0.6478370 total: 274us remaining: 27.2ms
1: learn: 0.6050464 total: 515us remaining: 25.2ms
2: learn: 0.5715635 total: 686us remaining: 22.2ms
3: learn: 0.5388645 total: 870us remaining: 20.9ms
4: learn: 0.5127410 total: 995us remaining: 18.9ms
5: learn: 0.4894968 total: 1.21ms remaining: 18.9ms
6: learn: 0.4690824 total: 1.65ms remaining: 21.9ms
7: learn: 0.4527488 total: 2.27ms remaining: 26.1ms
8: learn: 0.4367211 total: 2.51ms remaining: 25.4ms
9: learn: 0.4197791 total: 2.95ms remaining: 26.6ms
10: learn: 0.4076903 total: 3.36ms remaining: 27.2ms
11: learn: 0.3953699 total: 3.73ms remaining: 27.4ms
12: learn: 0.3835805 total: 4.14ms remaining: 27.7ms
13: learn: 0.3706580 total: 4.62ms remaining: 28.4ms
14: learn: 0.3632837 total: 5.02ms remaining: 28.4ms
15: learn: 0.3557629 total: 5.56ms remaining: 29.2ms
16: learn: 0.3465254 total: 5.92ms remaining: 28.9ms
17: learn: 0.3424635 total: 6.17ms remaining: 28.1ms
18: learn: 0.3340110 total: 6.6ms remaining: 28.2ms
19: learn: 0.3290275 total: 7.5ms remaining: 30ms
20: learn: 0.3221192 total: 8.06ms remaining: 30.3ms
21: learn: 0.3160905 total: 8.4ms remaining: 29.8ms
22: learn: 0.3106294 total: 8.63ms remaining: 28.9ms
23: learn: 0.3056046 total: 9.23ms remaining: 29.2ms
24: learn: 0.3024425 total: 10.1ms remaining: 30.2ms
25: learn: 0.2989832 total: 10.7ms remaining: 30.6ms
26: learn: 0.2944404 total: 11.3ms remaining: 30.4ms
27: learn: 0.2902619 total: 11.5ms remaining: 29.5ms
28: learn: 0.2872351 total: 11.9ms remaining: 29.1ms
29: learn: 0.2832049 total: 12.5ms remaining: 29.2ms
30: learn: 0.2811090 total: 12.8ms remaining: 28.5ms
31: learn: 0.2773095 total: 13.2ms remaining: 28.1ms
32: learn: 0.2748773 total: 13.7ms remaining: 27.7ms
33: learn: 0.2728913 total: 14ms remaining: 27.1ms
34: learn: 0.2704194 total: 14.4ms remaining: 26.8ms
35: learn: 0.2677007 total: 14.7ms remaining: 26.1ms
36: learn: 0.2654716 total: 15.2ms remaining: 25.8ms
37: learn: 0.2625555 total: 16.1ms remaining: 26.2ms
38: learn: 0.2588682 total: 16.7ms remaining: 26.1ms
39: learn: 0.2557431 total: 17.2ms remaining: 25.8ms
40: learn: 0.2541972 total: 17.5ms remaining: 25.2ms
41: learn: 0.2533046 total: 18.1ms remaining: 25ms
42: learn: 0.2508947 total: 19.1ms remaining: 25.3ms
43: learn: 0.2488413 total: 19.7ms remaining: 25ms
44: learn: 0.2460522 total: 20.4ms remaining: 24.9ms
45: learn: 0.2437101 total: 21.4ms remaining: 25.2ms
46: learn: 0.2420143 total: 22.3ms remaining: 25.2ms
47: learn: 0.2394717 total: 22.9ms remaining: 24.8ms
48: learn: 0.2377849 total: 23.5ms remaining: 24.5ms
49: learn: 0.2356327 total: 23.9ms remaining: 23.9ms
50: learn: 0.2333147 total: 25ms remaining: 24ms
51: learn: 0.2314324 total: 25.5ms remaining: 23.6ms
52: learn: 0.2295085 total: 26ms remaining: 23ms
53: learn: 0.2276318 total: 26.2ms remaining: 22.3ms
54: learn: 0.2268899 total: 26.4ms remaining: 21.6ms
55: learn: 0.2237974 total: 26.6ms remaining: 20.9ms
56: learn: 0.2224561 total: 26.8ms remaining: 20.2ms
57: learn: 0.2192132 total: 27.2ms remaining: 19.7ms
58: learn: 0.2173932 total: 27.5ms remaining: 19.1ms
59: learn: 0.2150435 total: 28.4ms remaining: 18.9ms
60: learn: 0.2132824 total: 28.8ms remaining: 18.4ms
61: learn: 0.2119521 total: 29ms remaining: 17.8ms
62: learn: 0.2102608 total: 29.2ms remaining: 17.2ms
63: learn: 0.2068338 total: 29.5ms remaining: 16.6ms
64: learn: 0.2054391 total: 30ms remaining: 16.2ms
65: learn: 0.2037807 total: 30.3ms remaining: 15.6ms
66: learn: 0.2022487 total: 30.5ms remaining: 15ms
67: learn: 0.1999168 total: 30.9ms remaining: 14.5ms
68: learn: 0.1984479 total: 31.6ms remaining: 14.2ms
69: learn: 0.1970045 total: 32ms remaining: 13.7ms
70: learn: 0.1956535 total: 32.7ms remaining: 13.3ms
71: learn: 0.1942315 total: 33.3ms remaining: 12.9ms
72: learn: 0.1932206 total: 33.6ms remaining: 12.4ms
73: learn: 0.1917156 total: 33.8ms remaining: 11.9ms
74: learn: 0.1897797 total: 34ms remaining: 11.3ms
75: learn: 0.1888298 total: 34.3ms remaining: 10.8ms
76: learn: 0.1875736 total: 34.8ms remaining: 10.4ms
77: learn: 0.1861071 total: 35.7ms remaining: 10.1ms
78: learn: 0.1849552 total: 35.9ms remaining: 9.56ms
79: learn: 0.1830319 total: 36.2ms remaining: 9.05ms
80: learn: 0.1811466 total: 36.5ms remaining: 8.55ms
81: learn: 0.1805334 total: 37.3ms remaining: 8.19ms
82: learn: 0.1784587 total: 38ms remaining: 7.79ms
83: learn: 0.1777659 total: 38.4ms remaining: 7.32ms
84: learn: 0.1770975 total: 38.8ms remaining: 6.85ms
85: learn: 0.1754173 total: 39.3ms remaining: 6.39ms
86: learn: 0.1737798 total: 39.7ms remaining: 5.94ms
87: learn: 0.1726413 total: 41ms remaining: 5.59ms
88: learn: 0.1712179 total: 41.3ms remaining: 5.11ms
89: learn: 0.1706006 total: 41.6ms remaining: 4.62ms
90: learn: 0.1688359 total: 41.8ms remaining: 4.13ms
91: learn: 0.1683922 total: 42ms remaining: 3.65ms
92: learn: 0.1679554 total: 43.1ms remaining: 3.24ms
93: learn: 0.1668017 total: 43.3ms remaining: 2.77ms
94: learn: 0.1659074 total: 43.5ms remaining: 2.29ms
95: learn: 0.1646166 total: 44ms remaining: 1.83ms
96: learn: 0.1642003 total: 44.4ms remaining: 1.37ms
97: learn: 0.1631870 total: 44.8ms remaining: 913us
98: learn: 0.1620912 total: 45.3ms remaining: 458us
99: learn: 0.1612658 total: 46ms remaining: 0us
0: learn: 0.6489461 total: 345us remaining: 34.2ms
1: learn: 0.6064240 total: 547us remaining: 26.8ms
2: learn: 0.5700715 total: 748us remaining: 24.2ms
3: learn: 0.5400758 total: 958us remaining: 23ms
4: learn: 0.5145103 total: 1.21ms remaining: 23ms
5: learn: 0.4866673 total: 1.63ms remaining: 25.6ms
6: learn: 0.4673804 total: 2.12ms remaining: 28.1ms
7: learn: 0.4481874 total: 2.59ms remaining: 29.8ms
8: learn: 0.4300384 total: 3.17ms remaining: 32ms
9: learn: 0.4134195 total: 3.51ms remaining: 31.6ms
10: learn: 0.3962502 total: 4.02ms remaining: 32.6ms
11: learn: 0.3811386 total: 4.42ms remaining: 32.4ms
12: learn: 0.3693335 total: 4.89ms remaining: 32.7ms
13: learn: 0.3598914 total: 5.1ms remaining: 31.3ms
14: learn: 0.3516374 total: 5.54ms remaining: 31.4ms
15: learn: 0.3406200 total: 6.12ms remaining: 32.1ms
16: learn: 0.3323188 total: 6.66ms remaining: 32.5ms
17: learn: 0.3250314 total: 7ms remaining: 31.9ms
18: learn: 0.3192270 total: 7.73ms remaining: 32.9ms
19: learn: 0.3128178 total: 8.54ms remaining: 34.2ms
20: learn: 0.3059293 total: 9.41ms remaining: 35.4ms
21: learn: 0.2987314 total: 9.99ms remaining: 35.4ms
22: learn: 0.2940187 total: 10.4ms remaining: 34.9ms
23: learn: 0.2900778 total: 11.1ms remaining: 35.2ms
24: learn: 0.2872265 total: 11.7ms remaining: 35.2ms
25: learn: 0.2842832 total: 12.5ms remaining: 35.5ms
26: learn: 0.2804576 total: 13.1ms remaining: 35.4ms
27: learn: 0.2770150 total: 13.7ms remaining: 35.3ms
28: learn: 0.2748851 total: 14.2ms remaining: 34.7ms
29: learn: 0.2718988 total: 14.7ms remaining: 34.3ms
30: learn: 0.2689076 total: 15ms remaining: 33.4ms
31: learn: 0.2655543 total: 15.5ms remaining: 32.9ms
32: learn: 0.2633803 total: 16.1ms remaining: 32.6ms
33: learn: 0.2607521 total: 16.6ms remaining: 32.3ms
34: learn: 0.2586816 total: 17.1ms remaining: 31.7ms
35: learn: 0.2561481 total: 17.5ms remaining: 31.2ms
36: learn: 0.2535555 total: 18.1ms remaining: 30.8ms
37: learn: 0.2498320 total: 18.7ms remaining: 30.4ms
38: learn: 0.2465524 total: 19.2ms remaining: 30ms
39: learn: 0.2453466 total: 19.6ms remaining: 29.3ms
40: learn: 0.2437222 total: 19.9ms remaining: 28.7ms
41: learn: 0.2416669 total: 20.4ms remaining: 28.2ms
42: learn: 0.2391285 total: 20.8ms remaining: 27.5ms
43: learn: 0.2365899 total: 21ms remaining: 26.7ms
44: learn: 0.2348016 total: 21.6ms remaining: 26.4ms
45: learn: 0.2321103 total: 22ms remaining: 25.8ms
46: learn: 0.2307285 total: 22.2ms remaining: 25ms
47: learn: 0.2292365 total: 22.6ms remaining: 24.5ms
48: learn: 0.2281501 total: 22.8ms remaining: 23.7ms
49: learn: 0.2266474 total: 23.1ms remaining: 23.1ms
50: learn: 0.2254592 total: 23.5ms remaining: 22.6ms
51: learn: 0.2225412 total: 23.9ms remaining: 22.1ms
52: learn: 0.2209814 total: 24.4ms remaining: 21.7ms
53: learn: 0.2188452 total: 25ms remaining: 21.3ms
54: learn: 0.2176260 total: 25.3ms remaining: 20.7ms
55: learn: 0.2160541 total: 25.8ms remaining: 20.3ms
56: learn: 0.2145296 total: 26.3ms remaining: 19.9ms
57: learn: 0.2131576 total: 26.9ms remaining: 19.5ms
58: learn: 0.2112919 total: 27.4ms remaining: 19.1ms
59: learn: 0.2099507 total: 27.9ms remaining: 18.6ms
60: learn: 0.2078440 total: 28.5ms remaining: 18.2ms
61: learn: 0.2063141 total: 29ms remaining: 17.8ms
62: learn: 0.2047629 total: 29.7ms remaining: 17.5ms
63: learn: 0.2038695 total: 30.5ms remaining: 17.2ms
64: learn: 0.2023354 total: 31.1ms remaining: 16.8ms
65: learn: 0.1996688 total: 31.5ms remaining: 16.2ms
66: learn: 0.1973198 total: 32ms remaining: 15.8ms
67: learn: 0.1959162 total: 32.3ms remaining: 15.2ms
68: learn: 0.1951752 total: 32.8ms remaining: 14.7ms
69: learn: 0.1940283 total: 33.5ms remaining: 14.4ms
70: learn: 0.1930343 total: 33.9ms remaining: 13.9ms
71: learn: 0.1907450 total: 34.4ms remaining: 13.4ms
72: learn: 0.1887311 total: 34.8ms remaining: 12.9ms
73: learn: 0.1874915 total: 35.3ms remaining: 12.4ms
74: learn: 0.1860438 total: 35.8ms remaining: 11.9ms
75: learn: 0.1851532 total: 36.2ms remaining: 11.4ms
76: learn: 0.1843621 total: 36.5ms remaining: 10.9ms
77: learn: 0.1828291 total: 36.9ms remaining: 10.4ms
78: learn: 0.1819787 total: 37.4ms remaining: 9.95ms
79: learn: 0.1800943 total: 38.1ms remaining: 9.52ms
80: learn: 0.1789557 total: 38.5ms remaining: 9.04ms
81: learn: 0.1783137 total: 39ms remaining: 8.57ms
82: learn: 0.1776746 total: 39.5ms remaining: 8.1ms
83: learn: 0.1766376 total: 40ms remaining: 7.62ms
84: learn: 0.1754587 total: 40.3ms remaining: 7.11ms
85: learn: 0.1747746 total: 40.6ms remaining: 6.61ms
86: learn: 0.1734613 total: 41ms remaining: 6.12ms
87: learn: 0.1723720 total: 41.4ms remaining: 5.65ms
88: learn: 0.1716154 total: 42ms remaining: 5.2ms
89: learn: 0.1705611 total: 42.6ms remaining: 4.73ms
90: learn: 0.1699904 total: 42.8ms remaining: 4.24ms
91: learn: 0.1692174 total: 43.5ms remaining: 3.78ms
92: learn: 0.1685566 total: 44ms remaining: 3.31ms
93: learn: 0.1679728 total: 44.8ms remaining: 2.86ms
94: learn: 0.1674829 total: 45.4ms remaining: 2.39ms
95: learn: 0.1669377 total: 46.3ms remaining: 1.93ms
96: learn: 0.1663326 total: 46.8ms remaining: 1.45ms
97: learn: 0.1654778 total: 47.1ms remaining: 960us
98: learn: 0.1643119 total: 47.7ms remaining: 481us
99: learn: 0.1631584 total: 48.6ms remaining: 0us
0: learn: 0.6590402 total: 325us remaining: 32.2ms
1: learn: 0.6114009 total: 533us remaining: 26.1ms
2: learn: 0.5810295 total: 738us remaining: 23.9ms
3: learn: 0.5494286 total: 1.12ms remaining: 26.9ms
4: learn: 0.5218481 total: 2.1ms remaining: 39.9ms
5: learn: 0.4961409 total: 2.57ms remaining: 40.3ms
6: learn: 0.4825933 total: 3.03ms remaining: 40.3ms
7: learn: 0.4612439 total: 3.37ms remaining: 38.8ms
8: learn: 0.4405597 total: 3.77ms remaining: 38.1ms
9: learn: 0.4236230 total: 4.12ms remaining: 37.1ms
10: learn: 0.4086284 total: 4.43ms remaining: 35.8ms
11: learn: 0.3966478 total: 4.72ms remaining: 34.6ms
12: learn: 0.3845862 total: 5.07ms remaining: 33.9ms
13: learn: 0.3734868 total: 5.41ms remaining: 33.2ms
14: learn: 0.3612373 total: 5.65ms remaining: 32ms
15: learn: 0.3563945 total: 5.84ms remaining: 30.7ms
16: learn: 0.3520736 total: 6.02ms remaining: 29.4ms
17: learn: 0.3437252 total: 6.37ms remaining: 29ms
18: learn: 0.3366232 total: 6.7ms remaining: 28.6ms
19: learn: 0.3305014 total: 7.02ms remaining: 28.1ms
20: learn: 0.3237168 total: 7.42ms remaining: 27.9ms
21: learn: 0.3165086 total: 7.72ms remaining: 27.4ms
22: learn: 0.3115073 total: 8.01ms remaining: 26.8ms
23: learn: 0.3070963 total: 8.42ms remaining: 26.7ms
24: learn: 0.3018950 total: 8.83ms remaining: 26.5ms
25: learn: 0.2978404 total: 9.19ms remaining: 26.2ms
26: learn: 0.2944708 total: 9.46ms remaining: 25.6ms
27: learn: 0.2896176 total: 9.76ms remaining: 25.1ms
28: learn: 0.2862532 total: 10.1ms remaining: 24.8ms
29: learn: 0.2832470 total: 10.3ms remaining: 24.1ms
30: learn: 0.2798789 total: 10.6ms remaining: 23.7ms
31: learn: 0.2760300 total: 10.9ms remaining: 23.1ms
32: learn: 0.2725581 total: 11.1ms remaining: 22.6ms
33: learn: 0.2704228 total: 11.4ms remaining: 22.2ms
34: learn: 0.2677841 total: 11.7ms remaining: 21.7ms
35: learn: 0.2653151 total: 12ms remaining: 21.4ms
36: learn: 0.2633276 total: 12.3ms remaining: 21ms
37: learn: 0.2602114 total: 12.6ms remaining: 20.6ms
38: learn: 0.2575564 total: 13ms remaining: 20.3ms
39: learn: 0.2550264 total: 13.3ms remaining: 20ms
40: learn: 0.2531548 total: 13.6ms remaining: 19.6ms
41: learn: 0.2507302 total: 13.9ms remaining: 19.2ms
42: learn: 0.2494902 total: 14.3ms remaining: 18.9ms
43: learn: 0.2463801 total: 14.5ms remaining: 18.4ms
44: learn: 0.2447248 total: 14.8ms remaining: 18.1ms
45: learn: 0.2424518 total: 15.2ms remaining: 17.9ms
46: learn: 0.2411762 total: 15.4ms remaining: 17.3ms
47: learn: 0.2387374 total: 15.8ms remaining: 17.1ms
48: learn: 0.2362627 total: 16ms remaining: 16.6ms
49: learn: 0.2354837 total: 16.3ms remaining: 16.3ms
50: learn: 0.2334826 total: 16.6ms remaining: 16ms
51: learn: 0.2320311 total: 17ms remaining: 15.7ms
52: learn: 0.2304605 total: 17.3ms remaining: 15.4ms
53: learn: 0.2283454 total: 17.6ms remaining: 15ms
54: learn: 0.2259308 total: 18.1ms remaining: 14.8ms
55: learn: 0.2243436 total: 18.4ms remaining: 14.4ms
56: learn: 0.2234152 total: 18.8ms remaining: 14.1ms
57: learn: 0.2222624 total: 19.3ms remaining: 13.9ms
58: learn: 0.2198506 total: 19.6ms remaining: 13.6ms
59: learn: 0.2180131 total: 19.8ms remaining: 13.2ms
60: learn: 0.2153596 total: 20.2ms remaining: 12.9ms
61: learn: 0.2135086 total: 20.6ms remaining: 12.6ms
62: learn: 0.2112580 total: 20.9ms remaining: 12.3ms
63: learn: 0.2097674 total: 21.2ms remaining: 11.9ms
64: learn: 0.2083025 total: 21.6ms remaining: 11.6ms
65: learn: 0.2067273 total: 21.9ms remaining: 11.3ms
66: learn: 0.2052166 total: 22.2ms remaining: 10.9ms
67: learn: 0.2045802 total: 22.5ms remaining: 10.6ms
68: learn: 0.2034975 total: 22.6ms remaining: 10.2ms
69: learn: 0.2027905 total: 22.9ms remaining: 9.81ms
70: learn: 0.2018563 total: 23.3ms remaining: 9.52ms
71: learn: 0.2000458 total: 23.6ms remaining: 9.19ms
72: learn: 0.1986833 total: 24.1ms remaining: 8.93ms
73: learn: 0.1981076 total: 24.6ms remaining: 8.64ms
74: learn: 0.1975341 total: 24.9ms remaining: 8.28ms
75: learn: 0.1966905 total: 25ms remaining: 7.91ms
76: learn: 0.1956072 total: 25.2ms remaining: 7.54ms
77: learn: 0.1944255 total: 25.6ms remaining: 7.22ms
78: learn: 0.1935610 total: 25.9ms remaining: 6.9ms
79: learn: 0.1927345 total: 26.3ms remaining: 6.57ms
80: learn: 0.1919683 total: 26.5ms remaining: 6.22ms
81: learn: 0.1910788 total: 26.7ms remaining: 5.87ms
82: learn: 0.1899076 total: 26.9ms remaining: 5.52ms
83: learn: 0.1886907 total: 27.1ms remaining: 5.17ms
84: learn: 0.1880866 total: 27.3ms remaining: 4.82ms
85: learn: 0.1877364 total: 27.6ms remaining: 4.5ms
86: learn: 0.1869338 total: 28ms remaining: 4.18ms
87: learn: 0.1860941 total: 28.3ms remaining: 3.86ms
88: learn: 0.1855313 total: 28.6ms remaining: 3.54ms
89: learn: 0.1846593 total: 29.1ms remaining: 3.23ms
90: learn: 0.1840153 total: 29.6ms remaining: 2.93ms
91: learn: 0.1833519 total: 30.1ms remaining: 2.62ms
92: learn: 0.1817814 total: 30.7ms remaining: 2.31ms
93: learn: 0.1806517 total: 31.2ms remaining: 1.99ms
94: learn: 0.1801285 total: 31.5ms remaining: 1.66ms
95: learn: 0.1795813 total: 31.7ms remaining: 1.32ms
96: learn: 0.1790231 total: 32.1ms remaining: 993us
97: learn: 0.1784280 total: 32.5ms remaining: 663us
98: learn: 0.1779345 total: 32.7ms remaining: 330us
99: learn: 0.1763333 total: 33.4ms remaining: 0us
0: learn: 0.6577557 total: 422us remaining: 41.8ms
1: learn: 0.6115590 total: 994us remaining: 48.7ms
2: learn: 0.5782661 total: 1.71ms remaining: 55.4ms
3: learn: 0.5504365 total: 2.08ms remaining: 49.9ms
4: learn: 0.5239138 total: 2.42ms remaining: 46.1ms
5: learn: 0.4960141 total: 3.19ms remaining: 49.9ms
6: learn: 0.4751299 total: 4.38ms remaining: 58.2ms
7: learn: 0.4522825 total: 4.86ms remaining: 55.9ms
8: learn: 0.4350004 total: 5.99ms remaining: 60.6ms
9: learn: 0.4172532 total: 6.27ms remaining: 56.4ms
10: learn: 0.4031272 total: 7.85ms remaining: 63.5ms
11: learn: 0.3894778 total: 8.62ms remaining: 63.2ms
12: learn: 0.3759268 total: 9.39ms remaining: 62.8ms
13: learn: 0.3646396 total: 10.4ms remaining: 63.8ms
14: learn: 0.3550212 total: 10.7ms remaining: 60.6ms
15: learn: 0.3459478 total: 11.1ms remaining: 58ms
16: learn: 0.3387418 total: 11.3ms remaining: 55.2ms
17: learn: 0.3317651 total: 11.5ms remaining: 52.5ms
18: learn: 0.3245328 total: 12ms remaining: 51.1ms
19: learn: 0.3186379 total: 13.3ms remaining: 53.3ms
20: learn: 0.3123720 total: 13.7ms remaining: 51.4ms
21: learn: 0.3066610 total: 14.1ms remaining: 50ms
22: learn: 0.3021609 total: 14.5ms remaining: 48.4ms
23: learn: 0.2981775 total: 14.9ms remaining: 47.3ms
24: learn: 0.2947302 total: 16ms remaining: 48ms
25: learn: 0.2894158 total: 17.6ms remaining: 50.1ms
26: learn: 0.2849985 total: 19.3ms remaining: 52.1ms
27: learn: 0.2816687 total: 21.1ms remaining: 54.3ms
28: learn: 0.2784744 total: 22.7ms remaining: 55.6ms
29: learn: 0.2753041 total: 24.2ms remaining: 56.4ms
30: learn: 0.2704793 total: 25.7ms remaining: 57.3ms
31: learn: 0.2655361 total: 27.1ms remaining: 57.7ms
32: learn: 0.2598502 total: 28.6ms remaining: 58.1ms
33: learn: 0.2568552 total: 29.5ms remaining: 57.2ms
34: learn: 0.2540340 total: 30.7ms remaining: 56.9ms
35: learn: 0.2503535 total: 31.6ms remaining: 56.2ms
36: learn: 0.2479424 total: 32.6ms remaining: 55.6ms
37: learn: 0.2453326 total: 34ms remaining: 55.4ms
38: learn: 0.2425282 total: 35.2ms remaining: 55.1ms
39: learn: 0.2398649 total: 36.7ms remaining: 55ms
40: learn: 0.2385556 total: 37.8ms remaining: 54.4ms
41: learn: 0.2365283 total: 38.9ms remaining: 53.7ms
42: learn: 0.2331381 total: 39.8ms remaining: 52.7ms
43: learn: 0.2300152 total: 40.7ms remaining: 51.8ms
44: learn: 0.2279245 total: 41.4ms remaining: 50.6ms
45: learn: 0.2263903 total: 41.8ms remaining: 49ms
46: learn: 0.2238257 total: 42.6ms remaining: 48ms
47: learn: 0.2216545 total: 43.4ms remaining: 47ms
48: learn: 0.2196501 total: 43.7ms remaining: 45.5ms
49: learn: 0.2168023 total: 43.9ms remaining: 43.9ms
50: learn: 0.2145388 total: 44.1ms remaining: 42.4ms
51: learn: 0.2132267 total: 44.4ms remaining: 40.9ms
52: learn: 0.2103353 total: 44.6ms remaining: 39.5ms
53: learn: 0.2089741 total: 44.9ms remaining: 38.2ms
54: learn: 0.2074504 total: 45.4ms remaining: 37.2ms
55: learn: 0.2053822 total: 45.7ms remaining: 35.9ms
56: learn: 0.2031189 total: 46ms remaining: 34.7ms
57: learn: 0.2017266 total: 46.5ms remaining: 33.6ms
58: learn: 0.1996794 total: 46.9ms remaining: 32.6ms
59: learn: 0.1972371 total: 47.4ms remaining: 31.6ms
60: learn: 0.1949181 total: 48ms remaining: 30.7ms
61: learn: 0.1912835 total: 48.4ms remaining: 29.7ms
62: learn: 0.1902439 total: 48.8ms remaining: 28.7ms
63: learn: 0.1884128 total: 49.2ms remaining: 27.7ms
64: learn: 0.1868456 total: 49.5ms remaining: 26.6ms
65: learn: 0.1856003 total: 49.7ms remaining: 25.6ms
66: learn: 0.1835379 total: 49.9ms remaining: 24.6ms
67: learn: 0.1825284 total: 50.1ms remaining: 23.6ms
68: learn: 0.1804093 total: 50.4ms remaining: 22.6ms
69: learn: 0.1791994 total: 50.6ms remaining: 21.7ms
70: learn: 0.1783252 total: 50.7ms remaining: 20.7ms
71: learn: 0.1765206 total: 51ms remaining: 19.8ms
72: learn: 0.1748911 total: 51.2ms remaining: 18.9ms
73: learn: 0.1736684 total: 51.4ms remaining: 18.1ms
74: learn: 0.1727610 total: 51.7ms remaining: 17.2ms
75: learn: 0.1704809 total: 52ms remaining: 16.4ms
76: learn: 0.1688713 total: 52.2ms remaining: 15.6ms
77: learn: 0.1679488 total: 52.4ms remaining: 14.8ms
78: learn: 0.1667283 total: 52.8ms remaining: 14ms
79: learn: 0.1657982 total: 53.1ms remaining: 13.3ms
80: learn: 0.1642468 total: 53.3ms remaining: 12.5ms
81: learn: 0.1631564 total: 53.5ms remaining: 11.7ms
82: learn: 0.1624591 total: 53.7ms remaining: 11ms
83: learn: 0.1611993 total: 53.9ms remaining: 10.3ms
84: learn: 0.1597753 total: 54.1ms remaining: 9.55ms
85: learn: 0.1589474 total: 54.3ms remaining: 8.84ms
86: learn: 0.1580195 total: 54.5ms remaining: 8.15ms
87: learn: 0.1575410 total: 54.7ms remaining: 7.46ms
88: learn: 0.1563160 total: 55ms remaining: 6.8ms
89: learn: 0.1557749 total: 55.2ms remaining: 6.13ms
90: learn: 0.1549278 total: 55.4ms remaining: 5.48ms
91: learn: 0.1534022 total: 55.6ms remaining: 4.83ms
92: learn: 0.1514222 total: 55.8ms remaining: 4.2ms
93: learn: 0.1508935 total: 56ms remaining: 3.58ms
94: learn: 0.1492736 total: 56.2ms remaining: 2.96ms
95: learn: 0.1484335 total: 56.4ms remaining: 2.35ms
96: learn: 0.1476766 total: 56.6ms remaining: 1.75ms
97: learn: 0.1466290 total: 56.8ms remaining: 1.16ms
98: learn: 0.1455973 total: 57ms remaining: 575us
99: learn: 0.1440446 total: 57.3ms remaining: 0us
0: learn: 0.6583442 total: 867us remaining: 85.9ms
1: learn: 0.6160693 total: 1.76ms remaining: 86.2ms
2: learn: 0.5795190 total: 2.4ms remaining: 77.7ms
3: learn: 0.5466177 total: 3.37ms remaining: 80.8ms
4: learn: 0.5207080 total: 4.21ms remaining: 80ms
5: learn: 0.4950930 total: 4.88ms remaining: 76.5ms
6: learn: 0.4717440 total: 5.78ms remaining: 76.8ms
7: learn: 0.4536730 total: 6.56ms remaining: 75.5ms
8: learn: 0.4349721 total: 7.34ms remaining: 74.2ms
9: learn: 0.4202796 total: 8.22ms remaining: 74ms
10: learn: 0.4077376 total: 8.8ms remaining: 71.2ms
11: learn: 0.3960184 total: 9.7ms remaining: 71.2ms
12: learn: 0.3826414 total: 10.4ms remaining: 69.7ms
13: learn: 0.3722208 total: 11.1ms remaining: 68.3ms
14: learn: 0.3627836 total: 12.4ms remaining: 70.1ms
15: learn: 0.3526643 total: 12.9ms remaining: 67.7ms
16: learn: 0.3433313 total: 13.7ms remaining: 66.8ms
17: learn: 0.3337858 total: 14.3ms remaining: 65ms
18: learn: 0.3281039 total: 15.1ms remaining: 64.2ms
19: learn: 0.3204933 total: 15.6ms remaining: 62.4ms
20: learn: 0.3172523 total: 16.5ms remaining: 62.2ms
21: learn: 0.3112378 total: 17.6ms remaining: 62.5ms
22: learn: 0.3055845 total: 17.9ms remaining: 59.9ms
23: learn: 0.3017372 total: 18.6ms remaining: 58.9ms
24: learn: 0.2956202 total: 19.6ms remaining: 58.7ms
25: learn: 0.2904468 total: 21.9ms remaining: 62.3ms
26: learn: 0.2863467 total: 22.8ms remaining: 61.5ms
27: learn: 0.2831806 total: 24.1ms remaining: 62ms
28: learn: 0.2799353 total: 24.6ms remaining: 60.3ms
29: learn: 0.2769338 total: 25.1ms remaining: 58.7ms
30: learn: 0.2725319 total: 25.8ms remaining: 57.4ms
31: learn: 0.2690325 total: 26.4ms remaining: 56.1ms
32: learn: 0.2653722 total: 27ms remaining: 54.8ms
33: learn: 0.2626298 total: 27.6ms remaining: 53.6ms
34: learn: 0.2613544 total: 29.4ms remaining: 54.7ms
35: learn: 0.2590320 total: 29.9ms remaining: 53.1ms
36: learn: 0.2566611 total: 30.4ms remaining: 51.8ms
37: learn: 0.2535962 total: 31.9ms remaining: 52.1ms
38: learn: 0.2497372 total: 32.5ms remaining: 50.8ms
39: learn: 0.2470181 total: 32.8ms remaining: 49.3ms
40: learn: 0.2445742 total: 33.4ms remaining: 48.1ms
41: learn: 0.2430848 total: 33.9ms remaining: 46.9ms
42: learn: 0.2422891 total: 34.4ms remaining: 45.6ms
43: learn: 0.2397990 total: 34.9ms remaining: 44.5ms
44: learn: 0.2371706 total: 35.7ms remaining: 43.6ms
45: learn: 0.2352512 total: 36.3ms remaining: 42.6ms
46: learn: 0.2327897 total: 36.7ms remaining: 41.4ms
47: learn: 0.2308399 total: 37ms remaining: 40.1ms
48: learn: 0.2296719 total: 37.4ms remaining: 38.9ms
49: learn: 0.2277718 total: 37.9ms remaining: 37.9ms
50: learn: 0.2263527 total: 38.2ms remaining: 36.7ms
51: learn: 0.2248207 total: 38.6ms remaining: 35.6ms
52: learn: 0.2232927 total: 39.5ms remaining: 35.1ms
53: learn: 0.2217390 total: 40.2ms remaining: 34.3ms
54: learn: 0.2188630 total: 40.7ms remaining: 33.3ms
55: learn: 0.2174118 total: 41.4ms remaining: 32.5ms
56: learn: 0.2154217 total: 41.8ms remaining: 31.5ms
57: learn: 0.2132996 total: 42.6ms remaining: 30.8ms
58: learn: 0.2119245 total: 43ms remaining: 29.9ms
59: learn: 0.2092403 total: 43.9ms remaining: 29.3ms
60: learn: 0.2081820 total: 44.4ms remaining: 28.4ms
61: learn: 0.2069303 total: 45.2ms remaining: 27.7ms
62: learn: 0.2057730 total: 45.8ms remaining: 26.9ms
63: learn: 0.2043888 total: 46.6ms remaining: 26.2ms
64: learn: 0.2033712 total: 47.2ms remaining: 25.4ms
65: learn: 0.2023271 total: 47.9ms remaining: 24.7ms
66: learn: 0.2007830 total: 48.7ms remaining: 24ms
67: learn: 0.1984698 total: 49.7ms remaining: 23.4ms
68: learn: 0.1971153 total: 50.5ms remaining: 22.7ms
69: learn: 0.1960784 total: 51.4ms remaining: 22ms
70: learn: 0.1949722 total: 52.2ms remaining: 21.3ms
71: learn: 0.1941931 total: 53ms remaining: 20.6ms
72: learn: 0.1930148 total: 53.5ms remaining: 19.8ms
73: learn: 0.1922700 total: 54.2ms remaining: 19ms
74: learn: 0.1912845 total: 54.7ms remaining: 18.2ms
75: learn: 0.1902818 total: 55.3ms remaining: 17.5ms
76: learn: 0.1879536 total: 56.2ms remaining: 16.8ms
77: learn: 0.1870270 total: 56.7ms remaining: 16ms
78: learn: 0.1848320 total: 57.1ms remaining: 15.2ms
79: learn: 0.1838008 total: 57.6ms remaining: 14.4ms
80: learn: 0.1828718 total: 58ms remaining: 13.6ms
81: learn: 0.1812732 total: 58.2ms remaining: 12.8ms
82: learn: 0.1803925 total: 58.5ms remaining: 12ms
83: learn: 0.1797710 total: 58.7ms remaining: 11.2ms
84: learn: 0.1789050 total: 59ms remaining: 10.4ms
85: learn: 0.1783196 total: 59.5ms remaining: 9.69ms
86: learn: 0.1777546 total: 60.3ms remaining: 9.01ms
87: learn: 0.1765896 total: 60.8ms remaining: 8.3ms
88: learn: 0.1757566 total: 61ms remaining: 7.54ms
89: learn: 0.1753380 total: 61.2ms remaining: 6.8ms
90: learn: 0.1738739 total: 61.4ms remaining: 6.08ms
91: learn: 0.1732464 total: 62.3ms remaining: 5.42ms
92: learn: 0.1719087 total: 62.8ms remaining: 4.73ms
93: learn: 0.1714328 total: 63ms remaining: 4.02ms
94: learn: 0.1701988 total: 63.2ms remaining: 3.33ms
95: learn: 0.1696867 total: 63.4ms remaining: 2.64ms
96: learn: 0.1691051 total: 63.7ms remaining: 1.97ms
97: learn: 0.1682508 total: 63.9ms remaining: 1.3ms
98: learn: 0.1678090 total: 64.1ms remaining: 647us
99: learn: 0.1672186 total: 64.4ms remaining: 0us
0: learn: 0.6475054 total: 365us remaining: 36.2ms
1: learn: 0.6071370 total: 583us remaining: 28.6ms
2: learn: 0.5688928 total: 761us remaining: 24.6ms
3: learn: 0.5371729 total: 913us remaining: 21.9ms
4: learn: 0.5086493 total: 1.14ms remaining: 21.6ms
5: learn: 0.4795564 total: 1.42ms remaining: 22.2ms
6: learn: 0.4579098 total: 1.65ms remaining: 21.9ms
7: learn: 0.4308290 total: 1.85ms remaining: 21.2ms
8: learn: 0.4093250 total: 2.02ms remaining: 20.4ms
9: learn: 0.3941826 total: 2.17ms remaining: 19.5ms
10: learn: 0.3800904 total: 2.36ms remaining: 19.1ms
11: learn: 0.3639103 total: 2.53ms remaining: 18.6ms
12: learn: 0.3496290 total: 2.81ms remaining: 18.8ms
13: learn: 0.3407169 total: 3ms remaining: 18.4ms
14: learn: 0.3297829 total: 3.2ms remaining: 18.1ms
15: learn: 0.3189842 total: 3.42ms remaining: 17.9ms
16: learn: 0.3114926 total: 3.57ms remaining: 17.4ms
17: learn: 0.3031443 total: 3.71ms remaining: 16.9ms
18: learn: 0.2955707 total: 3.9ms remaining: 16.6ms
19: learn: 0.2880411 total: 4.09ms remaining: 16.4ms
20: learn: 0.2805200 total: 4.28ms remaining: 16.1ms
21: learn: 0.2726320 total: 4.49ms remaining: 15.9ms
22: learn: 0.2675664 total: 4.66ms remaining: 15.6ms
23: learn: 0.2634753 total: 4.85ms remaining: 15.4ms
24: learn: 0.2596825 total: 5.05ms remaining: 15.1ms
25: learn: 0.2559484 total: 5.23ms remaining: 14.9ms
26: learn: 0.2515160 total: 5.45ms remaining: 14.7ms
27: learn: 0.2483297 total: 5.8ms remaining: 14.9ms
28: learn: 0.2462752 total: 5.95ms remaining: 14.6ms
29: learn: 0.2429713 total: 6.15ms remaining: 14.4ms
30: learn: 0.2395684 total: 6.26ms remaining: 13.9ms
31: learn: 0.2364600 total: 6.49ms remaining: 13.8ms
32: learn: 0.2339638 total: 6.64ms remaining: 13.5ms
33: learn: 0.2316042 total: 6.78ms remaining: 13.2ms
34: learn: 0.2292298 total: 7ms remaining: 13ms
35: learn: 0.2264514 total: 7.23ms remaining: 12.9ms
36: learn: 0.2223391 total: 7.75ms remaining: 13.2ms
37: learn: 0.2194596 total: 8ms remaining: 13.1ms
38: learn: 0.2165141 total: 8.2ms remaining: 12.8ms
39: learn: 0.2141335 total: 8.5ms remaining: 12.7ms
40: learn: 0.2121124 total: 8.96ms remaining: 12.9ms
41: learn: 0.2102779 total: 9.2ms remaining: 12.7ms
42: learn: 0.2082953 total: 9.41ms remaining: 12.5ms
43: learn: 0.2072916 total: 9.82ms remaining: 12.5ms
44: learn: 0.2049074 total: 10.5ms remaining: 12.8ms
45: learn: 0.2036420 total: 11.1ms remaining: 13.1ms
46: learn: 0.2014209 total: 11.3ms remaining: 12.8ms
47: learn: 0.1999690 total: 11.7ms remaining: 12.6ms
48: learn: 0.1990182 total: 12ms remaining: 12.5ms
49: learn: 0.1973890 total: 12.3ms remaining: 12.3ms
50: learn: 0.1938346 total: 12.5ms remaining: 12ms
51: learn: 0.1927559 total: 12.8ms remaining: 11.8ms
52: learn: 0.1916926 total: 13.2ms remaining: 11.7ms
53: learn: 0.1904656 total: 13.7ms remaining: 11.7ms
54: learn: 0.1892644 total: 14.2ms remaining: 11.6ms
55: learn: 0.1881377 total: 14.4ms remaining: 11.3ms
56: learn: 0.1865936 total: 14.6ms remaining: 11ms
57: learn: 0.1847520 total: 14.8ms remaining: 10.7ms
58: learn: 0.1834779 total: 15.2ms remaining: 10.5ms
59: learn: 0.1821843 total: 15.4ms remaining: 10.3ms
60: learn: 0.1803011 total: 15.7ms remaining: 10.1ms
61: learn: 0.1777742 total: 16ms remaining: 9.81ms
62: learn: 0.1772984 total: 16.2ms remaining: 9.54ms
63: learn: 0.1764391 total: 16.8ms remaining: 9.46ms
64: learn: 0.1757728 total: 17.4ms remaining: 9.38ms
65: learn: 0.1743333 total: 18.1ms remaining: 9.32ms
66: learn: 0.1731950 total: 18.5ms remaining: 9.12ms
67: learn: 0.1703715 total: 19.1ms remaining: 8.97ms
68: learn: 0.1688254 total: 19.3ms remaining: 8.67ms
69: learn: 0.1671513 total: 19.6ms remaining: 8.41ms
70: learn: 0.1655474 total: 20.1ms remaining: 8.2ms
71: learn: 0.1646720 total: 20.4ms remaining: 7.92ms
72: learn: 0.1632495 total: 20.6ms remaining: 7.62ms
73: learn: 0.1618558 total: 20.9ms remaining: 7.35ms
74: learn: 0.1613448 total: 21.3ms remaining: 7.1ms
75: learn: 0.1605755 total: 21.7ms remaining: 6.86ms
76: learn: 0.1590612 total: 21.9ms remaining: 6.55ms
77: learn: 0.1576873 total: 22.2ms remaining: 6.25ms
78: learn: 0.1568384 total: 22.9ms remaining: 6.09ms
79: learn: 0.1560417 total: 23.4ms remaining: 5.86ms
80: learn: 0.1553088 total: 24ms remaining: 5.63ms
81: learn: 0.1543515 total: 24.3ms remaining: 5.34ms
82: learn: 0.1536344 total: 24.7ms remaining: 5.07ms
83: learn: 0.1528763 total: 25ms remaining: 4.76ms
84: learn: 0.1522848 total: 25.2ms remaining: 4.45ms
85: learn: 0.1503108 total: 25.5ms remaining: 4.16ms
86: learn: 0.1489568 total: 25.8ms remaining: 3.85ms
87: learn: 0.1480627 total: 26ms remaining: 3.54ms
88: learn: 0.1472803 total: 26.3ms remaining: 3.25ms
89: learn: 0.1463411 total: 26.5ms remaining: 2.95ms
90: learn: 0.1457018 total: 26.8ms remaining: 2.65ms
91: learn: 0.1451322 total: 27ms remaining: 2.35ms
92: learn: 0.1439193 total: 27.3ms remaining: 2.05ms
93: learn: 0.1426374 total: 27.6ms remaining: 1.76ms
94: learn: 0.1411973 total: 27.8ms remaining: 1.47ms
95: learn: 0.1407247 total: 28.1ms remaining: 1.17ms
96: learn: 0.1402650 total: 28.4ms remaining: 877us
97: learn: 0.1398238 total: 28.7ms remaining: 585us
98: learn: 0.1394731 total: 29.2ms remaining: 294us
99: learn: 0.1391102 total: 29.5ms remaining: 0us
0: learn: 0.6426234 total: 295us remaining: 29.2ms
1: learn: 0.5966344 total: 601us remaining: 29.5ms
2: learn: 0.5610290 total: 767us remaining: 24.8ms
3: learn: 0.5301488 total: 1.15ms remaining: 27.7ms
4: learn: 0.5029961 total: 1.49ms remaining: 28.4ms
5: learn: 0.4793889 total: 1.84ms remaining: 28.8ms
6: learn: 0.4547454 total: 2ms remaining: 26.6ms
7: learn: 0.4372305 total: 2.29ms remaining: 26.3ms
8: learn: 0.4185281 total: 2.45ms remaining: 24.8ms
9: learn: 0.4020008 total: 2.67ms remaining: 24ms
10: learn: 0.3875013 total: 2.85ms remaining: 23.1ms
11: learn: 0.3742145 total: 3.02ms remaining: 22.1ms
12: learn: 0.3622586 total: 3.26ms remaining: 21.8ms
13: learn: 0.3518018 total: 3.64ms remaining: 22.3ms
14: learn: 0.3435048 total: 3.84ms remaining: 21.8ms
15: learn: 0.3326371 total: 4.03ms remaining: 21.1ms
16: learn: 0.3257278 total: 4.25ms remaining: 20.8ms
17: learn: 0.3167057 total: 4.56ms remaining: 20.8ms
18: learn: 0.3097649 total: 4.8ms remaining: 20.5ms
19: learn: 0.3042157 total: 5.11ms remaining: 20.4ms
20: learn: 0.2982697 total: 5.4ms remaining: 20.3ms
21: learn: 0.2921152 total: 5.71ms remaining: 20.2ms
22: learn: 0.2875303 total: 5.97ms remaining: 20ms
23: learn: 0.2834689 total: 6.37ms remaining: 20.2ms
24: learn: 0.2785533 total: 6.52ms remaining: 19.6ms
25: learn: 0.2762674 total: 6.84ms remaining: 19.5ms
26: learn: 0.2729498 total: 7.08ms remaining: 19.1ms
27: learn: 0.2685974 total: 7.25ms remaining: 18.7ms
28: learn: 0.2648926 total: 7.67ms remaining: 18.8ms
29: learn: 0.2629856 total: 7.91ms remaining: 18.4ms
30: learn: 0.2605379 total: 8.16ms remaining: 18.2ms
31: learn: 0.2583238 total: 8.28ms remaining: 17.6ms
32: learn: 0.2549441 total: 8.56ms remaining: 17.4ms
33: learn: 0.2528828 total: 8.74ms remaining: 17ms
34: learn: 0.2500136 total: 9.06ms remaining: 16.8ms
35: learn: 0.2472779 total: 9.39ms remaining: 16.7ms
36: learn: 0.2443514 total: 9.66ms remaining: 16.4ms
37: learn: 0.2422131 total: 10ms remaining: 16.3ms
38: learn: 0.2400465 total: 10.3ms remaining: 16.1ms
39: learn: 0.2378067 total: 10.7ms remaining: 16ms
40: learn: 0.2362807 total: 11.1ms remaining: 16ms
41: learn: 0.2342833 total: 11.5ms remaining: 15.9ms
42: learn: 0.2325844 total: 11.9ms remaining: 15.7ms
43: learn: 0.2309996 total: 12.2ms remaining: 15.5ms
44: learn: 0.2297499 total: 12.6ms remaining: 15.4ms
45: learn: 0.2277609 total: 13.1ms remaining: 15.4ms
46: learn: 0.2248966 total: 13.5ms remaining: 15.2ms
47: learn: 0.2233626 total: 13.8ms remaining: 15ms
48: learn: 0.2210270 total: 14.1ms remaining: 14.6ms
49: learn: 0.2200753 total: 14.5ms remaining: 14.5ms
50: learn: 0.2187332 total: 14.9ms remaining: 14.4ms
51: learn: 0.2163816 total: 15.3ms remaining: 14.2ms
52: learn: 0.2149486 total: 15.9ms remaining: 14.1ms
53: learn: 0.2138791 total: 16.2ms remaining: 13.8ms
54: learn: 0.2127221 total: 16.7ms remaining: 13.6ms
55: learn: 0.2106814 total: 17ms remaining: 13.4ms
56: learn: 0.2094962 total: 17.3ms remaining: 13ms
57: learn: 0.2069710 total: 17.7ms remaining: 12.8ms
58: learn: 0.2056220 total: 18.1ms remaining: 12.6ms
59: learn: 0.2035830 total: 18.7ms remaining: 12.5ms
60: learn: 0.2030326 total: 19ms remaining: 12.2ms
61: learn: 0.2013024 total: 19.4ms remaining: 11.9ms
62: learn: 0.1998871 total: 19.7ms remaining: 11.6ms
63: learn: 0.1983454 total: 20ms remaining: 11.3ms
64: learn: 0.1963874 total: 20.4ms remaining: 11ms
65: learn: 0.1944519 total: 20.8ms remaining: 10.7ms
66: learn: 0.1928225 total: 21.2ms remaining: 10.4ms
67: learn: 0.1914112 total: 21.6ms remaining: 10.2ms
68: learn: 0.1897957 total: 21.9ms remaining: 9.86ms
69: learn: 0.1890089 total: 22.3ms remaining: 9.57ms
70: learn: 0.1879103 total: 22.8ms remaining: 9.31ms
71: learn: 0.1863096 total: 23.2ms remaining: 9.01ms
72: learn: 0.1852996 total: 23.5ms remaining: 8.7ms
73: learn: 0.1844780 total: 24ms remaining: 8.44ms
74: learn: 0.1826719 total: 24.3ms remaining: 8.12ms
75: learn: 0.1818801 total: 25ms remaining: 7.91ms
76: learn: 0.1810469 total: 25.4ms remaining: 7.59ms
77: learn: 0.1801707 total: 25.8ms remaining: 7.29ms
78: learn: 0.1788266 total: 26.3ms remaining: 6.99ms
79: learn: 0.1774748 total: 26.9ms remaining: 6.74ms
80: learn: 0.1769838 total: 27.4ms remaining: 6.42ms
81: learn: 0.1755796 total: 27.9ms remaining: 6.11ms
82: learn: 0.1747006 total: 28.3ms remaining: 5.8ms
83: learn: 0.1733876 total: 28.6ms remaining: 5.44ms
84: learn: 0.1723565 total: 28.8ms remaining: 5.08ms
85: learn: 0.1717806 total: 29ms remaining: 4.72ms
86: learn: 0.1710007 total: 29.2ms remaining: 4.36ms
87: learn: 0.1701884 total: 29.5ms remaining: 4.03ms
88: learn: 0.1691883 total: 29.9ms remaining: 3.69ms
89: learn: 0.1687620 total: 30.5ms remaining: 3.39ms
90: learn: 0.1679230 total: 31.1ms remaining: 3.07ms
91: learn: 0.1669217 total: 31.6ms remaining: 2.75ms
92: learn: 0.1661735 total: 32.2ms remaining: 2.42ms
93: learn: 0.1652063 total: 32.6ms remaining: 2.08ms
94: learn: 0.1641294 total: 33.1ms remaining: 1.74ms
95: learn: 0.1635903 total: 33.4ms remaining: 1.39ms
96: learn: 0.1629956 total: 33.6ms remaining: 1.04ms
97: learn: 0.1625569 total: 33.8ms remaining: 689us
98: learn: 0.1621201 total: 34ms remaining: 343us
99: learn: 0.1617013 total: 34.2ms remaining: 0us
0: learn: 0.6457342 total: 487us remaining: 48.3ms
1: learn: 0.6010041 total: 827us remaining: 40.6ms
2: learn: 0.5607770 total: 1.07ms remaining: 34.6ms
3: learn: 0.5254774 total: 1.3ms remaining: 31.2ms
4: learn: 0.4970300 total: 1.53ms remaining: 29.1ms
5: learn: 0.4712240 total: 1.66ms remaining: 26ms
6: learn: 0.4472541 total: 1.84ms remaining: 24.4ms
7: learn: 0.4302047 total: 2.02ms remaining: 23.2ms
8: learn: 0.4096284 total: 2.21ms remaining: 22.4ms
9: learn: 0.3943730 total: 2.43ms remaining: 21.9ms
10: learn: 0.3825996 total: 2.69ms remaining: 21.7ms
11: learn: 0.3682381 total: 2.9ms remaining: 21.2ms
12: learn: 0.3542027 total: 3.11ms remaining: 20.8ms
13: learn: 0.3416179 total: 3.36ms remaining: 20.7ms
14: learn: 0.3315787 total: 3.56ms remaining: 20.2ms
15: learn: 0.3215391 total: 3.85ms remaining: 20.2ms
16: learn: 0.3128192 total: 4.22ms remaining: 20.6ms
17: learn: 0.3060670 total: 4.41ms remaining: 20.1ms
18: learn: 0.2996001 total: 4.9ms remaining: 20.9ms
19: learn: 0.2933228 total: 5.18ms remaining: 20.7ms
20: learn: 0.2865127 total: 5.55ms remaining: 20.9ms
21: learn: 0.2823357 total: 5.76ms remaining: 20.4ms
22: learn: 0.2769019 total: 5.96ms remaining: 19.9ms
23: learn: 0.2713227 total: 6.16ms remaining: 19.5ms
24: learn: 0.2655899 total: 6.45ms remaining: 19.3ms
25: learn: 0.2602218 total: 6.69ms remaining: 19ms
26: learn: 0.2562777 total: 6.91ms remaining: 18.7ms
27: learn: 0.2537223 total: 7.63ms remaining: 19.6ms
28: learn: 0.2511636 total: 7.84ms remaining: 19.2ms
29: learn: 0.2464586 total: 8.21ms remaining: 19.2ms
30: learn: 0.2430363 total: 8.53ms remaining: 19ms
31: learn: 0.2402076 total: 8.74ms remaining: 18.6ms
32: learn: 0.2372463 total: 8.96ms remaining: 18.2ms
33: learn: 0.2343819 total: 9.17ms remaining: 17.8ms
34: learn: 0.2322473 total: 9.69ms remaining: 18ms
35: learn: 0.2294492 total: 10.1ms remaining: 17.9ms
36: learn: 0.2274605 total: 10.4ms remaining: 17.8ms
37: learn: 0.2247957 total: 10.8ms remaining: 17.6ms
38: learn: 0.2220142 total: 11.1ms remaining: 17.4ms
39: learn: 0.2205209 total: 11.3ms remaining: 17ms
40: learn: 0.2186829 total: 11.6ms remaining: 16.7ms
41: learn: 0.2163716 total: 11.9ms remaining: 16.4ms
42: learn: 0.2141476 total: 12.2ms remaining: 16.2ms
43: learn: 0.2117608 total: 12.6ms remaining: 16.1ms
44: learn: 0.2093530 total: 12.9ms remaining: 15.8ms
45: learn: 0.2075321 total: 13.3ms remaining: 15.6ms
46: learn: 0.2060894 total: 13.7ms remaining: 15.4ms
47: learn: 0.2051089 total: 13.9ms remaining: 15ms
48: learn: 0.2037288 total: 14.1ms remaining: 14.7ms
49: learn: 0.2020560 total: 14.5ms remaining: 14.5ms
50: learn: 0.2012479 total: 15ms remaining: 14.4ms
51: learn: 0.2000097 total: 15.5ms remaining: 14.3ms
52: learn: 0.1985358 total: 15.8ms remaining: 14ms
53: learn: 0.1966868 total: 16ms remaining: 13.6ms
54: learn: 0.1953039 total: 16.2ms remaining: 13.3ms
55: learn: 0.1942397 total: 16.4ms remaining: 12.9ms
56: learn: 0.1930919 total: 16.8ms remaining: 12.7ms
57: learn: 0.1918476 total: 17.1ms remaining: 12.4ms
58: learn: 0.1905491 total: 17.4ms remaining: 12.1ms
59: learn: 0.1895913 total: 17.8ms remaining: 11.8ms
60: learn: 0.1876756 total: 18.1ms remaining: 11.6ms
61: learn: 0.1864385 total: 18.3ms remaining: 11.2ms
62: learn: 0.1857486 total: 18.6ms remaining: 10.9ms
63: learn: 0.1843478 total: 18.8ms remaining: 10.6ms
64: learn: 0.1832449 total: 19ms remaining: 10.2ms
65: learn: 0.1824715 total: 19.3ms remaining: 9.92ms
66: learn: 0.1813436 total: 19.6ms remaining: 9.66ms
67: learn: 0.1802899 total: 19.9ms remaining: 9.36ms
68: learn: 0.1795508 total: 20.4ms remaining: 9.14ms
69: learn: 0.1785407 total: 20.7ms remaining: 8.89ms
70: learn: 0.1773668 total: 21.1ms remaining: 8.6ms
71: learn: 0.1753779 total: 21.4ms remaining: 8.32ms
72: learn: 0.1744473 total: 21.7ms remaining: 8.03ms
73: learn: 0.1731742 total: 22.4ms remaining: 7.85ms
74: learn: 0.1719759 total: 23.2ms remaining: 7.72ms
75: learn: 0.1708276 total: 23.5ms remaining: 7.41ms
76: learn: 0.1700271 total: 23.7ms remaining: 7.08ms
77: learn: 0.1691305 total: 24ms remaining: 6.77ms
78: learn: 0.1681001 total: 24.9ms remaining: 6.61ms
79: learn: 0.1671839 total: 25.4ms remaining: 6.36ms
80: learn: 0.1660925 total: 26ms remaining: 6.1ms
81: learn: 0.1651765 total: 26.7ms remaining: 5.87ms
82: learn: 0.1640415 total: 27.1ms remaining: 5.56ms
83: learn: 0.1632295 total: 27.4ms remaining: 5.22ms
84: learn: 0.1629375 total: 27.7ms remaining: 4.88ms
85: learn: 0.1618696 total: 28ms remaining: 4.56ms
86: learn: 0.1616127 total: 28.5ms remaining: 4.25ms
87: learn: 0.1608334 total: 28.8ms remaining: 3.93ms
88: learn: 0.1599306 total: 29.4ms remaining: 3.63ms
89: learn: 0.1594392 total: 29.7ms remaining: 3.29ms
90: learn: 0.1584703 total: 30.4ms remaining: 3ms
91: learn: 0.1577935 total: 30.6ms remaining: 2.66ms
92: learn: 0.1573067 total: 31.1ms remaining: 2.34ms
93: learn: 0.1565912 total: 31.4ms remaining: 2ms
94: learn: 0.1559672 total: 31.7ms remaining: 1.67ms
95: learn: 0.1553522 total: 32.2ms remaining: 1.34ms
96: learn: 0.1543906 total: 32.5ms remaining: 1ms
97: learn: 0.1535468 total: 32.8ms remaining: 670us
98: learn: 0.1528294 total: 33.4ms remaining: 337us
99: learn: 0.1522443 total: 33.6ms remaining: 0us
0: learn: 0.6495311 total: 305us remaining: 30.3ms
1: learn: 0.6040737 total: 552us remaining: 27.1ms
2: learn: 0.5672379 total: 738us remaining: 23.9ms
3: learn: 0.5406197 total: 1.95ms remaining: 46.9ms
4: learn: 0.5151644 total: 2.75ms remaining: 52.3ms
5: learn: 0.4914903 total: 2.94ms remaining: 46ms
6: learn: 0.4692605 total: 3.14ms remaining: 41.7ms
7: learn: 0.4519017 total: 3.45ms remaining: 39.6ms
8: learn: 0.4380025 total: 3.68ms remaining: 37.2ms
9: learn: 0.4235189 total: 3.85ms remaining: 34.7ms
10: learn: 0.4113278 total: 4.21ms remaining: 34.1ms
11: learn: 0.3991034 total: 5.14ms remaining: 37.7ms
12: learn: 0.3858848 total: 5.44ms remaining: 36.4ms
13: learn: 0.3728550 total: 6.64ms remaining: 40.8ms
14: learn: 0.3636348 total: 7.06ms remaining: 40ms
15: learn: 0.3534055 total: 7.51ms remaining: 39.4ms
16: learn: 0.3436837 total: 7.88ms remaining: 38.5ms
17: learn: 0.3372644 total: 8.05ms remaining: 36.7ms
18: learn: 0.3313884 total: 8.21ms remaining: 35ms
19: learn: 0.3262682 total: 8.42ms remaining: 33.7ms
20: learn: 0.3172674 total: 8.65ms remaining: 32.5ms
21: learn: 0.3110035 total: 9.03ms remaining: 32ms
22: learn: 0.3032744 total: 9.27ms remaining: 31ms
23: learn: 0.2978424 total: 9.57ms remaining: 30.3ms
24: learn: 0.2923408 total: 9.93ms remaining: 29.8ms
25: learn: 0.2896119 total: 10.2ms remaining: 29.1ms
26: learn: 0.2859146 total: 10.5ms remaining: 28.4ms
27: learn: 0.2821476 total: 10.8ms remaining: 27.8ms
28: learn: 0.2781989 total: 11.2ms remaining: 27.4ms
29: learn: 0.2739802 total: 11.8ms remaining: 27.4ms
30: learn: 0.2710510 total: 12.5ms remaining: 27.9ms
31: learn: 0.2675356 total: 13.1ms remaining: 27.8ms
32: learn: 0.2641957 total: 13.9ms remaining: 28.1ms
33: learn: 0.2593319 total: 14.2ms remaining: 27.6ms
34: learn: 0.2557795 total: 15.5ms remaining: 28.8ms
35: learn: 0.2535475 total: 15.9ms remaining: 28.3ms
36: learn: 0.2505609 total: 16.1ms remaining: 27.4ms
37: learn: 0.2480422 total: 16.3ms remaining: 26.6ms
38: learn: 0.2455669 total: 16.6ms remaining: 25.9ms
39: learn: 0.2439872 total: 16.8ms remaining: 25.1ms
40: learn: 0.2422787 total: 17ms remaining: 24.5ms
41: learn: 0.2409530 total: 17.3ms remaining: 23.8ms
42: learn: 0.2383699 total: 18.1ms remaining: 24ms
43: learn: 0.2357869 total: 18.3ms remaining: 23.3ms
44: learn: 0.2337662 total: 18.9ms remaining: 23.1ms
45: learn: 0.2326745 total: 19.1ms remaining: 22.4ms
46: learn: 0.2309611 total: 20ms remaining: 22.5ms
47: learn: 0.2293255 total: 21.1ms remaining: 22.9ms
48: learn: 0.2276355 total: 21.3ms remaining: 22.2ms
49: learn: 0.2256338 total: 21.6ms remaining: 21.6ms
50: learn: 0.2230134 total: 21.8ms remaining: 20.9ms
51: learn: 0.2215946 total: 22ms remaining: 20.3ms
52: learn: 0.2190297 total: 22.2ms remaining: 19.7ms
53: learn: 0.2169878 total: 22.6ms remaining: 19.3ms
54: learn: 0.2153076 total: 23ms remaining: 18.8ms
55: learn: 0.2139220 total: 23.3ms remaining: 18.3ms
56: learn: 0.2124221 total: 23.7ms remaining: 17.8ms
57: learn: 0.2108960 total: 23.8ms remaining: 17.3ms
58: learn: 0.2088332 total: 24.1ms remaining: 16.8ms
59: learn: 0.2074714 total: 24.6ms remaining: 16.4ms
60: learn: 0.2060640 total: 24.9ms remaining: 15.9ms
61: learn: 0.2055411 total: 25.2ms remaining: 15.4ms
62: learn: 0.2045437 total: 26.4ms remaining: 15.5ms
63: learn: 0.2027296 total: 26.6ms remaining: 15ms
64: learn: 0.2002059 total: 27ms remaining: 14.5ms
65: learn: 0.1987645 total: 27.4ms remaining: 14.1ms
66: learn: 0.1965463 total: 28.1ms remaining: 13.8ms
67: learn: 0.1950846 total: 28.7ms remaining: 13.5ms
68: learn: 0.1938663 total: 29ms remaining: 13ms
69: learn: 0.1921205 total: 29.3ms remaining: 12.5ms
70: learn: 0.1898144 total: 29.6ms remaining: 12.1ms
71: learn: 0.1890448 total: 30.1ms remaining: 11.7ms
72: learn: 0.1877640 total: 30.5ms remaining: 11.3ms
73: learn: 0.1861304 total: 31ms remaining: 10.9ms
74: learn: 0.1838183 total: 31.3ms remaining: 10.4ms
75: learn: 0.1817840 total: 32.1ms remaining: 10.1ms
76: learn: 0.1811012 total: 33.3ms remaining: 9.96ms
77: learn: 0.1801302 total: 33.6ms remaining: 9.48ms
78: learn: 0.1788358 total: 33.9ms remaining: 9.01ms
79: learn: 0.1771961 total: 34.2ms remaining: 8.55ms
80: learn: 0.1765127 total: 34.5ms remaining: 8.1ms
81: learn: 0.1752301 total: 34.9ms remaining: 7.65ms
82: learn: 0.1746056 total: 35.5ms remaining: 7.26ms
83: learn: 0.1732466 total: 35.9ms remaining: 6.84ms
84: learn: 0.1724781 total: 36.3ms remaining: 6.4ms
85: learn: 0.1711030 total: 36.6ms remaining: 5.96ms
86: learn: 0.1702846 total: 37.3ms remaining: 5.57ms
87: learn: 0.1697629 total: 37.7ms remaining: 5.15ms
88: learn: 0.1691562 total: 38.2ms remaining: 4.73ms
89: learn: 0.1683050 total: 38.7ms remaining: 4.3ms
90: learn: 0.1675551 total: 39.2ms remaining: 3.87ms
91: learn: 0.1668608 total: 39.6ms remaining: 3.44ms
92: learn: 0.1659986 total: 39.9ms remaining: 3ms
93: learn: 0.1652068 total: 40.3ms remaining: 2.57ms
94: learn: 0.1642219 total: 41.5ms remaining: 2.18ms
95: learn: 0.1635650 total: 41.8ms remaining: 1.74ms
96: learn: 0.1630106 total: 42.9ms remaining: 1.33ms
97: learn: 0.1618195 total: 43.9ms remaining: 896us
98: learn: 0.1612227 total: 44.2ms remaining: 446us
99: learn: 0.1607275 total: 44.4ms remaining: 0us
Evaluating param combo 15/16: {'boosted_cb_model__iterations': 100, 'boosted_cb_model__learning_rate': 0.1, 'boosted_cb_model__max_depth': 6, 'boosted_cb_model__num_leaves': 8}
Duplicate found for this combo (hash=46ceb846705e28850644f75b1e681bf4). Removing old run: 76d8f16f4efc467dbd93153b3c6d3b12
0: learn: 0.6460229 total: 423us remaining: 42ms
1: learn: 0.5986942 total: 677us remaining: 33.2ms
2: learn: 0.5698636 total: 957us remaining: 31ms
3: learn: 0.5334742 total: 1.33ms remaining: 32.1ms
4: learn: 0.5066109 total: 1.78ms remaining: 33.7ms
5: learn: 0.4843498 total: 2.06ms remaining: 32.3ms
6: learn: 0.4615797 total: 2.41ms remaining: 32ms
7: learn: 0.4449571 total: 2.97ms remaining: 34.1ms
8: learn: 0.4229021 total: 4.11ms remaining: 41.6ms
9: learn: 0.4101460 total: 5.36ms remaining: 48.3ms
10: learn: 0.3974463 total: 5.71ms remaining: 46.2ms
11: learn: 0.3876826 total: 6.43ms remaining: 47.1ms
12: learn: 0.3757080 total: 6.85ms remaining: 45.8ms
13: learn: 0.3620377 total: 7.21ms remaining: 44.3ms
14: learn: 0.3517450 total: 7.87ms remaining: 44.6ms
15: learn: 0.3423458 total: 8.51ms remaining: 44.7ms
16: learn: 0.3313032 total: 9.15ms remaining: 44.7ms
17: learn: 0.3212979 total: 9.8ms remaining: 44.7ms
18: learn: 0.3162087 total: 10.7ms remaining: 45.8ms
19: learn: 0.3091345 total: 11.3ms remaining: 45.4ms
20: learn: 0.3015900 total: 12ms remaining: 45.2ms
21: learn: 0.2984053 total: 12.8ms remaining: 45.3ms
22: learn: 0.2936107 total: 13.5ms remaining: 45.1ms
23: learn: 0.2878470 total: 14.3ms remaining: 45.2ms
24: learn: 0.2836974 total: 14.9ms remaining: 44.7ms
25: learn: 0.2780346 total: 15.3ms remaining: 43.6ms
26: learn: 0.2743370 total: 15.9ms remaining: 43ms
27: learn: 0.2683212 total: 16.3ms remaining: 42ms
28: learn: 0.2662361 total: 16.5ms remaining: 40.5ms
29: learn: 0.2639247 total: 17ms remaining: 39.6ms
30: learn: 0.2606274 total: 17.8ms remaining: 39.6ms
31: learn: 0.2570408 total: 18.5ms remaining: 39.2ms
32: learn: 0.2553281 total: 18.6ms remaining: 37.8ms
33: learn: 0.2520468 total: 19.1ms remaining: 37.2ms
34: learn: 0.2495426 total: 19.8ms remaining: 36.8ms
35: learn: 0.2475286 total: 20.4ms remaining: 36.3ms
36: learn: 0.2457805 total: 21ms remaining: 35.8ms
37: learn: 0.2425179 total: 21.5ms remaining: 35ms
38: learn: 0.2409787 total: 22.1ms remaining: 34.6ms
39: learn: 0.2392860 total: 22.6ms remaining: 33.8ms
40: learn: 0.2368910 total: 23ms remaining: 33.1ms
41: learn: 0.2336958 total: 23.5ms remaining: 32.5ms
42: learn: 0.2302500 total: 24.2ms remaining: 32ms
43: learn: 0.2295681 total: 24.8ms remaining: 31.6ms
44: learn: 0.2256848 total: 25.3ms remaining: 31ms
45: learn: 0.2228299 total: 26ms remaining: 30.5ms
46: learn: 0.2209182 total: 26.7ms remaining: 30.1ms
47: learn: 0.2189152 total: 27.3ms remaining: 29.6ms
48: learn: 0.2158229 total: 28ms remaining: 29.2ms
49: learn: 0.2140103 total: 28.4ms remaining: 28.4ms
50: learn: 0.2119451 total: 29ms remaining: 27.9ms
51: learn: 0.2102723 total: 29.5ms remaining: 27.2ms
52: learn: 0.2093145 total: 30ms remaining: 26.6ms
53: learn: 0.2081931 total: 30.5ms remaining: 26ms
54: learn: 0.2054612 total: 30.9ms remaining: 25.3ms
55: learn: 0.2033785 total: 31.6ms remaining: 24.8ms
56: learn: 0.2022919 total: 32.2ms remaining: 24.3ms
57: learn: 0.1999491 total: 32.7ms remaining: 23.7ms
58: learn: 0.1988943 total: 33.2ms remaining: 23.1ms
59: learn: 0.1969574 total: 33.7ms remaining: 22.5ms
60: learn: 0.1958633 total: 34.3ms remaining: 21.9ms
61: learn: 0.1937584 total: 34.9ms remaining: 21.4ms
62: learn: 0.1912862 total: 35.4ms remaining: 20.8ms
63: learn: 0.1893931 total: 35.9ms remaining: 20.2ms
64: learn: 0.1880613 total: 36.4ms remaining: 19.6ms
65: learn: 0.1861488 total: 36.9ms remaining: 19ms
66: learn: 0.1845152 total: 37.4ms remaining: 18.4ms
67: learn: 0.1826215 total: 37.9ms remaining: 17.8ms
68: learn: 0.1810028 total: 38.3ms remaining: 17.2ms
69: learn: 0.1799899 total: 38.6ms remaining: 16.6ms
70: learn: 0.1783062 total: 39ms remaining: 15.9ms
71: learn: 0.1768881 total: 39.4ms remaining: 15.3ms
72: learn: 0.1759256 total: 39.8ms remaining: 14.7ms
73: learn: 0.1744777 total: 40.3ms remaining: 14.1ms
74: learn: 0.1734800 total: 40.7ms remaining: 13.6ms
75: learn: 0.1728562 total: 41.2ms remaining: 13ms
76: learn: 0.1716425 total: 41.5ms remaining: 12.4ms
77: learn: 0.1705949 total: 42ms remaining: 11.8ms
78: learn: 0.1693543 total: 42.6ms remaining: 11.3ms
79: learn: 0.1683177 total: 43ms remaining: 10.7ms
80: learn: 0.1672269 total: 43.5ms remaining: 10.2ms
81: learn: 0.1663139 total: 43.9ms remaining: 9.63ms
82: learn: 0.1654929 total: 44.4ms remaining: 9.1ms
83: learn: 0.1642729 total: 44.8ms remaining: 8.53ms
84: learn: 0.1633882 total: 45.2ms remaining: 7.97ms
85: learn: 0.1627040 total: 45.7ms remaining: 7.44ms
86: learn: 0.1612942 total: 46.5ms remaining: 6.94ms
87: learn: 0.1605489 total: 47.1ms remaining: 6.42ms
88: learn: 0.1597711 total: 47.6ms remaining: 5.88ms
89: learn: 0.1590690 total: 48.2ms remaining: 5.35ms
90: learn: 0.1586275 total: 48.6ms remaining: 4.81ms
91: learn: 0.1575598 total: 49.2ms remaining: 4.28ms
92: learn: 0.1563936 total: 49.6ms remaining: 3.74ms
93: learn: 0.1552779 total: 50.5ms remaining: 3.22ms
94: learn: 0.1544500 total: 50.9ms remaining: 2.68ms
95: learn: 0.1536301 total: 51.3ms remaining: 2.14ms
96: learn: 0.1528251 total: 51.7ms remaining: 1.6ms
97: learn: 0.1518105 total: 52.1ms remaining: 1.06ms
98: learn: 0.1508414 total: 52.5ms remaining: 530us
99: learn: 0.1501001 total: 53.2ms remaining: 0us
0: learn: 0.6434696 total: 488us remaining: 48.3ms
1: learn: 0.5908671 total: 807us remaining: 39.6ms
2: learn: 0.5540161 total: 1.25ms remaining: 40.4ms
3: learn: 0.5260698 total: 2.07ms remaining: 49.7ms
4: learn: 0.4972882 total: 2.51ms remaining: 47.7ms
5: learn: 0.4743007 total: 2.9ms remaining: 45.5ms
6: learn: 0.4529556 total: 3.33ms remaining: 44.2ms
7: learn: 0.4365312 total: 3.73ms remaining: 42.9ms
8: learn: 0.4267163 total: 3.98ms remaining: 40.2ms
9: learn: 0.4125750 total: 4.7ms remaining: 42.3ms
10: learn: 0.3975618 total: 5.26ms remaining: 42.6ms
11: learn: 0.3840776 total: 5.67ms remaining: 41.6ms
12: learn: 0.3664514 total: 5.83ms remaining: 39ms
13: learn: 0.3545502 total: 6.28ms remaining: 38.6ms
14: learn: 0.3431669 total: 6.69ms remaining: 37.9ms
15: learn: 0.3336461 total: 7.08ms remaining: 37.2ms
16: learn: 0.3257986 total: 7.7ms remaining: 37.6ms
17: learn: 0.3165228 total: 8.64ms remaining: 39.4ms
18: learn: 0.3073494 total: 9ms remaining: 38.4ms
19: learn: 0.3013560 total: 9.49ms remaining: 38ms
20: learn: 0.2931992 total: 10.7ms remaining: 40.4ms
21: learn: 0.2889907 total: 11.5ms remaining: 40.7ms
22: learn: 0.2834065 total: 13.7ms remaining: 45.7ms
23: learn: 0.2772113 total: 14.1ms remaining: 44.7ms
24: learn: 0.2703179 total: 15.4ms remaining: 46.3ms
25: learn: 0.2673409 total: 16.2ms remaining: 46.1ms
26: learn: 0.2622606 total: 16.7ms remaining: 45.2ms
27: learn: 0.2564959 total: 17.5ms remaining: 45.1ms
28: learn: 0.2518273 total: 18.4ms remaining: 45.1ms
29: learn: 0.2477439 total: 19.1ms remaining: 44.5ms
30: learn: 0.2441075 total: 19.6ms remaining: 43.6ms
31: learn: 0.2419642 total: 20.2ms remaining: 42.9ms
32: learn: 0.2381178 total: 21.6ms remaining: 43.8ms
33: learn: 0.2347685 total: 22ms remaining: 42.8ms
34: learn: 0.2324366 total: 22.6ms remaining: 42ms
35: learn: 0.2309254 total: 23.1ms remaining: 41.1ms
36: learn: 0.2282484 total: 23.7ms remaining: 40.3ms
37: learn: 0.2263869 total: 24.2ms remaining: 39.5ms
38: learn: 0.2241798 total: 25.4ms remaining: 39.7ms
39: learn: 0.2218812 total: 25.7ms remaining: 38.6ms
40: learn: 0.2187091 total: 26.4ms remaining: 37.9ms
41: learn: 0.2167929 total: 28.3ms remaining: 39.1ms
42: learn: 0.2157566 total: 29.1ms remaining: 38.5ms
43: learn: 0.2137873 total: 30ms remaining: 38.1ms
44: learn: 0.2117076 total: 31.9ms remaining: 38.9ms
45: learn: 0.2088020 total: 32.7ms remaining: 38.4ms
46: learn: 0.2060638 total: 33.3ms remaining: 37.5ms
47: learn: 0.2018728 total: 34.1ms remaining: 37ms
48: learn: 0.2001262 total: 35.6ms remaining: 37ms
49: learn: 0.1994772 total: 35.9ms remaining: 35.9ms
50: learn: 0.1959672 total: 36.4ms remaining: 35ms
51: learn: 0.1943815 total: 37.1ms remaining: 34.2ms
52: learn: 0.1934081 total: 37.8ms remaining: 33.5ms
53: learn: 0.1914274 total: 38.3ms remaining: 32.6ms
54: learn: 0.1902557 total: 38.7ms remaining: 31.7ms
55: learn: 0.1883236 total: 39.6ms remaining: 31.1ms
56: learn: 0.1859214 total: 40.2ms remaining: 30.3ms
57: learn: 0.1843155 total: 40.9ms remaining: 29.6ms
58: learn: 0.1821992 total: 41.8ms remaining: 29.1ms
59: learn: 0.1800120 total: 42.3ms remaining: 28.2ms
60: learn: 0.1786133 total: 42.8ms remaining: 27.4ms
61: learn: 0.1766987 total: 44.3ms remaining: 27.1ms
62: learn: 0.1740381 total: 44.6ms remaining: 26.2ms
63: learn: 0.1724637 total: 45ms remaining: 25.3ms
64: learn: 0.1701975 total: 45.6ms remaining: 24.6ms
65: learn: 0.1682817 total: 46.4ms remaining: 23.9ms
66: learn: 0.1660708 total: 47.1ms remaining: 23.2ms
67: learn: 0.1655515 total: 47.5ms remaining: 22.3ms
68: learn: 0.1634683 total: 48.4ms remaining: 21.7ms
69: learn: 0.1616501 total: 48.9ms remaining: 21ms
70: learn: 0.1600388 total: 49.9ms remaining: 20.4ms
71: learn: 0.1587681 total: 50.3ms remaining: 19.6ms
72: learn: 0.1579250 total: 50.7ms remaining: 18.8ms
73: learn: 0.1574547 total: 51.1ms remaining: 18ms
74: learn: 0.1556420 total: 51.8ms remaining: 17.3ms
75: learn: 0.1540917 total: 52.6ms remaining: 16.6ms
76: learn: 0.1527257 total: 53.2ms remaining: 15.9ms
77: learn: 0.1510435 total: 53.6ms remaining: 15.1ms
78: learn: 0.1501632 total: 54ms remaining: 14.3ms
79: learn: 0.1488908 total: 54.4ms remaining: 13.6ms
80: learn: 0.1479702 total: 54.9ms remaining: 12.9ms
81: learn: 0.1465679 total: 55.9ms remaining: 12.3ms
82: learn: 0.1457778 total: 56.3ms remaining: 11.5ms
83: learn: 0.1449621 total: 56.9ms remaining: 10.8ms
84: learn: 0.1434229 total: 57.6ms remaining: 10.2ms
85: learn: 0.1425868 total: 58.3ms remaining: 9.48ms
86: learn: 0.1417698 total: 59ms remaining: 8.82ms
87: learn: 0.1403898 total: 59.6ms remaining: 8.13ms
88: learn: 0.1397810 total: 60.2ms remaining: 7.44ms
89: learn: 0.1391092 total: 60.7ms remaining: 6.75ms
90: learn: 0.1382155 total: 61.2ms remaining: 6.05ms
91: learn: 0.1377273 total: 61.7ms remaining: 5.37ms
92: learn: 0.1374231 total: 62.4ms remaining: 4.69ms
93: learn: 0.1363552 total: 63ms remaining: 4.02ms
94: learn: 0.1353232 total: 63.5ms remaining: 3.34ms
95: learn: 0.1343840 total: 63.9ms remaining: 2.66ms
96: learn: 0.1337788 total: 64.6ms remaining: 2ms
97: learn: 0.1333743 total: 65.4ms remaining: 1.33ms
98: learn: 0.1323607 total: 65.9ms remaining: 665us
99: learn: 0.1317172 total: 66.4ms remaining: 0us
0: learn: 0.6422518 total: 516us remaining: 51.1ms
1: learn: 0.5993870 total: 825us remaining: 40.5ms
2: learn: 0.5628584 total: 1.12ms remaining: 36.4ms
3: learn: 0.5386216 total: 1.42ms remaining: 34ms
4: learn: 0.5066031 total: 1.83ms remaining: 34.8ms
5: learn: 0.4820305 total: 2.17ms remaining: 34ms
6: learn: 0.4639540 total: 2.47ms remaining: 32.8ms
7: learn: 0.4407244 total: 2.72ms remaining: 31.3ms
8: learn: 0.4206859 total: 3.05ms remaining: 30.9ms
9: learn: 0.4032046 total: 3.66ms remaining: 33ms
10: learn: 0.3882634 total: 4.01ms remaining: 32.5ms
11: learn: 0.3760172 total: 4.25ms remaining: 31.2ms
12: learn: 0.3631583 total: 4.63ms remaining: 31ms
13: learn: 0.3521457 total: 4.96ms remaining: 30.5ms
14: learn: 0.3431454 total: 5.34ms remaining: 30.3ms
15: learn: 0.3315988 total: 5.67ms remaining: 29.8ms
16: learn: 0.3202798 total: 5.99ms remaining: 29.2ms
17: learn: 0.3134526 total: 6.49ms remaining: 29.6ms
18: learn: 0.3069279 total: 6.84ms remaining: 29.2ms
19: learn: 0.3004042 total: 7.07ms remaining: 28.3ms
20: learn: 0.2938632 total: 7.48ms remaining: 28.1ms
21: learn: 0.2883339 total: 7.92ms remaining: 28.1ms
22: learn: 0.2812017 total: 8.26ms remaining: 27.6ms
23: learn: 0.2786116 total: 8.58ms remaining: 27.2ms
24: learn: 0.2723876 total: 8.91ms remaining: 26.7ms
25: learn: 0.2681037 total: 9.21ms remaining: 26.2ms
26: learn: 0.2635183 total: 9.5ms remaining: 25.7ms
27: learn: 0.2580335 total: 9.76ms remaining: 25.1ms
28: learn: 0.2537586 total: 10.1ms remaining: 24.7ms
29: learn: 0.2492840 total: 10.5ms remaining: 24.4ms
30: learn: 0.2446996 total: 10.8ms remaining: 24.1ms
31: learn: 0.2426673 total: 11.2ms remaining: 23.7ms
32: learn: 0.2403391 total: 11.5ms remaining: 23.4ms
33: learn: 0.2370377 total: 11.8ms remaining: 23ms
34: learn: 0.2346893 total: 12.1ms remaining: 22.5ms
35: learn: 0.2319904 total: 12.4ms remaining: 22.1ms
36: learn: 0.2293336 total: 12.8ms remaining: 21.9ms
37: learn: 0.2261521 total: 13.5ms remaining: 22ms
38: learn: 0.2221190 total: 13.9ms remaining: 21.8ms
39: learn: 0.2200094 total: 14.3ms remaining: 21.4ms
40: learn: 0.2169681 total: 14.6ms remaining: 21ms
41: learn: 0.2148130 total: 14.9ms remaining: 20.6ms
42: learn: 0.2138348 total: 15.4ms remaining: 20.4ms
43: learn: 0.2114684 total: 15.7ms remaining: 20ms
44: learn: 0.2087580 total: 16.1ms remaining: 19.7ms
45: learn: 0.2072098 total: 16.5ms remaining: 19.3ms
46: learn: 0.2049209 total: 16.9ms remaining: 19.1ms
47: learn: 0.2036974 total: 17.3ms remaining: 18.7ms
48: learn: 0.2021657 total: 17.7ms remaining: 18.5ms
49: learn: 0.1997194 total: 18.2ms remaining: 18.2ms
50: learn: 0.1980052 total: 18.5ms remaining: 17.8ms
51: learn: 0.1966425 total: 18.8ms remaining: 17.4ms
52: learn: 0.1935867 total: 19.5ms remaining: 17.3ms
53: learn: 0.1924080 total: 20ms remaining: 17.1ms
54: learn: 0.1899832 total: 20.6ms remaining: 16.8ms
55: learn: 0.1874622 total: 21.1ms remaining: 16.5ms
56: learn: 0.1848535 total: 22.2ms remaining: 16.7ms
57: learn: 0.1838008 total: 22.9ms remaining: 16.6ms
58: learn: 0.1828902 total: 23.9ms remaining: 16.6ms
59: learn: 0.1820268 total: 24.2ms remaining: 16.2ms
60: learn: 0.1795715 total: 25ms remaining: 16ms
61: learn: 0.1771220 total: 25.8ms remaining: 15.8ms
62: learn: 0.1758677 total: 26.6ms remaining: 15.6ms
63: learn: 0.1738837 total: 27.4ms remaining: 15.4ms
64: learn: 0.1717833 total: 28ms remaining: 15.1ms
65: learn: 0.1700289 total: 28.6ms remaining: 14.7ms
66: learn: 0.1692278 total: 29ms remaining: 14.3ms
67: learn: 0.1682873 total: 29.6ms remaining: 13.9ms
68: learn: 0.1666555 total: 30.3ms remaining: 13.6ms
69: learn: 0.1649986 total: 30.9ms remaining: 13.2ms
70: learn: 0.1629487 total: 31.6ms remaining: 12.9ms
71: learn: 0.1619495 total: 32.3ms remaining: 12.6ms
72: learn: 0.1605822 total: 33ms remaining: 12.2ms
73: learn: 0.1592303 total: 33.6ms remaining: 11.8ms
74: learn: 0.1578588 total: 34.1ms remaining: 11.4ms
75: learn: 0.1567866 total: 34.5ms remaining: 10.9ms
76: learn: 0.1560535 total: 34.9ms remaining: 10.4ms
77: learn: 0.1545536 total: 35.5ms remaining: 10ms
78: learn: 0.1537759 total: 35.8ms remaining: 9.52ms
79: learn: 0.1525453 total: 36.2ms remaining: 9.06ms
80: learn: 0.1515362 total: 37.1ms remaining: 8.7ms
81: learn: 0.1504165 total: 37.6ms remaining: 8.25ms
82: learn: 0.1488280 total: 38.2ms remaining: 7.82ms
83: learn: 0.1472733 total: 38.8ms remaining: 7.39ms
84: learn: 0.1462674 total: 39.4ms remaining: 6.95ms
85: learn: 0.1451319 total: 40.1ms remaining: 6.52ms
86: learn: 0.1443509 total: 40.8ms remaining: 6.1ms
87: learn: 0.1431014 total: 41.2ms remaining: 5.62ms
88: learn: 0.1423838 total: 41.7ms remaining: 5.15ms
89: learn: 0.1411516 total: 42.2ms remaining: 4.69ms
90: learn: 0.1403972 total: 42.6ms remaining: 4.22ms
91: learn: 0.1392421 total: 43.3ms remaining: 3.76ms
92: learn: 0.1386712 total: 44ms remaining: 3.31ms
93: learn: 0.1374421 total: 44.6ms remaining: 2.85ms
94: learn: 0.1364334 total: 45.2ms remaining: 2.38ms
95: learn: 0.1355836 total: 45.7ms remaining: 1.9ms
96: learn: 0.1343640 total: 46.1ms remaining: 1.43ms
97: learn: 0.1333499 total: 46.8ms remaining: 954us
98: learn: 0.1327076 total: 47.3ms remaining: 478us
99: learn: 0.1316487 total: 47.7ms remaining: 0us
0: learn: 0.6416822 total: 501us remaining: 49.6ms
1: learn: 0.6019648 total: 817us remaining: 40.1ms
2: learn: 0.5687979 total: 1.21ms remaining: 39ms
3: learn: 0.5302771 total: 1.58ms remaining: 38ms
4: learn: 0.5040872 total: 1.72ms remaining: 32.6ms
5: learn: 0.4812169 total: 1.97ms remaining: 30.9ms
6: learn: 0.4567565 total: 2.29ms remaining: 30.5ms
7: learn: 0.4352898 total: 2.58ms remaining: 29.6ms
8: learn: 0.4165105 total: 2.86ms remaining: 28.9ms
9: learn: 0.3972385 total: 3.23ms remaining: 29.1ms
10: learn: 0.3820132 total: 3.95ms remaining: 32ms
11: learn: 0.3688061 total: 4.63ms remaining: 33.9ms
12: learn: 0.3540180 total: 4.9ms remaining: 32.8ms
13: learn: 0.3438991 total: 5.36ms remaining: 32.9ms
14: learn: 0.3371155 total: 5.76ms remaining: 32.6ms
15: learn: 0.3263958 total: 6.26ms remaining: 32.9ms
16: learn: 0.3163591 total: 6.77ms remaining: 33ms
17: learn: 0.3080109 total: 7.27ms remaining: 33.1ms
18: learn: 0.3011187 total: 7.74ms remaining: 33ms
19: learn: 0.2948551 total: 8.11ms remaining: 32.4ms
20: learn: 0.2862073 total: 8.8ms remaining: 33.1ms
21: learn: 0.2797604 total: 9.56ms remaining: 33.9ms
22: learn: 0.2738126 total: 10.3ms remaining: 34.3ms
23: learn: 0.2686730 total: 11.1ms remaining: 35.1ms
24: learn: 0.2626725 total: 11.9ms remaining: 35.8ms
25: learn: 0.2583266 total: 12.5ms remaining: 35.7ms
26: learn: 0.2549457 total: 12.9ms remaining: 35ms
27: learn: 0.2516413 total: 13.3ms remaining: 34.2ms
28: learn: 0.2460561 total: 13.9ms remaining: 33.9ms
29: learn: 0.2425478 total: 14.6ms remaining: 34ms
30: learn: 0.2389663 total: 15.4ms remaining: 34.2ms
31: learn: 0.2359938 total: 15.8ms remaining: 33.6ms
32: learn: 0.2322823 total: 16.4ms remaining: 33.4ms
33: learn: 0.2297660 total: 17.1ms remaining: 33.1ms
34: learn: 0.2275496 total: 17.8ms remaining: 33ms
35: learn: 0.2255628 total: 18.3ms remaining: 32.6ms
36: learn: 0.2230388 total: 18.9ms remaining: 32.2ms
37: learn: 0.2198693 total: 19.4ms remaining: 31.6ms
38: learn: 0.2175725 total: 19.8ms remaining: 30.9ms
39: learn: 0.2148144 total: 20.3ms remaining: 30.4ms
40: learn: 0.2123082 total: 21ms remaining: 30.2ms
41: learn: 0.2110802 total: 21.7ms remaining: 29.9ms
42: learn: 0.2096018 total: 22.3ms remaining: 29.6ms
43: learn: 0.2060006 total: 22.9ms remaining: 29.1ms
44: learn: 0.2032244 total: 23.3ms remaining: 28.5ms
45: learn: 0.2007813 total: 24ms remaining: 28.2ms
46: learn: 0.1999685 total: 24.5ms remaining: 27.6ms
47: learn: 0.1972742 total: 24.9ms remaining: 27ms
48: learn: 0.1961341 total: 25.2ms remaining: 26.2ms
49: learn: 0.1941166 total: 25.6ms remaining: 25.6ms
50: learn: 0.1922646 total: 26ms remaining: 25ms
51: learn: 0.1902090 total: 26.5ms remaining: 24.4ms
52: learn: 0.1887889 total: 27ms remaining: 24ms
53: learn: 0.1868298 total: 27.6ms remaining: 23.5ms
54: learn: 0.1843770 total: 27.9ms remaining: 22.9ms
55: learn: 0.1827855 total: 28.3ms remaining: 22.3ms
56: learn: 0.1816540 total: 28.7ms remaining: 21.7ms
57: learn: 0.1789795 total: 29.1ms remaining: 21.1ms
58: learn: 0.1772942 total: 29.6ms remaining: 20.5ms
59: learn: 0.1756055 total: 30.2ms remaining: 20.1ms
60: learn: 0.1737985 total: 30.7ms remaining: 19.6ms
61: learn: 0.1722969 total: 31.3ms remaining: 19.2ms
62: learn: 0.1705087 total: 32.1ms remaining: 18.8ms
63: learn: 0.1697700 total: 32.8ms remaining: 18.4ms
64: learn: 0.1682499 total: 33.2ms remaining: 17.9ms
65: learn: 0.1670629 total: 33.9ms remaining: 17.5ms
66: learn: 0.1653816 total: 34.5ms remaining: 17ms
67: learn: 0.1648115 total: 34.9ms remaining: 16.4ms
68: learn: 0.1640280 total: 35.2ms remaining: 15.8ms
69: learn: 0.1627162 total: 35.6ms remaining: 15.3ms
70: learn: 0.1608247 total: 35.9ms remaining: 14.7ms
71: learn: 0.1596687 total: 36.3ms remaining: 14.1ms
72: learn: 0.1589635 total: 36.7ms remaining: 13.6ms
73: learn: 0.1574077 total: 37.1ms remaining: 13ms
74: learn: 0.1563082 total: 37.5ms remaining: 12.5ms
75: learn: 0.1551933 total: 37.8ms remaining: 11.9ms
76: learn: 0.1537988 total: 38.3ms remaining: 11.4ms
77: learn: 0.1527140 total: 38.8ms remaining: 10.9ms
78: learn: 0.1512282 total: 39.2ms remaining: 10.4ms
79: learn: 0.1497531 total: 39.6ms remaining: 9.9ms
80: learn: 0.1488077 total: 40.2ms remaining: 9.42ms
81: learn: 0.1477021 total: 40.5ms remaining: 8.9ms
82: learn: 0.1464634 total: 41.1ms remaining: 8.41ms
83: learn: 0.1452284 total: 41.5ms remaining: 7.91ms
84: learn: 0.1445371 total: 42ms remaining: 7.42ms
85: learn: 0.1441144 total: 42.4ms remaining: 6.9ms
86: learn: 0.1429034 total: 42.8ms remaining: 6.4ms
87: learn: 0.1417126 total: 43.3ms remaining: 5.9ms
88: learn: 0.1410298 total: 43.9ms remaining: 5.43ms
89: learn: 0.1401819 total: 44.5ms remaining: 4.95ms
90: learn: 0.1390797 total: 44.9ms remaining: 4.44ms
91: learn: 0.1375786 total: 45.4ms remaining: 3.95ms
92: learn: 0.1361731 total: 46.1ms remaining: 3.47ms
93: learn: 0.1354231 total: 46.7ms remaining: 2.98ms
94: learn: 0.1347815 total: 47.3ms remaining: 2.49ms
95: learn: 0.1337366 total: 47.6ms remaining: 1.98ms
96: learn: 0.1333003 total: 48.1ms remaining: 1.49ms
97: learn: 0.1328509 total: 48.5ms remaining: 989us
98: learn: 0.1319694 total: 48.8ms remaining: 493us
99: learn: 0.1313437 total: 49.2ms remaining: 0us
0: learn: 0.6454307 total: 706us remaining: 70ms
1: learn: 0.6039107 total: 1.17ms remaining: 57.3ms
2: learn: 0.5680897 total: 1.53ms remaining: 49.5ms
3: learn: 0.5412015 total: 1.93ms remaining: 46.2ms
4: learn: 0.5182422 total: 2.34ms remaining: 44.5ms
5: learn: 0.4899378 total: 2.91ms remaining: 45.5ms
6: learn: 0.4712526 total: 3.32ms remaining: 44.1ms
7: learn: 0.4534745 total: 3.7ms remaining: 42.6ms
8: learn: 0.4335757 total: 4.14ms remaining: 41.8ms
9: learn: 0.4179533 total: 4.51ms remaining: 40.6ms
10: learn: 0.4047999 total: 5ms remaining: 40.4ms
11: learn: 0.3926710 total: 5.48ms remaining: 40.2ms
12: learn: 0.3830573 total: 5.83ms remaining: 39ms
13: learn: 0.3704344 total: 6.21ms remaining: 38.1ms
14: learn: 0.3596217 total: 6.64ms remaining: 37.6ms
15: learn: 0.3512870 total: 6.96ms remaining: 36.5ms
16: learn: 0.3442628 total: 7.42ms remaining: 36.2ms
17: learn: 0.3356590 total: 7.94ms remaining: 36.2ms
18: learn: 0.3259936 total: 8.5ms remaining: 36.2ms
19: learn: 0.3201304 total: 8.67ms remaining: 34.7ms
20: learn: 0.3140818 total: 9.11ms remaining: 34.3ms
21: learn: 0.3092937 total: 9.57ms remaining: 33.9ms
22: learn: 0.3023720 total: 10ms remaining: 33.6ms
23: learn: 0.2960557 total: 10.4ms remaining: 33ms
24: learn: 0.2915371 total: 10.8ms remaining: 32.4ms
25: learn: 0.2871922 total: 11.2ms remaining: 32ms
26: learn: 0.2829314 total: 11.6ms remaining: 31.3ms
27: learn: 0.2767304 total: 11.9ms remaining: 30.6ms
28: learn: 0.2737164 total: 12ms remaining: 29.5ms
29: learn: 0.2700901 total: 12.4ms remaining: 28.9ms
30: learn: 0.2675257 total: 12.7ms remaining: 28.3ms
31: learn: 0.2635270 total: 13.1ms remaining: 27.8ms
32: learn: 0.2598304 total: 13.5ms remaining: 27.5ms
33: learn: 0.2557064 total: 13.9ms remaining: 27ms
34: learn: 0.2518669 total: 14.4ms remaining: 26.7ms
35: learn: 0.2491863 total: 14.7ms remaining: 26.1ms
36: learn: 0.2459948 total: 15.1ms remaining: 25.7ms
37: learn: 0.2429728 total: 15.4ms remaining: 25.1ms
38: learn: 0.2407984 total: 15.7ms remaining: 24.6ms
39: learn: 0.2391175 total: 16.2ms remaining: 24.3ms
40: learn: 0.2358972 total: 16.5ms remaining: 23.8ms
41: learn: 0.2344892 total: 16.7ms remaining: 23.1ms
42: learn: 0.2323901 total: 17.1ms remaining: 22.6ms
43: learn: 0.2306705 total: 17.5ms remaining: 22.2ms
44: learn: 0.2291525 total: 17.8ms remaining: 21.8ms
45: learn: 0.2285173 total: 18.2ms remaining: 21.4ms
46: learn: 0.2262623 total: 18.6ms remaining: 21ms
47: learn: 0.2230465 total: 19ms remaining: 20.6ms
48: learn: 0.2225959 total: 19.3ms remaining: 20.1ms
49: learn: 0.2210729 total: 19.8ms remaining: 19.8ms
50: learn: 0.2188844 total: 20.3ms remaining: 19.5ms
51: learn: 0.2175699 total: 20.6ms remaining: 19ms
52: learn: 0.2132824 total: 21.1ms remaining: 18.7ms
53: learn: 0.2117432 total: 21.4ms remaining: 18.3ms
54: learn: 0.2086707 total: 21.9ms remaining: 17.9ms
55: learn: 0.2067342 total: 22.2ms remaining: 17.5ms
56: learn: 0.2044487 total: 22.7ms remaining: 17.1ms
57: learn: 0.2022364 total: 23.1ms remaining: 16.7ms
58: learn: 0.2006899 total: 23.6ms remaining: 16.4ms
59: learn: 0.1981092 total: 24ms remaining: 16ms
60: learn: 0.1972342 total: 24.3ms remaining: 15.6ms
61: learn: 0.1955354 total: 24.7ms remaining: 15.1ms
62: learn: 0.1929683 total: 25.1ms remaining: 14.8ms
63: learn: 0.1908658 total: 25.6ms remaining: 14.4ms
64: learn: 0.1893034 total: 26ms remaining: 14ms
65: learn: 0.1863986 total: 26.7ms remaining: 13.7ms
66: learn: 0.1838074 total: 27.1ms remaining: 13.3ms
67: learn: 0.1824642 total: 27.5ms remaining: 12.9ms
68: learn: 0.1799343 total: 28ms remaining: 12.6ms
69: learn: 0.1779507 total: 28.5ms remaining: 12.2ms
70: learn: 0.1766070 total: 29ms remaining: 11.8ms
71: learn: 0.1751538 total: 29.5ms remaining: 11.5ms
72: learn: 0.1730211 total: 30ms remaining: 11.1ms
73: learn: 0.1714725 total: 30.4ms remaining: 10.7ms
74: learn: 0.1705358 total: 30.8ms remaining: 10.3ms
75: learn: 0.1688950 total: 31.3ms remaining: 9.89ms
76: learn: 0.1680273 total: 31.7ms remaining: 9.46ms
77: learn: 0.1666096 total: 32.2ms remaining: 9.09ms
78: learn: 0.1651738 total: 32.8ms remaining: 8.71ms
79: learn: 0.1639785 total: 33.1ms remaining: 8.28ms
80: learn: 0.1620317 total: 33.5ms remaining: 7.87ms
81: learn: 0.1615176 total: 33.9ms remaining: 7.44ms
82: learn: 0.1596799 total: 34.3ms remaining: 7.03ms
83: learn: 0.1585117 total: 34.7ms remaining: 6.61ms
84: learn: 0.1569362 total: 35.1ms remaining: 6.2ms
85: learn: 0.1552066 total: 35.7ms remaining: 5.82ms
86: learn: 0.1536697 total: 36.3ms remaining: 5.42ms
87: learn: 0.1528888 total: 36.8ms remaining: 5.02ms
88: learn: 0.1514037 total: 37.3ms remaining: 4.61ms
89: learn: 0.1504573 total: 37.8ms remaining: 4.2ms
90: learn: 0.1497236 total: 38.4ms remaining: 3.8ms
91: learn: 0.1488347 total: 38.8ms remaining: 3.38ms
92: learn: 0.1480035 total: 39.3ms remaining: 2.96ms
93: learn: 0.1474358 total: 39.8ms remaining: 2.54ms
94: learn: 0.1460922 total: 40.3ms remaining: 2.12ms
95: learn: 0.1449649 total: 40.8ms remaining: 1.7ms
96: learn: 0.1440412 total: 41.2ms remaining: 1.27ms
97: learn: 0.1431665 total: 41.5ms remaining: 847us
98: learn: 0.1420706 total: 42ms remaining: 424us
99: learn: 0.1411751 total: 42.5ms remaining: 0us
0: learn: 0.6482460 total: 483us remaining: 47.9ms
1: learn: 0.5994620 total: 783us remaining: 38.4ms
2: learn: 0.5597606 total: 1.04ms remaining: 33.7ms
3: learn: 0.5297838 total: 1.65ms remaining: 39.7ms
4: learn: 0.4998076 total: 1.95ms remaining: 37ms
5: learn: 0.4730549 total: 2.22ms remaining: 34.8ms
6: learn: 0.4488273 total: 2.55ms remaining: 33.9ms
7: learn: 0.4308397 total: 2.84ms remaining: 32.7ms
8: learn: 0.4166495 total: 3.1ms remaining: 31.4ms
9: learn: 0.3991651 total: 3.35ms remaining: 30.1ms
10: learn: 0.3836696 total: 3.65ms remaining: 29.6ms
11: learn: 0.3709195 total: 3.94ms remaining: 28.9ms
12: learn: 0.3607502 total: 4.2ms remaining: 28.1ms
13: learn: 0.3534224 total: 4.55ms remaining: 28ms
14: learn: 0.3415889 total: 4.83ms remaining: 27.4ms
15: learn: 0.3315429 total: 5.23ms remaining: 27.5ms
16: learn: 0.3227477 total: 5.52ms remaining: 26.9ms
17: learn: 0.3143145 total: 5.79ms remaining: 26.4ms
18: learn: 0.3076283 total: 6.09ms remaining: 26ms
19: learn: 0.3030449 total: 6.37ms remaining: 25.5ms
20: learn: 0.2978045 total: 6.63ms remaining: 24.9ms
21: learn: 0.2912842 total: 6.9ms remaining: 24.5ms
22: learn: 0.2878362 total: 7.26ms remaining: 24.3ms
23: learn: 0.2841128 total: 7.53ms remaining: 23.8ms
24: learn: 0.2788583 total: 7.91ms remaining: 23.7ms
25: learn: 0.2749961 total: 8.29ms remaining: 23.6ms
26: learn: 0.2695994 total: 8.63ms remaining: 23.3ms
27: learn: 0.2649796 total: 9.09ms remaining: 23.4ms
28: learn: 0.2618379 total: 9.42ms remaining: 23.1ms
29: learn: 0.2589920 total: 9.75ms remaining: 22.7ms
30: learn: 0.2569354 total: 10ms remaining: 22.3ms
31: learn: 0.2524786 total: 10.3ms remaining: 21.9ms
32: learn: 0.2489823 total: 10.7ms remaining: 21.6ms
33: learn: 0.2444734 total: 11ms remaining: 21.4ms
34: learn: 0.2422778 total: 11.4ms remaining: 21.2ms
35: learn: 0.2410345 total: 11.7ms remaining: 20.7ms
36: learn: 0.2357798 total: 12ms remaining: 20.5ms
37: learn: 0.2320050 total: 12.4ms remaining: 20.2ms
38: learn: 0.2296439 total: 12.8ms remaining: 20.1ms
39: learn: 0.2275001 total: 13.2ms remaining: 19.7ms
40: learn: 0.2265651 total: 13.5ms remaining: 19.4ms
41: learn: 0.2250516 total: 13.8ms remaining: 19ms
42: learn: 0.2221955 total: 14.1ms remaining: 18.7ms
43: learn: 0.2200962 total: 14.6ms remaining: 18.6ms
44: learn: 0.2183577 total: 15ms remaining: 18.3ms
45: learn: 0.2171256 total: 15.1ms remaining: 17.7ms
46: learn: 0.2154213 total: 15.5ms remaining: 17.5ms
47: learn: 0.2144414 total: 15.9ms remaining: 17.2ms
48: learn: 0.2114539 total: 16.3ms remaining: 16.9ms
49: learn: 0.2087620 total: 16.7ms remaining: 16.7ms
50: learn: 0.2073633 total: 17.2ms remaining: 16.5ms
51: learn: 0.2062790 total: 17.6ms remaining: 16.2ms
52: learn: 0.2046617 total: 17.9ms remaining: 15.9ms
53: learn: 0.2019813 total: 18.3ms remaining: 15.6ms
54: learn: 0.2000729 total: 18.8ms remaining: 15.4ms
55: learn: 0.1980745 total: 19.2ms remaining: 15.1ms
56: learn: 0.1963953 total: 19.8ms remaining: 14.9ms
57: learn: 0.1954676 total: 20.2ms remaining: 14.6ms
58: learn: 0.1927920 total: 20.6ms remaining: 14.3ms
59: learn: 0.1919992 total: 21ms remaining: 14ms
60: learn: 0.1897627 total: 21.4ms remaining: 13.7ms
61: learn: 0.1880689 total: 21.9ms remaining: 13.4ms
62: learn: 0.1872845 total: 22.3ms remaining: 13.1ms
63: learn: 0.1859812 total: 22.9ms remaining: 12.9ms
64: learn: 0.1847340 total: 23.3ms remaining: 12.6ms
65: learn: 0.1834789 total: 23.9ms remaining: 12.3ms
66: learn: 0.1815676 total: 24.4ms remaining: 12ms
67: learn: 0.1796680 total: 24.9ms remaining: 11.7ms
68: learn: 0.1784089 total: 25.3ms remaining: 11.4ms
69: learn: 0.1767732 total: 25.8ms remaining: 11ms
70: learn: 0.1752524 total: 26.3ms remaining: 10.8ms
71: learn: 0.1736631 total: 26.7ms remaining: 10.4ms
72: learn: 0.1720111 total: 27.2ms remaining: 10.1ms
73: learn: 0.1698143 total: 27.7ms remaining: 9.73ms
74: learn: 0.1679986 total: 28.3ms remaining: 9.42ms
75: learn: 0.1664287 total: 28.8ms remaining: 9.08ms
76: learn: 0.1651468 total: 29.2ms remaining: 8.72ms
77: learn: 0.1640673 total: 29.7ms remaining: 8.38ms
78: learn: 0.1629770 total: 30.2ms remaining: 8.04ms
79: learn: 0.1617743 total: 30.7ms remaining: 7.68ms
80: learn: 0.1604518 total: 31.2ms remaining: 7.33ms
81: learn: 0.1594080 total: 31.7ms remaining: 6.95ms
82: learn: 0.1583856 total: 32.2ms remaining: 6.6ms
83: learn: 0.1578502 total: 32.7ms remaining: 6.23ms
84: learn: 0.1573959 total: 33.2ms remaining: 5.86ms
85: learn: 0.1562034 total: 33.7ms remaining: 5.49ms
86: learn: 0.1554047 total: 34.2ms remaining: 5.11ms
87: learn: 0.1543375 total: 34.6ms remaining: 4.72ms
88: learn: 0.1529024 total: 35.1ms remaining: 4.33ms
89: learn: 0.1519306 total: 35.6ms remaining: 3.95ms
90: learn: 0.1516271 total: 36ms remaining: 3.56ms
91: learn: 0.1512948 total: 36.5ms remaining: 3.17ms
92: learn: 0.1501188 total: 37.1ms remaining: 2.79ms
93: learn: 0.1485239 total: 37.4ms remaining: 2.39ms
94: learn: 0.1477443 total: 37.8ms remaining: 1.99ms
95: learn: 0.1460301 total: 38.2ms remaining: 1.59ms
96: learn: 0.1447156 total: 38.7ms remaining: 1.2ms
97: learn: 0.1434485 total: 39.2ms remaining: 800us
98: learn: 0.1428498 total: 39.7ms remaining: 400us
99: learn: 0.1416213 total: 40.1ms remaining: 0us
0: learn: 0.6459251 total: 877us remaining: 86.9ms
1: learn: 0.5967732 total: 1.67ms remaining: 81.8ms
2: learn: 0.5604266 total: 1.99ms remaining: 64.3ms
3: learn: 0.5301914 total: 2.28ms remaining: 54.7ms
4: learn: 0.5001033 total: 2.85ms remaining: 54.2ms
5: learn: 0.4757285 total: 3.83ms remaining: 60ms
6: learn: 0.4534027 total: 4.51ms remaining: 60ms
7: learn: 0.4332717 total: 5.3ms remaining: 61ms
8: learn: 0.4169403 total: 6.11ms remaining: 61.8ms
9: learn: 0.4024786 total: 6.83ms remaining: 61.5ms
10: learn: 0.3870025 total: 7.54ms remaining: 61ms
11: learn: 0.3708718 total: 8.1ms remaining: 59.4ms
12: learn: 0.3621849 total: 8.44ms remaining: 56.5ms
13: learn: 0.3487949 total: 8.84ms remaining: 54.3ms
14: learn: 0.3388521 total: 9.23ms remaining: 52.3ms
15: learn: 0.3304012 total: 9.35ms remaining: 49.1ms
16: learn: 0.3191993 total: 9.79ms remaining: 47.8ms
17: learn: 0.3131216 total: 10.2ms remaining: 46.5ms
18: learn: 0.3060119 total: 10.5ms remaining: 44.9ms
19: learn: 0.2994195 total: 10.8ms remaining: 43.4ms
20: learn: 0.2932420 total: 11.2ms remaining: 42.1ms
21: learn: 0.2870467 total: 11.8ms remaining: 41.7ms
22: learn: 0.2822750 total: 12.1ms remaining: 40.5ms
23: learn: 0.2740107 total: 12.6ms remaining: 40ms
24: learn: 0.2716019 total: 13ms remaining: 39ms
25: learn: 0.2674096 total: 13.3ms remaining: 38ms
26: learn: 0.2645899 total: 13.6ms remaining: 36.7ms
27: learn: 0.2602843 total: 13.8ms remaining: 35.5ms
28: learn: 0.2526958 total: 14.1ms remaining: 34.5ms
29: learn: 0.2493028 total: 14.4ms remaining: 33.7ms
30: learn: 0.2472036 total: 15.1ms remaining: 33.5ms
31: learn: 0.2430176 total: 15.7ms remaining: 33.5ms
32: learn: 0.2391329 total: 16.2ms remaining: 32.8ms
33: learn: 0.2358302 total: 16.5ms remaining: 32ms
34: learn: 0.2310165 total: 17.2ms remaining: 31.9ms
35: learn: 0.2277757 total: 17.5ms remaining: 31.1ms
36: learn: 0.2253298 total: 17.8ms remaining: 30.3ms
37: learn: 0.2233005 total: 18.1ms remaining: 29.5ms
38: learn: 0.2212937 total: 18.4ms remaining: 28.8ms
39: learn: 0.2194811 total: 18.8ms remaining: 28.2ms
40: learn: 0.2147043 total: 19.4ms remaining: 27.9ms
41: learn: 0.2138031 total: 19.8ms remaining: 27.4ms
42: learn: 0.2100221 total: 20.2ms remaining: 26.8ms
43: learn: 0.2084343 total: 20.6ms remaining: 26.2ms
44: learn: 0.2054505 total: 21ms remaining: 25.7ms
45: learn: 0.2016074 total: 21.5ms remaining: 25.3ms
46: learn: 0.1987326 total: 22ms remaining: 24.8ms
47: learn: 0.1961880 total: 22.3ms remaining: 24.2ms
48: learn: 0.1951000 total: 22.7ms remaining: 23.6ms
49: learn: 0.1916939 total: 23.1ms remaining: 23.1ms
50: learn: 0.1890724 total: 24ms remaining: 23.1ms
51: learn: 0.1868853 total: 24.7ms remaining: 22.8ms
52: learn: 0.1852767 total: 25.3ms remaining: 22.5ms
53: learn: 0.1833287 total: 25.8ms remaining: 22ms
54: learn: 0.1810154 total: 26.3ms remaining: 21.5ms
55: learn: 0.1787887 total: 26.9ms remaining: 21.1ms
56: learn: 0.1768586 total: 27.4ms remaining: 20.7ms
57: learn: 0.1756359 total: 28.1ms remaining: 20.3ms
58: learn: 0.1747836 total: 28.5ms remaining: 19.8ms
59: learn: 0.1728584 total: 28.9ms remaining: 19.3ms
60: learn: 0.1715426 total: 29.3ms remaining: 18.7ms
61: learn: 0.1691059 total: 29.7ms remaining: 18.2ms
62: learn: 0.1675363 total: 30.3ms remaining: 17.8ms
63: learn: 0.1651880 total: 30.8ms remaining: 17.3ms
64: learn: 0.1640820 total: 31.2ms remaining: 16.8ms
65: learn: 0.1629242 total: 31.6ms remaining: 16.3ms
66: learn: 0.1605127 total: 31.9ms remaining: 15.7ms
67: learn: 0.1598041 total: 32.2ms remaining: 15.2ms
68: learn: 0.1591679 total: 32.6ms remaining: 14.6ms
69: learn: 0.1575901 total: 32.9ms remaining: 14.1ms
70: learn: 0.1561751 total: 33.5ms remaining: 13.7ms
71: learn: 0.1544019 total: 33.9ms remaining: 13.2ms
72: learn: 0.1531977 total: 34.3ms remaining: 12.7ms
73: learn: 0.1512375 total: 35.8ms remaining: 12.6ms
74: learn: 0.1504852 total: 36.3ms remaining: 12.1ms
75: learn: 0.1488729 total: 37.1ms remaining: 11.7ms
76: learn: 0.1471366 total: 37.5ms remaining: 11.2ms
77: learn: 0.1461028 total: 37.9ms remaining: 10.7ms
78: learn: 0.1449585 total: 38.4ms remaining: 10.2ms
79: learn: 0.1431747 total: 38.8ms remaining: 9.7ms
80: learn: 0.1421230 total: 39.2ms remaining: 9.18ms
81: learn: 0.1411879 total: 39.6ms remaining: 8.69ms
82: learn: 0.1396318 total: 40.3ms remaining: 8.26ms
83: learn: 0.1381443 total: 40.7ms remaining: 7.75ms
84: learn: 0.1373405 total: 41ms remaining: 7.23ms
85: learn: 0.1357847 total: 41.3ms remaining: 6.72ms
86: learn: 0.1349063 total: 41.7ms remaining: 6.24ms
87: learn: 0.1341299 total: 42.2ms remaining: 5.76ms
88: learn: 0.1330911 total: 42.7ms remaining: 5.28ms
89: learn: 0.1320781 total: 43.2ms remaining: 4.8ms
90: learn: 0.1311151 total: 43.9ms remaining: 4.34ms
91: learn: 0.1300194 total: 44.2ms remaining: 3.84ms
92: learn: 0.1287699 total: 44.5ms remaining: 3.35ms
93: learn: 0.1276108 total: 44.9ms remaining: 2.87ms
94: learn: 0.1263275 total: 45.3ms remaining: 2.38ms
95: learn: 0.1255466 total: 45.8ms remaining: 1.91ms
96: learn: 0.1250082 total: 46.3ms remaining: 1.43ms
97: learn: 0.1242084 total: 46.7ms remaining: 952us
98: learn: 0.1236661 total: 47ms remaining: 474us
99: learn: 0.1229932 total: 47.4ms remaining: 0us
0: learn: 0.6481220 total: 485us remaining: 48ms
1: learn: 0.6111293 total: 1.07ms remaining: 52.3ms
2: learn: 0.5802804 total: 1.85ms remaining: 59.7ms
3: learn: 0.5439726 total: 2.59ms remaining: 62.2ms
4: learn: 0.5119527 total: 3.27ms remaining: 62ms
5: learn: 0.4901590 total: 4.06ms remaining: 63.7ms
6: learn: 0.4674184 total: 4.64ms remaining: 61.6ms
7: learn: 0.4472015 total: 5.32ms remaining: 61.2ms
8: learn: 0.4320357 total: 5.83ms remaining: 58.9ms
9: learn: 0.4137385 total: 6.4ms remaining: 57.6ms
10: learn: 0.4027291 total: 7ms remaining: 56.7ms
11: learn: 0.3875597 total: 7.16ms remaining: 52.5ms
12: learn: 0.3749398 total: 7.95ms remaining: 53.2ms
13: learn: 0.3680215 total: 8.6ms remaining: 52.8ms
14: learn: 0.3587988 total: 9.31ms remaining: 52.8ms
15: learn: 0.3490724 total: 10ms remaining: 52.5ms
16: learn: 0.3430159 total: 10.7ms remaining: 52.1ms
17: learn: 0.3346041 total: 11.5ms remaining: 52.3ms
18: learn: 0.3270005 total: 12ms remaining: 51.2ms
19: learn: 0.3193122 total: 12.4ms remaining: 49.4ms
20: learn: 0.3131098 total: 12.8ms remaining: 48.1ms
21: learn: 0.3077639 total: 13.2ms remaining: 46.9ms
22: learn: 0.3023597 total: 13.8ms remaining: 46.1ms
23: learn: 0.2988230 total: 14.2ms remaining: 44.8ms
24: learn: 0.2948105 total: 15ms remaining: 44.9ms
25: learn: 0.2903789 total: 16.1ms remaining: 45.8ms
26: learn: 0.2834294 total: 16.6ms remaining: 44.9ms
27: learn: 0.2779859 total: 17.5ms remaining: 45.1ms
28: learn: 0.2730893 total: 18.1ms remaining: 44.2ms
29: learn: 0.2678348 total: 18.5ms remaining: 43.3ms
30: learn: 0.2641220 total: 19.3ms remaining: 43ms
31: learn: 0.2609596 total: 19.7ms remaining: 41.8ms
32: learn: 0.2563068 total: 20ms remaining: 40.6ms
33: learn: 0.2530053 total: 21ms remaining: 40.7ms
34: learn: 0.2495580 total: 21.8ms remaining: 40.5ms
35: learn: 0.2460362 total: 22ms remaining: 39.2ms
36: learn: 0.2422195 total: 22.4ms remaining: 38.2ms
37: learn: 0.2397035 total: 23.3ms remaining: 38ms
38: learn: 0.2375111 total: 24.1ms remaining: 37.7ms
39: learn: 0.2351878 total: 24.5ms remaining: 36.8ms
40: learn: 0.2322549 total: 25.1ms remaining: 36.2ms
41: learn: 0.2296954 total: 25.5ms remaining: 35.2ms
42: learn: 0.2271196 total: 25.9ms remaining: 34.3ms
43: learn: 0.2242304 total: 26.8ms remaining: 34.1ms
44: learn: 0.2222700 total: 27.4ms remaining: 33.4ms
45: learn: 0.2202307 total: 28.2ms remaining: 33.1ms
46: learn: 0.2180234 total: 28.8ms remaining: 32.5ms
47: learn: 0.2162309 total: 29.1ms remaining: 31.6ms
48: learn: 0.2143964 total: 30ms remaining: 31.2ms
49: learn: 0.2133614 total: 30.4ms remaining: 30.4ms
50: learn: 0.2108225 total: 30.8ms remaining: 29.6ms
51: learn: 0.2081551 total: 31.2ms remaining: 28.8ms
52: learn: 0.2062048 total: 31.5ms remaining: 28ms
53: learn: 0.2047539 total: 32.1ms remaining: 27.3ms
54: learn: 0.2029284 total: 32.5ms remaining: 26.6ms
55: learn: 0.2014443 total: 33ms remaining: 26ms
56: learn: 0.1989066 total: 33.6ms remaining: 25.4ms
57: learn: 0.1978963 total: 34.2ms remaining: 24.8ms
58: learn: 0.1967623 total: 35.1ms remaining: 24.4ms
59: learn: 0.1944301 total: 35.6ms remaining: 23.7ms
60: learn: 0.1927543 total: 36.1ms remaining: 23.1ms
61: learn: 0.1897249 total: 38.1ms remaining: 23.3ms
62: learn: 0.1881984 total: 39.3ms remaining: 23.1ms
63: learn: 0.1860998 total: 39.7ms remaining: 22.3ms
64: learn: 0.1847722 total: 40.5ms remaining: 21.8ms
65: learn: 0.1831115 total: 41ms remaining: 21.1ms
66: learn: 0.1812653 total: 41.4ms remaining: 20.4ms
67: learn: 0.1797829 total: 41.8ms remaining: 19.7ms
68: learn: 0.1785188 total: 42.1ms remaining: 18.9ms
69: learn: 0.1775237 total: 42.5ms remaining: 18.2ms
70: learn: 0.1756976 total: 42.8ms remaining: 17.5ms
71: learn: 0.1742396 total: 43.2ms remaining: 16.8ms
72: learn: 0.1726201 total: 43.6ms remaining: 16.1ms
73: learn: 0.1710115 total: 44.6ms remaining: 15.7ms
74: learn: 0.1701401 total: 45.5ms remaining: 15.2ms
75: learn: 0.1686296 total: 46.2ms remaining: 14.6ms
76: learn: 0.1670643 total: 46.7ms remaining: 13.9ms
77: learn: 0.1649674 total: 48ms remaining: 13.5ms
78: learn: 0.1635317 total: 48.6ms remaining: 12.9ms
79: learn: 0.1622078 total: 49.1ms remaining: 12.3ms
80: learn: 0.1612491 total: 49.5ms remaining: 11.6ms
81: learn: 0.1606524 total: 49.9ms remaining: 11ms
82: learn: 0.1595372 total: 50.3ms remaining: 10.3ms
83: learn: 0.1582192 total: 51ms remaining: 9.71ms
84: learn: 0.1567791 total: 51.6ms remaining: 9.11ms
85: learn: 0.1553771 total: 52.5ms remaining: 8.54ms
86: learn: 0.1546524 total: 53.1ms remaining: 7.94ms
87: learn: 0.1530995 total: 53.5ms remaining: 7.3ms
88: learn: 0.1520824 total: 54.2ms remaining: 6.7ms
89: learn: 0.1510000 total: 54.5ms remaining: 6.05ms
90: learn: 0.1504653 total: 54.8ms remaining: 5.42ms
91: learn: 0.1500492 total: 55.7ms remaining: 4.84ms
92: learn: 0.1490266 total: 56.1ms remaining: 4.22ms
93: learn: 0.1483426 total: 56.7ms remaining: 3.62ms
94: learn: 0.1471332 total: 57.2ms remaining: 3.01ms
95: learn: 0.1462601 total: 58ms remaining: 2.42ms
96: learn: 0.1450494 total: 58.6ms remaining: 1.81ms
97: learn: 0.1437640 total: 59.3ms remaining: 1.21ms
98: learn: 0.1431763 total: 59.7ms remaining: 602us
99: learn: 0.1424434 total: 60.1ms remaining: 0us
0: learn: 0.6438231 total: 445us remaining: 44.1ms
1: learn: 0.5942339 total: 938us remaining: 46ms
2: learn: 0.5577436 total: 1.43ms remaining: 46.2ms
3: learn: 0.5227298 total: 1.74ms remaining: 41.7ms
4: learn: 0.4957887 total: 1.99ms remaining: 37.7ms
5: learn: 0.4699463 total: 2.3ms remaining: 36.1ms
6: learn: 0.4473918 total: 2.76ms remaining: 36.7ms
7: learn: 0.4301896 total: 3ms remaining: 34.5ms
8: learn: 0.4101801 total: 3.31ms remaining: 33.4ms
9: learn: 0.3951413 total: 3.63ms remaining: 32.7ms
10: learn: 0.3797073 total: 3.96ms remaining: 32ms
11: learn: 0.3670122 total: 4.27ms remaining: 31.3ms
12: learn: 0.3548199 total: 4.65ms remaining: 31.1ms
13: learn: 0.3447084 total: 5.07ms remaining: 31.2ms
14: learn: 0.3357773 total: 5.65ms remaining: 32ms
15: learn: 0.3263992 total: 5.77ms remaining: 30.3ms
16: learn: 0.3192555 total: 6.05ms remaining: 29.5ms
17: learn: 0.3126032 total: 6.35ms remaining: 28.9ms
18: learn: 0.3073360 total: 6.62ms remaining: 28.2ms
19: learn: 0.2996768 total: 6.95ms remaining: 27.8ms
20: learn: 0.2937950 total: 7.32ms remaining: 27.5ms
21: learn: 0.2894520 total: 7.75ms remaining: 27.5ms
22: learn: 0.2850523 total: 8.36ms remaining: 28ms
23: learn: 0.2786365 total: 8.91ms remaining: 28.2ms
24: learn: 0.2739428 total: 9.49ms remaining: 28.5ms
25: learn: 0.2704767 total: 10ms remaining: 28.5ms
26: learn: 0.2672330 total: 10.7ms remaining: 28.9ms
27: learn: 0.2617352 total: 11.4ms remaining: 29.3ms
28: learn: 0.2572261 total: 11.8ms remaining: 28.9ms
29: learn: 0.2552006 total: 12.4ms remaining: 29ms
30: learn: 0.2514638 total: 12.7ms remaining: 28.3ms
31: learn: 0.2491248 total: 13ms remaining: 27.7ms
32: learn: 0.2460170 total: 13.4ms remaining: 27.2ms
33: learn: 0.2432369 total: 13.8ms remaining: 26.8ms
34: learn: 0.2395334 total: 14.2ms remaining: 26.4ms
35: learn: 0.2375372 total: 14.7ms remaining: 26.2ms
36: learn: 0.2354697 total: 15.3ms remaining: 26.1ms
37: learn: 0.2335946 total: 15.8ms remaining: 25.7ms
38: learn: 0.2303347 total: 16ms remaining: 25.1ms
39: learn: 0.2283544 total: 16.3ms remaining: 24.5ms
40: learn: 0.2269470 total: 16.7ms remaining: 24ms
41: learn: 0.2246784 total: 17.5ms remaining: 24.1ms
42: learn: 0.2233697 total: 18.1ms remaining: 24ms
43: learn: 0.2206222 total: 18.5ms remaining: 23.6ms
44: learn: 0.2184053 total: 18.8ms remaining: 23ms
45: learn: 0.2162331 total: 19.3ms remaining: 22.7ms
46: learn: 0.2135035 total: 19.8ms remaining: 22.4ms
47: learn: 0.2124312 total: 20.6ms remaining: 22.3ms
48: learn: 0.2112126 total: 20.9ms remaining: 21.8ms
49: learn: 0.2093780 total: 21.5ms remaining: 21.5ms
50: learn: 0.2069938 total: 22.2ms remaining: 21.3ms
51: learn: 0.2051590 total: 22.9ms remaining: 21.1ms
52: learn: 0.2026844 total: 23.4ms remaining: 20.8ms
53: learn: 0.2013613 total: 23.8ms remaining: 20.3ms
54: learn: 0.1988510 total: 24.3ms remaining: 19.9ms
55: learn: 0.1982063 total: 24.6ms remaining: 19.4ms
56: learn: 0.1961221 total: 25.4ms remaining: 19.2ms
57: learn: 0.1954671 total: 25.8ms remaining: 18.7ms
58: learn: 0.1940105 total: 26.2ms remaining: 18.2ms
59: learn: 0.1919199 total: 26.6ms remaining: 17.7ms
60: learn: 0.1891903 total: 27ms remaining: 17.3ms
61: learn: 0.1875479 total: 27.5ms remaining: 16.8ms
62: learn: 0.1855201 total: 28.1ms remaining: 16.5ms
63: learn: 0.1838181 total: 28.4ms remaining: 16ms
64: learn: 0.1817402 total: 28.7ms remaining: 15.5ms
65: learn: 0.1799330 total: 29.2ms remaining: 15ms
66: learn: 0.1776151 total: 29.6ms remaining: 14.6ms
67: learn: 0.1764638 total: 30.4ms remaining: 14.3ms
68: learn: 0.1750410 total: 30.9ms remaining: 13.9ms
69: learn: 0.1739522 total: 31.2ms remaining: 13.4ms
70: learn: 0.1725858 total: 31.8ms remaining: 13ms
71: learn: 0.1713943 total: 32.5ms remaining: 12.6ms
72: learn: 0.1701219 total: 33.2ms remaining: 12.3ms
73: learn: 0.1691621 total: 33.7ms remaining: 11.9ms
74: learn: 0.1678395 total: 34.6ms remaining: 11.5ms
75: learn: 0.1668259 total: 35ms remaining: 11.1ms
76: learn: 0.1657933 total: 35.5ms remaining: 10.6ms
77: learn: 0.1649136 total: 36ms remaining: 10.1ms
78: learn: 0.1630982 total: 37.4ms remaining: 9.94ms
79: learn: 0.1623115 total: 37.8ms remaining: 9.45ms
80: learn: 0.1616336 total: 38.1ms remaining: 8.95ms
81: learn: 0.1602423 total: 38.5ms remaining: 8.45ms
82: learn: 0.1586111 total: 39ms remaining: 8ms
83: learn: 0.1577384 total: 39.6ms remaining: 7.54ms
84: learn: 0.1569119 total: 40.1ms remaining: 7.08ms
85: learn: 0.1563218 total: 41ms remaining: 6.67ms
86: learn: 0.1556317 total: 41.9ms remaining: 6.26ms
87: learn: 0.1545180 total: 42.3ms remaining: 5.76ms
88: learn: 0.1536534 total: 42.7ms remaining: 5.28ms
89: learn: 0.1527228 total: 43.6ms remaining: 4.85ms
90: learn: 0.1516318 total: 44.4ms remaining: 4.39ms
91: learn: 0.1506953 total: 45ms remaining: 3.91ms
92: learn: 0.1498776 total: 45.7ms remaining: 3.44ms
93: learn: 0.1489490 total: 47.2ms remaining: 3.01ms
94: learn: 0.1482286 total: 47.7ms remaining: 2.51ms
95: learn: 0.1475212 total: 48ms remaining: 2ms
96: learn: 0.1464758 total: 48.3ms remaining: 1.5ms
97: learn: 0.1454925 total: 48.7ms remaining: 993us
98: learn: 0.1446619 total: 49.2ms remaining: 497us
99: learn: 0.1440331 total: 49.5ms remaining: 0us
0: learn: 0.6495864 total: 493us remaining: 48.9ms
1: learn: 0.6173131 total: 845us remaining: 41.4ms
2: learn: 0.5809182 total: 1.19ms remaining: 38.5ms
3: learn: 0.5592879 total: 1.53ms remaining: 36.7ms
4: learn: 0.5333497 total: 1.66ms remaining: 31.6ms
5: learn: 0.5100357 total: 2ms remaining: 31.3ms
6: learn: 0.4871572 total: 2.48ms remaining: 32.9ms
7: learn: 0.4656432 total: 3.04ms remaining: 35ms
8: learn: 0.4488647 total: 3.65ms remaining: 36.9ms
9: learn: 0.4359401 total: 4.14ms remaining: 37.3ms
10: learn: 0.4213552 total: 4.49ms remaining: 36.3ms
11: learn: 0.4112009 total: 4.76ms remaining: 34.9ms
12: learn: 0.3990163 total: 4.9ms remaining: 32.8ms
13: learn: 0.3899301 total: 5.34ms remaining: 32.8ms
14: learn: 0.3778913 total: 5.74ms remaining: 32.5ms
15: learn: 0.3704319 total: 6.1ms remaining: 32ms
16: learn: 0.3637520 total: 6.43ms remaining: 31.4ms
17: learn: 0.3549960 total: 6.81ms remaining: 31ms
18: learn: 0.3469922 total: 7.19ms remaining: 30.6ms
19: learn: 0.3421919 total: 7.55ms remaining: 30.2ms
20: learn: 0.3359789 total: 7.88ms remaining: 29.6ms
21: learn: 0.3289882 total: 8.28ms remaining: 29.3ms
22: learn: 0.3247038 total: 8.72ms remaining: 29.2ms
23: learn: 0.3173687 total: 9.35ms remaining: 29.6ms
24: learn: 0.3117996 total: 9.91ms remaining: 29.7ms
25: learn: 0.3062690 total: 10.3ms remaining: 29.2ms
26: learn: 0.3010276 total: 10.6ms remaining: 28.8ms
27: learn: 0.2966909 total: 11ms remaining: 28.3ms
28: learn: 0.2916840 total: 11.4ms remaining: 27.9ms
29: learn: 0.2884667 total: 12.1ms remaining: 28.3ms
30: learn: 0.2841525 total: 12.5ms remaining: 27.8ms
31: learn: 0.2803788 total: 12.8ms remaining: 27.3ms
32: learn: 0.2780639 total: 13.1ms remaining: 26.6ms
33: learn: 0.2750141 total: 13.4ms remaining: 26.1ms
34: learn: 0.2714735 total: 13.8ms remaining: 25.7ms
35: learn: 0.2683093 total: 14.2ms remaining: 25.3ms
36: learn: 0.2664962 total: 14.8ms remaining: 25.2ms
37: learn: 0.2626528 total: 15.4ms remaining: 25.2ms
38: learn: 0.2595677 total: 15.9ms remaining: 24.8ms
39: learn: 0.2553350 total: 16.5ms remaining: 24.8ms
40: learn: 0.2529060 total: 16.9ms remaining: 24.3ms
41: learn: 0.2497261 total: 17.2ms remaining: 23.8ms
42: learn: 0.2471071 total: 17.8ms remaining: 23.6ms
43: learn: 0.2460101 total: 18.1ms remaining: 23.1ms
44: learn: 0.2438296 total: 18.5ms remaining: 22.6ms
45: learn: 0.2411975 total: 18.8ms remaining: 22.1ms
46: learn: 0.2397885 total: 19.4ms remaining: 21.9ms
47: learn: 0.2384377 total: 20ms remaining: 21.6ms
48: learn: 0.2360721 total: 20.7ms remaining: 21.6ms
49: learn: 0.2334278 total: 21.5ms remaining: 21.5ms
50: learn: 0.2321367 total: 22.5ms remaining: 21.6ms
51: learn: 0.2294726 total: 23ms remaining: 21.2ms
52: learn: 0.2264535 total: 23.8ms remaining: 21.1ms
53: learn: 0.2249275 total: 24.7ms remaining: 21ms
54: learn: 0.2225142 total: 25.2ms remaining: 20.6ms
55: learn: 0.2203405 total: 26.1ms remaining: 20.5ms
56: learn: 0.2185270 total: 26.5ms remaining: 20ms
57: learn: 0.2173819 total: 27.5ms remaining: 19.9ms
58: learn: 0.2143567 total: 28.1ms remaining: 19.5ms
59: learn: 0.2115298 total: 28.6ms remaining: 19ms
60: learn: 0.2092046 total: 29.2ms remaining: 18.7ms
61: learn: 0.2076664 total: 29.8ms remaining: 18.3ms
62: learn: 0.2053319 total: 30.4ms remaining: 17.8ms
63: learn: 0.2034479 total: 31.5ms remaining: 17.7ms
64: learn: 0.2020957 total: 32ms remaining: 17.2ms
65: learn: 0.2003246 total: 33.6ms remaining: 17.3ms
66: learn: 0.1983271 total: 34.1ms remaining: 16.8ms
67: learn: 0.1968854 total: 34.5ms remaining: 16.2ms
68: learn: 0.1956967 total: 35ms remaining: 15.7ms
69: learn: 0.1932236 total: 35.8ms remaining: 15.4ms
70: learn: 0.1924645 total: 36.7ms remaining: 15ms
71: learn: 0.1905839 total: 37.6ms remaining: 14.6ms
72: learn: 0.1889518 total: 38ms remaining: 14.1ms
73: learn: 0.1879034 total: 38.5ms remaining: 13.5ms
74: learn: 0.1862801 total: 39.2ms remaining: 13.1ms
75: learn: 0.1843327 total: 39.7ms remaining: 12.5ms
76: learn: 0.1831375 total: 40.2ms remaining: 12ms
77: learn: 0.1812536 total: 40.9ms remaining: 11.5ms
78: learn: 0.1800181 total: 41.3ms remaining: 11ms
79: learn: 0.1783145 total: 42ms remaining: 10.5ms
80: learn: 0.1767922 total: 42.6ms remaining: 10ms
81: learn: 0.1755307 total: 43.2ms remaining: 9.48ms
82: learn: 0.1741847 total: 43.6ms remaining: 8.92ms
83: learn: 0.1729547 total: 44ms remaining: 8.38ms
84: learn: 0.1715502 total: 45.2ms remaining: 7.97ms
85: learn: 0.1705513 total: 45.6ms remaining: 7.43ms
86: learn: 0.1691987 total: 46.5ms remaining: 6.95ms
87: learn: 0.1680186 total: 47ms remaining: 6.4ms
88: learn: 0.1669470 total: 47.4ms remaining: 5.85ms
89: learn: 0.1659363 total: 48.1ms remaining: 5.34ms
90: learn: 0.1650771 total: 48.9ms remaining: 4.84ms
91: learn: 0.1639152 total: 50.2ms remaining: 4.37ms
92: learn: 0.1629719 total: 50.6ms remaining: 3.81ms
93: learn: 0.1618433 total: 51.1ms remaining: 3.26ms
94: learn: 0.1608519 total: 52ms remaining: 2.73ms
95: learn: 0.1599917 total: 52.7ms remaining: 2.19ms
96: learn: 0.1590048 total: 53ms remaining: 1.64ms
97: learn: 0.1584523 total: 53.5ms remaining: 1.09ms
98: learn: 0.1570499 total: 53.9ms remaining: 544us
99: learn: 0.1561710 total: 54.8ms remaining: 0us
0: learn: 0.6511846 total: 455us remaining: 45.1ms
1: learn: 0.6076570 total: 802us remaining: 39.3ms
2: learn: 0.5723418 total: 1.05ms remaining: 33.9ms
3: learn: 0.5438088 total: 1.36ms remaining: 32.6ms
4: learn: 0.5133112 total: 1.65ms remaining: 31.4ms
5: learn: 0.4856231 total: 2.11ms remaining: 33.1ms
6: learn: 0.4656300 total: 2.44ms remaining: 32.4ms
7: learn: 0.4409271 total: 2.75ms remaining: 31.6ms
8: learn: 0.4197159 total: 3.04ms remaining: 30.7ms
9: learn: 0.3996955 total: 3.33ms remaining: 30ms
10: learn: 0.3883977 total: 3.81ms remaining: 30.8ms
11: learn: 0.3761112 total: 4.3ms remaining: 31.6ms
12: learn: 0.3651722 total: 4.9ms remaining: 32.8ms
13: learn: 0.3541517 total: 5.48ms remaining: 33.7ms
14: learn: 0.3505325 total: 6.1ms remaining: 34.6ms
15: learn: 0.3446431 total: 6.22ms remaining: 32.7ms
16: learn: 0.3309643 total: 6.82ms remaining: 33.3ms
17: learn: 0.3234425 total: 7.28ms remaining: 33.2ms
18: learn: 0.3167732 total: 7.84ms remaining: 33.4ms
19: learn: 0.3116347 total: 8.3ms remaining: 33.2ms
20: learn: 0.3032964 total: 8.84ms remaining: 33.3ms
21: learn: 0.2968374 total: 9.38ms remaining: 33.2ms
22: learn: 0.2927937 total: 9.97ms remaining: 33.4ms
23: learn: 0.2860013 total: 10.6ms remaining: 33.5ms
24: learn: 0.2835729 total: 11.2ms remaining: 33.5ms
25: learn: 0.2788450 total: 11.8ms remaining: 33.5ms
26: learn: 0.2749840 total: 12.3ms remaining: 33.4ms
27: learn: 0.2704904 total: 13.2ms remaining: 34ms
28: learn: 0.2651415 total: 13.8ms remaining: 33.8ms
29: learn: 0.2608183 total: 14.5ms remaining: 33.9ms
30: learn: 0.2566741 total: 15ms remaining: 33.4ms
31: learn: 0.2537151 total: 15.5ms remaining: 32.8ms
32: learn: 0.2517625 total: 15.9ms remaining: 32.4ms
33: learn: 0.2481143 total: 16.4ms remaining: 31.8ms
34: learn: 0.2460842 total: 17ms remaining: 31.5ms
35: learn: 0.2418761 total: 17.4ms remaining: 30.9ms
36: learn: 0.2385708 total: 18.2ms remaining: 31.1ms
37: learn: 0.2351499 total: 18.8ms remaining: 30.7ms
38: learn: 0.2325407 total: 19.9ms remaining: 31.2ms
39: learn: 0.2308443 total: 21.1ms remaining: 31.7ms
40: learn: 0.2286408 total: 22.3ms remaining: 32.1ms
41: learn: 0.2263965 total: 22.9ms remaining: 31.6ms
42: learn: 0.2249888 total: 23.3ms remaining: 30.9ms
43: learn: 0.2228905 total: 23.7ms remaining: 30.2ms
44: learn: 0.2200580 total: 24.1ms remaining: 29.4ms
45: learn: 0.2176117 total: 24.6ms remaining: 28.9ms
46: learn: 0.2158570 total: 25.4ms remaining: 28.7ms
47: learn: 0.2132175 total: 26.3ms remaining: 28.5ms
48: learn: 0.2113158 total: 26.7ms remaining: 27.8ms
49: learn: 0.2085489 total: 27.1ms remaining: 27.1ms
50: learn: 0.2071999 total: 27.6ms remaining: 26.5ms
51: learn: 0.2045916 total: 28.8ms remaining: 26.6ms
52: learn: 0.2023669 total: 30ms remaining: 26.6ms
53: learn: 0.1995389 total: 30.5ms remaining: 26ms
54: learn: 0.1975750 total: 31.2ms remaining: 25.5ms
55: learn: 0.1961345 total: 32ms remaining: 25.1ms
56: learn: 0.1939941 total: 32.8ms remaining: 24.7ms
57: learn: 0.1927492 total: 33.5ms remaining: 24.3ms
58: learn: 0.1906379 total: 34.1ms remaining: 23.7ms
59: learn: 0.1891975 total: 34.8ms remaining: 23.2ms
60: learn: 0.1881798 total: 35.8ms remaining: 22.9ms
61: learn: 0.1865213 total: 36.3ms remaining: 22.2ms
62: learn: 0.1852302 total: 36.6ms remaining: 21.5ms
63: learn: 0.1833525 total: 37.1ms remaining: 20.9ms
64: learn: 0.1814688 total: 37.7ms remaining: 20.3ms
65: learn: 0.1802001 total: 38.5ms remaining: 19.8ms
66: learn: 0.1785566 total: 39.1ms remaining: 19.3ms
67: learn: 0.1776592 total: 39.8ms remaining: 18.7ms
68: learn: 0.1764516 total: 40.3ms remaining: 18.1ms
69: learn: 0.1750527 total: 40.8ms remaining: 17.5ms
70: learn: 0.1736182 total: 41.6ms remaining: 17ms
71: learn: 0.1721287 total: 42.5ms remaining: 16.5ms
72: learn: 0.1709128 total: 42.9ms remaining: 15.9ms
73: learn: 0.1697592 total: 43.3ms remaining: 15.2ms
74: learn: 0.1678313 total: 43.7ms remaining: 14.6ms
75: learn: 0.1670008 total: 44.4ms remaining: 14ms
76: learn: 0.1657926 total: 44.8ms remaining: 13.4ms
77: learn: 0.1644537 total: 45.5ms remaining: 12.8ms
78: learn: 0.1636771 total: 46ms remaining: 12.2ms
79: learn: 0.1621901 total: 46.8ms remaining: 11.7ms
80: learn: 0.1612640 total: 47.9ms remaining: 11.2ms
81: learn: 0.1603972 total: 48.4ms remaining: 10.6ms
82: learn: 0.1595154 total: 48.9ms remaining: 10ms
83: learn: 0.1582315 total: 49.2ms remaining: 9.38ms
84: learn: 0.1575468 total: 49.7ms remaining: 8.76ms
85: learn: 0.1563534 total: 50.1ms remaining: 8.15ms
86: learn: 0.1551965 total: 50.9ms remaining: 7.61ms
87: learn: 0.1540434 total: 51.8ms remaining: 7.06ms
88: learn: 0.1526571 total: 52.5ms remaining: 6.49ms
89: learn: 0.1511779 total: 53.4ms remaining: 5.93ms
90: learn: 0.1500077 total: 54.1ms remaining: 5.35ms
91: learn: 0.1488126 total: 54.6ms remaining: 4.75ms
92: learn: 0.1481620 total: 55.1ms remaining: 4.14ms
93: learn: 0.1473607 total: 55.6ms remaining: 3.55ms
94: learn: 0.1462452 total: 55.9ms remaining: 2.94ms
95: learn: 0.1455584 total: 56.4ms remaining: 2.35ms
96: learn: 0.1445430 total: 57.2ms remaining: 1.77ms
97: learn: 0.1434738 total: 58.1ms remaining: 1.19ms
98: learn: 0.1429231 total: 58.6ms remaining: 592us
99: learn: 0.1423377 total: 59.1ms remaining: 0us
0: learn: 0.6488410 total: 487us remaining: 48.3ms
1: learn: 0.6041830 total: 913us remaining: 44.7ms
2: learn: 0.5666197 total: 1.21ms remaining: 39ms
3: learn: 0.5371348 total: 1.57ms remaining: 37.7ms
4: learn: 0.5069919 total: 1.91ms remaining: 36.2ms
5: learn: 0.4873744 total: 2.31ms remaining: 36.1ms
6: learn: 0.4635549 total: 2.76ms remaining: 36.7ms
7: learn: 0.4477490 total: 3.19ms remaining: 36.7ms
8: learn: 0.4331475 total: 3.65ms remaining: 36.9ms
9: learn: 0.4162288 total: 4.06ms remaining: 36.6ms
10: learn: 0.4022050 total: 4.47ms remaining: 36.1ms
11: learn: 0.3883253 total: 4.68ms remaining: 34.3ms
12: learn: 0.3768763 total: 5.12ms remaining: 34.2ms
13: learn: 0.3671179 total: 5.61ms remaining: 34.4ms
14: learn: 0.3574952 total: 5.91ms remaining: 33.5ms
15: learn: 0.3486668 total: 6.28ms remaining: 33ms
16: learn: 0.3413200 total: 6.68ms remaining: 32.6ms
17: learn: 0.3349934 total: 7.32ms remaining: 33.4ms
18: learn: 0.3267828 total: 7.84ms remaining: 33.4ms
19: learn: 0.3197310 total: 8.19ms remaining: 32.8ms
20: learn: 0.3149930 total: 8.49ms remaining: 31.9ms
21: learn: 0.3076532 total: 8.83ms remaining: 31.3ms
22: learn: 0.3028168 total: 9.22ms remaining: 30.9ms
23: learn: 0.2967363 total: 9.64ms remaining: 30.5ms
24: learn: 0.2906921 total: 10.1ms remaining: 30.3ms
25: learn: 0.2859462 total: 10.5ms remaining: 30ms
26: learn: 0.2822333 total: 10.9ms remaining: 29.3ms
27: learn: 0.2779236 total: 11.2ms remaining: 28.9ms
28: learn: 0.2729841 total: 11.6ms remaining: 28.5ms
29: learn: 0.2699343 total: 11.9ms remaining: 27.8ms
30: learn: 0.2659386 total: 12.3ms remaining: 27.3ms
31: learn: 0.2629158 total: 12.6ms remaining: 26.8ms
32: learn: 0.2604045 total: 13.2ms remaining: 26.9ms
33: learn: 0.2582302 total: 13.9ms remaining: 26.9ms
34: learn: 0.2554016 total: 14.4ms remaining: 26.8ms
35: learn: 0.2539357 total: 14.7ms remaining: 26.1ms
36: learn: 0.2511286 total: 15.4ms remaining: 26.2ms
37: learn: 0.2493651 total: 15.9ms remaining: 25.9ms
38: learn: 0.2467166 total: 16.6ms remaining: 26ms
39: learn: 0.2437269 total: 17.2ms remaining: 25.9ms
40: learn: 0.2427853 total: 17.9ms remaining: 25.7ms
41: learn: 0.2427569 total: 18ms remaining: 24.8ms
42: learn: 0.2406668 total: 18.4ms remaining: 24.3ms
43: learn: 0.2381068 total: 18.8ms remaining: 23.9ms
44: learn: 0.2354987 total: 19.1ms remaining: 23.3ms
45: learn: 0.2324478 total: 19.6ms remaining: 23ms
46: learn: 0.2301235 total: 20.2ms remaining: 22.7ms
47: learn: 0.2284376 total: 20.6ms remaining: 22.4ms
48: learn: 0.2268030 total: 21.1ms remaining: 22ms
49: learn: 0.2256024 total: 21.5ms remaining: 21.5ms
50: learn: 0.2239383 total: 21.9ms remaining: 21ms
51: learn: 0.2216564 total: 22.3ms remaining: 20.6ms
52: learn: 0.2192051 total: 22.8ms remaining: 20.2ms
53: learn: 0.2179268 total: 23.2ms remaining: 19.7ms
54: learn: 0.2164056 total: 23.5ms remaining: 19.2ms
55: learn: 0.2143912 total: 23.9ms remaining: 18.8ms
56: learn: 0.2111898 total: 24.6ms remaining: 18.6ms
57: learn: 0.2086606 total: 25.1ms remaining: 18.2ms
58: learn: 0.2067375 total: 25.7ms remaining: 17.9ms
59: learn: 0.2057138 total: 26.2ms remaining: 17.5ms
60: learn: 0.2039027 total: 26.6ms remaining: 17ms
61: learn: 0.2021739 total: 27ms remaining: 16.5ms
62: learn: 0.2008954 total: 27.5ms remaining: 16.1ms
63: learn: 0.1987454 total: 28ms remaining: 15.7ms
64: learn: 0.1964593 total: 28.4ms remaining: 15.3ms
65: learn: 0.1954175 total: 29ms remaining: 14.9ms
66: learn: 0.1944282 total: 29.5ms remaining: 14.6ms
67: learn: 0.1927253 total: 30.2ms remaining: 14.2ms
68: learn: 0.1918243 total: 30.7ms remaining: 13.8ms
69: learn: 0.1904622 total: 31.1ms remaining: 13.3ms
70: learn: 0.1885280 total: 31.6ms remaining: 12.9ms
71: learn: 0.1871618 total: 32.1ms remaining: 12.5ms
72: learn: 0.1860210 total: 32.7ms remaining: 12.1ms
73: learn: 0.1839733 total: 33.1ms remaining: 11.6ms
74: learn: 0.1825771 total: 33.7ms remaining: 11.2ms
75: learn: 0.1810811 total: 34.2ms remaining: 10.8ms
76: learn: 0.1795348 total: 34.7ms remaining: 10.4ms
77: learn: 0.1782247 total: 35.4ms remaining: 9.98ms
78: learn: 0.1771325 total: 36.3ms remaining: 9.64ms
79: learn: 0.1755010 total: 37.1ms remaining: 9.28ms
80: learn: 0.1741631 total: 37.8ms remaining: 8.87ms
81: learn: 0.1731037 total: 38.6ms remaining: 8.48ms
82: learn: 0.1722395 total: 39.3ms remaining: 8.04ms
83: learn: 0.1707454 total: 39.8ms remaining: 7.59ms
84: learn: 0.1697471 total: 40.2ms remaining: 7.09ms
85: learn: 0.1686024 total: 40.8ms remaining: 6.65ms
86: learn: 0.1676882 total: 41.2ms remaining: 6.16ms
87: learn: 0.1665142 total: 41.6ms remaining: 5.67ms
88: learn: 0.1658191 total: 42.5ms remaining: 5.25ms
89: learn: 0.1652434 total: 43.3ms remaining: 4.81ms
90: learn: 0.1640851 total: 44ms remaining: 4.35ms
91: learn: 0.1635332 total: 44.4ms remaining: 3.86ms
92: learn: 0.1625477 total: 45.1ms remaining: 3.4ms
93: learn: 0.1617999 total: 45.9ms remaining: 2.93ms
94: learn: 0.1608273 total: 46.7ms remaining: 2.46ms
95: learn: 0.1597764 total: 47.2ms remaining: 1.97ms
96: learn: 0.1593550 total: 47.7ms remaining: 1.48ms
97: learn: 0.1587727 total: 48.2ms remaining: 984us
98: learn: 0.1581202 total: 49.1ms remaining: 496us
99: learn: 0.1569767 total: 49.6ms remaining: 0us
0: learn: 0.6474641 total: 431us remaining: 42.8ms
1: learn: 0.6077883 total: 758us remaining: 37.1ms
2: learn: 0.5742449 total: 906us remaining: 29.3ms
3: learn: 0.5421671 total: 1.22ms remaining: 29.4ms
4: learn: 0.5167581 total: 1.88ms remaining: 35.8ms
5: learn: 0.4928078 total: 2.26ms remaining: 35.4ms
6: learn: 0.4707335 total: 2.64ms remaining: 35ms
7: learn: 0.4500471 total: 2.99ms remaining: 34.4ms
8: learn: 0.4322172 total: 3.31ms remaining: 33.5ms
9: learn: 0.4154537 total: 3.77ms remaining: 33.9ms
10: learn: 0.4026877 total: 4.19ms remaining: 33.9ms
11: learn: 0.3917246 total: 4.6ms remaining: 33.8ms
12: learn: 0.3834524 total: 5.18ms remaining: 34.7ms
13: learn: 0.3745413 total: 5.85ms remaining: 36ms
14: learn: 0.3678016 total: 6.56ms remaining: 37.2ms
15: learn: 0.3580243 total: 7.38ms remaining: 38.7ms
16: learn: 0.3506033 total: 7.64ms remaining: 37.3ms
17: learn: 0.3415945 total: 7.99ms remaining: 36.4ms
18: learn: 0.3352856 total: 8.37ms remaining: 35.7ms
19: learn: 0.3292979 total: 8.75ms remaining: 35ms
20: learn: 0.3232338 total: 9.1ms remaining: 34.2ms
21: learn: 0.3176816 total: 9.48ms remaining: 33.6ms
22: learn: 0.3105225 total: 9.85ms remaining: 33ms
23: learn: 0.3060575 total: 10.3ms remaining: 32.7ms
24: learn: 0.3022308 total: 10.8ms remaining: 32.4ms
25: learn: 0.2980852 total: 11.3ms remaining: 32.1ms
26: learn: 0.2926775 total: 11.7ms remaining: 31.6ms
27: learn: 0.2897220 total: 12.1ms remaining: 31.1ms
28: learn: 0.2849458 total: 12.9ms remaining: 31.6ms
29: learn: 0.2816172 total: 13.3ms remaining: 31.1ms
30: learn: 0.2802763 total: 13.9ms remaining: 31ms
31: learn: 0.2776989 total: 14.5ms remaining: 30.9ms
32: learn: 0.2734024 total: 15.4ms remaining: 31.2ms
33: learn: 0.2700002 total: 16.1ms remaining: 31.3ms
34: learn: 0.2674873 total: 16.8ms remaining: 31.1ms
35: learn: 0.2658062 total: 17.4ms remaining: 30.9ms
36: learn: 0.2625090 total: 18ms remaining: 30.6ms
37: learn: 0.2614340 total: 18.3ms remaining: 29.9ms
38: learn: 0.2592705 total: 18.8ms remaining: 29.4ms
39: learn: 0.2550580 total: 19.7ms remaining: 29.6ms
40: learn: 0.2526475 total: 20.6ms remaining: 29.6ms
41: learn: 0.2498844 total: 21.3ms remaining: 29.5ms
42: learn: 0.2485077 total: 21.7ms remaining: 28.8ms
43: learn: 0.2473002 total: 22.3ms remaining: 28.3ms
44: learn: 0.2447949 total: 23ms remaining: 28.1ms
45: learn: 0.2438473 total: 23.4ms remaining: 27.5ms
46: learn: 0.2420054 total: 23.8ms remaining: 26.9ms
47: learn: 0.2399732 total: 24.4ms remaining: 26.4ms
48: learn: 0.2368189 total: 24.8ms remaining: 25.8ms
49: learn: 0.2339327 total: 25.2ms remaining: 25.2ms
50: learn: 0.2321580 total: 26ms remaining: 25ms
51: learn: 0.2303962 total: 26.6ms remaining: 24.6ms
52: learn: 0.2280310 total: 27.6ms remaining: 24.4ms
53: learn: 0.2269895 total: 28.2ms remaining: 24ms
54: learn: 0.2248960 total: 29ms remaining: 23.8ms
55: learn: 0.2223448 total: 29.9ms remaining: 23.5ms
56: learn: 0.2197124 total: 30.4ms remaining: 22.9ms
57: learn: 0.2183733 total: 31ms remaining: 22.4ms
58: learn: 0.2158602 total: 31.9ms remaining: 22.2ms
59: learn: 0.2142115 total: 32.4ms remaining: 21.6ms
60: learn: 0.2120369 total: 33.2ms remaining: 21.2ms
61: learn: 0.2100350 total: 33.6ms remaining: 20.6ms
62: learn: 0.2076103 total: 34ms remaining: 20ms
63: learn: 0.2055892 total: 34.5ms remaining: 19.4ms
64: learn: 0.2040218 total: 35.3ms remaining: 19ms
65: learn: 0.2015573 total: 36.2ms remaining: 18.7ms
66: learn: 0.1997539 total: 36.8ms remaining: 18.1ms
67: learn: 0.1986287 total: 37.3ms remaining: 17.5ms
68: learn: 0.1962710 total: 38.2ms remaining: 17.2ms
69: learn: 0.1936191 total: 39.3ms remaining: 16.8ms
70: learn: 0.1918479 total: 39.7ms remaining: 16.2ms
71: learn: 0.1906414 total: 40.4ms remaining: 15.7ms
72: learn: 0.1897030 total: 40.9ms remaining: 15.1ms
73: learn: 0.1881570 total: 41.5ms remaining: 14.6ms
74: learn: 0.1857148 total: 42ms remaining: 14ms
75: learn: 0.1840591 total: 42.5ms remaining: 13.4ms
76: learn: 0.1822852 total: 42.9ms remaining: 12.8ms
77: learn: 0.1799874 total: 43.3ms remaining: 12.2ms
78: learn: 0.1785394 total: 43.8ms remaining: 11.6ms
79: learn: 0.1770115 total: 44.7ms remaining: 11.2ms
80: learn: 0.1763398 total: 45.3ms remaining: 10.6ms
81: learn: 0.1747446 total: 45.9ms remaining: 10.1ms
82: learn: 0.1732753 total: 46.4ms remaining: 9.51ms
83: learn: 0.1724768 total: 46.9ms remaining: 8.93ms
84: learn: 0.1715662 total: 47.5ms remaining: 8.37ms
85: learn: 0.1692734 total: 48.3ms remaining: 7.86ms
86: learn: 0.1682219 total: 48.7ms remaining: 7.28ms
87: learn: 0.1670159 total: 49.2ms remaining: 6.7ms
88: learn: 0.1657418 total: 49.9ms remaining: 6.16ms
89: learn: 0.1646984 total: 50.3ms remaining: 5.59ms
90: learn: 0.1637992 total: 51.3ms remaining: 5.08ms
91: learn: 0.1624310 total: 51.8ms remaining: 4.51ms
92: learn: 0.1614818 total: 52.4ms remaining: 3.94ms
93: learn: 0.1608375 total: 53.3ms remaining: 3.4ms
94: learn: 0.1601106 total: 53.8ms remaining: 2.83ms
95: learn: 0.1591783 total: 55ms remaining: 2.29ms
96: learn: 0.1581593 total: 55.3ms remaining: 1.71ms
97: learn: 0.1574560 total: 55.8ms remaining: 1.14ms
98: learn: 0.1569974 total: 56.2ms remaining: 567us
99: learn: 0.1558268 total: 56.7ms remaining: 0us
0: learn: 0.6477647 total: 403us remaining: 39.9ms
1: learn: 0.6127518 total: 700us remaining: 34.3ms
2: learn: 0.5886244 total: 1.01ms remaining: 32.6ms
3: learn: 0.5523313 total: 1.31ms remaining: 31.5ms
4: learn: 0.5216426 total: 1.72ms remaining: 32.7ms
5: learn: 0.4985728 total: 2.4ms remaining: 37.6ms
6: learn: 0.4714201 total: 3.14ms remaining: 41.7ms
7: learn: 0.4519625 total: 3.87ms remaining: 44.6ms
8: learn: 0.4321295 total: 4.56ms remaining: 46.1ms
9: learn: 0.4141023 total: 5.33ms remaining: 47.9ms
10: learn: 0.4001725 total: 6.14ms remaining: 49.7ms
11: learn: 0.3879811 total: 6.92ms remaining: 50.7ms
12: learn: 0.3739023 total: 7.64ms remaining: 51.1ms
13: learn: 0.3611474 total: 8.25ms remaining: 50.7ms
14: learn: 0.3500715 total: 8.85ms remaining: 50.1ms
15: learn: 0.3390426 total: 9.36ms remaining: 49.2ms
16: learn: 0.3304780 total: 9.91ms remaining: 48.4ms
17: learn: 0.3243833 total: 10.6ms remaining: 48.2ms
18: learn: 0.3174226 total: 11.2ms remaining: 47.6ms
19: learn: 0.3090807 total: 11.9ms remaining: 47.5ms
20: learn: 0.3010340 total: 12.5ms remaining: 47.2ms
21: learn: 0.2940049 total: 13.2ms remaining: 46.7ms
22: learn: 0.2890414 total: 13.8ms remaining: 46.2ms
23: learn: 0.2838094 total: 14.4ms remaining: 45.5ms
24: learn: 0.2793484 total: 15.1ms remaining: 45.3ms
25: learn: 0.2745319 total: 15.8ms remaining: 44.9ms
26: learn: 0.2711734 total: 16.5ms remaining: 44.5ms
27: learn: 0.2679744 total: 17.1ms remaining: 43.9ms
28: learn: 0.2612836 total: 17.7ms remaining: 43.3ms
29: learn: 0.2591571 total: 17.9ms remaining: 41.7ms
30: learn: 0.2573990 total: 18.2ms remaining: 40.5ms
31: learn: 0.2536914 total: 19.4ms remaining: 41.2ms
32: learn: 0.2520893 total: 19.8ms remaining: 40.1ms
33: learn: 0.2475817 total: 20.2ms remaining: 39.2ms
34: learn: 0.2444968 total: 20.8ms remaining: 38.7ms
35: learn: 0.2418765 total: 21.5ms remaining: 38.2ms
36: learn: 0.2382324 total: 22.2ms remaining: 37.7ms
37: learn: 0.2348878 total: 23.1ms remaining: 37.7ms
38: learn: 0.2327583 total: 23.7ms remaining: 37.1ms
39: learn: 0.2315955 total: 24.3ms remaining: 36.4ms
40: learn: 0.2295426 total: 25ms remaining: 36ms
41: learn: 0.2272825 total: 25.7ms remaining: 35.5ms
42: learn: 0.2246312 total: 26.4ms remaining: 35ms
43: learn: 0.2213682 total: 27.1ms remaining: 34.5ms
44: learn: 0.2189293 total: 27.8ms remaining: 33.9ms
45: learn: 0.2179273 total: 28.2ms remaining: 33.1ms
46: learn: 0.2159202 total: 28.7ms remaining: 32.3ms
47: learn: 0.2133740 total: 29.3ms remaining: 31.7ms
48: learn: 0.2098472 total: 30.1ms remaining: 31.4ms
49: learn: 0.2068154 total: 30.8ms remaining: 30.8ms
50: learn: 0.2061113 total: 31.4ms remaining: 30.2ms
51: learn: 0.2037805 total: 32.2ms remaining: 29.7ms
52: learn: 0.2026674 total: 33.2ms remaining: 29.4ms
53: learn: 0.2010886 total: 33.6ms remaining: 28.6ms
54: learn: 0.1994794 total: 33.9ms remaining: 27.7ms
55: learn: 0.1984573 total: 34.5ms remaining: 27.1ms
56: learn: 0.1959629 total: 35.4ms remaining: 26.7ms
57: learn: 0.1939351 total: 37.1ms remaining: 26.8ms
58: learn: 0.1915941 total: 37.5ms remaining: 26.1ms
59: learn: 0.1898296 total: 37.9ms remaining: 25.3ms
60: learn: 0.1881750 total: 38.5ms remaining: 24.6ms
61: learn: 0.1844752 total: 39.1ms remaining: 24ms
62: learn: 0.1821605 total: 39.8ms remaining: 23.4ms
63: learn: 0.1796806 total: 40.2ms remaining: 22.6ms
64: learn: 0.1776474 total: 40.6ms remaining: 21.9ms
65: learn: 0.1750052 total: 41ms remaining: 21.1ms
66: learn: 0.1731621 total: 41.5ms remaining: 20.4ms
67: learn: 0.1719016 total: 42.2ms remaining: 19.9ms
68: learn: 0.1695609 total: 42.7ms remaining: 19.2ms
69: learn: 0.1691065 total: 43.6ms remaining: 18.7ms
70: learn: 0.1679148 total: 44ms remaining: 18ms
71: learn: 0.1663014 total: 44.5ms remaining: 17.3ms
72: learn: 0.1641277 total: 45.1ms remaining: 16.7ms
73: learn: 0.1630318 total: 46ms remaining: 16.2ms
74: learn: 0.1612617 total: 46.6ms remaining: 15.5ms
75: learn: 0.1590640 total: 47.1ms remaining: 14.9ms
76: learn: 0.1579279 total: 48.7ms remaining: 14.5ms
77: learn: 0.1567992 total: 49.2ms remaining: 13.9ms
78: learn: 0.1547183 total: 49.6ms remaining: 13.2ms
79: learn: 0.1532674 total: 50.4ms remaining: 12.6ms
80: learn: 0.1520756 total: 50.8ms remaining: 11.9ms
81: learn: 0.1507511 total: 51.9ms remaining: 11.4ms
82: learn: 0.1495220 total: 52.4ms remaining: 10.7ms
83: learn: 0.1487438 total: 52.8ms remaining: 10.1ms
84: learn: 0.1479020 total: 53.4ms remaining: 9.43ms
85: learn: 0.1462077 total: 55.1ms remaining: 8.97ms
86: learn: 0.1452365 total: 55.7ms remaining: 8.32ms
87: learn: 0.1436059 total: 56.3ms remaining: 7.68ms
88: learn: 0.1422820 total: 56.8ms remaining: 7.02ms
89: learn: 0.1412853 total: 57.7ms remaining: 6.41ms
90: learn: 0.1398929 total: 58.2ms remaining: 5.75ms
91: learn: 0.1389708 total: 59.1ms remaining: 5.14ms
92: learn: 0.1376155 total: 59.8ms remaining: 4.5ms
93: learn: 0.1369376 total: 60.7ms remaining: 3.88ms
94: learn: 0.1360997 total: 61.2ms remaining: 3.22ms
95: learn: 0.1353171 total: 61.9ms remaining: 2.58ms
96: learn: 0.1347899 total: 63.5ms remaining: 1.96ms
97: learn: 0.1333686 total: 64.9ms remaining: 1.32ms
98: learn: 0.1321242 total: 65.3ms remaining: 659us
99: learn: 0.1311064 total: 66.3ms remaining: 0us
0: learn: 0.6522701 total: 890us remaining: 88.1ms
1: learn: 0.6036416 total: 1.57ms remaining: 76.8ms
2: learn: 0.5640355 total: 2.18ms remaining: 70.5ms
3: learn: 0.5322241 total: 2.79ms remaining: 66.9ms
4: learn: 0.5010241 total: 3.41ms remaining: 64.8ms
5: learn: 0.4775931 total: 4.23ms remaining: 66.3ms
6: learn: 0.4563549 total: 5ms remaining: 66.5ms
7: learn: 0.4387096 total: 5.97ms remaining: 68.7ms
8: learn: 0.4244739 total: 6.52ms remaining: 66ms
9: learn: 0.4063645 total: 7.11ms remaining: 64ms
10: learn: 0.3924550 total: 7.61ms remaining: 61.6ms
11: learn: 0.3783817 total: 8.05ms remaining: 59ms
12: learn: 0.3647220 total: 8.82ms remaining: 59.1ms
13: learn: 0.3577826 total: 9.23ms remaining: 56.7ms
14: learn: 0.3494593 total: 9.89ms remaining: 56.1ms
15: learn: 0.3400580 total: 10.7ms remaining: 56.4ms
16: learn: 0.3285507 total: 11.6ms remaining: 56.6ms
17: learn: 0.3217872 total: 12.1ms remaining: 55.1ms
18: learn: 0.3149769 total: 12.6ms remaining: 53.8ms
19: learn: 0.3077400 total: 13.1ms remaining: 52.5ms
20: learn: 0.3042261 total: 13.5ms remaining: 50.9ms
21: learn: 0.2983474 total: 14.1ms remaining: 49.9ms
22: learn: 0.2942262 total: 14.8ms remaining: 49.4ms
23: learn: 0.2882372 total: 15.4ms remaining: 48.8ms
24: learn: 0.2851206 total: 16ms remaining: 48ms
25: learn: 0.2815644 total: 16.5ms remaining: 47ms
26: learn: 0.2776790 total: 17.3ms remaining: 46.7ms
27: learn: 0.2734958 total: 17.9ms remaining: 46.1ms
28: learn: 0.2701786 total: 18.5ms remaining: 45.3ms
29: learn: 0.2667674 total: 19.1ms remaining: 44.7ms
30: learn: 0.2630019 total: 19.7ms remaining: 43.8ms
31: learn: 0.2581845 total: 20.3ms remaining: 43.1ms
32: learn: 0.2556435 total: 20.7ms remaining: 42.1ms
33: learn: 0.2538021 total: 21.4ms remaining: 41.6ms
34: learn: 0.2517359 total: 21.9ms remaining: 40.8ms
35: learn: 0.2485945 total: 22.5ms remaining: 40ms
36: learn: 0.2465127 total: 22.7ms remaining: 38.7ms
37: learn: 0.2438727 total: 23.4ms remaining: 38.2ms
38: learn: 0.2420634 total: 24ms remaining: 37.5ms
39: learn: 0.2404796 total: 24.6ms remaining: 36.8ms
40: learn: 0.2388046 total: 25ms remaining: 36ms
41: learn: 0.2371937 total: 25.4ms remaining: 35.1ms
42: learn: 0.2338179 total: 26ms remaining: 34.5ms
43: learn: 0.2308034 total: 26.5ms remaining: 33.7ms
44: learn: 0.2280634 total: 26.9ms remaining: 32.8ms
45: learn: 0.2253802 total: 27.4ms remaining: 32.2ms
46: learn: 0.2243953 total: 27.9ms remaining: 31.4ms
47: learn: 0.2231214 total: 28.3ms remaining: 30.6ms
48: learn: 0.2208584 total: 28.9ms remaining: 30ms
49: learn: 0.2188559 total: 29.4ms remaining: 29.4ms
50: learn: 0.2175474 total: 29.9ms remaining: 28.7ms
51: learn: 0.2166235 total: 30.5ms remaining: 28.1ms
52: learn: 0.2144388 total: 31ms remaining: 27.5ms
53: learn: 0.2121554 total: 31.5ms remaining: 26.8ms
54: learn: 0.2098647 total: 32ms remaining: 26.2ms
55: learn: 0.2086553 total: 32.5ms remaining: 25.5ms
56: learn: 0.2069467 total: 33ms remaining: 24.9ms
57: learn: 0.2057201 total: 33.6ms remaining: 24.3ms
58: learn: 0.2027529 total: 34.2ms remaining: 23.8ms
59: learn: 0.2004828 total: 34.8ms remaining: 23.2ms
60: learn: 0.1988509 total: 35.4ms remaining: 22.7ms
61: learn: 0.1970354 total: 36ms remaining: 22.1ms
62: learn: 0.1947051 total: 36.5ms remaining: 21.4ms
63: learn: 0.1939672 total: 36.8ms remaining: 20.7ms
64: learn: 0.1920435 total: 37.3ms remaining: 20.1ms
65: learn: 0.1903993 total: 37.7ms remaining: 19.4ms
66: learn: 0.1887878 total: 38.3ms remaining: 18.9ms
67: learn: 0.1874798 total: 38.9ms remaining: 18.3ms
68: learn: 0.1854338 total: 39.4ms remaining: 17.7ms
69: learn: 0.1844063 total: 39.9ms remaining: 17.1ms
70: learn: 0.1834861 total: 40.5ms remaining: 16.6ms
71: learn: 0.1815721 total: 41ms remaining: 16ms
72: learn: 0.1802420 total: 41.6ms remaining: 15.4ms
73: learn: 0.1790963 total: 42.1ms remaining: 14.8ms
74: learn: 0.1770389 total: 42.5ms remaining: 14.2ms
75: learn: 0.1754908 total: 43ms remaining: 13.6ms
76: learn: 0.1745791 total: 43.6ms remaining: 13ms
77: learn: 0.1729327 total: 44.1ms remaining: 12.4ms
78: learn: 0.1715906 total: 44.6ms remaining: 11.9ms
79: learn: 0.1699566 total: 45.1ms remaining: 11.3ms
80: learn: 0.1686836 total: 45.6ms remaining: 10.7ms
81: learn: 0.1681575 total: 46.1ms remaining: 10.1ms
82: learn: 0.1671318 total: 46.6ms remaining: 9.54ms
83: learn: 0.1660038 total: 47ms remaining: 8.94ms
84: learn: 0.1645107 total: 47.5ms remaining: 8.37ms
85: learn: 0.1633423 total: 47.9ms remaining: 7.8ms
86: learn: 0.1622611 total: 48.3ms remaining: 7.22ms
87: learn: 0.1613205 total: 48.7ms remaining: 6.64ms
88: learn: 0.1600571 total: 49.2ms remaining: 6.09ms
89: learn: 0.1587184 total: 49.7ms remaining: 5.52ms
90: learn: 0.1577681 total: 50ms remaining: 4.95ms
91: learn: 0.1567456 total: 50.6ms remaining: 4.4ms
92: learn: 0.1557140 total: 52.6ms remaining: 3.96ms
93: learn: 0.1546785 total: 53.1ms remaining: 3.39ms
94: learn: 0.1538591 total: 53.6ms remaining: 2.82ms
95: learn: 0.1531077 total: 54ms remaining: 2.25ms
96: learn: 0.1520755 total: 54.5ms remaining: 1.68ms
97: learn: 0.1512373 total: 55.1ms remaining: 1.12ms
98: learn: 0.1502051 total: 57ms remaining: 575us
99: learn: 0.1496630 total: 58.1ms remaining: 0us
0: learn: 0.6451451 total: 755us remaining: 74.8ms
1: learn: 0.6042992 total: 1.68ms remaining: 82.5ms
2: learn: 0.5723961 total: 2.29ms remaining: 74ms
3: learn: 0.5505362 total: 2.93ms remaining: 70.4ms
4: learn: 0.5203715 total: 3.4ms remaining: 64.5ms
5: learn: 0.4949788 total: 4.11ms remaining: 64.4ms
6: learn: 0.4705068 total: 4.63ms remaining: 61.5ms
7: learn: 0.4529208 total: 5.18ms remaining: 59.6ms
8: learn: 0.4367037 total: 5.77ms remaining: 58.4ms
9: learn: 0.4220916 total: 6.49ms remaining: 58.5ms
10: learn: 0.4072312 total: 7.04ms remaining: 56.9ms
11: learn: 0.3918750 total: 7.61ms remaining: 55.8ms
12: learn: 0.3797389 total: 8.41ms remaining: 56.3ms
13: learn: 0.3713153 total: 9.01ms remaining: 55.4ms
14: learn: 0.3621565 total: 9.65ms remaining: 54.7ms
15: learn: 0.3513609 total: 10.1ms remaining: 53ms
16: learn: 0.3437247 total: 10.9ms remaining: 53.4ms
17: learn: 0.3380817 total: 11.6ms remaining: 52.9ms
18: learn: 0.3312335 total: 12.2ms remaining: 52ms
19: learn: 0.3250979 total: 13ms remaining: 52.2ms
20: learn: 0.3185907 total: 13.9ms remaining: 52.3ms
21: learn: 0.3126966 total: 14.7ms remaining: 52ms
22: learn: 0.3073101 total: 15.3ms remaining: 51.4ms
23: learn: 0.3028373 total: 15.9ms remaining: 50.4ms
24: learn: 0.2980124 total: 16.5ms remaining: 49.6ms
25: learn: 0.2929595 total: 17.4ms remaining: 49.6ms
26: learn: 0.2896203 total: 18.5ms remaining: 50.1ms
27: learn: 0.2835319 total: 19.2ms remaining: 49.4ms
28: learn: 0.2797981 total: 19.9ms remaining: 48.7ms
29: learn: 0.2771595 total: 20.5ms remaining: 47.9ms
30: learn: 0.2729367 total: 21.1ms remaining: 47ms
31: learn: 0.2688679 total: 21.8ms remaining: 46.2ms
32: learn: 0.2653477 total: 22.5ms remaining: 45.8ms
33: learn: 0.2611816 total: 23.2ms remaining: 45ms
34: learn: 0.2576321 total: 23.7ms remaining: 44.1ms
35: learn: 0.2548372 total: 24.4ms remaining: 43.4ms
36: learn: 0.2529773 total: 24.8ms remaining: 42.2ms
37: learn: 0.2487806 total: 25.4ms remaining: 41.5ms
38: learn: 0.2461394 total: 26ms remaining: 40.7ms
39: learn: 0.2428244 total: 26.5ms remaining: 39.8ms
40: learn: 0.2400398 total: 27ms remaining: 38.9ms
41: learn: 0.2380082 total: 27.6ms remaining: 38.1ms
42: learn: 0.2364277 total: 28.5ms remaining: 37.7ms
43: learn: 0.2332589 total: 29ms remaining: 36.9ms
44: learn: 0.2325072 total: 29.5ms remaining: 36.1ms
45: learn: 0.2297279 total: 30.1ms remaining: 35.3ms
46: learn: 0.2262849 total: 30.7ms remaining: 34.7ms
47: learn: 0.2240035 total: 31.3ms remaining: 33.9ms
48: learn: 0.2228309 total: 31.9ms remaining: 33.2ms
49: learn: 0.2212052 total: 32.6ms remaining: 32.6ms
50: learn: 0.2185792 total: 33.2ms remaining: 31.9ms
51: learn: 0.2170915 total: 33.8ms remaining: 31.2ms
52: learn: 0.2151489 total: 34.6ms remaining: 30.7ms
53: learn: 0.2126566 total: 35.2ms remaining: 30ms
54: learn: 0.2119384 total: 36ms remaining: 29.4ms
55: learn: 0.2099443 total: 36.6ms remaining: 28.8ms
56: learn: 0.2089844 total: 37.3ms remaining: 28.1ms
57: learn: 0.2064028 total: 38.2ms remaining: 27.6ms
58: learn: 0.2045883 total: 39ms remaining: 27.1ms
59: learn: 0.2026679 total: 40.2ms remaining: 26.8ms
60: learn: 0.2010630 total: 41ms remaining: 26.2ms
61: learn: 0.1989599 total: 41.7ms remaining: 25.5ms
62: learn: 0.1978796 total: 42.5ms remaining: 25ms
63: learn: 0.1962388 total: 43.3ms remaining: 24.4ms
64: learn: 0.1941716 total: 44.2ms remaining: 23.8ms
65: learn: 0.1931220 total: 45.1ms remaining: 23.2ms
66: learn: 0.1917293 total: 45.9ms remaining: 22.6ms
67: learn: 0.1903946 total: 46.8ms remaining: 22ms
68: learn: 0.1885611 total: 47.6ms remaining: 21.4ms
69: learn: 0.1865977 total: 48.2ms remaining: 20.7ms
70: learn: 0.1850735 total: 48.9ms remaining: 20ms
71: learn: 0.1830569 total: 49.8ms remaining: 19.4ms
72: learn: 0.1815147 total: 50.4ms remaining: 18.6ms
73: learn: 0.1789950 total: 51.3ms remaining: 18ms
74: learn: 0.1781168 total: 51.9ms remaining: 17.3ms
75: learn: 0.1769358 total: 52.5ms remaining: 16.6ms
76: learn: 0.1752666 total: 53.4ms remaining: 15.9ms
77: learn: 0.1739681 total: 53.9ms remaining: 15.2ms
78: learn: 0.1730544 total: 55ms remaining: 14.6ms
79: learn: 0.1718053 total: 55.6ms remaining: 13.9ms
80: learn: 0.1707527 total: 57ms remaining: 13.4ms
81: learn: 0.1699461 total: 57.7ms remaining: 12.7ms
82: learn: 0.1690114 total: 58.3ms remaining: 11.9ms
83: learn: 0.1677074 total: 59ms remaining: 11.2ms
84: learn: 0.1667715 total: 59.6ms remaining: 10.5ms
85: learn: 0.1659646 total: 60.2ms remaining: 9.8ms
86: learn: 0.1648152 total: 61ms remaining: 9.11ms
87: learn: 0.1637020 total: 61.8ms remaining: 8.42ms
88: learn: 0.1627011 total: 62.5ms remaining: 7.72ms
89: learn: 0.1614639 total: 63.6ms remaining: 7.06ms
90: learn: 0.1603660 total: 64.9ms remaining: 6.41ms
91: learn: 0.1590917 total: 65.7ms remaining: 5.71ms
92: learn: 0.1582291 total: 66.6ms remaining: 5.01ms
93: learn: 0.1576251 total: 67.6ms remaining: 4.31ms
94: learn: 0.1566021 total: 68.3ms remaining: 3.6ms
95: learn: 0.1558731 total: 69ms remaining: 2.87ms
96: learn: 0.1550613 total: 69.9ms remaining: 2.16ms
97: learn: 0.1539704 total: 70.8ms remaining: 1.44ms
98: learn: 0.1531952 total: 71.5ms remaining: 721us
99: learn: 0.1519390 total: 72.2ms remaining: 0us
0: learn: 0.6447701 total: 764us remaining: 75.7ms
1: learn: 0.6056301 total: 1.07ms remaining: 52.3ms
2: learn: 0.5733978 total: 1.53ms remaining: 49.3ms
3: learn: 0.5441743 total: 1.98ms remaining: 47.4ms
4: learn: 0.5177609 total: 2.49ms remaining: 47.3ms
5: learn: 0.4945935 total: 3.38ms remaining: 53ms
6: learn: 0.4688536 total: 3.98ms remaining: 52.9ms
7: learn: 0.4504243 total: 4.58ms remaining: 52.6ms
8: learn: 0.4310441 total: 5.21ms remaining: 52.6ms
9: learn: 0.4211420 total: 6.06ms remaining: 54.6ms
10: learn: 0.4040153 total: 6.8ms remaining: 55ms
11: learn: 0.3899990 total: 7.42ms remaining: 54.4ms
12: learn: 0.3811531 total: 7.86ms remaining: 52.6ms
13: learn: 0.3741547 total: 8.51ms remaining: 52.3ms
14: learn: 0.3608915 total: 9.13ms remaining: 51.7ms
15: learn: 0.3514996 total: 9.73ms remaining: 51.1ms
16: learn: 0.3453344 total: 9.99ms remaining: 48.8ms
17: learn: 0.3362904 total: 10.7ms remaining: 48.9ms
18: learn: 0.3315063 total: 11.3ms remaining: 48ms
19: learn: 0.3234206 total: 11.8ms remaining: 47.2ms
20: learn: 0.3207093 total: 12.6ms remaining: 47.3ms
21: learn: 0.3149737 total: 12.9ms remaining: 45.8ms
22: learn: 0.3079959 total: 14ms remaining: 46.9ms
23: learn: 0.3030842 total: 14.6ms remaining: 46.1ms
24: learn: 0.2982382 total: 15.2ms remaining: 45.7ms
25: learn: 0.2940517 total: 15.9ms remaining: 45.2ms
26: learn: 0.2880557 total: 16.5ms remaining: 44.7ms
27: learn: 0.2842384 total: 17.2ms remaining: 44.2ms
28: learn: 0.2790874 total: 17.8ms remaining: 43.6ms
29: learn: 0.2742982 total: 18.6ms remaining: 43.5ms
30: learn: 0.2681622 total: 19.2ms remaining: 42.8ms
31: learn: 0.2664259 total: 19.8ms remaining: 42.1ms
32: learn: 0.2647500 total: 20.3ms remaining: 41.3ms
33: learn: 0.2628096 total: 20.9ms remaining: 40.7ms
34: learn: 0.2588150 total: 21.6ms remaining: 40.2ms
35: learn: 0.2561726 total: 22.2ms remaining: 39.5ms
36: learn: 0.2541944 total: 22.9ms remaining: 39ms
37: learn: 0.2513833 total: 23.6ms remaining: 38.4ms
38: learn: 0.2476279 total: 24ms remaining: 37.6ms
39: learn: 0.2456028 total: 24.5ms remaining: 36.7ms
40: learn: 0.2430387 total: 24.9ms remaining: 35.9ms
41: learn: 0.2411154 total: 25.4ms remaining: 35.1ms
42: learn: 0.2384670 total: 26ms remaining: 34.5ms
43: learn: 0.2342631 total: 26.5ms remaining: 33.7ms
44: learn: 0.2306784 total: 27ms remaining: 33ms
45: learn: 0.2253848 total: 27.5ms remaining: 32.3ms
46: learn: 0.2227968 total: 27.9ms remaining: 31.5ms
47: learn: 0.2208583 total: 28.4ms remaining: 30.7ms
48: learn: 0.2187812 total: 28.8ms remaining: 29.9ms
49: learn: 0.2158569 total: 29.2ms remaining: 29.2ms
50: learn: 0.2130382 total: 29.6ms remaining: 28.4ms
51: learn: 0.2102869 total: 30.1ms remaining: 27.7ms
52: learn: 0.2087435 total: 30.5ms remaining: 27ms
53: learn: 0.2074826 total: 30.9ms remaining: 26.3ms
54: learn: 0.2061974 total: 31.4ms remaining: 25.6ms
55: learn: 0.2044226 total: 31.7ms remaining: 24.9ms
56: learn: 0.2032657 total: 32.1ms remaining: 24.2ms
57: learn: 0.2015775 total: 32.6ms remaining: 23.6ms
58: learn: 0.1991958 total: 33.1ms remaining: 23ms
59: learn: 0.1965103 total: 33.6ms remaining: 22.4ms
60: learn: 0.1941532 total: 34.1ms remaining: 21.8ms
61: learn: 0.1912086 total: 34.5ms remaining: 21.2ms
62: learn: 0.1891089 total: 34.9ms remaining: 20.5ms
63: learn: 0.1861982 total: 35.3ms remaining: 19.9ms
64: learn: 0.1842951 total: 35.8ms remaining: 19.3ms
65: learn: 0.1812502 total: 36.1ms remaining: 18.6ms
66: learn: 0.1785166 total: 36.6ms remaining: 18ms
67: learn: 0.1767820 total: 37.3ms remaining: 17.5ms
68: learn: 0.1749276 total: 37.9ms remaining: 17ms
69: learn: 0.1737319 total: 38.3ms remaining: 16.4ms
70: learn: 0.1717897 total: 38.8ms remaining: 15.8ms
71: learn: 0.1699087 total: 39.3ms remaining: 15.3ms
72: learn: 0.1678962 total: 39.8ms remaining: 14.7ms
73: learn: 0.1662050 total: 40.4ms remaining: 14.2ms
74: learn: 0.1653409 total: 40.8ms remaining: 13.6ms
75: learn: 0.1645030 total: 41.3ms remaining: 13ms
76: learn: 0.1634063 total: 41.7ms remaining: 12.5ms
77: learn: 0.1620137 total: 42.1ms remaining: 11.9ms
78: learn: 0.1606836 total: 42.5ms remaining: 11.3ms
79: learn: 0.1587245 total: 43ms remaining: 10.7ms
80: learn: 0.1565554 total: 43.4ms remaining: 10.2ms
81: learn: 0.1546546 total: 43.8ms remaining: 9.62ms
82: learn: 0.1530069 total: 44.3ms remaining: 9.06ms
83: learn: 0.1512483 total: 44.8ms remaining: 8.52ms
84: learn: 0.1493381 total: 45.2ms remaining: 7.97ms
85: learn: 0.1485220 total: 45.6ms remaining: 7.42ms
86: learn: 0.1467454 total: 46ms remaining: 6.88ms
87: learn: 0.1459096 total: 46.4ms remaining: 6.33ms
88: learn: 0.1450653 total: 46.9ms remaining: 5.79ms
89: learn: 0.1434247 total: 47.3ms remaining: 5.26ms
90: learn: 0.1424792 total: 47.8ms remaining: 4.72ms
91: learn: 0.1413129 total: 48.2ms remaining: 4.19ms
92: learn: 0.1402986 total: 48.6ms remaining: 3.66ms
93: learn: 0.1381899 total: 49.2ms remaining: 3.14ms
94: learn: 0.1369743 total: 51ms remaining: 2.68ms
95: learn: 0.1360709 total: 51.5ms remaining: 2.15ms
96: learn: 0.1344922 total: 52ms remaining: 1.61ms
97: learn: 0.1337746 total: 52.6ms remaining: 1.07ms
98: learn: 0.1324802 total: 53.1ms remaining: 536us
99: learn: 0.1313830 total: 53.7ms remaining: 0us
0: learn: 0.6458849 total: 510us remaining: 50.5ms
1: learn: 0.6133520 total: 865us remaining: 42.4ms
2: learn: 0.5788935 total: 1.18ms remaining: 38.3ms
3: learn: 0.5506121 total: 1.6ms remaining: 38.3ms
4: learn: 0.5188294 total: 1.91ms remaining: 36.3ms
5: learn: 0.4954000 total: 2.28ms remaining: 35.7ms
6: learn: 0.4734426 total: 2.76ms remaining: 36.7ms
7: learn: 0.4556870 total: 3.14ms remaining: 36.1ms
8: learn: 0.4339884 total: 3.51ms remaining: 35.5ms
9: learn: 0.4206988 total: 4.13ms remaining: 37.2ms
10: learn: 0.4079334 total: 4.55ms remaining: 36.8ms
11: learn: 0.3911875 total: 4.96ms remaining: 36.3ms
12: learn: 0.3788150 total: 5.51ms remaining: 36.8ms
13: learn: 0.3691222 total: 6.02ms remaining: 37ms
14: learn: 0.3599406 total: 6.43ms remaining: 36.4ms
15: learn: 0.3492508 total: 6.56ms remaining: 34.5ms
16: learn: 0.3372736 total: 7.01ms remaining: 34.2ms
17: learn: 0.3290084 total: 7.28ms remaining: 33.2ms
18: learn: 0.3184555 total: 7.84ms remaining: 33.4ms
19: learn: 0.3135967 total: 8.22ms remaining: 32.9ms
20: learn: 0.3097415 total: 8.74ms remaining: 32.9ms
21: learn: 0.3035698 total: 9.2ms remaining: 32.6ms
22: learn: 0.3004913 total: 9.51ms remaining: 31.8ms
23: learn: 0.2958960 total: 9.96ms remaining: 31.5ms
24: learn: 0.2910836 total: 10.6ms remaining: 31.7ms
25: learn: 0.2872216 total: 10.9ms remaining: 31ms
26: learn: 0.2824063 total: 11.3ms remaining: 30.5ms
27: learn: 0.2793386 total: 11.7ms remaining: 30.1ms
28: learn: 0.2753716 total: 12ms remaining: 29.4ms
29: learn: 0.2718755 total: 12.4ms remaining: 28.8ms
30: learn: 0.2690465 total: 12.7ms remaining: 28.4ms
31: learn: 0.2671851 total: 13.1ms remaining: 27.8ms
32: learn: 0.2623224 total: 13.5ms remaining: 27.5ms
33: learn: 0.2594564 total: 13.9ms remaining: 27ms
34: learn: 0.2556683 total: 14.4ms remaining: 26.8ms
35: learn: 0.2514976 total: 15ms remaining: 26.7ms
36: learn: 0.2476577 total: 15.4ms remaining: 26.3ms
37: learn: 0.2463112 total: 15.8ms remaining: 25.8ms
38: learn: 0.2431250 total: 16.2ms remaining: 25.4ms
39: learn: 0.2414802 total: 16.6ms remaining: 24.8ms
40: learn: 0.2386433 total: 17.1ms remaining: 24.6ms
41: learn: 0.2367208 total: 17.5ms remaining: 24.1ms
42: learn: 0.2339689 total: 17.9ms remaining: 23.7ms
43: learn: 0.2313215 total: 18.3ms remaining: 23.3ms
44: learn: 0.2287363 total: 18.6ms remaining: 22.8ms
45: learn: 0.2278756 total: 19ms remaining: 22.3ms
46: learn: 0.2254814 total: 19.3ms remaining: 21.8ms
47: learn: 0.2239846 total: 19.7ms remaining: 21.4ms
48: learn: 0.2226685 total: 20.1ms remaining: 21ms
49: learn: 0.2202107 total: 20.6ms remaining: 20.6ms
50: learn: 0.2171960 total: 20.9ms remaining: 20.1ms
51: learn: 0.2151221 total: 21.2ms remaining: 19.6ms
52: learn: 0.2140390 total: 21.5ms remaining: 19.1ms
53: learn: 0.2115634 total: 21.9ms remaining: 18.6ms
54: learn: 0.2094185 total: 22.2ms remaining: 18.2ms
55: learn: 0.2070625 total: 22.6ms remaining: 17.8ms
56: learn: 0.2052080 total: 23.1ms remaining: 17.4ms
57: learn: 0.2029810 total: 23.4ms remaining: 17ms
58: learn: 0.2012912 total: 23.7ms remaining: 16.5ms
59: learn: 0.1993082 total: 24.1ms remaining: 16.1ms
60: learn: 0.1981063 total: 24.6ms remaining: 15.7ms
61: learn: 0.1951256 total: 24.9ms remaining: 15.3ms
62: learn: 0.1943817 total: 25.2ms remaining: 14.8ms
63: learn: 0.1926787 total: 25.7ms remaining: 14.5ms
64: learn: 0.1911710 total: 26.1ms remaining: 14.1ms
65: learn: 0.1886861 total: 26.5ms remaining: 13.6ms
66: learn: 0.1863373 total: 26.9ms remaining: 13.2ms
67: learn: 0.1850114 total: 27.4ms remaining: 12.9ms
68: learn: 0.1831335 total: 27.8ms remaining: 12.5ms
69: learn: 0.1817557 total: 28.1ms remaining: 12.1ms
70: learn: 0.1810677 total: 28.5ms remaining: 11.6ms
71: learn: 0.1794123 total: 29.1ms remaining: 11.3ms
72: learn: 0.1781799 total: 29.6ms remaining: 10.9ms
73: learn: 0.1765436 total: 29.9ms remaining: 10.5ms
74: learn: 0.1751594 total: 30.3ms remaining: 10.1ms
75: learn: 0.1738585 total: 30.6ms remaining: 9.67ms
76: learn: 0.1727655 total: 31.1ms remaining: 9.3ms
77: learn: 0.1714937 total: 31.6ms remaining: 8.91ms
78: learn: 0.1703010 total: 32.1ms remaining: 8.53ms
79: learn: 0.1691029 total: 32.5ms remaining: 8.14ms
80: learn: 0.1675268 total: 32.9ms remaining: 7.71ms
81: learn: 0.1657979 total: 33.4ms remaining: 7.32ms
82: learn: 0.1642409 total: 33.7ms remaining: 6.9ms
83: learn: 0.1622945 total: 34.1ms remaining: 6.49ms
84: learn: 0.1614470 total: 34.7ms remaining: 6.12ms
85: learn: 0.1604327 total: 35.1ms remaining: 5.71ms
86: learn: 0.1595932 total: 35.5ms remaining: 5.3ms
87: learn: 0.1583188 total: 36ms remaining: 4.91ms
88: learn: 0.1567842 total: 36.6ms remaining: 4.52ms
89: learn: 0.1552409 total: 37.1ms remaining: 4.12ms
90: learn: 0.1535638 total: 37.7ms remaining: 3.73ms
91: learn: 0.1526073 total: 38.5ms remaining: 3.35ms
92: learn: 0.1519107 total: 39ms remaining: 2.93ms
93: learn: 0.1511137 total: 39.3ms remaining: 2.51ms
94: learn: 0.1506922 total: 39.8ms remaining: 2.09ms
95: learn: 0.1491531 total: 40.2ms remaining: 1.68ms
96: learn: 0.1478150 total: 40.7ms remaining: 1.26ms
97: learn: 0.1467851 total: 41.2ms remaining: 840us
98: learn: 0.1456745 total: 41.7ms remaining: 421us
99: learn: 0.1451717 total: 42.1ms remaining: 0us
0: learn: 0.6561469 total: 548us remaining: 54.3ms
1: learn: 0.6141428 total: 1.02ms remaining: 49.8ms
2: learn: 0.5806953 total: 1.41ms remaining: 45.5ms
3: learn: 0.5532450 total: 1.8ms remaining: 43.3ms
4: learn: 0.5284830 total: 2.31ms remaining: 44ms
5: learn: 0.5031883 total: 2.79ms remaining: 43.8ms
6: learn: 0.4813054 total: 3.08ms remaining: 40.9ms
7: learn: 0.4639089 total: 3.47ms remaining: 39.9ms
8: learn: 0.4440443 total: 3.83ms remaining: 38.7ms
9: learn: 0.4251806 total: 4.25ms remaining: 38.3ms
10: learn: 0.4131648 total: 4.58ms remaining: 37ms
11: learn: 0.3995829 total: 4.92ms remaining: 36.1ms
12: learn: 0.3895909 total: 5.24ms remaining: 35.1ms
13: learn: 0.3790789 total: 5.63ms remaining: 34.6ms
14: learn: 0.3702250 total: 5.94ms remaining: 33.7ms
15: learn: 0.3628179 total: 6.3ms remaining: 33.1ms
16: learn: 0.3521130 total: 6.68ms remaining: 32.6ms
17: learn: 0.3455921 total: 7.01ms remaining: 31.9ms
18: learn: 0.3399362 total: 7.29ms remaining: 31.1ms
19: learn: 0.3315420 total: 7.67ms remaining: 30.7ms
20: learn: 0.3265923 total: 8.01ms remaining: 30.1ms
21: learn: 0.3224172 total: 8.4ms remaining: 29.8ms
22: learn: 0.3211601 total: 8.7ms remaining: 29.1ms
23: learn: 0.3154900 total: 9ms remaining: 28.5ms
24: learn: 0.3131349 total: 9.44ms remaining: 28.3ms
25: learn: 0.3068798 total: 9.79ms remaining: 27.9ms
26: learn: 0.3016954 total: 10.2ms remaining: 27.5ms
27: learn: 0.2983040 total: 10.5ms remaining: 27ms
28: learn: 0.2934145 total: 10.8ms remaining: 26.5ms
29: learn: 0.2892725 total: 11.2ms remaining: 26.2ms
30: learn: 0.2831620 total: 11.6ms remaining: 25.8ms
31: learn: 0.2787553 total: 11.9ms remaining: 25.2ms
32: learn: 0.2756261 total: 12.3ms remaining: 24.9ms
33: learn: 0.2716675 total: 12.7ms remaining: 24.7ms
34: learn: 0.2672679 total: 13.2ms remaining: 24.4ms
35: learn: 0.2655949 total: 13.4ms remaining: 23.9ms
36: learn: 0.2626896 total: 13.8ms remaining: 23.4ms
37: learn: 0.2603952 total: 14.1ms remaining: 23.1ms
38: learn: 0.2570534 total: 14.5ms remaining: 22.6ms
39: learn: 0.2542723 total: 14.9ms remaining: 22.3ms
40: learn: 0.2506732 total: 15.4ms remaining: 22.2ms
41: learn: 0.2464743 total: 15.9ms remaining: 21.9ms
42: learn: 0.2426782 total: 16.3ms remaining: 21.5ms
43: learn: 0.2401948 total: 16.7ms remaining: 21.3ms
44: learn: 0.2385713 total: 17.3ms remaining: 21.1ms
45: learn: 0.2362299 total: 17.8ms remaining: 20.9ms
46: learn: 0.2326157 total: 18.3ms remaining: 20.6ms
47: learn: 0.2316490 total: 18.8ms remaining: 20.3ms
48: learn: 0.2282742 total: 19.2ms remaining: 20ms
49: learn: 0.2270968 total: 19.8ms remaining: 19.8ms
50: learn: 0.2259638 total: 20ms remaining: 19.2ms
51: learn: 0.2247657 total: 20.2ms remaining: 18.6ms
52: learn: 0.2233193 total: 20.6ms remaining: 18.3ms
53: learn: 0.2205222 total: 21ms remaining: 17.9ms
54: learn: 0.2192851 total: 21.5ms remaining: 17.6ms
55: learn: 0.2182013 total: 22ms remaining: 17.3ms
56: learn: 0.2167010 total: 22.5ms remaining: 17ms
57: learn: 0.2147984 total: 23.1ms remaining: 16.7ms
58: learn: 0.2128245 total: 23.5ms remaining: 16.3ms
59: learn: 0.2109271 total: 24ms remaining: 16ms
60: learn: 0.2088641 total: 24.6ms remaining: 15.7ms
61: learn: 0.2064593 total: 25.1ms remaining: 15.4ms
62: learn: 0.2051058 total: 25.5ms remaining: 15ms
63: learn: 0.2024714 total: 26ms remaining: 14.6ms
64: learn: 0.2005225 total: 26.6ms remaining: 14.3ms
65: learn: 0.1987558 total: 27.1ms remaining: 14ms
66: learn: 0.1968005 total: 27.8ms remaining: 13.7ms
67: learn: 0.1948838 total: 28.3ms remaining: 13.3ms
68: learn: 0.1932047 total: 28.7ms remaining: 12.9ms
69: learn: 0.1912417 total: 29.2ms remaining: 12.5ms
70: learn: 0.1887524 total: 29.9ms remaining: 12.2ms
71: learn: 0.1874792 total: 30.5ms remaining: 11.9ms
72: learn: 0.1859013 total: 31ms remaining: 11.5ms
73: learn: 0.1848770 total: 31.5ms remaining: 11.1ms
74: learn: 0.1837501 total: 31.9ms remaining: 10.6ms
75: learn: 0.1819594 total: 32.4ms remaining: 10.2ms
76: learn: 0.1812631 total: 32.9ms remaining: 9.82ms
77: learn: 0.1801700 total: 33.3ms remaining: 9.39ms
78: learn: 0.1784495 total: 33.8ms remaining: 8.98ms
79: learn: 0.1778314 total: 34.4ms remaining: 8.59ms
80: learn: 0.1764189 total: 34.9ms remaining: 8.19ms
81: learn: 0.1747795 total: 35.4ms remaining: 7.76ms
82: learn: 0.1740287 total: 36.2ms remaining: 7.41ms
83: learn: 0.1725595 total: 36.7ms remaining: 7ms
84: learn: 0.1714423 total: 37.2ms remaining: 6.57ms
85: learn: 0.1699679 total: 37.7ms remaining: 6.14ms
86: learn: 0.1691006 total: 38.2ms remaining: 5.71ms
87: learn: 0.1683647 total: 38.6ms remaining: 5.27ms
88: learn: 0.1679061 total: 39.1ms remaining: 4.83ms
89: learn: 0.1671002 total: 39.5ms remaining: 4.39ms
90: learn: 0.1654977 total: 40ms remaining: 3.95ms
91: learn: 0.1646972 total: 40.5ms remaining: 3.52ms
92: learn: 0.1640330 total: 40.9ms remaining: 3.08ms
93: learn: 0.1629333 total: 41.3ms remaining: 2.64ms
94: learn: 0.1616853 total: 41.8ms remaining: 2.2ms
95: learn: 0.1606318 total: 42.2ms remaining: 1.76ms
96: learn: 0.1601456 total: 42.6ms remaining: 1.32ms
97: learn: 0.1593405 total: 42.9ms remaining: 876us
98: learn: 0.1586283 total: 43.5ms remaining: 439us
99: learn: 0.1572542 total: 43.9ms remaining: 0us
0: learn: 0.6616346 total: 538us remaining: 53.3ms
1: learn: 0.6109973 total: 884us remaining: 43.3ms
2: learn: 0.5722396 total: 1.19ms remaining: 38.4ms
3: learn: 0.5408827 total: 1.56ms remaining: 37.5ms
4: learn: 0.5094934 total: 1.91ms remaining: 36.2ms
5: learn: 0.4906131 total: 2.29ms remaining: 35.8ms
6: learn: 0.4645653 total: 2.7ms remaining: 35.9ms
7: learn: 0.4444164 total: 2.83ms remaining: 32.5ms
8: learn: 0.4272058 total: 3.15ms remaining: 31.9ms
9: learn: 0.4115717 total: 3.46ms remaining: 31.2ms
10: learn: 0.3950856 total: 3.79ms remaining: 30.6ms
11: learn: 0.3835814 total: 4.17ms remaining: 30.6ms
12: learn: 0.3704893 total: 4.33ms remaining: 29ms
13: learn: 0.3607335 total: 4.71ms remaining: 28.9ms
14: learn: 0.3545259 total: 5.11ms remaining: 28.9ms
15: learn: 0.3461680 total: 5.5ms remaining: 28.9ms
16: learn: 0.3363363 total: 5.83ms remaining: 28.5ms
17: learn: 0.3290670 total: 6.19ms remaining: 28.2ms
18: learn: 0.3213655 total: 6.5ms remaining: 27.7ms
19: learn: 0.3164629 total: 6.9ms remaining: 27.6ms
20: learn: 0.3102984 total: 7.21ms remaining: 27.1ms
21: learn: 0.3031531 total: 7.59ms remaining: 26.9ms
22: learn: 0.2953113 total: 7.89ms remaining: 26.4ms
23: learn: 0.2912851 total: 8.19ms remaining: 25.9ms
24: learn: 0.2842226 total: 8.69ms remaining: 26.1ms
25: learn: 0.2815780 total: 9.05ms remaining: 25.7ms
26: learn: 0.2782092 total: 9.39ms remaining: 25.4ms
27: learn: 0.2749687 total: 9.62ms remaining: 24.7ms
28: learn: 0.2712762 total: 9.87ms remaining: 24.2ms
29: learn: 0.2651409 total: 10.2ms remaining: 23.8ms
30: learn: 0.2625774 total: 10.5ms remaining: 23.3ms
31: learn: 0.2607545 total: 10.7ms remaining: 22.8ms
32: learn: 0.2581615 total: 10.9ms remaining: 22.1ms
33: learn: 0.2530686 total: 11.2ms remaining: 21.8ms
34: learn: 0.2482771 total: 11.6ms remaining: 21.6ms
35: learn: 0.2443547 total: 12ms remaining: 21.3ms
36: learn: 0.2390791 total: 12.4ms remaining: 21.1ms
37: learn: 0.2359834 total: 12.7ms remaining: 20.7ms
38: learn: 0.2340394 total: 13ms remaining: 20.3ms
39: learn: 0.2312033 total: 13.3ms remaining: 20ms
40: learn: 0.2302284 total: 13.7ms remaining: 19.8ms
41: learn: 0.2263697 total: 14.2ms remaining: 19.6ms
42: learn: 0.2228828 total: 14.6ms remaining: 19.3ms
43: learn: 0.2206554 total: 15.1ms remaining: 19.2ms
44: learn: 0.2177805 total: 15.5ms remaining: 19ms
45: learn: 0.2166365 total: 15.7ms remaining: 18.4ms
46: learn: 0.2147782 total: 16.2ms remaining: 18.3ms
47: learn: 0.2117934 total: 16.6ms remaining: 18ms
48: learn: 0.2099487 total: 17.1ms remaining: 17.8ms
49: learn: 0.2071163 total: 17.4ms remaining: 17.4ms
50: learn: 0.2046263 total: 17.8ms remaining: 17.1ms
51: learn: 0.2024450 total: 18.4ms remaining: 17ms
52: learn: 0.2005754 total: 19ms remaining: 16.8ms
53: learn: 0.1981363 total: 19.4ms remaining: 16.5ms
54: learn: 0.1961990 total: 19.8ms remaining: 16.2ms
55: learn: 0.1937006 total: 20.2ms remaining: 15.8ms
56: learn: 0.1910742 total: 20.5ms remaining: 15.5ms
57: learn: 0.1882669 total: 21ms remaining: 15.2ms
58: learn: 0.1860093 total: 21.5ms remaining: 14.9ms
59: learn: 0.1837245 total: 22ms remaining: 14.7ms
60: learn: 0.1810336 total: 22.4ms remaining: 14.3ms
61: learn: 0.1795047 total: 22.8ms remaining: 14ms
62: learn: 0.1769841 total: 23.3ms remaining: 13.7ms
63: learn: 0.1749374 total: 23.8ms remaining: 13.4ms
64: learn: 0.1731419 total: 24.3ms remaining: 13.1ms
65: learn: 0.1711514 total: 24.7ms remaining: 12.7ms
66: learn: 0.1691857 total: 25.2ms remaining: 12.4ms
67: learn: 0.1674547 total: 25.6ms remaining: 12.1ms
68: learn: 0.1654834 total: 26.1ms remaining: 11.7ms
69: learn: 0.1642283 total: 26.6ms remaining: 11.4ms
70: learn: 0.1625059 total: 27.2ms remaining: 11.1ms
71: learn: 0.1601459 total: 27.8ms remaining: 10.8ms
72: learn: 0.1581288 total: 28.2ms remaining: 10.4ms
73: learn: 0.1575355 total: 28.6ms remaining: 10.1ms
74: learn: 0.1563107 total: 29ms remaining: 9.68ms
75: learn: 0.1549076 total: 29.6ms remaining: 9.34ms
76: learn: 0.1526753 total: 30.3ms remaining: 9.04ms
77: learn: 0.1515277 total: 30.9ms remaining: 8.71ms
78: learn: 0.1496484 total: 31.4ms remaining: 8.34ms
79: learn: 0.1485007 total: 31.8ms remaining: 7.96ms
80: learn: 0.1461037 total: 32.3ms remaining: 7.58ms
81: learn: 0.1445475 total: 32.9ms remaining: 7.23ms
82: learn: 0.1432127 total: 33.4ms remaining: 6.85ms
83: learn: 0.1414773 total: 34.1ms remaining: 6.49ms
84: learn: 0.1398074 total: 34.5ms remaining: 6.09ms
85: learn: 0.1385001 total: 35ms remaining: 5.7ms
86: learn: 0.1373486 total: 35.4ms remaining: 5.3ms
87: learn: 0.1362773 total: 36ms remaining: 4.9ms
88: learn: 0.1355164 total: 36.5ms remaining: 4.51ms
89: learn: 0.1343375 total: 37ms remaining: 4.11ms
90: learn: 0.1337581 total: 37.5ms remaining: 3.71ms
91: learn: 0.1325804 total: 38ms remaining: 3.3ms
92: learn: 0.1316402 total: 38.4ms remaining: 2.89ms
93: learn: 0.1304202 total: 38.8ms remaining: 2.48ms
94: learn: 0.1298267 total: 39.3ms remaining: 2.07ms
95: learn: 0.1285233 total: 39.9ms remaining: 1.66ms
96: learn: 0.1271916 total: 40.5ms remaining: 1.25ms
97: learn: 0.1264605 total: 41ms remaining: 836us
98: learn: 0.1252049 total: 41.5ms remaining: 419us
99: learn: 0.1244674 total: 42ms remaining: 0us
0: learn: 0.6589850 total: 479us remaining: 47.5ms
1: learn: 0.6173268 total: 815us remaining: 40ms
2: learn: 0.5819228 total: 1.09ms remaining: 35.4ms
3: learn: 0.5574915 total: 1.46ms remaining: 34.9ms
4: learn: 0.5332271 total: 1.77ms remaining: 33.7ms
5: learn: 0.5109981 total: 2.09ms remaining: 32.7ms
6: learn: 0.4817409 total: 2.41ms remaining: 32ms
7: learn: 0.4578100 total: 2.65ms remaining: 30.5ms
8: learn: 0.4400720 total: 3.31ms remaining: 33.5ms
9: learn: 0.4247016 total: 3.83ms remaining: 34.5ms
10: learn: 0.4074585 total: 4.42ms remaining: 35.7ms
11: learn: 0.3976352 total: 4.81ms remaining: 35.3ms
12: learn: 0.3866986 total: 5.24ms remaining: 35ms
13: learn: 0.3755725 total: 5.75ms remaining: 35.3ms
14: learn: 0.3623562 total: 6.23ms remaining: 35.3ms
15: learn: 0.3535372 total: 6.65ms remaining: 34.9ms
16: learn: 0.3441112 total: 7.17ms remaining: 35ms
17: learn: 0.3373060 total: 7.58ms remaining: 34.5ms
18: learn: 0.3273168 total: 8.59ms remaining: 36.6ms
19: learn: 0.3180001 total: 9.04ms remaining: 36.2ms
20: learn: 0.3132445 total: 9.39ms remaining: 35.3ms
21: learn: 0.3055298 total: 9.8ms remaining: 34.7ms
22: learn: 0.3002391 total: 10.2ms remaining: 34.2ms
23: learn: 0.2953327 total: 10.6ms remaining: 33.7ms
24: learn: 0.2890278 total: 11ms remaining: 33ms
25: learn: 0.2838117 total: 11.5ms remaining: 32.7ms
26: learn: 0.2787583 total: 12ms remaining: 32.5ms
27: learn: 0.2758002 total: 12.4ms remaining: 31.8ms
28: learn: 0.2709398 total: 12.8ms remaining: 31.4ms
29: learn: 0.2657668 total: 13.2ms remaining: 30.9ms
30: learn: 0.2618131 total: 13.6ms remaining: 30.3ms
31: learn: 0.2588815 total: 14.2ms remaining: 30.1ms
32: learn: 0.2559394 total: 14.6ms remaining: 29.7ms
33: learn: 0.2534781 total: 15.2ms remaining: 29.4ms
34: learn: 0.2504927 total: 15.6ms remaining: 29ms
35: learn: 0.2487456 total: 16ms remaining: 28.5ms
36: learn: 0.2457432 total: 16.4ms remaining: 28ms
37: learn: 0.2436861 total: 16.9ms remaining: 27.6ms
38: learn: 0.2404680 total: 17.4ms remaining: 27.2ms
39: learn: 0.2375523 total: 17.8ms remaining: 26.8ms
40: learn: 0.2342454 total: 18.4ms remaining: 26.4ms
41: learn: 0.2318136 total: 18.8ms remaining: 26ms
42: learn: 0.2288749 total: 19.3ms remaining: 25.6ms
43: learn: 0.2265562 total: 19.7ms remaining: 25.1ms
44: learn: 0.2232778 total: 20.1ms remaining: 24.5ms
45: learn: 0.2213184 total: 20.4ms remaining: 24ms
46: learn: 0.2187823 total: 20.9ms remaining: 23.5ms
47: learn: 0.2166353 total: 21.3ms remaining: 23ms
48: learn: 0.2147026 total: 21.8ms remaining: 22.7ms
49: learn: 0.2128187 total: 22.2ms remaining: 22.2ms
50: learn: 0.2101182 total: 22.8ms remaining: 21.9ms
51: learn: 0.2092353 total: 23.2ms remaining: 21.4ms
52: learn: 0.2075377 total: 23.6ms remaining: 20.9ms
53: learn: 0.2066530 total: 24ms remaining: 20.5ms
54: learn: 0.2044811 total: 24.5ms remaining: 20ms
55: learn: 0.2026730 total: 24.9ms remaining: 19.5ms
56: learn: 0.2000305 total: 25.3ms remaining: 19.1ms
57: learn: 0.1984260 total: 25.7ms remaining: 18.6ms
58: learn: 0.1976436 total: 26ms remaining: 18.1ms
59: learn: 0.1950815 total: 26.5ms remaining: 17.7ms
60: learn: 0.1929250 total: 27ms remaining: 17.3ms
61: learn: 0.1915891 total: 27.5ms remaining: 16.8ms
62: learn: 0.1893576 total: 27.9ms remaining: 16.4ms
63: learn: 0.1881031 total: 28.3ms remaining: 15.9ms
64: learn: 0.1867330 total: 28.8ms remaining: 15.5ms
65: learn: 0.1853646 total: 29.3ms remaining: 15.1ms
66: learn: 0.1837686 total: 29.9ms remaining: 14.7ms
67: learn: 0.1826417 total: 30.4ms remaining: 14.3ms
68: learn: 0.1813746 total: 30.9ms remaining: 13.9ms
69: learn: 0.1800007 total: 31.3ms remaining: 13.4ms
70: learn: 0.1777328 total: 31.7ms remaining: 13ms
71: learn: 0.1767782 total: 32.2ms remaining: 12.5ms
72: learn: 0.1754490 total: 32.8ms remaining: 12.1ms
73: learn: 0.1736202 total: 33.3ms remaining: 11.7ms
74: learn: 0.1723229 total: 33.7ms remaining: 11.2ms
75: learn: 0.1711417 total: 34.2ms remaining: 10.8ms
76: learn: 0.1696771 total: 34.7ms remaining: 10.4ms
77: learn: 0.1685262 total: 35.3ms remaining: 9.94ms
78: learn: 0.1676439 total: 35.7ms remaining: 9.48ms
79: learn: 0.1667190 total: 36.2ms remaining: 9.04ms
80: learn: 0.1653152 total: 36.7ms remaining: 8.61ms
81: learn: 0.1644322 total: 37.1ms remaining: 8.15ms
82: learn: 0.1635745 total: 37.6ms remaining: 7.69ms
83: learn: 0.1622066 total: 37.9ms remaining: 7.22ms
84: learn: 0.1609589 total: 38.3ms remaining: 6.75ms
85: learn: 0.1598233 total: 38.6ms remaining: 6.29ms
86: learn: 0.1592661 total: 39.1ms remaining: 5.84ms
87: learn: 0.1584185 total: 39.6ms remaining: 5.4ms
88: learn: 0.1572580 total: 40.1ms remaining: 4.95ms
89: learn: 0.1561033 total: 40.4ms remaining: 4.49ms
90: learn: 0.1553269 total: 41ms remaining: 4.05ms
91: learn: 0.1538309 total: 41.4ms remaining: 3.6ms
92: learn: 0.1524678 total: 41.9ms remaining: 3.15ms
93: learn: 0.1516504 total: 42.4ms remaining: 2.7ms
94: learn: 0.1512200 total: 42.8ms remaining: 2.25ms
95: learn: 0.1499684 total: 43.1ms remaining: 1.8ms
96: learn: 0.1494016 total: 43.7ms remaining: 1.35ms
97: learn: 0.1488526 total: 44.2ms remaining: 901us
98: learn: 0.1482695 total: 44.6ms remaining: 450us
99: learn: 0.1475814 total: 45ms remaining: 0us
0: learn: 0.6452274 total: 661us remaining: 65.5ms
1: learn: 0.6007395 total: 1.22ms remaining: 59.6ms
2: learn: 0.5606860 total: 1.7ms remaining: 54.9ms
3: learn: 0.5356117 total: 2.21ms remaining: 53ms
4: learn: 0.5040476 total: 2.94ms remaining: 55.8ms
5: learn: 0.4782473 total: 3.61ms remaining: 56.5ms
6: learn: 0.4579484 total: 4.29ms remaining: 57ms
7: learn: 0.4339063 total: 4.87ms remaining: 56ms
8: learn: 0.4136929 total: 5.6ms remaining: 56.6ms
9: learn: 0.3964760 total: 6.1ms remaining: 54.9ms
10: learn: 0.3776982 total: 6.55ms remaining: 53ms
11: learn: 0.3631631 total: 7.08ms remaining: 52ms
12: learn: 0.3538404 total: 7.66ms remaining: 51.3ms
13: learn: 0.3402280 total: 8.54ms remaining: 52.5ms
14: learn: 0.3279063 total: 8.97ms remaining: 50.8ms
15: learn: 0.3190464 total: 9.26ms remaining: 48.6ms
16: learn: 0.3131871 total: 9.47ms remaining: 46.3ms
17: learn: 0.3048667 total: 9.78ms remaining: 44.5ms
18: learn: 0.2987186 total: 10.1ms remaining: 43ms
19: learn: 0.2904161 total: 11.1ms remaining: 44.5ms
20: learn: 0.2830408 total: 11.6ms remaining: 43.6ms
21: learn: 0.2757885 total: 12.6ms remaining: 44.6ms
22: learn: 0.2687860 total: 13ms remaining: 43.4ms
23: learn: 0.2623586 total: 13.3ms remaining: 42.2ms
24: learn: 0.2581471 total: 13.8ms remaining: 41.5ms
25: learn: 0.2539348 total: 14.5ms remaining: 41.4ms
26: learn: 0.2503573 total: 14.8ms remaining: 40ms
27: learn: 0.2460689 total: 15.1ms remaining: 38.8ms
28: learn: 0.2428545 total: 15.2ms remaining: 37.2ms
29: learn: 0.2395749 total: 15.5ms remaining: 36.1ms
30: learn: 0.2357687 total: 16ms remaining: 35.6ms
31: learn: 0.2310186 total: 16.6ms remaining: 35.3ms
32: learn: 0.2269526 total: 17ms remaining: 34.5ms
33: learn: 0.2230483 total: 17.6ms remaining: 34.2ms
34: learn: 0.2196256 total: 18.2ms remaining: 33.8ms
35: learn: 0.2164008 total: 18.8ms remaining: 33.4ms
36: learn: 0.2136322 total: 19.1ms remaining: 32.6ms
37: learn: 0.2115325 total: 19.7ms remaining: 32.1ms
38: learn: 0.2080654 total: 20.2ms remaining: 31.6ms
39: learn: 0.2058333 total: 20.6ms remaining: 30.9ms
40: learn: 0.2037221 total: 21ms remaining: 30.2ms
41: learn: 0.2015290 total: 21.3ms remaining: 29.4ms
42: learn: 0.1996680 total: 21.7ms remaining: 28.7ms
43: learn: 0.1985694 total: 22ms remaining: 28.1ms
44: learn: 0.1962434 total: 22.6ms remaining: 27.6ms
45: learn: 0.1949811 total: 23ms remaining: 27ms
46: learn: 0.1933688 total: 23.3ms remaining: 26.3ms
47: learn: 0.1904367 total: 23.7ms remaining: 25.7ms
48: learn: 0.1886177 total: 24.1ms remaining: 25.1ms
49: learn: 0.1877138 total: 24.5ms remaining: 24.5ms
50: learn: 0.1855138 total: 24.9ms remaining: 23.9ms
51: learn: 0.1845785 total: 25.3ms remaining: 23.4ms
52: learn: 0.1824223 total: 25.7ms remaining: 22.8ms
53: learn: 0.1799806 total: 26ms remaining: 22.2ms
54: learn: 0.1788668 total: 26.3ms remaining: 21.5ms
55: learn: 0.1779603 total: 26.6ms remaining: 20.9ms
56: learn: 0.1759896 total: 27.1ms remaining: 20.5ms
57: learn: 0.1748177 total: 27.8ms remaining: 20.1ms
58: learn: 0.1727186 total: 28.5ms remaining: 19.8ms
59: learn: 0.1711060 total: 29.1ms remaining: 19.4ms
60: learn: 0.1698123 total: 29.9ms remaining: 19.1ms
61: learn: 0.1689338 total: 30.7ms remaining: 18.8ms
62: learn: 0.1674283 total: 31.5ms remaining: 18.5ms
63: learn: 0.1656366 total: 32.5ms remaining: 18.3ms
64: learn: 0.1640729 total: 33.1ms remaining: 17.8ms
65: learn: 0.1620183 total: 34ms remaining: 17.5ms
66: learn: 0.1608405 total: 34.7ms remaining: 17.1ms
67: learn: 0.1600532 total: 35.7ms remaining: 16.8ms
68: learn: 0.1585397 total: 36.9ms remaining: 16.6ms
69: learn: 0.1574716 total: 37.5ms remaining: 16.1ms
70: learn: 0.1562461 total: 38.6ms remaining: 15.8ms
71: learn: 0.1551565 total: 39.3ms remaining: 15.3ms
72: learn: 0.1541272 total: 39.9ms remaining: 14.8ms
73: learn: 0.1530555 total: 40.7ms remaining: 14.3ms
74: learn: 0.1517833 total: 42ms remaining: 14ms
75: learn: 0.1507274 total: 42.4ms remaining: 13.4ms
76: learn: 0.1502550 total: 42.8ms remaining: 12.8ms
77: learn: 0.1492409 total: 43.1ms remaining: 12.2ms
78: learn: 0.1470354 total: 43.6ms remaining: 11.6ms
79: learn: 0.1461855 total: 44.2ms remaining: 11.1ms
80: learn: 0.1450661 total: 45.2ms remaining: 10.6ms
81: learn: 0.1441868 total: 45.8ms remaining: 10ms
82: learn: 0.1428044 total: 46.5ms remaining: 9.53ms
83: learn: 0.1411136 total: 47.4ms remaining: 9.03ms
84: learn: 0.1399459 total: 48.3ms remaining: 8.52ms
85: learn: 0.1384995 total: 48.8ms remaining: 7.94ms
86: learn: 0.1375867 total: 49.2ms remaining: 7.36ms
87: learn: 0.1366461 total: 49.9ms remaining: 6.8ms
88: learn: 0.1352229 total: 50.3ms remaining: 6.22ms
89: learn: 0.1337635 total: 50.9ms remaining: 5.66ms
90: learn: 0.1328132 total: 51.5ms remaining: 5.09ms
91: learn: 0.1321318 total: 52.1ms remaining: 4.53ms
92: learn: 0.1312117 total: 52.6ms remaining: 3.96ms
93: learn: 0.1300240 total: 53ms remaining: 3.38ms
94: learn: 0.1292502 total: 53.6ms remaining: 2.82ms
95: learn: 0.1285517 total: 54.2ms remaining: 2.26ms
96: learn: 0.1278014 total: 54.6ms remaining: 1.69ms
97: learn: 0.1267940 total: 55.2ms remaining: 1.13ms
98: learn: 0.1255733 total: 55.7ms remaining: 562us
99: learn: 0.1247385 total: 56.1ms remaining: 0us
0: learn: 0.6413457 total: 539us remaining: 53.4ms
1: learn: 0.5900578 total: 813us remaining: 39.9ms
2: learn: 0.5519438 total: 1.06ms remaining: 34.3ms
3: learn: 0.5243187 total: 1.33ms remaining: 32.1ms
4: learn: 0.4950511 total: 1.59ms remaining: 30.2ms
5: learn: 0.4709211 total: 1.81ms remaining: 28.4ms
6: learn: 0.4495564 total: 2.1ms remaining: 27.9ms
7: learn: 0.4249429 total: 2.47ms remaining: 28.4ms
8: learn: 0.4057863 total: 2.74ms remaining: 27.7ms
9: learn: 0.3912050 total: 3.08ms remaining: 27.7ms
10: learn: 0.3781296 total: 3.5ms remaining: 28.3ms
11: learn: 0.3678321 total: 3.77ms remaining: 27.7ms
12: learn: 0.3538277 total: 4.24ms remaining: 28.4ms
13: learn: 0.3401897 total: 4.62ms remaining: 28.4ms
14: learn: 0.3271568 total: 5.05ms remaining: 28.6ms
15: learn: 0.3166289 total: 5.38ms remaining: 28.3ms
16: learn: 0.3094882 total: 5.57ms remaining: 27.2ms
17: learn: 0.3004746 total: 6.01ms remaining: 27.4ms
18: learn: 0.2952407 total: 6.36ms remaining: 27.1ms
19: learn: 0.2889081 total: 6.67ms remaining: 26.7ms
20: learn: 0.2864670 total: 6.95ms remaining: 26.1ms
21: learn: 0.2819508 total: 7.28ms remaining: 25.8ms
22: learn: 0.2767701 total: 7.61ms remaining: 25.5ms
23: learn: 0.2728949 total: 8.01ms remaining: 25.4ms
24: learn: 0.2690209 total: 8.44ms remaining: 25.3ms
25: learn: 0.2661912 total: 8.75ms remaining: 24.9ms
26: learn: 0.2623100 total: 9.07ms remaining: 24.5ms
27: learn: 0.2602555 total: 9.36ms remaining: 24.1ms
28: learn: 0.2581525 total: 9.58ms remaining: 23.5ms
29: learn: 0.2538615 total: 9.98ms remaining: 23.3ms
30: learn: 0.2516232 total: 10.4ms remaining: 23.2ms
31: learn: 0.2491004 total: 10.8ms remaining: 22.9ms
32: learn: 0.2458602 total: 11.2ms remaining: 22.7ms
33: learn: 0.2437416 total: 11.5ms remaining: 22.3ms
34: learn: 0.2418580 total: 12ms remaining: 22.2ms
35: learn: 0.2394861 total: 12.6ms remaining: 22.4ms
36: learn: 0.2365999 total: 13.2ms remaining: 22.4ms
37: learn: 0.2328293 total: 13.7ms remaining: 22.4ms
38: learn: 0.2304114 total: 14.3ms remaining: 22.3ms
39: learn: 0.2277712 total: 14.8ms remaining: 22.2ms
40: learn: 0.2248847 total: 16ms remaining: 23ms
41: learn: 0.2237335 total: 16.6ms remaining: 23ms
42: learn: 0.2203033 total: 17.3ms remaining: 22.9ms
43: learn: 0.2170414 total: 17.9ms remaining: 22.8ms
44: learn: 0.2140231 total: 18.7ms remaining: 22.9ms
45: learn: 0.2131041 total: 19.3ms remaining: 22.6ms
46: learn: 0.2109699 total: 19.9ms remaining: 22.4ms
47: learn: 0.2098659 total: 20.6ms remaining: 22.3ms
48: learn: 0.2092036 total: 21.2ms remaining: 22.1ms
49: learn: 0.2079761 total: 21.8ms remaining: 21.8ms
50: learn: 0.2055578 total: 22.4ms remaining: 21.6ms
51: learn: 0.2030421 total: 23.1ms remaining: 21.3ms
52: learn: 0.2003143 total: 23.8ms remaining: 21.1ms
53: learn: 0.1980229 total: 24.5ms remaining: 20.8ms
54: learn: 0.1971450 total: 25.1ms remaining: 20.5ms
55: learn: 0.1962788 total: 25.7ms remaining: 20.2ms
56: learn: 0.1938905 total: 26.3ms remaining: 19.8ms
57: learn: 0.1926346 total: 26.9ms remaining: 19.4ms
58: learn: 0.1902046 total: 27.4ms remaining: 19ms
59: learn: 0.1894448 total: 27.9ms remaining: 18.6ms
60: learn: 0.1873198 total: 28.6ms remaining: 18.3ms
61: learn: 0.1860127 total: 29.2ms remaining: 17.9ms
62: learn: 0.1846397 total: 29.7ms remaining: 17.4ms
63: learn: 0.1834666 total: 30.2ms remaining: 17ms
64: learn: 0.1822810 total: 30.8ms remaining: 16.6ms
65: learn: 0.1805458 total: 31.4ms remaining: 16.2ms
66: learn: 0.1784709 total: 31.9ms remaining: 15.7ms
67: learn: 0.1770346 total: 32.3ms remaining: 15.2ms
68: learn: 0.1752650 total: 32.7ms remaining: 14.7ms
69: learn: 0.1733905 total: 33.1ms remaining: 14.2ms
70: learn: 0.1718145 total: 33.8ms remaining: 13.8ms
71: learn: 0.1708119 total: 34.3ms remaining: 13.3ms
72: learn: 0.1687135 total: 34.7ms remaining: 12.8ms
73: learn: 0.1670206 total: 35.2ms remaining: 12.4ms
74: learn: 0.1656156 total: 35.7ms remaining: 11.9ms
75: learn: 0.1649156 total: 36.1ms remaining: 11.4ms
76: learn: 0.1632530 total: 36.5ms remaining: 10.9ms
77: learn: 0.1616361 total: 37ms remaining: 10.4ms
78: learn: 0.1608543 total: 37.4ms remaining: 9.95ms
79: learn: 0.1597547 total: 37.7ms remaining: 9.44ms
80: learn: 0.1590644 total: 38.1ms remaining: 8.95ms
81: learn: 0.1584403 total: 38.5ms remaining: 8.45ms
82: learn: 0.1571011 total: 38.9ms remaining: 7.96ms
83: learn: 0.1560923 total: 39.3ms remaining: 7.49ms
84: learn: 0.1551411 total: 39.8ms remaining: 7.03ms
85: learn: 0.1545136 total: 40.5ms remaining: 6.6ms
86: learn: 0.1540241 total: 41.1ms remaining: 6.14ms
87: learn: 0.1533892 total: 41.8ms remaining: 5.7ms
88: learn: 0.1521288 total: 42.4ms remaining: 5.24ms
89: learn: 0.1509581 total: 43ms remaining: 4.78ms
90: learn: 0.1502992 total: 43.5ms remaining: 4.3ms
91: learn: 0.1489227 total: 43.9ms remaining: 3.81ms
92: learn: 0.1479268 total: 44.2ms remaining: 3.33ms
93: learn: 0.1473020 total: 44.6ms remaining: 2.85ms
94: learn: 0.1458495 total: 45ms remaining: 2.37ms
95: learn: 0.1451447 total: 45.3ms remaining: 1.89ms
96: learn: 0.1444871 total: 45.6ms remaining: 1.41ms
97: learn: 0.1434088 total: 46ms remaining: 939us
98: learn: 0.1425895 total: 46.4ms remaining: 469us
99: learn: 0.1417209 total: 46.7ms remaining: 0us
0: learn: 0.6452260 total: 550us remaining: 54.5ms
1: learn: 0.5951641 total: 866us remaining: 42.5ms
2: learn: 0.5576028 total: 1.18ms remaining: 38.1ms
3: learn: 0.5242696 total: 1.44ms remaining: 34.6ms
4: learn: 0.4943754 total: 1.7ms remaining: 32.3ms
5: learn: 0.4725212 total: 1.8ms remaining: 28.3ms
6: learn: 0.4484628 total: 2.14ms remaining: 28.5ms
7: learn: 0.4263461 total: 2.42ms remaining: 27.8ms
8: learn: 0.4072501 total: 3.32ms remaining: 33.6ms
9: learn: 0.3935810 total: 3.62ms remaining: 32.6ms
10: learn: 0.3762641 total: 4.06ms remaining: 32.8ms
11: learn: 0.3641626 total: 4.61ms remaining: 33.8ms
12: learn: 0.3522660 total: 5ms remaining: 33.5ms
13: learn: 0.3433186 total: 5.45ms remaining: 33.5ms
14: learn: 0.3338644 total: 5.96ms remaining: 33.8ms
15: learn: 0.3241537 total: 6.46ms remaining: 33.9ms
16: learn: 0.3143320 total: 6.85ms remaining: 33.5ms
17: learn: 0.3102218 total: 7.27ms remaining: 33.1ms
18: learn: 0.3024684 total: 7.8ms remaining: 33.3ms
19: learn: 0.2942581 total: 8.28ms remaining: 33.1ms
20: learn: 0.2864357 total: 8.84ms remaining: 33.2ms
21: learn: 0.2801061 total: 9.28ms remaining: 32.9ms
22: learn: 0.2754716 total: 9.61ms remaining: 32.2ms
23: learn: 0.2698246 total: 10ms remaining: 31.8ms
24: learn: 0.2647569 total: 10.6ms remaining: 31.7ms
25: learn: 0.2603520 total: 11ms remaining: 31.4ms
26: learn: 0.2562941 total: 11.3ms remaining: 30.6ms
27: learn: 0.2523354 total: 11.7ms remaining: 30ms
28: learn: 0.2471671 total: 12.1ms remaining: 29.7ms
29: learn: 0.2439060 total: 12.6ms remaining: 29.3ms
30: learn: 0.2400371 total: 13ms remaining: 29ms
31: learn: 0.2367053 total: 13.5ms remaining: 28.6ms
32: learn: 0.2346123 total: 13.8ms remaining: 28.1ms
33: learn: 0.2307711 total: 14.2ms remaining: 27.6ms
34: learn: 0.2275036 total: 14.8ms remaining: 27.4ms
35: learn: 0.2242418 total: 15.2ms remaining: 27ms
36: learn: 0.2214850 total: 15.6ms remaining: 26.6ms
37: learn: 0.2196511 total: 16ms remaining: 26.1ms
38: learn: 0.2176325 total: 16.5ms remaining: 25.8ms
39: learn: 0.2149849 total: 16.9ms remaining: 25.4ms
40: learn: 0.2134714 total: 17.3ms remaining: 24.9ms
41: learn: 0.2107307 total: 17.8ms remaining: 24.6ms
42: learn: 0.2093829 total: 18.3ms remaining: 24.2ms
43: learn: 0.2062526 total: 18.7ms remaining: 23.8ms
44: learn: 0.2041734 total: 19.2ms remaining: 23.5ms
45: learn: 0.2034350 total: 19.7ms remaining: 23.2ms
46: learn: 0.2008729 total: 20.2ms remaining: 22.8ms
47: learn: 0.1982945 total: 20.9ms remaining: 22.6ms
48: learn: 0.1964192 total: 21.3ms remaining: 22.2ms
49: learn: 0.1941753 total: 21.9ms remaining: 21.9ms
50: learn: 0.1922923 total: 22.4ms remaining: 21.5ms
51: learn: 0.1915179 total: 23ms remaining: 21.2ms
52: learn: 0.1899475 total: 23.6ms remaining: 20.9ms
53: learn: 0.1887676 total: 24.1ms remaining: 20.5ms
54: learn: 0.1866542 total: 24.5ms remaining: 20.1ms
55: learn: 0.1849546 total: 25.1ms remaining: 19.7ms
56: learn: 0.1835797 total: 25.6ms remaining: 19.3ms
57: learn: 0.1814780 total: 26.3ms remaining: 19ms
58: learn: 0.1797862 total: 26.9ms remaining: 18.7ms
59: learn: 0.1782307 total: 27.5ms remaining: 18.3ms
60: learn: 0.1764140 total: 28.1ms remaining: 18ms
61: learn: 0.1750435 total: 28.8ms remaining: 17.7ms
62: learn: 0.1731903 total: 29.5ms remaining: 17.3ms
63: learn: 0.1718950 total: 30ms remaining: 16.9ms
64: learn: 0.1705192 total: 30.9ms remaining: 16.6ms
65: learn: 0.1694197 total: 31.4ms remaining: 16.2ms
66: learn: 0.1684674 total: 32.2ms remaining: 15.8ms
67: learn: 0.1671599 total: 32.8ms remaining: 15.4ms
68: learn: 0.1660256 total: 33.3ms remaining: 15ms
69: learn: 0.1648235 total: 34ms remaining: 14.6ms
70: learn: 0.1637161 total: 34.5ms remaining: 14.1ms
71: learn: 0.1624292 total: 35.1ms remaining: 13.6ms
72: learn: 0.1611962 total: 35.6ms remaining: 13.2ms
73: learn: 0.1598332 total: 36.1ms remaining: 12.7ms
74: learn: 0.1582758 total: 36.5ms remaining: 12.2ms
75: learn: 0.1562274 total: 37.1ms remaining: 11.7ms
76: learn: 0.1556210 total: 37.6ms remaining: 11.2ms
77: learn: 0.1546046 total: 38.1ms remaining: 10.7ms
78: learn: 0.1530285 total: 38.5ms remaining: 10.2ms
79: learn: 0.1521724 total: 39.1ms remaining: 9.77ms
80: learn: 0.1513836 total: 39.8ms remaining: 9.33ms
81: learn: 0.1498292 total: 40.5ms remaining: 8.88ms
82: learn: 0.1485866 total: 41.1ms remaining: 8.41ms
83: learn: 0.1478196 total: 41.9ms remaining: 7.97ms
84: learn: 0.1466833 total: 42.5ms remaining: 7.5ms
85: learn: 0.1457152 total: 43.3ms remaining: 7.04ms
86: learn: 0.1447497 total: 43.7ms remaining: 6.53ms
87: learn: 0.1438988 total: 44ms remaining: 6ms
88: learn: 0.1430282 total: 44.5ms remaining: 5.49ms
89: learn: 0.1422060 total: 44.8ms remaining: 4.98ms
90: learn: 0.1415122 total: 45.2ms remaining: 4.47ms
91: learn: 0.1401087 total: 45.6ms remaining: 3.97ms
92: learn: 0.1393979 total: 46.1ms remaining: 3.47ms
93: learn: 0.1386233 total: 46.5ms remaining: 2.97ms
94: learn: 0.1372819 total: 46.8ms remaining: 2.46ms
95: learn: 0.1363838 total: 47.3ms remaining: 1.97ms
96: learn: 0.1352899 total: 47.8ms remaining: 1.48ms
97: learn: 0.1346396 total: 48.2ms remaining: 984us
98: learn: 0.1336550 total: 48.7ms remaining: 492us
99: learn: 0.1330828 total: 49.1ms remaining: 0us
0: learn: 0.6489590 total: 1.15ms remaining: 114ms
1: learn: 0.6054729 total: 2.14ms remaining: 105ms
2: learn: 0.5713667 total: 2.89ms remaining: 93.4ms
3: learn: 0.5478161 total: 4.01ms remaining: 96.3ms
4: learn: 0.5216653 total: 5.55ms remaining: 106ms
5: learn: 0.4966537 total: 6.33ms remaining: 99.2ms
6: learn: 0.4782736 total: 6.98ms remaining: 92.7ms
7: learn: 0.4638061 total: 7.65ms remaining: 88ms
8: learn: 0.4486561 total: 8.5ms remaining: 86ms
9: learn: 0.4323845 total: 9.3ms remaining: 83.7ms
10: learn: 0.4182749 total: 9.97ms remaining: 80.7ms
11: learn: 0.4006948 total: 10.3ms remaining: 75.2ms
12: learn: 0.3868310 total: 11.3ms remaining: 75.7ms
13: learn: 0.3766702 total: 12.4ms remaining: 76.2ms
14: learn: 0.3630724 total: 13.4ms remaining: 75.9ms
15: learn: 0.3532909 total: 14.6ms remaining: 76.4ms
16: learn: 0.3404533 total: 15.4ms remaining: 75.1ms
17: learn: 0.3326679 total: 16.1ms remaining: 73.5ms
18: learn: 0.3261320 total: 16.7ms remaining: 71.2ms
19: learn: 0.3193806 total: 17.6ms remaining: 70.3ms
20: learn: 0.3129853 total: 18.9ms remaining: 70.9ms
21: learn: 0.3052955 total: 19.7ms remaining: 70ms
22: learn: 0.2995204 total: 20.4ms remaining: 68.3ms
23: learn: 0.2938979 total: 21.2ms remaining: 67.1ms
24: learn: 0.2905536 total: 22.5ms remaining: 67.5ms
25: learn: 0.2864712 total: 23.2ms remaining: 65.9ms
26: learn: 0.2807762 total: 24ms remaining: 64.9ms
27: learn: 0.2749710 total: 24.9ms remaining: 64.1ms
28: learn: 0.2681340 total: 25.6ms remaining: 62.8ms
29: learn: 0.2656587 total: 26.3ms remaining: 61.4ms
30: learn: 0.2628564 total: 26.9ms remaining: 59.8ms
31: learn: 0.2568914 total: 27.2ms remaining: 57.9ms
32: learn: 0.2537556 total: 27.7ms remaining: 56.2ms
33: learn: 0.2519176 total: 28.2ms remaining: 54.7ms
34: learn: 0.2473640 total: 28.7ms remaining: 53.2ms
35: learn: 0.2450667 total: 29.4ms remaining: 52.2ms
36: learn: 0.2436387 total: 29.8ms remaining: 50.8ms
37: learn: 0.2417667 total: 30.5ms remaining: 49.8ms
38: learn: 0.2383129 total: 32.5ms remaining: 50.9ms
39: learn: 0.2367566 total: 34.9ms remaining: 52.3ms
40: learn: 0.2354297 total: 36.4ms remaining: 52.4ms
41: learn: 0.2339993 total: 38.3ms remaining: 52.9ms
42: learn: 0.2309912 total: 41.3ms remaining: 54.8ms
43: learn: 0.2284946 total: 44ms remaining: 55.9ms
44: learn: 0.2269643 total: 46.4ms remaining: 56.8ms
45: learn: 0.2244732 total: 48ms remaining: 56.4ms
46: learn: 0.2206652 total: 49.1ms remaining: 55.4ms
47: learn: 0.2180727 total: 50.7ms remaining: 55ms
48: learn: 0.2169677 total: 51.2ms remaining: 53.3ms
49: learn: 0.2159717 total: 52.3ms remaining: 52.3ms
50: learn: 0.2147252 total: 53.3ms remaining: 51.2ms
51: learn: 0.2126697 total: 53.8ms remaining: 49.7ms
52: learn: 0.2094491 total: 54.4ms remaining: 48.3ms
53: learn: 0.2059811 total: 55.6ms remaining: 47.3ms
54: learn: 0.2036358 total: 56.2ms remaining: 46ms
55: learn: 0.2030384 total: 57.1ms remaining: 44.9ms
56: learn: 0.2014926 total: 58ms remaining: 43.7ms
57: learn: 0.2003530 total: 58.8ms remaining: 42.6ms
58: learn: 0.1973485 total: 59.6ms remaining: 41.4ms
59: learn: 0.1945387 total: 60.3ms remaining: 40.2ms
60: learn: 0.1923427 total: 61.1ms remaining: 39.1ms
61: learn: 0.1899936 total: 61.7ms remaining: 37.8ms
62: learn: 0.1888055 total: 62.1ms remaining: 36.5ms
63: learn: 0.1868306 total: 62.5ms remaining: 35.2ms
64: learn: 0.1842215 total: 63ms remaining: 33.9ms
65: learn: 0.1830473 total: 63.5ms remaining: 32.7ms
66: learn: 0.1814550 total: 64.3ms remaining: 31.7ms
67: learn: 0.1798772 total: 65.3ms remaining: 30.7ms
68: learn: 0.1777053 total: 66ms remaining: 29.7ms
69: learn: 0.1751818 total: 66.9ms remaining: 28.7ms
70: learn: 0.1733743 total: 67.6ms remaining: 27.6ms
71: learn: 0.1718073 total: 68.1ms remaining: 26.5ms
72: learn: 0.1711001 total: 68.4ms remaining: 25.3ms
73: learn: 0.1692128 total: 68.7ms remaining: 24.2ms
74: learn: 0.1676232 total: 69.1ms remaining: 23ms
75: learn: 0.1664282 total: 69.6ms remaining: 22ms
76: learn: 0.1640009 total: 70ms remaining: 20.9ms
77: learn: 0.1621925 total: 70.3ms remaining: 19.8ms
78: learn: 0.1608005 total: 70.7ms remaining: 18.8ms
79: learn: 0.1593342 total: 71.3ms remaining: 17.8ms
80: learn: 0.1585114 total: 71.9ms remaining: 16.9ms
81: learn: 0.1573162 total: 72.4ms remaining: 15.9ms
82: learn: 0.1564807 total: 72.9ms remaining: 14.9ms
83: learn: 0.1554987 total: 73.3ms remaining: 14ms
84: learn: 0.1542502 total: 74ms remaining: 13.1ms
85: learn: 0.1529965 total: 74.3ms remaining: 12.1ms
86: learn: 0.1517095 total: 74.7ms remaining: 11.2ms
87: learn: 0.1505284 total: 75.2ms remaining: 10.3ms
88: learn: 0.1492240 total: 75.9ms remaining: 9.39ms
89: learn: 0.1480363 total: 76.5ms remaining: 8.51ms
90: learn: 0.1467339 total: 77.3ms remaining: 7.65ms
91: learn: 0.1454709 total: 78.1ms remaining: 6.79ms
92: learn: 0.1444853 total: 78.5ms remaining: 5.91ms
93: learn: 0.1429242 total: 79ms remaining: 5.04ms
94: learn: 0.1421633 total: 79.5ms remaining: 4.18ms
95: learn: 0.1413099 total: 80ms remaining: 3.33ms
96: learn: 0.1404696 total: 80.7ms remaining: 2.49ms
97: learn: 0.1397359 total: 81.1ms remaining: 1.66ms
98: learn: 0.1387876 total: 81.7ms remaining: 824us
99: learn: 0.1382322 total: 82.1ms remaining: 0us
Evaluating param combo 16/16: {'boosted_cb_model__iterations': 100, 'boosted_cb_model__learning_rate': 0.1, 'boosted_cb_model__max_depth': 6, 'boosted_cb_model__num_leaves': 16}
Duplicate found for this combo (hash=9d5b729e51135eafed880be835f768be). Removing old run: eb5ef7998d82471f9d250ba40646e2e2
0: learn: 0.6460564 total: 1.62ms remaining: 161ms
1: learn: 0.6115491 total: 3.82ms remaining: 187ms
2: learn: 0.5773897 total: 6.94ms remaining: 224ms
3: learn: 0.5478549 total: 8.65ms remaining: 208ms
4: learn: 0.5182940 total: 9.87ms remaining: 188ms
5: learn: 0.4881346 total: 10.6ms remaining: 166ms
6: learn: 0.4636411 total: 11.2ms remaining: 149ms
7: learn: 0.4414647 total: 11.8ms remaining: 136ms
8: learn: 0.4212306 total: 12.5ms remaining: 126ms
9: learn: 0.4061705 total: 13.2ms remaining: 119ms
10: learn: 0.3929099 total: 14.1ms remaining: 114ms
11: learn: 0.3795535 total: 16.5ms remaining: 121ms
12: learn: 0.3676262 total: 17ms remaining: 114ms
13: learn: 0.3571708 total: 18.1ms remaining: 111ms
14: learn: 0.3438041 total: 19.4ms remaining: 110ms
15: learn: 0.3339876 total: 20.4ms remaining: 107ms
16: learn: 0.3256147 total: 21.2ms remaining: 104ms
17: learn: 0.3237170 total: 21.5ms remaining: 97.7ms
18: learn: 0.3154076 total: 22.1ms remaining: 94.4ms
19: learn: 0.3071222 total: 22.6ms remaining: 90.5ms
20: learn: 0.2995403 total: 23.5ms remaining: 88.5ms
21: learn: 0.2945462 total: 23.8ms remaining: 84.3ms
22: learn: 0.2896742 total: 24.5ms remaining: 82.1ms
23: learn: 0.2845711 total: 25ms remaining: 79.2ms
24: learn: 0.2800764 total: 25.7ms remaining: 77.1ms
25: learn: 0.2747643 total: 26ms remaining: 74.1ms
26: learn: 0.2706259 total: 26.6ms remaining: 71.9ms
27: learn: 0.2647070 total: 27.4ms remaining: 70.4ms
28: learn: 0.2612732 total: 28.1ms remaining: 68.8ms
29: learn: 0.2569599 total: 29.5ms remaining: 68.9ms
30: learn: 0.2539537 total: 30.3ms remaining: 67.4ms
31: learn: 0.2499569 total: 30.9ms remaining: 65.7ms
32: learn: 0.2464926 total: 31.9ms remaining: 64.8ms
33: learn: 0.2425367 total: 32.6ms remaining: 63.3ms
34: learn: 0.2380646 total: 33.2ms remaining: 61.6ms
35: learn: 0.2359067 total: 34.1ms remaining: 60.6ms
36: learn: 0.2324772 total: 35.4ms remaining: 60.2ms
37: learn: 0.2296507 total: 36ms remaining: 58.7ms
38: learn: 0.2279374 total: 36.3ms remaining: 56.8ms
39: learn: 0.2260783 total: 37.3ms remaining: 55.9ms
40: learn: 0.2234158 total: 37.9ms remaining: 54.5ms
41: learn: 0.2204111 total: 38.5ms remaining: 53.2ms
42: learn: 0.2189710 total: 39.5ms remaining: 52.4ms
43: learn: 0.2167972 total: 40.5ms remaining: 51.5ms
44: learn: 0.2140913 total: 41.3ms remaining: 50.4ms
45: learn: 0.2114060 total: 42ms remaining: 49.3ms
46: learn: 0.2096511 total: 43.1ms remaining: 48.6ms
47: learn: 0.2066832 total: 44.4ms remaining: 48.1ms
48: learn: 0.2035646 total: 44.9ms remaining: 46.8ms
49: learn: 0.2009341 total: 45.6ms remaining: 45.6ms
50: learn: 0.1990334 total: 46.1ms remaining: 44.3ms
51: learn: 0.1965160 total: 47ms remaining: 43.4ms
52: learn: 0.1941812 total: 47.6ms remaining: 42.3ms
53: learn: 0.1922171 total: 49.6ms remaining: 42.3ms
54: learn: 0.1898604 total: 50.3ms remaining: 41.1ms
55: learn: 0.1876379 total: 51.2ms remaining: 40.2ms
56: learn: 0.1858364 total: 51.9ms remaining: 39.1ms
57: learn: 0.1845777 total: 52.6ms remaining: 38.1ms
58: learn: 0.1838015 total: 53.3ms remaining: 37ms
59: learn: 0.1822539 total: 56ms remaining: 37.4ms
60: learn: 0.1816199 total: 56.2ms remaining: 35.9ms
61: learn: 0.1793068 total: 56.8ms remaining: 34.8ms
62: learn: 0.1766040 total: 59ms remaining: 34.6ms
63: learn: 0.1750871 total: 59.7ms remaining: 33.6ms
64: learn: 0.1729869 total: 60.3ms remaining: 32.5ms
65: learn: 0.1714244 total: 61.4ms remaining: 31.6ms
66: learn: 0.1692713 total: 62.7ms remaining: 30.9ms
67: learn: 0.1672872 total: 63.3ms remaining: 29.8ms
68: learn: 0.1656736 total: 64.4ms remaining: 28.9ms
69: learn: 0.1640504 total: 65.2ms remaining: 27.9ms
70: learn: 0.1625983 total: 66.1ms remaining: 27ms
71: learn: 0.1605809 total: 67ms remaining: 26ms
72: learn: 0.1593449 total: 68ms remaining: 25.1ms
73: learn: 0.1577407 total: 69.6ms remaining: 24.5ms
74: learn: 0.1560696 total: 70.5ms remaining: 23.5ms
75: learn: 0.1543792 total: 72.3ms remaining: 22.8ms
76: learn: 0.1530379 total: 74.6ms remaining: 22.3ms
77: learn: 0.1519702 total: 76.1ms remaining: 21.5ms
78: learn: 0.1504854 total: 77.9ms remaining: 20.7ms
79: learn: 0.1488243 total: 79.4ms remaining: 19.8ms
80: learn: 0.1471932 total: 80.3ms remaining: 18.8ms
81: learn: 0.1456928 total: 81.1ms remaining: 17.8ms
82: learn: 0.1448080 total: 83.6ms remaining: 17.1ms
83: learn: 0.1432590 total: 84.8ms remaining: 16.1ms
84: learn: 0.1420120 total: 88.1ms remaining: 15.5ms
85: learn: 0.1411562 total: 89.6ms remaining: 14.6ms
86: learn: 0.1397316 total: 91.5ms remaining: 13.7ms
87: learn: 0.1388008 total: 92.3ms remaining: 12.6ms
88: learn: 0.1375409 total: 94.3ms remaining: 11.7ms
89: learn: 0.1367936 total: 95ms remaining: 10.6ms
90: learn: 0.1356756 total: 96ms remaining: 9.49ms
91: learn: 0.1345972 total: 96.8ms remaining: 8.42ms
92: learn: 0.1338271 total: 97.8ms remaining: 7.36ms
93: learn: 0.1332789 total: 98.5ms remaining: 6.29ms
94: learn: 0.1324934 total: 99.2ms remaining: 5.22ms
95: learn: 0.1314454 total: 100ms remaining: 4.17ms
96: learn: 0.1306761 total: 101ms remaining: 3.11ms
97: learn: 0.1298523 total: 102ms remaining: 2.07ms
98: learn: 0.1284715 total: 103ms remaining: 1.04ms
99: learn: 0.1275061 total: 104ms remaining: 0us
0: learn: 0.6424726 total: 697us remaining: 69.1ms
1: learn: 0.5994503 total: 1.01ms remaining: 49.6ms
2: learn: 0.5650641 total: 1.5ms remaining: 48.6ms
3: learn: 0.5330205 total: 1.62ms remaining: 39ms
4: learn: 0.5040646 total: 2.13ms remaining: 40.5ms
5: learn: 0.4756208 total: 2.62ms remaining: 41ms
6: learn: 0.4556726 total: 3.61ms remaining: 48ms
7: learn: 0.4312891 total: 4.63ms remaining: 53.3ms
8: learn: 0.4153181 total: 5.14ms remaining: 52ms
9: learn: 0.4013725 total: 6.43ms remaining: 57.9ms
10: learn: 0.3841636 total: 7.12ms remaining: 57.6ms
11: learn: 0.3713251 total: 7.3ms remaining: 53.5ms
12: learn: 0.3553684 total: 7.83ms remaining: 52.4ms
13: learn: 0.3468555 total: 9ms remaining: 55.3ms
14: learn: 0.3368721 total: 10ms remaining: 56.8ms
15: learn: 0.3250555 total: 10.7ms remaining: 56.4ms
16: learn: 0.3135326 total: 12ms remaining: 58.6ms
17: learn: 0.3068917 total: 12.6ms remaining: 57.4ms
18: learn: 0.2983682 total: 13.3ms remaining: 56.7ms
19: learn: 0.2903195 total: 15.3ms remaining: 61ms
20: learn: 0.2825822 total: 16.1ms remaining: 60.4ms
21: learn: 0.2786885 total: 16.5ms remaining: 58.7ms
22: learn: 0.2713611 total: 17.4ms remaining: 58.4ms
23: learn: 0.2655422 total: 18.3ms remaining: 58.1ms
24: learn: 0.2595446 total: 18.9ms remaining: 56.8ms
25: learn: 0.2553193 total: 19.5ms remaining: 55.6ms
26: learn: 0.2485124 total: 20.3ms remaining: 54.9ms
27: learn: 0.2449750 total: 21.5ms remaining: 55.3ms
28: learn: 0.2407880 total: 22.1ms remaining: 54.2ms
29: learn: 0.2379387 total: 22.7ms remaining: 53ms
30: learn: 0.2331728 total: 23.1ms remaining: 51.5ms
31: learn: 0.2301211 total: 23.6ms remaining: 50.2ms
32: learn: 0.2274073 total: 24.4ms remaining: 49.6ms
33: learn: 0.2239642 total: 25.2ms remaining: 48.9ms
34: learn: 0.2218696 total: 25.7ms remaining: 47.8ms
35: learn: 0.2190630 total: 26.9ms remaining: 47.8ms
36: learn: 0.2162074 total: 27.5ms remaining: 46.9ms
37: learn: 0.2142116 total: 28.3ms remaining: 46.2ms
38: learn: 0.2122932 total: 29.6ms remaining: 46.3ms
39: learn: 0.2092948 total: 30.4ms remaining: 45.6ms
40: learn: 0.2081077 total: 30.9ms remaining: 44.5ms
41: learn: 0.2036558 total: 31.5ms remaining: 43.5ms
42: learn: 0.2020587 total: 32.5ms remaining: 43.1ms
43: learn: 0.1991714 total: 33.4ms remaining: 42.6ms
44: learn: 0.1963465 total: 35.4ms remaining: 43.3ms
45: learn: 0.1949122 total: 36.2ms remaining: 42.5ms
46: learn: 0.1922773 total: 37.4ms remaining: 42.2ms
47: learn: 0.1906400 total: 38.4ms remaining: 41.6ms
48: learn: 0.1875899 total: 39.4ms remaining: 41ms
49: learn: 0.1859991 total: 40.6ms remaining: 40.6ms
50: learn: 0.1847537 total: 41.3ms remaining: 39.7ms
51: learn: 0.1824626 total: 42.5ms remaining: 39.3ms
52: learn: 0.1810482 total: 44.5ms remaining: 39.5ms
53: learn: 0.1780986 total: 45.5ms remaining: 38.7ms
54: learn: 0.1764889 total: 46.3ms remaining: 37.9ms
55: learn: 0.1745581 total: 47.4ms remaining: 37.3ms
56: learn: 0.1730598 total: 48.4ms remaining: 36.5ms
57: learn: 0.1719532 total: 49ms remaining: 35.5ms
58: learn: 0.1693763 total: 50.5ms remaining: 35.1ms
59: learn: 0.1678983 total: 51.4ms remaining: 34.3ms
60: learn: 0.1661835 total: 52.2ms remaining: 33.4ms
61: learn: 0.1639334 total: 53.1ms remaining: 32.6ms
62: learn: 0.1623231 total: 54ms remaining: 31.7ms
63: learn: 0.1600026 total: 54.7ms remaining: 30.8ms
64: learn: 0.1575103 total: 55.5ms remaining: 29.9ms
65: learn: 0.1565985 total: 56.2ms remaining: 28.9ms
66: learn: 0.1538045 total: 57ms remaining: 28.1ms
67: learn: 0.1513697 total: 58.8ms remaining: 27.7ms
68: learn: 0.1496827 total: 61ms remaining: 27.4ms
69: learn: 0.1476345 total: 61.7ms remaining: 26.4ms
70: learn: 0.1463918 total: 62.6ms remaining: 25.6ms
71: learn: 0.1449643 total: 63.4ms remaining: 24.7ms
72: learn: 0.1432436 total: 64.2ms remaining: 23.8ms
73: learn: 0.1416623 total: 65.2ms remaining: 22.9ms
74: learn: 0.1405137 total: 66.9ms remaining: 22.3ms
75: learn: 0.1395736 total: 68.4ms remaining: 21.6ms
76: learn: 0.1381913 total: 70.8ms remaining: 21.1ms
77: learn: 0.1359855 total: 72.2ms remaining: 20.4ms
78: learn: 0.1344471 total: 74.2ms remaining: 19.7ms
79: learn: 0.1329802 total: 76.1ms remaining: 19ms
80: learn: 0.1317570 total: 77.9ms remaining: 18.3ms
81: learn: 0.1305140 total: 79.8ms remaining: 17.5ms
82: learn: 0.1289247 total: 81.8ms remaining: 16.7ms
83: learn: 0.1281610 total: 83.9ms remaining: 16ms
84: learn: 0.1267920 total: 86.2ms remaining: 15.2ms
85: learn: 0.1253611 total: 87.5ms remaining: 14.3ms
86: learn: 0.1240218 total: 89.2ms remaining: 13.3ms
87: learn: 0.1234992 total: 90.6ms remaining: 12.4ms
88: learn: 0.1227209 total: 92.4ms remaining: 11.4ms
89: learn: 0.1220948 total: 93.5ms remaining: 10.4ms
90: learn: 0.1213376 total: 94.7ms remaining: 9.37ms
91: learn: 0.1204634 total: 96ms remaining: 8.35ms
92: learn: 0.1193109 total: 96.9ms remaining: 7.3ms
93: learn: 0.1182654 total: 98.3ms remaining: 6.28ms
94: learn: 0.1175628 total: 99.4ms remaining: 5.23ms
95: learn: 0.1165376 total: 101ms remaining: 4.2ms
96: learn: 0.1155220 total: 103ms remaining: 3.19ms
97: learn: 0.1147181 total: 105ms remaining: 2.14ms
98: learn: 0.1139527 total: 107ms remaining: 1.08ms
99: learn: 0.1134197 total: 108ms remaining: 0us
0: learn: 0.6425807 total: 2.1ms remaining: 208ms
1: learn: 0.6095934 total: 4.68ms remaining: 229ms
2: learn: 0.5724604 total: 7.46ms remaining: 241ms
3: learn: 0.5389820 total: 9.82ms remaining: 236ms
4: learn: 0.5086857 total: 11.7ms remaining: 222ms
5: learn: 0.4798514 total: 13.1ms remaining: 205ms
6: learn: 0.4553136 total: 15.8ms remaining: 210ms
7: learn: 0.4379000 total: 16.6ms remaining: 191ms
8: learn: 0.4197578 total: 18ms remaining: 182ms
9: learn: 0.4020864 total: 18.7ms remaining: 168ms
10: learn: 0.3874267 total: 21ms remaining: 170ms
11: learn: 0.3753395 total: 23.5ms remaining: 172ms
12: learn: 0.3618715 total: 25.3ms remaining: 169ms
13: learn: 0.3506784 total: 27.1ms remaining: 166ms
14: learn: 0.3434627 total: 28.5ms remaining: 161ms
15: learn: 0.3329389 total: 30.2ms remaining: 159ms
16: learn: 0.3242477 total: 31.1ms remaining: 152ms
17: learn: 0.3125269 total: 32.3ms remaining: 147ms
18: learn: 0.3053817 total: 32.5ms remaining: 139ms
19: learn: 0.2966711 total: 33.6ms remaining: 135ms
20: learn: 0.2902346 total: 37.2ms remaining: 140ms
21: learn: 0.2821835 total: 38.4ms remaining: 136ms
22: learn: 0.2768605 total: 39.2ms remaining: 131ms
23: learn: 0.2723296 total: 39.4ms remaining: 125ms
24: learn: 0.2672881 total: 40.2ms remaining: 121ms
25: learn: 0.2625101 total: 41.2ms remaining: 117ms
26: learn: 0.2578836 total: 41.7ms remaining: 113ms
27: learn: 0.2509535 total: 43.5ms remaining: 112ms
28: learn: 0.2465789 total: 45.3ms remaining: 111ms
29: learn: 0.2428655 total: 47.6ms remaining: 111ms
30: learn: 0.2391947 total: 48.6ms remaining: 108ms
31: learn: 0.2370659 total: 48.9ms remaining: 104ms
32: learn: 0.2336989 total: 49.8ms remaining: 101ms
33: learn: 0.2316409 total: 50.8ms remaining: 98.5ms
34: learn: 0.2276796 total: 51.5ms remaining: 95.6ms
35: learn: 0.2242121 total: 52.4ms remaining: 93.1ms
36: learn: 0.2216698 total: 53.2ms remaining: 90.5ms
37: learn: 0.2202014 total: 53.7ms remaining: 87.6ms
38: learn: 0.2165851 total: 54.8ms remaining: 85.7ms
39: learn: 0.2155305 total: 55.6ms remaining: 83.4ms
40: learn: 0.2132991 total: 56.8ms remaining: 81.8ms
41: learn: 0.2118226 total: 59.2ms remaining: 81.7ms
42: learn: 0.2098110 total: 60.4ms remaining: 80.1ms
43: learn: 0.2078951 total: 62ms remaining: 78.9ms
44: learn: 0.2053172 total: 63.4ms remaining: 77.5ms
45: learn: 0.2028354 total: 65ms remaining: 76.3ms
46: learn: 0.2013843 total: 66.5ms remaining: 74.9ms
47: learn: 0.1991441 total: 67.9ms remaining: 73.5ms
48: learn: 0.1973173 total: 69.5ms remaining: 72.3ms
49: learn: 0.1958110 total: 71.9ms remaining: 71.9ms
50: learn: 0.1935166 total: 73.8ms remaining: 70.9ms
51: learn: 0.1910786 total: 75.1ms remaining: 69.3ms
52: learn: 0.1888082 total: 77.5ms remaining: 68.8ms
53: learn: 0.1864464 total: 79.7ms remaining: 67.9ms
54: learn: 0.1841618 total: 82.1ms remaining: 67.2ms
55: learn: 0.1829305 total: 83.9ms remaining: 66ms
56: learn: 0.1813734 total: 86.1ms remaining: 64.9ms
57: learn: 0.1787215 total: 87.9ms remaining: 63.6ms
58: learn: 0.1769624 total: 88.9ms remaining: 61.8ms
59: learn: 0.1738250 total: 90.2ms remaining: 60.1ms
60: learn: 0.1721127 total: 91.6ms remaining: 58.6ms
61: learn: 0.1694950 total: 93.7ms remaining: 57.4ms
62: learn: 0.1674560 total: 94.6ms remaining: 55.5ms
63: learn: 0.1651146 total: 95.3ms remaining: 53.6ms
64: learn: 0.1636673 total: 96.3ms remaining: 51.8ms
65: learn: 0.1613912 total: 97.6ms remaining: 50.3ms
66: learn: 0.1596330 total: 99.6ms remaining: 49.1ms
67: learn: 0.1575805 total: 101ms remaining: 47.4ms
68: learn: 0.1552873 total: 103ms remaining: 46.2ms
69: learn: 0.1534061 total: 104ms remaining: 44.4ms
70: learn: 0.1521509 total: 105ms remaining: 42.7ms
71: learn: 0.1504389 total: 105ms remaining: 41ms
72: learn: 0.1482221 total: 107ms remaining: 39.5ms
73: learn: 0.1468191 total: 108ms remaining: 38ms
74: learn: 0.1450395 total: 109ms remaining: 36.3ms
75: learn: 0.1438418 total: 110ms remaining: 34.8ms
76: learn: 0.1415306 total: 111ms remaining: 33.3ms
77: learn: 0.1406844 total: 113ms remaining: 31.8ms
78: learn: 0.1391775 total: 114ms remaining: 30.3ms
79: learn: 0.1372589 total: 116ms remaining: 29.1ms
80: learn: 0.1362480 total: 118ms remaining: 27.6ms
81: learn: 0.1349545 total: 120ms remaining: 26.3ms
82: learn: 0.1339132 total: 121ms remaining: 24.8ms
83: learn: 0.1326293 total: 123ms remaining: 23.4ms
84: learn: 0.1313642 total: 124ms remaining: 21.9ms
85: learn: 0.1303793 total: 126ms remaining: 20.5ms
86: learn: 0.1294905 total: 127ms remaining: 18.9ms
87: learn: 0.1276614 total: 128ms remaining: 17.4ms
88: learn: 0.1261972 total: 129ms remaining: 15.9ms
89: learn: 0.1249437 total: 130ms remaining: 14.4ms
90: learn: 0.1236735 total: 131ms remaining: 12.9ms
91: learn: 0.1226221 total: 132ms remaining: 11.5ms
92: learn: 0.1213811 total: 133ms remaining: 10ms
93: learn: 0.1205026 total: 134ms remaining: 8.56ms
94: learn: 0.1190857 total: 135ms remaining: 7.12ms
95: learn: 0.1177405 total: 136ms remaining: 5.67ms
96: learn: 0.1165438 total: 137ms remaining: 4.24ms
97: learn: 0.1157073 total: 138ms remaining: 2.82ms
98: learn: 0.1144604 total: 139ms remaining: 1.4ms
99: learn: 0.1134937 total: 140ms remaining: 0us
0: learn: 0.6414042 total: 1.08ms remaining: 107ms
1: learn: 0.6036809 total: 2.12ms remaining: 104ms
2: learn: 0.5672952 total: 3.45ms remaining: 111ms
3: learn: 0.5305474 total: 4.22ms remaining: 101ms
4: learn: 0.5054898 total: 4.97ms remaining: 94.4ms
5: learn: 0.4765417 total: 5.61ms remaining: 87.9ms
6: learn: 0.4545655 total: 6.2ms remaining: 82.4ms
7: learn: 0.4373139 total: 6.72ms remaining: 77.3ms
8: learn: 0.4180403 total: 7.38ms remaining: 74.6ms
9: learn: 0.4024810 total: 8.19ms remaining: 73.7ms
10: learn: 0.3849705 total: 8.95ms remaining: 72.4ms
11: learn: 0.3727187 total: 9.62ms remaining: 70.5ms
12: learn: 0.3586289 total: 10.3ms remaining: 69.1ms
13: learn: 0.3477836 total: 11.1ms remaining: 68ms
14: learn: 0.3371383 total: 11.9ms remaining: 67.6ms
15: learn: 0.3260366 total: 12.2ms remaining: 63.8ms
16: learn: 0.3151335 total: 12.8ms remaining: 62.4ms
17: learn: 0.3072938 total: 13.6ms remaining: 61.9ms
18: learn: 0.3010589 total: 13.8ms remaining: 59ms
19: learn: 0.2941139 total: 14.4ms remaining: 57.7ms
20: learn: 0.2866631 total: 15.3ms remaining: 57.6ms
21: learn: 0.2785329 total: 16ms remaining: 56.6ms
22: learn: 0.2711571 total: 16.5ms remaining: 55.3ms
23: learn: 0.2662387 total: 17.1ms remaining: 54.2ms
24: learn: 0.2614794 total: 17.8ms remaining: 53.5ms
25: learn: 0.2561155 total: 18.5ms remaining: 52.8ms
26: learn: 0.2525781 total: 18.9ms remaining: 51.2ms
27: learn: 0.2485784 total: 19.6ms remaining: 50.3ms
28: learn: 0.2443970 total: 20.5ms remaining: 50.3ms
29: learn: 0.2414316 total: 21.5ms remaining: 50.2ms
30: learn: 0.2370679 total: 22.5ms remaining: 50ms
31: learn: 0.2333710 total: 23.3ms remaining: 49.5ms
32: learn: 0.2297558 total: 24.1ms remaining: 48.9ms
33: learn: 0.2275985 total: 24.3ms remaining: 47.3ms
34: learn: 0.2261549 total: 25ms remaining: 46.4ms
35: learn: 0.2232823 total: 25.5ms remaining: 45.4ms
36: learn: 0.2214664 total: 26ms remaining: 44.3ms
37: learn: 0.2187996 total: 26.3ms remaining: 43ms
38: learn: 0.2167631 total: 27ms remaining: 42.2ms
39: learn: 0.2150543 total: 27.6ms remaining: 41.5ms
40: learn: 0.2123694 total: 28.1ms remaining: 40.5ms
41: learn: 0.2092893 total: 28.7ms remaining: 39.6ms
42: learn: 0.2077938 total: 29.9ms remaining: 39.6ms
43: learn: 0.2051896 total: 30.4ms remaining: 38.7ms
44: learn: 0.2020838 total: 31.2ms remaining: 38.1ms
45: learn: 0.1996024 total: 32ms remaining: 37.5ms
46: learn: 0.1977749 total: 32.9ms remaining: 37.1ms
47: learn: 0.1958991 total: 33.8ms remaining: 36.6ms
48: learn: 0.1944225 total: 34.2ms remaining: 35.6ms
49: learn: 0.1931922 total: 35.1ms remaining: 35.1ms
50: learn: 0.1913610 total: 35.7ms remaining: 34.3ms
51: learn: 0.1896323 total: 37.2ms remaining: 34.3ms
52: learn: 0.1878096 total: 38ms remaining: 33.7ms
53: learn: 0.1856469 total: 38.6ms remaining: 32.9ms
54: learn: 0.1843376 total: 39.2ms remaining: 32.1ms
55: learn: 0.1809689 total: 39.7ms remaining: 31.2ms
56: learn: 0.1787633 total: 40.4ms remaining: 30.5ms
57: learn: 0.1771388 total: 41.9ms remaining: 30.3ms
58: learn: 0.1754872 total: 42.6ms remaining: 29.6ms
59: learn: 0.1734412 total: 44.3ms remaining: 29.5ms
60: learn: 0.1718546 total: 45.9ms remaining: 29.3ms
61: learn: 0.1703756 total: 46.5ms remaining: 28.5ms
62: learn: 0.1686704 total: 47.5ms remaining: 27.9ms
63: learn: 0.1672408 total: 48.5ms remaining: 27.3ms
64: learn: 0.1656192 total: 49.1ms remaining: 26.4ms
65: learn: 0.1635327 total: 50.4ms remaining: 26ms
66: learn: 0.1617759 total: 51.3ms remaining: 25.3ms
67: learn: 0.1602687 total: 52ms remaining: 24.5ms
68: learn: 0.1587617 total: 52.8ms remaining: 23.7ms
69: learn: 0.1567076 total: 53.5ms remaining: 22.9ms
70: learn: 0.1550130 total: 54.4ms remaining: 22.2ms
71: learn: 0.1536892 total: 55.2ms remaining: 21.5ms
72: learn: 0.1522625 total: 55.9ms remaining: 20.7ms
73: learn: 0.1509576 total: 57.2ms remaining: 20.1ms
74: learn: 0.1488420 total: 58.3ms remaining: 19.4ms
75: learn: 0.1473430 total: 59.2ms remaining: 18.7ms
76: learn: 0.1454817 total: 60.2ms remaining: 18ms
77: learn: 0.1436339 total: 61.1ms remaining: 17.2ms
78: learn: 0.1420377 total: 62ms remaining: 16.5ms
79: learn: 0.1406279 total: 62.7ms remaining: 15.7ms
80: learn: 0.1391946 total: 64.2ms remaining: 15.1ms
81: learn: 0.1377607 total: 65ms remaining: 14.3ms
82: learn: 0.1366854 total: 66ms remaining: 13.5ms
83: learn: 0.1355168 total: 67.2ms remaining: 12.8ms
84: learn: 0.1345287 total: 68.2ms remaining: 12ms
85: learn: 0.1335772 total: 69.2ms remaining: 11.3ms
86: learn: 0.1324271 total: 70.2ms remaining: 10.5ms
87: learn: 0.1313599 total: 71.2ms remaining: 9.7ms
88: learn: 0.1302197 total: 72ms remaining: 8.9ms
89: learn: 0.1290454 total: 73.9ms remaining: 8.21ms
90: learn: 0.1274900 total: 74.8ms remaining: 7.4ms
91: learn: 0.1264243 total: 76.6ms remaining: 6.66ms
92: learn: 0.1252107 total: 77.3ms remaining: 5.82ms
93: learn: 0.1242705 total: 78.3ms remaining: 5ms
94: learn: 0.1232048 total: 80.4ms remaining: 4.23ms
95: learn: 0.1225731 total: 81.3ms remaining: 3.39ms
96: learn: 0.1215702 total: 82.1ms remaining: 2.54ms
97: learn: 0.1206040 total: 83ms remaining: 1.69ms
98: learn: 0.1196254 total: 84.7ms remaining: 855us
99: learn: 0.1183691 total: 85.4ms remaining: 0us
0: learn: 0.6483277 total: 505us remaining: 50.1ms
1: learn: 0.6155300 total: 958us remaining: 47ms
2: learn: 0.5801133 total: 2.52ms remaining: 81.6ms
3: learn: 0.5438671 total: 3.11ms remaining: 74.6ms
4: learn: 0.5153476 total: 3.47ms remaining: 65.9ms
5: learn: 0.4930093 total: 3.9ms remaining: 61.2ms
6: learn: 0.4716985 total: 4.32ms remaining: 57.4ms
7: learn: 0.4519960 total: 4.83ms remaining: 55.5ms
8: learn: 0.4380235 total: 5.19ms remaining: 52.5ms
9: learn: 0.4193870 total: 5.75ms remaining: 51.8ms
10: learn: 0.4050681 total: 6.02ms remaining: 48.7ms
11: learn: 0.3896630 total: 6.56ms remaining: 48.1ms
12: learn: 0.3783375 total: 7.14ms remaining: 47.8ms
13: learn: 0.3636949 total: 7.68ms remaining: 47.2ms
14: learn: 0.3514784 total: 8.27ms remaining: 46.9ms
15: learn: 0.3406462 total: 8.92ms remaining: 46.9ms
16: learn: 0.3328499 total: 9.38ms remaining: 45.8ms
17: learn: 0.3269265 total: 9.88ms remaining: 45ms
18: learn: 0.3204205 total: 10.3ms remaining: 44.1ms
19: learn: 0.3124206 total: 10.8ms remaining: 43.3ms
20: learn: 0.3037066 total: 11.5ms remaining: 43.3ms
21: learn: 0.2972371 total: 12.4ms remaining: 43.9ms
22: learn: 0.2900100 total: 13.2ms remaining: 44.3ms
23: learn: 0.2832710 total: 13.6ms remaining: 43.2ms
24: learn: 0.2786096 total: 14.2ms remaining: 42.6ms
25: learn: 0.2745835 total: 15.1ms remaining: 42.9ms
26: learn: 0.2714739 total: 15.9ms remaining: 43ms
27: learn: 0.2660829 total: 16.9ms remaining: 43.4ms
28: learn: 0.2649405 total: 17.3ms remaining: 42.3ms
29: learn: 0.2617376 total: 17.9ms remaining: 41.8ms
30: learn: 0.2580553 total: 19.3ms remaining: 43.1ms
31: learn: 0.2532095 total: 19.9ms remaining: 42.3ms
32: learn: 0.2507447 total: 20.2ms remaining: 41.1ms
33: learn: 0.2467854 total: 20.8ms remaining: 40.3ms
34: learn: 0.2443274 total: 21.2ms remaining: 39.3ms
35: learn: 0.2410986 total: 22.3ms remaining: 39.7ms
36: learn: 0.2381974 total: 23.8ms remaining: 40.5ms
37: learn: 0.2342700 total: 24.5ms remaining: 40ms
38: learn: 0.2309641 total: 25.3ms remaining: 39.6ms
39: learn: 0.2275861 total: 26.6ms remaining: 39.9ms
40: learn: 0.2227911 total: 27.7ms remaining: 39.9ms
41: learn: 0.2206702 total: 28.3ms remaining: 39ms
42: learn: 0.2178427 total: 28.9ms remaining: 38.3ms
43: learn: 0.2155070 total: 30.1ms remaining: 38.3ms
44: learn: 0.2124377 total: 30.8ms remaining: 37.7ms
45: learn: 0.2095209 total: 31.4ms remaining: 36.9ms
46: learn: 0.2068874 total: 32.9ms remaining: 37.1ms
47: learn: 0.2049574 total: 33.8ms remaining: 36.6ms
48: learn: 0.2033372 total: 34.3ms remaining: 35.7ms
49: learn: 0.2018192 total: 35.1ms remaining: 35.1ms
50: learn: 0.2003974 total: 35.8ms remaining: 34.4ms
51: learn: 0.1972085 total: 36.5ms remaining: 33.6ms
52: learn: 0.1940012 total: 37.2ms remaining: 33ms
53: learn: 0.1913792 total: 38ms remaining: 32.4ms
54: learn: 0.1900020 total: 39.1ms remaining: 32ms
55: learn: 0.1884141 total: 40.4ms remaining: 31.7ms
56: learn: 0.1857276 total: 41.1ms remaining: 31ms
57: learn: 0.1830402 total: 41.8ms remaining: 30.3ms
58: learn: 0.1802661 total: 42.8ms remaining: 29.7ms
59: learn: 0.1775120 total: 43.4ms remaining: 29ms
60: learn: 0.1743496 total: 44ms remaining: 28.1ms
61: learn: 0.1724312 total: 44.8ms remaining: 27.4ms
62: learn: 0.1697467 total: 45.4ms remaining: 26.7ms
63: learn: 0.1677510 total: 46.2ms remaining: 26ms
64: learn: 0.1651310 total: 47.1ms remaining: 25.4ms
65: learn: 0.1628205 total: 47.9ms remaining: 24.7ms
66: learn: 0.1608895 total: 49ms remaining: 24.1ms
67: learn: 0.1587382 total: 49.7ms remaining: 23.4ms
68: learn: 0.1571322 total: 50.5ms remaining: 22.7ms
69: learn: 0.1555672 total: 51.4ms remaining: 22ms
70: learn: 0.1540817 total: 52.1ms remaining: 21.3ms
71: learn: 0.1516280 total: 52.7ms remaining: 20.5ms
72: learn: 0.1492683 total: 54ms remaining: 20ms
73: learn: 0.1475752 total: 54.7ms remaining: 19.2ms
74: learn: 0.1459547 total: 55.7ms remaining: 18.6ms
75: learn: 0.1441532 total: 56.5ms remaining: 17.8ms
76: learn: 0.1426665 total: 57.6ms remaining: 17.2ms
77: learn: 0.1411967 total: 58.5ms remaining: 16.5ms
78: learn: 0.1397386 total: 59.5ms remaining: 15.8ms
79: learn: 0.1383296 total: 60.1ms remaining: 15ms
80: learn: 0.1366788 total: 61ms remaining: 14.3ms
81: learn: 0.1355422 total: 61.7ms remaining: 13.5ms
82: learn: 0.1338257 total: 62.8ms remaining: 12.9ms
83: learn: 0.1322048 total: 63.6ms remaining: 12.1ms
84: learn: 0.1308557 total: 64.3ms remaining: 11.3ms
85: learn: 0.1288535 total: 65.4ms remaining: 10.6ms
86: learn: 0.1273786 total: 66.1ms remaining: 9.88ms
87: learn: 0.1259338 total: 67.2ms remaining: 9.17ms
88: learn: 0.1248328 total: 68.2ms remaining: 8.43ms
89: learn: 0.1237220 total: 69.1ms remaining: 7.68ms
90: learn: 0.1226572 total: 70.1ms remaining: 6.93ms
91: learn: 0.1210698 total: 70.8ms remaining: 6.15ms
92: learn: 0.1197389 total: 71.9ms remaining: 5.41ms
93: learn: 0.1185527 total: 73ms remaining: 4.66ms
94: learn: 0.1173842 total: 73.7ms remaining: 3.88ms
95: learn: 0.1158753 total: 74.5ms remaining: 3.1ms
96: learn: 0.1148342 total: 75.3ms remaining: 2.33ms
97: learn: 0.1132491 total: 76.1ms remaining: 1.55ms
98: learn: 0.1117718 total: 77.1ms remaining: 778us
99: learn: 0.1105715 total: 77.8ms remaining: 0us
0: learn: 0.6478540 total: 655us remaining: 64.9ms
1: learn: 0.6188368 total: 1.36ms remaining: 66.7ms
2: learn: 0.5812869 total: 2.02ms remaining: 65.4ms
3: learn: 0.5494925 total: 2.62ms remaining: 62.9ms
4: learn: 0.5199929 total: 3.1ms remaining: 58.9ms
5: learn: 0.4941833 total: 3.88ms remaining: 60.7ms
6: learn: 0.4742569 total: 4.05ms remaining: 53.9ms
7: learn: 0.4530433 total: 4.7ms remaining: 54.1ms
8: learn: 0.4320795 total: 5.07ms remaining: 51.2ms
9: learn: 0.4138967 total: 5.56ms remaining: 50ms
10: learn: 0.3986699 total: 6.1ms remaining: 49.4ms
11: learn: 0.3873607 total: 6.62ms remaining: 48.6ms
12: learn: 0.3746075 total: 7.3ms remaining: 48.9ms
13: learn: 0.3635151 total: 7.88ms remaining: 48.4ms
14: learn: 0.3541650 total: 8.46ms remaining: 47.9ms
15: learn: 0.3423315 total: 8.95ms remaining: 47ms
16: learn: 0.3322791 total: 9.47ms remaining: 46.3ms
17: learn: 0.3245152 total: 10ms remaining: 45.6ms
18: learn: 0.3141369 total: 10.6ms remaining: 45.1ms
19: learn: 0.3077105 total: 11.2ms remaining: 44.8ms
20: learn: 0.2985603 total: 11.8ms remaining: 44.2ms
21: learn: 0.2914161 total: 12.3ms remaining: 43.5ms
22: learn: 0.2847772 total: 12.9ms remaining: 43ms
23: learn: 0.2771363 total: 13.4ms remaining: 42.3ms
24: learn: 0.2718045 total: 13.8ms remaining: 41.5ms
25: learn: 0.2658817 total: 14.3ms remaining: 40.8ms
26: learn: 0.2597141 total: 14.9ms remaining: 40.2ms
27: learn: 0.2534277 total: 15.5ms remaining: 39.8ms
28: learn: 0.2484247 total: 16ms remaining: 39.2ms
29: learn: 0.2437953 total: 16.5ms remaining: 38.6ms
30: learn: 0.2402339 total: 17.2ms remaining: 38.3ms
31: learn: 0.2374675 total: 18.2ms remaining: 38.7ms
32: learn: 0.2352614 total: 18.4ms remaining: 37.4ms
33: learn: 0.2301768 total: 19.1ms remaining: 37.1ms
34: learn: 0.2275918 total: 19.2ms remaining: 35.7ms
35: learn: 0.2240243 total: 19.8ms remaining: 35.2ms
36: learn: 0.2212741 total: 20.2ms remaining: 34.4ms
37: learn: 0.2188742 total: 20.8ms remaining: 33.9ms
38: learn: 0.2153598 total: 21.5ms remaining: 33.6ms
39: learn: 0.2115964 total: 21.9ms remaining: 32.9ms
40: learn: 0.2089791 total: 22.4ms remaining: 32.2ms
41: learn: 0.2068259 total: 22.9ms remaining: 31.7ms
42: learn: 0.2045457 total: 23.7ms remaining: 31.4ms
43: learn: 0.2025532 total: 24.3ms remaining: 30.9ms
44: learn: 0.1992884 total: 25.1ms remaining: 30.6ms
45: learn: 0.1963081 total: 25.8ms remaining: 30.3ms
46: learn: 0.1936464 total: 26.5ms remaining: 29.9ms
47: learn: 0.1918183 total: 27.3ms remaining: 29.5ms
48: learn: 0.1896215 total: 28ms remaining: 29.2ms
49: learn: 0.1881047 total: 28.7ms remaining: 28.7ms
50: learn: 0.1864221 total: 29.4ms remaining: 28.2ms
51: learn: 0.1840211 total: 30.1ms remaining: 27.8ms
52: learn: 0.1816152 total: 30.7ms remaining: 27.2ms
53: learn: 0.1806960 total: 31.3ms remaining: 26.7ms
54: learn: 0.1791919 total: 32ms remaining: 26.1ms
55: learn: 0.1770581 total: 32.6ms remaining: 25.7ms
56: learn: 0.1749057 total: 33.4ms remaining: 25.2ms
57: learn: 0.1723042 total: 34ms remaining: 24.6ms
58: learn: 0.1709858 total: 34.6ms remaining: 24ms
59: learn: 0.1694587 total: 35.2ms remaining: 23.5ms
60: learn: 0.1671948 total: 35.9ms remaining: 22.9ms
61: learn: 0.1645024 total: 36.5ms remaining: 22.4ms
62: learn: 0.1623542 total: 37.3ms remaining: 21.9ms
63: learn: 0.1609887 total: 38ms remaining: 21.4ms
64: learn: 0.1590499 total: 38.8ms remaining: 20.9ms
65: learn: 0.1561850 total: 39.6ms remaining: 20.4ms
66: learn: 0.1546082 total: 40.5ms remaining: 19.9ms
67: learn: 0.1530022 total: 41.4ms remaining: 19.5ms
68: learn: 0.1505926 total: 42.1ms remaining: 18.9ms
69: learn: 0.1488946 total: 42.9ms remaining: 18.4ms
70: learn: 0.1478330 total: 43.8ms remaining: 17.9ms
71: learn: 0.1462355 total: 44.5ms remaining: 17.3ms
72: learn: 0.1447594 total: 45.4ms remaining: 16.8ms
73: learn: 0.1435484 total: 46.1ms remaining: 16.2ms
74: learn: 0.1420605 total: 47ms remaining: 15.7ms
75: learn: 0.1399332 total: 47.8ms remaining: 15.1ms
76: learn: 0.1384936 total: 48.5ms remaining: 14.5ms
77: learn: 0.1371481 total: 49.3ms remaining: 13.9ms
78: learn: 0.1356753 total: 50ms remaining: 13.3ms
79: learn: 0.1344624 total: 50.8ms remaining: 12.7ms
80: learn: 0.1333094 total: 51.7ms remaining: 12.1ms
81: learn: 0.1319504 total: 52.4ms remaining: 11.5ms
82: learn: 0.1301410 total: 53.2ms remaining: 10.9ms
83: learn: 0.1289650 total: 54ms remaining: 10.3ms
84: learn: 0.1273603 total: 54.8ms remaining: 9.67ms
85: learn: 0.1256978 total: 55.8ms remaining: 9.08ms
86: learn: 0.1245942 total: 56.7ms remaining: 8.47ms
87: learn: 0.1234886 total: 57.5ms remaining: 7.84ms
88: learn: 0.1217552 total: 58.3ms remaining: 7.21ms
89: learn: 0.1206373 total: 59.1ms remaining: 6.57ms
90: learn: 0.1196699 total: 60ms remaining: 5.94ms
91: learn: 0.1186854 total: 60.6ms remaining: 5.27ms
92: learn: 0.1178456 total: 61.2ms remaining: 4.61ms
93: learn: 0.1170309 total: 61.9ms remaining: 3.95ms
94: learn: 0.1155022 total: 62.7ms remaining: 3.3ms
95: learn: 0.1142228 total: 63.4ms remaining: 2.64ms
96: learn: 0.1133006 total: 64.2ms remaining: 1.98ms
97: learn: 0.1118029 total: 64.9ms remaining: 1.32ms
98: learn: 0.1108799 total: 65.5ms remaining: 662us
99: learn: 0.1101087 total: 66.4ms remaining: 0us
0: learn: 0.6452232 total: 650us remaining: 64.4ms
1: learn: 0.6130854 total: 1.19ms remaining: 58.2ms
2: learn: 0.5785534 total: 1.8ms remaining: 58.2ms
3: learn: 0.5487896 total: 2.56ms remaining: 61.6ms
4: learn: 0.5235638 total: 3.16ms remaining: 60ms
5: learn: 0.4958000 total: 3.67ms remaining: 57.4ms
6: learn: 0.4707181 total: 4.43ms remaining: 58.8ms
7: learn: 0.4489693 total: 5.04ms remaining: 58ms
8: learn: 0.4264777 total: 5.54ms remaining: 56ms
9: learn: 0.4124217 total: 6.06ms remaining: 54.6ms
10: learn: 0.3945337 total: 7.14ms remaining: 57.8ms
11: learn: 0.3795706 total: 7.76ms remaining: 56.9ms
12: learn: 0.3660203 total: 8.54ms remaining: 57.1ms
13: learn: 0.3555878 total: 9.18ms remaining: 56.4ms
14: learn: 0.3450628 total: 9.63ms remaining: 54.6ms
15: learn: 0.3352606 total: 10.1ms remaining: 53ms
16: learn: 0.3276190 total: 10.7ms remaining: 52.3ms
17: learn: 0.3212609 total: 12.5ms remaining: 57.1ms
18: learn: 0.3123439 total: 13.1ms remaining: 55.9ms
19: learn: 0.3031837 total: 14.2ms remaining: 56.8ms
20: learn: 0.2938529 total: 14.8ms remaining: 55.8ms
21: learn: 0.2866061 total: 17ms remaining: 60.3ms
22: learn: 0.2828694 total: 18ms remaining: 60.2ms
23: learn: 0.2753056 total: 18.6ms remaining: 59ms
24: learn: 0.2711134 total: 19.3ms remaining: 57.9ms
25: learn: 0.2653193 total: 20ms remaining: 57ms
26: learn: 0.2603788 total: 21ms remaining: 56.7ms
27: learn: 0.2568278 total: 21.3ms remaining: 54.7ms
28: learn: 0.2506470 total: 22.2ms remaining: 54.3ms
29: learn: 0.2446159 total: 23.3ms remaining: 54.4ms
30: learn: 0.2402115 total: 24.1ms remaining: 53.6ms
31: learn: 0.2359320 total: 24.8ms remaining: 52.6ms
32: learn: 0.2322850 total: 25.8ms remaining: 52.3ms
33: learn: 0.2288307 total: 26.5ms remaining: 51.4ms
34: learn: 0.2263137 total: 27.5ms remaining: 51ms
35: learn: 0.2215138 total: 28.3ms remaining: 50.2ms
36: learn: 0.2196910 total: 29.5ms remaining: 50.3ms
37: learn: 0.2177473 total: 30ms remaining: 48.9ms
38: learn: 0.2123156 total: 32ms remaining: 50ms
39: learn: 0.2082721 total: 32.6ms remaining: 49ms
40: learn: 0.2048000 total: 33.7ms remaining: 48.5ms
41: learn: 0.2015699 total: 34.5ms remaining: 47.6ms
42: learn: 0.1971835 total: 35.5ms remaining: 47ms
43: learn: 0.1961963 total: 36.1ms remaining: 45.9ms
44: learn: 0.1947997 total: 38.3ms remaining: 46.9ms
45: learn: 0.1912582 total: 39.5ms remaining: 46.4ms
46: learn: 0.1887218 total: 40.9ms remaining: 46.1ms
47: learn: 0.1865246 total: 42.1ms remaining: 45.6ms
48: learn: 0.1839188 total: 43.3ms remaining: 45.1ms
49: learn: 0.1812008 total: 44.7ms remaining: 44.7ms
50: learn: 0.1793408 total: 46.3ms remaining: 44.5ms
51: learn: 0.1770457 total: 47.4ms remaining: 43.8ms
52: learn: 0.1737714 total: 48.4ms remaining: 43ms
53: learn: 0.1715542 total: 49.2ms remaining: 41.9ms
54: learn: 0.1690469 total: 50.3ms remaining: 41.1ms
55: learn: 0.1665154 total: 51.5ms remaining: 40.5ms
56: learn: 0.1647795 total: 52.4ms remaining: 39.5ms
57: learn: 0.1628518 total: 53.2ms remaining: 38.5ms
58: learn: 0.1603441 total: 54.1ms remaining: 37.6ms
59: learn: 0.1580241 total: 55.2ms remaining: 36.8ms
60: learn: 0.1555307 total: 55.8ms remaining: 35.7ms
61: learn: 0.1531722 total: 56.8ms remaining: 34.8ms
62: learn: 0.1513581 total: 57.7ms remaining: 33.9ms
63: learn: 0.1486551 total: 58.4ms remaining: 32.8ms
64: learn: 0.1473070 total: 59.4ms remaining: 32ms
65: learn: 0.1452171 total: 60.7ms remaining: 31.3ms
66: learn: 0.1433764 total: 61.7ms remaining: 30.4ms
67: learn: 0.1423939 total: 63.1ms remaining: 29.7ms
68: learn: 0.1411770 total: 64.3ms remaining: 28.9ms
69: learn: 0.1393073 total: 65.2ms remaining: 27.9ms
70: learn: 0.1368018 total: 66.8ms remaining: 27.3ms
71: learn: 0.1351955 total: 67.6ms remaining: 26.3ms
72: learn: 0.1334172 total: 69.4ms remaining: 25.7ms
73: learn: 0.1313265 total: 70.8ms remaining: 24.9ms
74: learn: 0.1294352 total: 71.6ms remaining: 23.9ms
75: learn: 0.1280175 total: 72.5ms remaining: 22.9ms
76: learn: 0.1261850 total: 73.5ms remaining: 21.9ms
77: learn: 0.1240069 total: 74.2ms remaining: 20.9ms
78: learn: 0.1227004 total: 76.8ms remaining: 20.4ms
79: learn: 0.1214925 total: 77.8ms remaining: 19.4ms
80: learn: 0.1200065 total: 78.6ms remaining: 18.4ms
81: learn: 0.1185123 total: 79.6ms remaining: 17.5ms
82: learn: 0.1172316 total: 81.2ms remaining: 16.6ms
83: learn: 0.1158209 total: 81.8ms remaining: 15.6ms
84: learn: 0.1146112 total: 82.7ms remaining: 14.6ms
85: learn: 0.1133338 total: 83.4ms remaining: 13.6ms
86: learn: 0.1121390 total: 86.1ms remaining: 12.9ms
87: learn: 0.1105949 total: 87ms remaining: 11.9ms
88: learn: 0.1096507 total: 87.9ms remaining: 10.9ms
89: learn: 0.1083503 total: 88.8ms remaining: 9.87ms
90: learn: 0.1072355 total: 89.6ms remaining: 8.87ms
91: learn: 0.1061779 total: 90.7ms remaining: 7.89ms
92: learn: 0.1051529 total: 91.6ms remaining: 6.89ms
93: learn: 0.1039878 total: 92.6ms remaining: 5.91ms
94: learn: 0.1031676 total: 93.7ms remaining: 4.93ms
95: learn: 0.1020187 total: 94.9ms remaining: 3.95ms
96: learn: 0.1012952 total: 95.5ms remaining: 2.95ms
97: learn: 0.1005362 total: 97ms remaining: 1.98ms
98: learn: 0.0995976 total: 98.1ms remaining: 991us
99: learn: 0.0987654 total: 98.9ms remaining: 0us
0: learn: 0.6482935 total: 571us remaining: 56.6ms
1: learn: 0.6097817 total: 1.03ms remaining: 50.7ms
2: learn: 0.5723488 total: 1.48ms remaining: 48ms
3: learn: 0.5391228 total: 1.82ms remaining: 43.7ms
4: learn: 0.5138453 total: 2.18ms remaining: 41.5ms
5: learn: 0.4910075 total: 2.71ms remaining: 42.4ms
6: learn: 0.4707257 total: 3.13ms remaining: 41.5ms
7: learn: 0.4519516 total: 3.49ms remaining: 40.2ms
8: learn: 0.4328789 total: 3.86ms remaining: 39.1ms
9: learn: 0.4164797 total: 4.26ms remaining: 38.3ms
10: learn: 0.3997077 total: 4.76ms remaining: 38.6ms
11: learn: 0.3883643 total: 5.07ms remaining: 37.1ms
12: learn: 0.3787946 total: 5.81ms remaining: 38.9ms
13: learn: 0.3660023 total: 6.33ms remaining: 38.9ms
14: learn: 0.3557177 total: 7.27ms remaining: 41.2ms
15: learn: 0.3464166 total: 7.9ms remaining: 41.5ms
16: learn: 0.3349490 total: 8.68ms remaining: 42.4ms
17: learn: 0.3258922 total: 9.33ms remaining: 42.5ms
18: learn: 0.3195433 total: 10.3ms remaining: 44ms
19: learn: 0.3100072 total: 10.9ms remaining: 43.5ms
20: learn: 0.3025717 total: 11.5ms remaining: 43.2ms
21: learn: 0.2956294 total: 12ms remaining: 42.5ms
22: learn: 0.2888096 total: 12.5ms remaining: 41.9ms
23: learn: 0.2839482 total: 13ms remaining: 41.3ms
24: learn: 0.2786366 total: 13.6ms remaining: 40.7ms
25: learn: 0.2740122 total: 14.2ms remaining: 40.4ms
26: learn: 0.2699392 total: 14.7ms remaining: 39.9ms
27: learn: 0.2675302 total: 15.3ms remaining: 39.3ms
28: learn: 0.2624445 total: 16.1ms remaining: 39.5ms
29: learn: 0.2568874 total: 16.8ms remaining: 39.3ms
30: learn: 0.2517272 total: 17.5ms remaining: 39ms
31: learn: 0.2486361 total: 18.1ms remaining: 38.5ms
32: learn: 0.2462152 total: 18.7ms remaining: 37.9ms
33: learn: 0.2436858 total: 19.5ms remaining: 37.8ms
34: learn: 0.2408060 total: 20.3ms remaining: 37.8ms
35: learn: 0.2372277 total: 21.3ms remaining: 37.8ms
36: learn: 0.2355722 total: 22.2ms remaining: 37.8ms
37: learn: 0.2322521 total: 22.9ms remaining: 37.4ms
38: learn: 0.2292490 total: 23.5ms remaining: 36.8ms
39: learn: 0.2252908 total: 24.2ms remaining: 36.2ms
40: learn: 0.2220657 total: 25.2ms remaining: 36.2ms
41: learn: 0.2207384 total: 25.6ms remaining: 35.3ms
42: learn: 0.2181120 total: 26.3ms remaining: 34.9ms
43: learn: 0.2155424 total: 27.3ms remaining: 34.8ms
44: learn: 0.2122211 total: 28.2ms remaining: 34.4ms
45: learn: 0.2101936 total: 29ms remaining: 34.1ms
46: learn: 0.2074187 total: 29.9ms remaining: 33.8ms
47: learn: 0.2059315 total: 30.8ms remaining: 33.4ms
48: learn: 0.2035496 total: 31.6ms remaining: 32.9ms
49: learn: 0.2002119 total: 32.4ms remaining: 32.4ms
50: learn: 0.1978201 total: 33.3ms remaining: 32ms
51: learn: 0.1954618 total: 34.2ms remaining: 31.6ms
52: learn: 0.1943978 total: 34.8ms remaining: 30.8ms
53: learn: 0.1921818 total: 35.4ms remaining: 30.2ms
54: learn: 0.1894813 total: 36.3ms remaining: 29.7ms
55: learn: 0.1863147 total: 37.1ms remaining: 29.1ms
56: learn: 0.1844038 total: 37.7ms remaining: 28.4ms
57: learn: 0.1817204 total: 38.4ms remaining: 27.8ms
58: learn: 0.1801922 total: 39.2ms remaining: 27.2ms
59: learn: 0.1782430 total: 40.8ms remaining: 27.2ms
60: learn: 0.1751776 total: 41.7ms remaining: 26.6ms
61: learn: 0.1719181 total: 42.7ms remaining: 26.1ms
62: learn: 0.1693431 total: 43.6ms remaining: 25.6ms
63: learn: 0.1677272 total: 44.6ms remaining: 25.1ms
64: learn: 0.1655813 total: 45.4ms remaining: 24.5ms
65: learn: 0.1635442 total: 46.4ms remaining: 23.9ms
66: learn: 0.1614970 total: 47.3ms remaining: 23.3ms
67: learn: 0.1597533 total: 48.1ms remaining: 22.6ms
68: learn: 0.1578856 total: 49.1ms remaining: 22.1ms
69: learn: 0.1561416 total: 50ms remaining: 21.4ms
70: learn: 0.1543075 total: 50.8ms remaining: 20.7ms
71: learn: 0.1524157 total: 51.6ms remaining: 20.1ms
72: learn: 0.1502702 total: 52.7ms remaining: 19.5ms
73: learn: 0.1489564 total: 53.8ms remaining: 18.9ms
74: learn: 0.1477989 total: 54.5ms remaining: 18.2ms
75: learn: 0.1463172 total: 55.9ms remaining: 17.7ms
76: learn: 0.1448132 total: 56.6ms remaining: 16.9ms
77: learn: 0.1439062 total: 57.5ms remaining: 16.2ms
78: learn: 0.1424043 total: 58.3ms remaining: 15.5ms
79: learn: 0.1409956 total: 60.4ms remaining: 15.1ms
80: learn: 0.1391833 total: 61.2ms remaining: 14.4ms
81: learn: 0.1377252 total: 62.2ms remaining: 13.6ms
82: learn: 0.1360079 total: 63ms remaining: 12.9ms
83: learn: 0.1347468 total: 64ms remaining: 12.2ms
84: learn: 0.1335413 total: 65.4ms remaining: 11.5ms
85: learn: 0.1317027 total: 66.4ms remaining: 10.8ms
86: learn: 0.1302597 total: 67.8ms remaining: 10.1ms
87: learn: 0.1289960 total: 68.5ms remaining: 9.34ms
88: learn: 0.1276287 total: 69.4ms remaining: 8.57ms
89: learn: 0.1263822 total: 70.5ms remaining: 7.84ms
90: learn: 0.1256466 total: 71.3ms remaining: 7.05ms
91: learn: 0.1244025 total: 72.2ms remaining: 6.28ms
92: learn: 0.1229601 total: 72.8ms remaining: 5.48ms
93: learn: 0.1213025 total: 73.9ms remaining: 4.71ms
94: learn: 0.1199285 total: 74.6ms remaining: 3.92ms
95: learn: 0.1184821 total: 75.5ms remaining: 3.14ms
96: learn: 0.1172785 total: 76.2ms remaining: 2.36ms
97: learn: 0.1162318 total: 78.8ms remaining: 1.61ms
98: learn: 0.1154944 total: 79.5ms remaining: 803us
99: learn: 0.1143362 total: 80.6ms remaining: 0us
0: learn: 0.6424092 total: 615us remaining: 61ms
1: learn: 0.6044739 total: 1.24ms remaining: 60.6ms
2: learn: 0.5658378 total: 1.64ms remaining: 53.1ms
3: learn: 0.5331348 total: 2.27ms remaining: 54.5ms
4: learn: 0.5020048 total: 2.71ms remaining: 51.5ms
5: learn: 0.4785496 total: 3.35ms remaining: 52.5ms
6: learn: 0.4548111 total: 4.42ms remaining: 58.7ms
7: learn: 0.4343862 total: 5.3ms remaining: 60.9ms
8: learn: 0.4164076 total: 6.59ms remaining: 66.7ms
9: learn: 0.3980669 total: 7.45ms remaining: 67ms
10: learn: 0.3842137 total: 8.42ms remaining: 68.2ms
11: learn: 0.3709775 total: 9.16ms remaining: 67.2ms
12: learn: 0.3556386 total: 9.81ms remaining: 65.6ms
13: learn: 0.3451018 total: 10.6ms remaining: 64.9ms
14: learn: 0.3348098 total: 12.5ms remaining: 70.8ms
15: learn: 0.3271334 total: 13.3ms remaining: 70ms
16: learn: 0.3190411 total: 14.2ms remaining: 69.1ms
17: learn: 0.3107571 total: 14.4ms remaining: 65.4ms
18: learn: 0.3020451 total: 15.2ms remaining: 64.6ms
19: learn: 0.2965826 total: 16.7ms remaining: 66.7ms
20: learn: 0.2903581 total: 17.5ms remaining: 66ms
21: learn: 0.2825763 total: 18.9ms remaining: 67ms
22: learn: 0.2779718 total: 19.8ms remaining: 66.3ms
23: learn: 0.2721132 total: 20.5ms remaining: 64.8ms
24: learn: 0.2690395 total: 21.3ms remaining: 63.9ms
25: learn: 0.2656442 total: 22.7ms remaining: 64.6ms
26: learn: 0.2595123 total: 24.2ms remaining: 65.3ms
27: learn: 0.2550912 total: 26.2ms remaining: 67.3ms
28: learn: 0.2509144 total: 27.9ms remaining: 68.4ms
29: learn: 0.2469739 total: 28.6ms remaining: 66.8ms
30: learn: 0.2433197 total: 29.8ms remaining: 66.4ms
31: learn: 0.2407384 total: 31.9ms remaining: 67.8ms
32: learn: 0.2381498 total: 32.8ms remaining: 66.5ms
33: learn: 0.2341570 total: 33.9ms remaining: 65.8ms
34: learn: 0.2301784 total: 35.3ms remaining: 65.5ms
35: learn: 0.2271857 total: 37ms remaining: 65.8ms
36: learn: 0.2245278 total: 39.5ms remaining: 67.3ms
37: learn: 0.2220755 total: 41.7ms remaining: 68ms
38: learn: 0.2187753 total: 44.4ms remaining: 69.4ms
39: learn: 0.2156415 total: 47ms remaining: 70.5ms
40: learn: 0.2132895 total: 48.1ms remaining: 69.3ms
41: learn: 0.2109095 total: 51.5ms remaining: 71.1ms
42: learn: 0.2088499 total: 53.7ms remaining: 71.2ms
43: learn: 0.2062905 total: 55.4ms remaining: 70.5ms
44: learn: 0.2043945 total: 56.9ms remaining: 69.5ms
45: learn: 0.2021090 total: 58.2ms remaining: 68.3ms
46: learn: 0.2012857 total: 59.5ms remaining: 67.1ms
47: learn: 0.1985814 total: 60.8ms remaining: 65.9ms
48: learn: 0.1967447 total: 62.5ms remaining: 65.1ms
49: learn: 0.1953459 total: 63.9ms remaining: 63.9ms
50: learn: 0.1929238 total: 65.7ms remaining: 63.2ms
51: learn: 0.1916273 total: 66.8ms remaining: 61.6ms
52: learn: 0.1893060 total: 67.8ms remaining: 60.2ms
53: learn: 0.1873613 total: 69ms remaining: 58.8ms
54: learn: 0.1860794 total: 70.3ms remaining: 57.5ms
55: learn: 0.1837366 total: 71.3ms remaining: 56ms
56: learn: 0.1810279 total: 73.8ms remaining: 55.7ms
57: learn: 0.1796809 total: 74.7ms remaining: 54.1ms
58: learn: 0.1772241 total: 76.1ms remaining: 52.9ms
59: learn: 0.1745327 total: 77ms remaining: 51.3ms
60: learn: 0.1726487 total: 79.7ms remaining: 51ms
61: learn: 0.1706294 total: 80.5ms remaining: 49.3ms
62: learn: 0.1692594 total: 81.5ms remaining: 47.9ms
63: learn: 0.1681378 total: 82.9ms remaining: 46.6ms
64: learn: 0.1667839 total: 84.2ms remaining: 45.4ms
65: learn: 0.1648415 total: 85.8ms remaining: 44.2ms
66: learn: 0.1622233 total: 87.9ms remaining: 43.3ms
67: learn: 0.1602809 total: 89.9ms remaining: 42.3ms
68: learn: 0.1592196 total: 91.5ms remaining: 41.1ms
69: learn: 0.1571752 total: 93.4ms remaining: 40ms
70: learn: 0.1558444 total: 94.9ms remaining: 38.8ms
71: learn: 0.1538037 total: 96.2ms remaining: 37.4ms
72: learn: 0.1523753 total: 98.2ms remaining: 36.3ms
73: learn: 0.1507850 total: 100ms remaining: 35.2ms
74: learn: 0.1490669 total: 102ms remaining: 34.1ms
75: learn: 0.1472015 total: 104ms remaining: 32.8ms
76: learn: 0.1457907 total: 105ms remaining: 31.5ms
77: learn: 0.1446345 total: 108ms remaining: 30.4ms
78: learn: 0.1438302 total: 109ms remaining: 29ms
79: learn: 0.1426836 total: 111ms remaining: 27.7ms
80: learn: 0.1414285 total: 113ms remaining: 26.6ms
81: learn: 0.1403873 total: 115ms remaining: 25.2ms
82: learn: 0.1388680 total: 117ms remaining: 23.9ms
83: learn: 0.1377954 total: 118ms remaining: 22.4ms
84: learn: 0.1369911 total: 120ms remaining: 21.3ms
85: learn: 0.1356640 total: 121ms remaining: 19.8ms
86: learn: 0.1346080 total: 123ms remaining: 18.3ms
87: learn: 0.1333840 total: 123ms remaining: 16.8ms
88: learn: 0.1323537 total: 124ms remaining: 15.4ms
89: learn: 0.1312237 total: 126ms remaining: 14ms
90: learn: 0.1299434 total: 127ms remaining: 12.6ms
91: learn: 0.1289586 total: 128ms remaining: 11.2ms
92: learn: 0.1281463 total: 129ms remaining: 9.75ms
93: learn: 0.1273422 total: 131ms remaining: 8.35ms
94: learn: 0.1262911 total: 133ms remaining: 6.99ms
95: learn: 0.1249987 total: 134ms remaining: 5.58ms
96: learn: 0.1236901 total: 135ms remaining: 4.19ms
97: learn: 0.1228552 total: 137ms remaining: 2.79ms
98: learn: 0.1222773 total: 138ms remaining: 1.4ms
99: learn: 0.1210190 total: 139ms remaining: 0us
0: learn: 0.6495789 total: 465us remaining: 46.1ms
1: learn: 0.6178464 total: 880us remaining: 43.1ms
2: learn: 0.5852578 total: 1.29ms remaining: 41.8ms
3: learn: 0.5533435 total: 1.8ms remaining: 43.3ms
4: learn: 0.5300644 total: 2.79ms remaining: 53.1ms
5: learn: 0.5104585 total: 3.6ms remaining: 56.4ms
6: learn: 0.4862911 total: 4.68ms remaining: 62.2ms
7: learn: 0.4666907 total: 5.27ms remaining: 60.6ms
8: learn: 0.4496751 total: 5.55ms remaining: 56.1ms
9: learn: 0.4323048 total: 6.48ms remaining: 58.3ms
10: learn: 0.4180174 total: 7.22ms remaining: 58.4ms
11: learn: 0.4056119 total: 7.71ms remaining: 56.6ms
12: learn: 0.3933982 total: 8.37ms remaining: 56ms
13: learn: 0.3860414 total: 9.04ms remaining: 55.5ms
14: learn: 0.3734601 total: 9.92ms remaining: 56.2ms
15: learn: 0.3647702 total: 10.5ms remaining: 55ms
16: learn: 0.3555684 total: 11.9ms remaining: 58.3ms
17: learn: 0.3451648 total: 12.7ms remaining: 58ms
18: learn: 0.3365500 total: 13.5ms remaining: 57.4ms
19: learn: 0.3307781 total: 14.2ms remaining: 56.7ms
20: learn: 0.3238906 total: 14.8ms remaining: 55.7ms
21: learn: 0.3172724 total: 15.6ms remaining: 55.4ms
22: learn: 0.3102313 total: 16.3ms remaining: 54.7ms
23: learn: 0.3047438 total: 17ms remaining: 53.7ms
24: learn: 0.2978030 total: 18.3ms remaining: 54.9ms
25: learn: 0.2927210 total: 18.9ms remaining: 53.8ms
26: learn: 0.2895587 total: 19.5ms remaining: 52.7ms
27: learn: 0.2840902 total: 20.3ms remaining: 52.3ms
28: learn: 0.2795832 total: 21.2ms remaining: 51.9ms
29: learn: 0.2755125 total: 21.9ms remaining: 51ms
30: learn: 0.2705809 total: 22.5ms remaining: 50ms
31: learn: 0.2674857 total: 23.3ms remaining: 49.4ms
32: learn: 0.2630010 total: 24.2ms remaining: 49.2ms
33: learn: 0.2567622 total: 25.3ms remaining: 49.2ms
34: learn: 0.2541048 total: 26.6ms remaining: 49.4ms
35: learn: 0.2515091 total: 27.4ms remaining: 48.7ms
36: learn: 0.2496420 total: 27.8ms remaining: 47.3ms
37: learn: 0.2462492 total: 28.3ms remaining: 46.2ms
38: learn: 0.2432209 total: 29.6ms remaining: 46.4ms
39: learn: 0.2400406 total: 30.3ms remaining: 45.4ms
40: learn: 0.2370158 total: 32ms remaining: 46ms
41: learn: 0.2339843 total: 32.6ms remaining: 45ms
42: learn: 0.2316020 total: 33.1ms remaining: 43.9ms
43: learn: 0.2285619 total: 33.7ms remaining: 42.9ms
44: learn: 0.2263588 total: 34.8ms remaining: 42.6ms
45: learn: 0.2230401 total: 35.4ms remaining: 41.6ms
46: learn: 0.2209444 total: 36.4ms remaining: 41ms
47: learn: 0.2184339 total: 37.4ms remaining: 40.5ms
48: learn: 0.2155951 total: 38ms remaining: 39.6ms
49: learn: 0.2125427 total: 38.7ms remaining: 38.7ms
50: learn: 0.2106964 total: 39.4ms remaining: 37.9ms
51: learn: 0.2087866 total: 40.2ms remaining: 37.1ms
52: learn: 0.2068120 total: 41.6ms remaining: 36.9ms
53: learn: 0.2045616 total: 42.2ms remaining: 35.9ms
54: learn: 0.2014660 total: 43.1ms remaining: 35.3ms
55: learn: 0.1989029 total: 43.8ms remaining: 34.4ms
56: learn: 0.1968948 total: 44.7ms remaining: 33.7ms
57: learn: 0.1940082 total: 45.5ms remaining: 32.9ms
58: learn: 0.1906136 total: 46.3ms remaining: 32.2ms
59: learn: 0.1890053 total: 47ms remaining: 31.4ms
60: learn: 0.1873394 total: 48ms remaining: 30.7ms
61: learn: 0.1853249 total: 49.4ms remaining: 30.3ms
62: learn: 0.1835254 total: 50.4ms remaining: 29.6ms
63: learn: 0.1815619 total: 51.6ms remaining: 29ms
64: learn: 0.1802908 total: 52.5ms remaining: 28.3ms
65: learn: 0.1778013 total: 53.8ms remaining: 27.7ms
66: learn: 0.1751446 total: 54.6ms remaining: 26.9ms
67: learn: 0.1725826 total: 55.7ms remaining: 26.2ms
68: learn: 0.1706549 total: 56.7ms remaining: 25.5ms
69: learn: 0.1692851 total: 57.8ms remaining: 24.8ms
70: learn: 0.1672780 total: 58.9ms remaining: 24.1ms
71: learn: 0.1645516 total: 59.8ms remaining: 23.3ms
72: learn: 0.1618093 total: 60.9ms remaining: 22.5ms
73: learn: 0.1599470 total: 62ms remaining: 21.8ms
74: learn: 0.1585582 total: 63.2ms remaining: 21.1ms
75: learn: 0.1559920 total: 64.4ms remaining: 20.3ms
76: learn: 0.1541175 total: 65.5ms remaining: 19.6ms
77: learn: 0.1526668 total: 66.4ms remaining: 18.7ms
78: learn: 0.1509238 total: 67.3ms remaining: 17.9ms
79: learn: 0.1493245 total: 68.3ms remaining: 17.1ms
80: learn: 0.1476028 total: 69.3ms remaining: 16.3ms
81: learn: 0.1451814 total: 70.3ms remaining: 15.4ms
82: learn: 0.1436671 total: 71.1ms remaining: 14.6ms
83: learn: 0.1424078 total: 72.1ms remaining: 13.7ms
84: learn: 0.1409789 total: 73ms remaining: 12.9ms
85: learn: 0.1389015 total: 73.6ms remaining: 12ms
86: learn: 0.1377363 total: 74.5ms remaining: 11.1ms
87: learn: 0.1365062 total: 75.4ms remaining: 10.3ms
88: learn: 0.1354969 total: 76.1ms remaining: 9.41ms
89: learn: 0.1344998 total: 76.9ms remaining: 8.54ms
90: learn: 0.1335139 total: 77.9ms remaining: 7.7ms
91: learn: 0.1320630 total: 78.7ms remaining: 6.85ms
92: learn: 0.1304506 total: 79.4ms remaining: 5.97ms
93: learn: 0.1291335 total: 80.2ms remaining: 5.12ms
94: learn: 0.1281305 total: 80.9ms remaining: 4.26ms
95: learn: 0.1272265 total: 81.8ms remaining: 3.41ms
96: learn: 0.1259320 total: 82.7ms remaining: 2.56ms
97: learn: 0.1245168 total: 83.6ms remaining: 1.71ms
98: learn: 0.1237258 total: 84.5ms remaining: 853us
99: learn: 0.1220054 total: 85.6ms remaining: 0us
0: learn: 0.6534941 total: 781us remaining: 77.4ms
1: learn: 0.6122449 total: 1.45ms remaining: 71ms
2: learn: 0.5769827 total: 2.06ms remaining: 66.7ms
3: learn: 0.5450784 total: 3.23ms remaining: 77.5ms
4: learn: 0.5199399 total: 3.94ms remaining: 75ms
5: learn: 0.4947532 total: 4.69ms remaining: 73.5ms
6: learn: 0.4703112 total: 5.76ms remaining: 76.5ms
7: learn: 0.4494416 total: 6.57ms remaining: 75.6ms
8: learn: 0.4298333 total: 7.44ms remaining: 75.3ms
9: learn: 0.4160616 total: 7.92ms remaining: 71.3ms
10: learn: 0.4025642 total: 8.94ms remaining: 72.4ms
11: learn: 0.3853415 total: 9.72ms remaining: 71.3ms
12: learn: 0.3727685 total: 11.5ms remaining: 76.8ms
13: learn: 0.3608699 total: 12.3ms remaining: 75.6ms
14: learn: 0.3496626 total: 12.8ms remaining: 72.8ms
15: learn: 0.3392487 total: 13.4ms remaining: 70.2ms
16: learn: 0.3287677 total: 14.9ms remaining: 72.8ms
17: learn: 0.3236872 total: 15.7ms remaining: 71.3ms
18: learn: 0.3140516 total: 16.2ms remaining: 69.2ms
19: learn: 0.3051473 total: 17.5ms remaining: 70.1ms
20: learn: 0.2994371 total: 18.1ms remaining: 68.3ms
21: learn: 0.2929577 total: 18.9ms remaining: 67ms
22: learn: 0.2855741 total: 19.8ms remaining: 66.3ms
23: learn: 0.2793941 total: 20.6ms remaining: 65.2ms
24: learn: 0.2750060 total: 21.7ms remaining: 65.1ms
25: learn: 0.2691104 total: 22.7ms remaining: 64.5ms
26: learn: 0.2622555 total: 23.6ms remaining: 63.7ms
27: learn: 0.2564873 total: 24.9ms remaining: 63.9ms
28: learn: 0.2530226 total: 25.6ms remaining: 62.6ms
29: learn: 0.2480208 total: 26.2ms remaining: 61.2ms
30: learn: 0.2438530 total: 26.6ms remaining: 59.2ms
31: learn: 0.2406571 total: 27.2ms remaining: 57.8ms
32: learn: 0.2361800 total: 27.9ms remaining: 56.7ms
33: learn: 0.2335486 total: 29.4ms remaining: 57.1ms
34: learn: 0.2296192 total: 30ms remaining: 55.7ms
35: learn: 0.2280074 total: 30.3ms remaining: 53.9ms
36: learn: 0.2268101 total: 31ms remaining: 52.8ms
37: learn: 0.2246386 total: 31.9ms remaining: 52ms
38: learn: 0.2218528 total: 32.4ms remaining: 50.7ms
39: learn: 0.2191333 total: 33.9ms remaining: 50.9ms
40: learn: 0.2161464 total: 34.6ms remaining: 49.8ms
41: learn: 0.2136653 total: 35.3ms remaining: 48.8ms
42: learn: 0.2110949 total: 36ms remaining: 47.7ms
43: learn: 0.2093502 total: 36.8ms remaining: 46.9ms
44: learn: 0.2073207 total: 38ms remaining: 46.4ms
45: learn: 0.2050560 total: 38.4ms remaining: 45.1ms
46: learn: 0.2020952 total: 39.2ms remaining: 44.3ms
47: learn: 0.2004267 total: 39.9ms remaining: 43.2ms
48: learn: 0.1976483 total: 41.6ms remaining: 43.3ms
49: learn: 0.1969053 total: 42.1ms remaining: 42.1ms
50: learn: 0.1941347 total: 42.7ms remaining: 41.1ms
51: learn: 0.1928375 total: 43.3ms remaining: 40ms
52: learn: 0.1907508 total: 44.8ms remaining: 39.7ms
53: learn: 0.1873079 total: 45.3ms remaining: 38.6ms
54: learn: 0.1846907 total: 46.7ms remaining: 38.2ms
55: learn: 0.1834587 total: 47.5ms remaining: 37.3ms
56: learn: 0.1825657 total: 48.4ms remaining: 36.5ms
57: learn: 0.1808045 total: 48.9ms remaining: 35.4ms
58: learn: 0.1789457 total: 49.5ms remaining: 34.4ms
59: learn: 0.1776847 total: 50.1ms remaining: 33.4ms
60: learn: 0.1747824 total: 51.2ms remaining: 32.7ms
61: learn: 0.1728133 total: 51.8ms remaining: 31.8ms
62: learn: 0.1707909 total: 52.4ms remaining: 30.8ms
63: learn: 0.1688952 total: 53.4ms remaining: 30.1ms
64: learn: 0.1670873 total: 54.3ms remaining: 29.2ms
65: learn: 0.1654977 total: 54.9ms remaining: 28.3ms
66: learn: 0.1634131 total: 55.7ms remaining: 27.4ms
67: learn: 0.1616935 total: 56.3ms remaining: 26.5ms
68: learn: 0.1598203 total: 57ms remaining: 25.6ms
69: learn: 0.1579050 total: 58.1ms remaining: 24.9ms
70: learn: 0.1560556 total: 58.9ms remaining: 24.1ms
71: learn: 0.1535245 total: 60.5ms remaining: 23.5ms
72: learn: 0.1521208 total: 61.2ms remaining: 22.6ms
73: learn: 0.1504559 total: 62.1ms remaining: 21.8ms
74: learn: 0.1490340 total: 63ms remaining: 21ms
75: learn: 0.1478127 total: 64.6ms remaining: 20.4ms
76: learn: 0.1461005 total: 65.3ms remaining: 19.5ms
77: learn: 0.1445503 total: 67.1ms remaining: 18.9ms
78: learn: 0.1431991 total: 67.9ms remaining: 18.1ms
79: learn: 0.1420370 total: 68.6ms remaining: 17.1ms
80: learn: 0.1403294 total: 69.6ms remaining: 16.3ms
81: learn: 0.1388279 total: 70.3ms remaining: 15.4ms
82: learn: 0.1379079 total: 71.5ms remaining: 14.6ms
83: learn: 0.1372589 total: 72.4ms remaining: 13.8ms
84: learn: 0.1359653 total: 73.3ms remaining: 12.9ms
85: learn: 0.1345054 total: 73.9ms remaining: 12ms
86: learn: 0.1332332 total: 74.8ms remaining: 11.2ms
87: learn: 0.1317270 total: 75.5ms remaining: 10.3ms
88: learn: 0.1300325 total: 76.4ms remaining: 9.44ms
89: learn: 0.1290779 total: 77.2ms remaining: 8.57ms
90: learn: 0.1273862 total: 78.2ms remaining: 7.73ms
91: learn: 0.1264447 total: 78.8ms remaining: 6.85ms
92: learn: 0.1254657 total: 79.5ms remaining: 5.98ms
93: learn: 0.1244671 total: 80.4ms remaining: 5.13ms
94: learn: 0.1230889 total: 81.3ms remaining: 4.28ms
95: learn: 0.1220404 total: 82.3ms remaining: 3.43ms
96: learn: 0.1210978 total: 83.2ms remaining: 2.57ms
97: learn: 0.1202150 total: 84ms remaining: 1.71ms
98: learn: 0.1189599 total: 85.1ms remaining: 859us
99: learn: 0.1180836 total: 85.8ms remaining: 0us
0: learn: 0.6476609 total: 525us remaining: 52ms
1: learn: 0.6078913 total: 1.48ms remaining: 72.6ms
2: learn: 0.5838627 total: 2.87ms remaining: 92.8ms
3: learn: 0.5587682 total: 4.88ms remaining: 117ms
4: learn: 0.5268380 total: 5.59ms remaining: 106ms
5: learn: 0.5053589 total: 6.22ms remaining: 97.4ms
6: learn: 0.4802092 total: 6.87ms remaining: 91.2ms
7: learn: 0.4607218 total: 8.17ms remaining: 93.9ms
8: learn: 0.4412163 total: 9.28ms remaining: 93.9ms
9: learn: 0.4208723 total: 10ms remaining: 90.1ms
10: learn: 0.4055317 total: 10.4ms remaining: 84.5ms
11: learn: 0.3942299 total: 12.2ms remaining: 89.4ms
12: learn: 0.3861527 total: 13.5ms remaining: 90.2ms
13: learn: 0.3773984 total: 14.4ms remaining: 88.5ms
14: learn: 0.3650244 total: 15.5ms remaining: 87.9ms
15: learn: 0.3543091 total: 16.3ms remaining: 85.4ms
16: learn: 0.3475590 total: 16.8ms remaining: 81.9ms
17: learn: 0.3398008 total: 17.2ms remaining: 78.4ms
18: learn: 0.3337312 total: 17.7ms remaining: 75.3ms
19: learn: 0.3257317 total: 18.2ms remaining: 72.9ms
20: learn: 0.3176647 total: 19ms remaining: 71.3ms
21: learn: 0.3113245 total: 19.7ms remaining: 69.8ms
22: learn: 0.3060822 total: 20.3ms remaining: 68.1ms
23: learn: 0.3013323 total: 20.5ms remaining: 64.8ms
24: learn: 0.2961287 total: 21.3ms remaining: 63.9ms
25: learn: 0.2900945 total: 22.1ms remaining: 62.8ms
26: learn: 0.2839752 total: 22.8ms remaining: 61.6ms
27: learn: 0.2776371 total: 23.4ms remaining: 60.2ms
28: learn: 0.2747837 total: 23.6ms remaining: 57.7ms
29: learn: 0.2714390 total: 24.1ms remaining: 56.3ms
30: learn: 0.2677248 total: 24.3ms remaining: 54.1ms
31: learn: 0.2649790 total: 24.8ms remaining: 52.8ms
32: learn: 0.2620417 total: 25.3ms remaining: 51.4ms
33: learn: 0.2575582 total: 25.9ms remaining: 50.3ms
34: learn: 0.2541548 total: 26.4ms remaining: 49.1ms
35: learn: 0.2520526 total: 26.9ms remaining: 47.9ms
36: learn: 0.2479510 total: 27.5ms remaining: 46.9ms
37: learn: 0.2449521 total: 28.4ms remaining: 46.3ms
38: learn: 0.2421768 total: 29.1ms remaining: 45.5ms
39: learn: 0.2390914 total: 29.9ms remaining: 44.8ms
40: learn: 0.2378562 total: 30.4ms remaining: 43.7ms
41: learn: 0.2356405 total: 31.1ms remaining: 43ms
42: learn: 0.2335215 total: 32.4ms remaining: 42.9ms
43: learn: 0.2311720 total: 33ms remaining: 42ms
44: learn: 0.2283697 total: 33.7ms remaining: 41.1ms
45: learn: 0.2243870 total: 34.5ms remaining: 40.5ms
46: learn: 0.2211732 total: 35.2ms remaining: 39.7ms
47: learn: 0.2177466 total: 35.7ms remaining: 38.7ms
48: learn: 0.2156051 total: 36.1ms remaining: 37.5ms
49: learn: 0.2122104 total: 36.8ms remaining: 36.8ms
50: learn: 0.2108903 total: 37.2ms remaining: 35.7ms
51: learn: 0.2092450 total: 38ms remaining: 35.1ms
52: learn: 0.2071529 total: 38.7ms remaining: 34.3ms
53: learn: 0.2036525 total: 39.4ms remaining: 33.6ms
54: learn: 0.2022467 total: 40ms remaining: 32.8ms
55: learn: 0.2004191 total: 41.3ms remaining: 32.5ms
56: learn: 0.1985497 total: 41.9ms remaining: 31.6ms
57: learn: 0.1962502 total: 42.7ms remaining: 30.9ms
58: learn: 0.1932903 total: 43.4ms remaining: 30.1ms
59: learn: 0.1912722 total: 44.5ms remaining: 29.7ms
60: learn: 0.1892666 total: 45.3ms remaining: 29ms
61: learn: 0.1871679 total: 46.5ms remaining: 28.5ms
62: learn: 0.1844720 total: 47.1ms remaining: 27.6ms
63: learn: 0.1820783 total: 48ms remaining: 27ms
64: learn: 0.1805908 total: 49.1ms remaining: 26.4ms
65: learn: 0.1790196 total: 50.4ms remaining: 26ms
66: learn: 0.1772076 total: 51ms remaining: 25.1ms
67: learn: 0.1752804 total: 52.4ms remaining: 24.7ms
68: learn: 0.1739981 total: 53ms remaining: 23.8ms
69: learn: 0.1716495 total: 53.8ms remaining: 23.1ms
70: learn: 0.1700248 total: 54.6ms remaining: 22.3ms
71: learn: 0.1679663 total: 56.1ms remaining: 21.8ms
72: learn: 0.1656447 total: 56.8ms remaining: 21ms
73: learn: 0.1641300 total: 57.5ms remaining: 20.2ms
74: learn: 0.1627951 total: 58.5ms remaining: 19.5ms
75: learn: 0.1612069 total: 59.2ms remaining: 18.7ms
76: learn: 0.1597159 total: 59.9ms remaining: 17.9ms
77: learn: 0.1582509 total: 61.4ms remaining: 17.3ms
78: learn: 0.1568882 total: 62.4ms remaining: 16.6ms
79: learn: 0.1557541 total: 63.5ms remaining: 15.9ms
80: learn: 0.1544303 total: 64.4ms remaining: 15.1ms
81: learn: 0.1532035 total: 65.3ms remaining: 14.3ms
82: learn: 0.1517390 total: 66.1ms remaining: 13.5ms
83: learn: 0.1505826 total: 67ms remaining: 12.8ms
84: learn: 0.1495833 total: 67.7ms remaining: 12ms
85: learn: 0.1480833 total: 68.5ms remaining: 11.2ms
86: learn: 0.1469503 total: 69.5ms remaining: 10.4ms
87: learn: 0.1461566 total: 71ms remaining: 9.68ms
88: learn: 0.1451075 total: 71.8ms remaining: 8.88ms
89: learn: 0.1441472 total: 72.9ms remaining: 8.1ms
90: learn: 0.1430349 total: 74.6ms remaining: 7.38ms
91: learn: 0.1417471 total: 75.8ms remaining: 6.59ms
92: learn: 0.1407590 total: 76.7ms remaining: 5.77ms
93: learn: 0.1395519 total: 78.3ms remaining: 5ms
94: learn: 0.1384687 total: 79.2ms remaining: 4.17ms
95: learn: 0.1372771 total: 80ms remaining: 3.33ms
96: learn: 0.1364788 total: 82.6ms remaining: 2.55ms
97: learn: 0.1356838 total: 83.5ms remaining: 1.7ms
98: learn: 0.1345136 total: 84.2ms remaining: 850us
99: learn: 0.1334763 total: 86.4ms remaining: 0us
0: learn: 0.6476069 total: 427us remaining: 42.3ms
1: learn: 0.6099705 total: 895us remaining: 43.9ms
2: learn: 0.5765838 total: 1.35ms remaining: 43.5ms
3: learn: 0.5427603 total: 1.78ms remaining: 42.8ms
4: learn: 0.5151544 total: 2.36ms remaining: 44.9ms
5: learn: 0.4924592 total: 2.86ms remaining: 44.8ms
6: learn: 0.4694414 total: 3.37ms remaining: 44.8ms
7: learn: 0.4535522 total: 3.97ms remaining: 45.7ms
8: learn: 0.4367589 total: 4.41ms remaining: 44.6ms
9: learn: 0.4192366 total: 4.75ms remaining: 42.7ms
10: learn: 0.4047310 total: 4.86ms remaining: 39.3ms
11: learn: 0.3939310 total: 5.17ms remaining: 37.9ms
12: learn: 0.3829631 total: 5.85ms remaining: 39.1ms
13: learn: 0.3716386 total: 6.5ms remaining: 39.9ms
14: learn: 0.3640746 total: 7.03ms remaining: 39.8ms
15: learn: 0.3538183 total: 7.5ms remaining: 39.4ms
16: learn: 0.3456950 total: 7.99ms remaining: 39ms
17: learn: 0.3368499 total: 8.51ms remaining: 38.8ms
18: learn: 0.3281077 total: 9.14ms remaining: 39ms
19: learn: 0.3206841 total: 9.65ms remaining: 38.6ms
20: learn: 0.3135813 total: 10.3ms remaining: 38.6ms
21: learn: 0.3073593 total: 10.8ms remaining: 38.2ms
22: learn: 0.3027511 total: 11.5ms remaining: 38.4ms
23: learn: 0.2978065 total: 12.6ms remaining: 40ms
24: learn: 0.2951878 total: 13.3ms remaining: 39.9ms
25: learn: 0.2918457 total: 14.2ms remaining: 40.3ms
26: learn: 0.2893576 total: 14.6ms remaining: 39.4ms
27: learn: 0.2841110 total: 15.3ms remaining: 39.2ms
28: learn: 0.2800618 total: 16.1ms remaining: 39.4ms
29: learn: 0.2749841 total: 16.8ms remaining: 39.3ms
30: learn: 0.2729425 total: 17.6ms remaining: 39.1ms
31: learn: 0.2681197 total: 18.3ms remaining: 39ms
32: learn: 0.2627012 total: 19.4ms remaining: 39.3ms
33: learn: 0.2591936 total: 20ms remaining: 38.8ms
34: learn: 0.2564770 total: 20.7ms remaining: 38.5ms
35: learn: 0.2527661 total: 21.7ms remaining: 38.6ms
36: learn: 0.2477277 total: 22.7ms remaining: 38.6ms
37: learn: 0.2461602 total: 23.3ms remaining: 38ms
38: learn: 0.2418578 total: 23.8ms remaining: 37.3ms
39: learn: 0.2390617 total: 24.9ms remaining: 37.3ms
40: learn: 0.2338773 total: 25.7ms remaining: 37ms
41: learn: 0.2302547 total: 26.3ms remaining: 36.3ms
42: learn: 0.2288855 total: 27.1ms remaining: 35.9ms
43: learn: 0.2262853 total: 27.8ms remaining: 35.4ms
44: learn: 0.2236002 total: 28.8ms remaining: 35.2ms
45: learn: 0.2200589 total: 29.9ms remaining: 35.1ms
46: learn: 0.2176929 total: 31ms remaining: 35ms
47: learn: 0.2143357 total: 31.8ms remaining: 34.4ms
48: learn: 0.2130031 total: 32.5ms remaining: 33.8ms
49: learn: 0.2103242 total: 32.7ms remaining: 32.7ms
50: learn: 0.2084953 total: 33.3ms remaining: 32ms
51: learn: 0.2063611 total: 34.1ms remaining: 31.4ms
52: learn: 0.2047310 total: 34.8ms remaining: 30.9ms
53: learn: 0.2026757 total: 35.5ms remaining: 30.3ms
54: learn: 0.1998299 total: 36.1ms remaining: 29.5ms
55: learn: 0.1979241 total: 36.7ms remaining: 28.9ms
56: learn: 0.1971645 total: 37.9ms remaining: 28.6ms
57: learn: 0.1951774 total: 38.7ms remaining: 28ms
58: learn: 0.1921831 total: 39.4ms remaining: 27.4ms
59: learn: 0.1898358 total: 40ms remaining: 26.7ms
60: learn: 0.1876472 total: 41.2ms remaining: 26.3ms
61: learn: 0.1858316 total: 42.4ms remaining: 26ms
62: learn: 0.1838957 total: 43.4ms remaining: 25.5ms
63: learn: 0.1822422 total: 44.5ms remaining: 25ms
64: learn: 0.1806515 total: 45.7ms remaining: 24.6ms
65: learn: 0.1786235 total: 46.6ms remaining: 24ms
66: learn: 0.1761166 total: 47.4ms remaining: 23.3ms
67: learn: 0.1752360 total: 48.2ms remaining: 22.7ms
68: learn: 0.1732268 total: 48.9ms remaining: 22ms
69: learn: 0.1710478 total: 49.6ms remaining: 21.2ms
70: learn: 0.1684948 total: 50.4ms remaining: 20.6ms
71: learn: 0.1669153 total: 51.9ms remaining: 20.2ms
72: learn: 0.1653334 total: 52.8ms remaining: 19.5ms
73: learn: 0.1637129 total: 53.7ms remaining: 18.9ms
74: learn: 0.1615837 total: 54.4ms remaining: 18.1ms
75: learn: 0.1597643 total: 55ms remaining: 17.4ms
76: learn: 0.1581688 total: 56.1ms remaining: 16.7ms
77: learn: 0.1570969 total: 56.8ms remaining: 16ms
78: learn: 0.1554987 total: 57.9ms remaining: 15.4ms
79: learn: 0.1538170 total: 58.9ms remaining: 14.7ms
80: learn: 0.1524636 total: 59.6ms remaining: 14ms
81: learn: 0.1514423 total: 60.6ms remaining: 13.3ms
82: learn: 0.1495602 total: 61.5ms remaining: 12.6ms
83: learn: 0.1476546 total: 62.2ms remaining: 11.8ms
84: learn: 0.1461042 total: 64.1ms remaining: 11.3ms
85: learn: 0.1447650 total: 64.7ms remaining: 10.5ms
86: learn: 0.1434032 total: 66.2ms remaining: 9.9ms
87: learn: 0.1418466 total: 67.1ms remaining: 9.15ms
88: learn: 0.1399695 total: 68ms remaining: 8.4ms
89: learn: 0.1386902 total: 68.8ms remaining: 7.64ms
90: learn: 0.1373321 total: 69.5ms remaining: 6.88ms
91: learn: 0.1363706 total: 70.2ms remaining: 6.11ms
92: learn: 0.1355587 total: 71.1ms remaining: 5.35ms
93: learn: 0.1343757 total: 71.7ms remaining: 4.58ms
94: learn: 0.1331809 total: 74ms remaining: 3.89ms
95: learn: 0.1322673 total: 75.3ms remaining: 3.14ms
96: learn: 0.1313192 total: 76.2ms remaining: 2.36ms
97: learn: 0.1305032 total: 77.1ms remaining: 1.57ms
98: learn: 0.1294217 total: 78.5ms remaining: 793us
99: learn: 0.1284574 total: 79.1ms remaining: 0us
0: learn: 0.6419363 total: 540us remaining: 53.5ms
1: learn: 0.6011176 total: 1.11ms remaining: 54.4ms
2: learn: 0.5650957 total: 1.76ms remaining: 56.8ms
3: learn: 0.5351090 total: 2.37ms remaining: 56.9ms
4: learn: 0.5040384 total: 2.92ms remaining: 55.6ms
5: learn: 0.4787169 total: 3.98ms remaining: 62.3ms
6: learn: 0.4553862 total: 4.58ms remaining: 60.8ms
7: learn: 0.4341118 total: 5.34ms remaining: 61.4ms
8: learn: 0.4170355 total: 5.91ms remaining: 59.8ms
9: learn: 0.4032727 total: 6.74ms remaining: 60.7ms
10: learn: 0.3912721 total: 7.67ms remaining: 62ms
11: learn: 0.3798536 total: 8.36ms remaining: 61.3ms
12: learn: 0.3686481 total: 9.78ms remaining: 65.4ms
13: learn: 0.3550926 total: 10.4ms remaining: 63.7ms
14: learn: 0.3444844 total: 11.3ms remaining: 64ms
15: learn: 0.3355957 total: 12ms remaining: 63ms
16: learn: 0.3291119 total: 12.7ms remaining: 61.8ms
17: learn: 0.3197235 total: 14.4ms remaining: 65.6ms
18: learn: 0.3112880 total: 14.9ms remaining: 63.7ms
19: learn: 0.3032755 total: 16.1ms remaining: 64.6ms
20: learn: 0.2962665 total: 17.3ms remaining: 65.1ms
21: learn: 0.2890773 total: 17.9ms remaining: 63.5ms
22: learn: 0.2838434 total: 18.4ms remaining: 61.6ms
23: learn: 0.2781875 total: 20.5ms remaining: 64.8ms
24: learn: 0.2731688 total: 21.8ms remaining: 65.3ms
25: learn: 0.2682044 total: 22.4ms remaining: 63.9ms
26: learn: 0.2636309 total: 23.1ms remaining: 62.4ms
27: learn: 0.2581415 total: 25.7ms remaining: 66ms
28: learn: 0.2540585 total: 26.7ms remaining: 65.4ms
29: learn: 0.2496847 total: 27.7ms remaining: 64.6ms
30: learn: 0.2484047 total: 28ms remaining: 62.4ms
31: learn: 0.2441142 total: 29.8ms remaining: 63.3ms
32: learn: 0.2380824 total: 31.4ms remaining: 63.8ms
33: learn: 0.2340262 total: 32.9ms remaining: 63.9ms
34: learn: 0.2297177 total: 34.4ms remaining: 63.9ms
35: learn: 0.2263603 total: 35.4ms remaining: 62.9ms
36: learn: 0.2233339 total: 37.1ms remaining: 63.2ms
37: learn: 0.2204648 total: 38.1ms remaining: 62.1ms
38: learn: 0.2164013 total: 39.5ms remaining: 61.8ms
39: learn: 0.2139482 total: 41.7ms remaining: 62.5ms
40: learn: 0.2108738 total: 43.8ms remaining: 63.1ms
41: learn: 0.2083536 total: 45.5ms remaining: 62.9ms
42: learn: 0.2045092 total: 47ms remaining: 62.3ms
43: learn: 0.2032248 total: 49.5ms remaining: 62.9ms
44: learn: 0.2018938 total: 50.4ms remaining: 61.6ms
45: learn: 0.1991045 total: 52.1ms remaining: 61.1ms
46: learn: 0.1973864 total: 53.5ms remaining: 60.3ms
47: learn: 0.1955079 total: 55ms remaining: 59.5ms
48: learn: 0.1931020 total: 56.6ms remaining: 58.9ms
49: learn: 0.1912296 total: 58.4ms remaining: 58.4ms
50: learn: 0.1891984 total: 60.1ms remaining: 57.7ms
51: learn: 0.1866213 total: 62ms remaining: 57.2ms
52: learn: 0.1836039 total: 64.6ms remaining: 57.2ms
53: learn: 0.1819193 total: 67ms remaining: 57.1ms
54: learn: 0.1801093 total: 68.6ms remaining: 56.2ms
55: learn: 0.1783823 total: 70.8ms remaining: 55.6ms
56: learn: 0.1769560 total: 72.4ms remaining: 54.6ms
57: learn: 0.1730717 total: 75ms remaining: 54.3ms
58: learn: 0.1697826 total: 76.7ms remaining: 53.3ms
59: learn: 0.1676643 total: 78.4ms remaining: 52.3ms
60: learn: 0.1664159 total: 79.9ms remaining: 51.1ms
61: learn: 0.1635990 total: 81.8ms remaining: 50.1ms
62: learn: 0.1607245 total: 83.6ms remaining: 49.1ms
63: learn: 0.1586759 total: 85ms remaining: 47.8ms
64: learn: 0.1571391 total: 86.6ms remaining: 46.6ms
65: learn: 0.1546529 total: 88.1ms remaining: 45.4ms
66: learn: 0.1525521 total: 89.4ms remaining: 44ms
67: learn: 0.1498612 total: 90.4ms remaining: 42.5ms
68: learn: 0.1476515 total: 91.5ms remaining: 41.1ms
69: learn: 0.1455110 total: 93ms remaining: 39.9ms
70: learn: 0.1434805 total: 94.7ms remaining: 38.7ms
71: learn: 0.1412362 total: 96.3ms remaining: 37.4ms
72: learn: 0.1390986 total: 97.7ms remaining: 36.1ms
73: learn: 0.1372169 total: 98.7ms remaining: 34.7ms
74: learn: 0.1349601 total: 100ms remaining: 33.4ms
75: learn: 0.1330158 total: 102ms remaining: 32.2ms
76: learn: 0.1308058 total: 105ms remaining: 31.3ms
77: learn: 0.1287715 total: 107ms remaining: 30.2ms
78: learn: 0.1272862 total: 109ms remaining: 29ms
79: learn: 0.1260736 total: 111ms remaining: 27.7ms
80: learn: 0.1253311 total: 112ms remaining: 26.4ms
81: learn: 0.1233086 total: 114ms remaining: 25ms
82: learn: 0.1214652 total: 116ms remaining: 23.8ms
83: learn: 0.1204899 total: 118ms remaining: 22.4ms
84: learn: 0.1188855 total: 120ms remaining: 21.1ms
85: learn: 0.1176528 total: 121ms remaining: 19.8ms
86: learn: 0.1161560 total: 124ms remaining: 18.5ms
87: learn: 0.1148655 total: 125ms remaining: 17ms
88: learn: 0.1133994 total: 126ms remaining: 15.6ms
89: learn: 0.1120974 total: 127ms remaining: 14.1ms
90: learn: 0.1108607 total: 128ms remaining: 12.7ms
91: learn: 0.1094853 total: 129ms remaining: 11.2ms
92: learn: 0.1081737 total: 130ms remaining: 9.82ms
93: learn: 0.1067023 total: 131ms remaining: 8.39ms
94: learn: 0.1054725 total: 132ms remaining: 6.96ms
95: learn: 0.1044273 total: 133ms remaining: 5.55ms
96: learn: 0.1032615 total: 134ms remaining: 4.16ms
97: learn: 0.1024604 total: 135ms remaining: 2.75ms
98: learn: 0.1011906 total: 136ms remaining: 1.38ms
99: learn: 0.1001702 total: 137ms remaining: 0us
0: learn: 0.6458911 total: 564us remaining: 55.9ms
1: learn: 0.6098312 total: 1.15ms remaining: 56.5ms
2: learn: 0.5719195 total: 1.7ms remaining: 55ms
3: learn: 0.5372780 total: 1.82ms remaining: 43.7ms
4: learn: 0.5094653 total: 2.25ms remaining: 42.7ms
5: learn: 0.4869324 total: 3ms remaining: 47ms
6: learn: 0.4639285 total: 3.45ms remaining: 45.8ms
7: learn: 0.4424897 total: 3.95ms remaining: 45.5ms
8: learn: 0.4271859 total: 4.24ms remaining: 42.9ms
9: learn: 0.4113984 total: 4.66ms remaining: 41.9ms
10: learn: 0.3951133 total: 5.06ms remaining: 41ms
11: learn: 0.3832256 total: 5.52ms remaining: 40.5ms
12: learn: 0.3720227 total: 5.96ms remaining: 39.9ms
13: learn: 0.3609613 total: 6.63ms remaining: 40.7ms
14: learn: 0.3515607 total: 7.11ms remaining: 40.3ms
15: learn: 0.3432732 total: 7.68ms remaining: 40.3ms
16: learn: 0.3364668 total: 8.14ms remaining: 39.7ms
17: learn: 0.3283772 total: 8.69ms remaining: 39.6ms
18: learn: 0.3221569 total: 9.38ms remaining: 40ms
19: learn: 0.3176788 total: 10.3ms remaining: 41.1ms
20: learn: 0.3096443 total: 11ms remaining: 41.5ms
21: learn: 0.3010229 total: 11.9ms remaining: 42ms
22: learn: 0.2947529 total: 12.8ms remaining: 42.7ms
23: learn: 0.2893152 total: 13.7ms remaining: 43.2ms
24: learn: 0.2865792 total: 14.5ms remaining: 43.6ms
25: learn: 0.2797334 total: 15.9ms remaining: 45.2ms
26: learn: 0.2745505 total: 16.8ms remaining: 45.3ms
27: learn: 0.2689394 total: 17.8ms remaining: 45.8ms
28: learn: 0.2629653 total: 19.1ms remaining: 46.9ms
29: learn: 0.2583618 total: 20ms remaining: 46.6ms
30: learn: 0.2532520 total: 21.5ms remaining: 47.9ms
31: learn: 0.2483569 total: 22.5ms remaining: 47.7ms
32: learn: 0.2449190 total: 23.7ms remaining: 48ms
33: learn: 0.2427465 total: 24.5ms remaining: 47.6ms
34: learn: 0.2399617 total: 25.8ms remaining: 47.8ms
35: learn: 0.2367692 total: 26.4ms remaining: 46.9ms
36: learn: 0.2343335 total: 27.5ms remaining: 46.8ms
37: learn: 0.2318076 total: 28.5ms remaining: 46.4ms
38: learn: 0.2295396 total: 29.2ms remaining: 45.7ms
39: learn: 0.2278173 total: 30.7ms remaining: 46ms
40: learn: 0.2238035 total: 31.3ms remaining: 45ms
41: learn: 0.2214597 total: 32.1ms remaining: 44.3ms
42: learn: 0.2183419 total: 32.7ms remaining: 43.3ms
43: learn: 0.2163152 total: 33.6ms remaining: 42.8ms
44: learn: 0.2142766 total: 34.5ms remaining: 42.2ms
45: learn: 0.2114595 total: 35.3ms remaining: 41.5ms
46: learn: 0.2094141 total: 35.9ms remaining: 40.5ms
47: learn: 0.2076379 total: 36.3ms remaining: 39.3ms
48: learn: 0.2062048 total: 37.3ms remaining: 38.8ms
49: learn: 0.2037692 total: 37.9ms remaining: 37.9ms
50: learn: 0.2012822 total: 38.9ms remaining: 37.4ms
51: learn: 0.1995988 total: 39.6ms remaining: 36.5ms
52: learn: 0.1974493 total: 40.2ms remaining: 35.6ms
53: learn: 0.1946770 total: 40.9ms remaining: 34.8ms
54: learn: 0.1930883 total: 41.7ms remaining: 34.1ms
55: learn: 0.1910026 total: 43.2ms remaining: 33.9ms
56: learn: 0.1896023 total: 44.1ms remaining: 33.3ms
57: learn: 0.1882956 total: 45ms remaining: 32.6ms
58: learn: 0.1864348 total: 46ms remaining: 32ms
59: learn: 0.1858779 total: 46.3ms remaining: 30.9ms
60: learn: 0.1834955 total: 46.9ms remaining: 30ms
61: learn: 0.1819352 total: 47.5ms remaining: 29.1ms
62: learn: 0.1800055 total: 48.6ms remaining: 28.5ms
63: learn: 0.1783279 total: 49.3ms remaining: 27.7ms
64: learn: 0.1757310 total: 50.1ms remaining: 27ms
65: learn: 0.1741460 total: 51.1ms remaining: 26.3ms
66: learn: 0.1725699 total: 52.2ms remaining: 25.7ms
67: learn: 0.1715953 total: 53.1ms remaining: 25ms
68: learn: 0.1698178 total: 54ms remaining: 24.3ms
69: learn: 0.1680054 total: 54.9ms remaining: 23.5ms
70: learn: 0.1667314 total: 55.9ms remaining: 22.8ms
71: learn: 0.1641113 total: 56.7ms remaining: 22ms
72: learn: 0.1626654 total: 57.6ms remaining: 21.3ms
73: learn: 0.1606705 total: 58.4ms remaining: 20.5ms
74: learn: 0.1586456 total: 59.5ms remaining: 19.8ms
75: learn: 0.1573660 total: 60.2ms remaining: 19ms
76: learn: 0.1555283 total: 60.9ms remaining: 18.2ms
77: learn: 0.1550416 total: 61.6ms remaining: 17.4ms
78: learn: 0.1533197 total: 62.6ms remaining: 16.6ms
79: learn: 0.1518345 total: 63.6ms remaining: 15.9ms
80: learn: 0.1501395 total: 64.4ms remaining: 15.1ms
81: learn: 0.1487209 total: 65.3ms remaining: 14.3ms
82: learn: 0.1478269 total: 66.3ms remaining: 13.6ms
83: learn: 0.1464552 total: 67.1ms remaining: 12.8ms
84: learn: 0.1451952 total: 67.8ms remaining: 12ms
85: learn: 0.1439841 total: 68.9ms remaining: 11.2ms
86: learn: 0.1425445 total: 70.4ms remaining: 10.5ms
87: learn: 0.1410727 total: 71.2ms remaining: 9.71ms
88: learn: 0.1394445 total: 71.8ms remaining: 8.88ms
89: learn: 0.1386525 total: 72.7ms remaining: 8.07ms
90: learn: 0.1378862 total: 74.5ms remaining: 7.37ms
91: learn: 0.1364339 total: 75.4ms remaining: 6.55ms
92: learn: 0.1356940 total: 76.1ms remaining: 5.72ms
93: learn: 0.1347577 total: 77.1ms remaining: 4.92ms
94: learn: 0.1337984 total: 78.1ms remaining: 4.11ms
95: learn: 0.1327860 total: 78.8ms remaining: 3.28ms
96: learn: 0.1318867 total: 79.7ms remaining: 2.47ms
97: learn: 0.1304096 total: 80.6ms remaining: 1.64ms
98: learn: 0.1294207 total: 82.2ms remaining: 830us
99: learn: 0.1284818 total: 83ms remaining: 0us
0: learn: 0.6449164 total: 707us remaining: 70.1ms
1: learn: 0.6085246 total: 1.39ms remaining: 68.2ms
2: learn: 0.5715641 total: 1.99ms remaining: 64.4ms
3: learn: 0.5404080 total: 2.47ms remaining: 59.4ms
4: learn: 0.5092282 total: 2.97ms remaining: 56.5ms
5: learn: 0.4858956 total: 3.56ms remaining: 55.7ms
6: learn: 0.4624937 total: 4.09ms remaining: 54.3ms
7: learn: 0.4465534 total: 4.77ms remaining: 54.8ms
8: learn: 0.4296513 total: 5.34ms remaining: 54ms
9: learn: 0.4162316 total: 5.82ms remaining: 52.4ms
10: learn: 0.4039378 total: 6.77ms remaining: 54.8ms
11: learn: 0.3929279 total: 7.36ms remaining: 53.9ms
12: learn: 0.3783309 total: 8.07ms remaining: 54ms
13: learn: 0.3689193 total: 8.59ms remaining: 52.8ms
14: learn: 0.3599247 total: 9.13ms remaining: 51.7ms
15: learn: 0.3482234 total: 9.61ms remaining: 50.5ms
16: learn: 0.3409746 total: 10.2ms remaining: 49.7ms
17: learn: 0.3310210 total: 10.7ms remaining: 48.8ms
18: learn: 0.3223923 total: 11.2ms remaining: 47.7ms
19: learn: 0.3150966 total: 11.7ms remaining: 46.9ms
20: learn: 0.3096095 total: 12.3ms remaining: 46.4ms
21: learn: 0.3043243 total: 12.9ms remaining: 45.8ms
22: learn: 0.2982197 total: 13.3ms remaining: 44.6ms
23: learn: 0.2937462 total: 13.9ms remaining: 43.9ms
24: learn: 0.2894376 total: 14.4ms remaining: 43.3ms
25: learn: 0.2841506 total: 15ms remaining: 42.7ms
26: learn: 0.2805311 total: 15.6ms remaining: 42.2ms
27: learn: 0.2743572 total: 16.2ms remaining: 41.6ms
28: learn: 0.2709245 total: 16.7ms remaining: 40.9ms
29: learn: 0.2672337 total: 17.4ms remaining: 40.6ms
30: learn: 0.2644080 total: 17.9ms remaining: 39.9ms
31: learn: 0.2585723 total: 18.5ms remaining: 39.3ms
32: learn: 0.2560024 total: 19ms remaining: 38.6ms
33: learn: 0.2526009 total: 19.7ms remaining: 38.2ms
34: learn: 0.2492032 total: 20.7ms remaining: 38.4ms
35: learn: 0.2470315 total: 21.1ms remaining: 37.5ms
36: learn: 0.2431673 total: 21.8ms remaining: 37.2ms
37: learn: 0.2408352 total: 22.5ms remaining: 36.6ms
38: learn: 0.2378579 total: 23ms remaining: 36ms
39: learn: 0.2358564 total: 23.7ms remaining: 35.5ms
40: learn: 0.2325985 total: 24.3ms remaining: 35ms
41: learn: 0.2308968 total: 24.9ms remaining: 34.3ms
42: learn: 0.2281575 total: 25.4ms remaining: 33.7ms
43: learn: 0.2270003 total: 25.9ms remaining: 33ms
44: learn: 0.2243739 total: 26.5ms remaining: 32.3ms
45: learn: 0.2216083 total: 27.1ms remaining: 31.8ms
46: learn: 0.2184681 total: 27.7ms remaining: 31.3ms
47: learn: 0.2168333 total: 28.3ms remaining: 30.6ms
48: learn: 0.2134635 total: 28.9ms remaining: 30.1ms
49: learn: 0.2105444 total: 29.7ms remaining: 29.7ms
50: learn: 0.2077680 total: 30.4ms remaining: 29.2ms
51: learn: 0.2062496 total: 31ms remaining: 28.6ms
52: learn: 0.2041004 total: 32ms remaining: 28.4ms
53: learn: 0.2016130 total: 32.8ms remaining: 28ms
54: learn: 0.1989750 total: 33.8ms remaining: 27.6ms
55: learn: 0.1967034 total: 34.7ms remaining: 27.2ms
56: learn: 0.1943877 total: 35.5ms remaining: 26.8ms
57: learn: 0.1909132 total: 36.4ms remaining: 26.3ms
58: learn: 0.1888175 total: 37.3ms remaining: 25.9ms
59: learn: 0.1858295 total: 38.1ms remaining: 25.4ms
60: learn: 0.1837096 total: 39ms remaining: 24.9ms
61: learn: 0.1807538 total: 39.8ms remaining: 24.4ms
62: learn: 0.1786693 total: 40.6ms remaining: 23.9ms
63: learn: 0.1761744 total: 41.7ms remaining: 23.5ms
64: learn: 0.1743178 total: 42.6ms remaining: 23ms
65: learn: 0.1721721 total: 43.5ms remaining: 22.4ms
66: learn: 0.1698389 total: 44.5ms remaining: 21.9ms
67: learn: 0.1683310 total: 45.3ms remaining: 21.3ms
68: learn: 0.1667973 total: 46.1ms remaining: 20.7ms
69: learn: 0.1655109 total: 47.1ms remaining: 20.2ms
70: learn: 0.1635133 total: 48ms remaining: 19.6ms
71: learn: 0.1613183 total: 48.9ms remaining: 19ms
72: learn: 0.1595464 total: 49.6ms remaining: 18.3ms
73: learn: 0.1578993 total: 50.5ms remaining: 17.7ms
74: learn: 0.1563150 total: 51.8ms remaining: 17.3ms
75: learn: 0.1548879 total: 53.2ms remaining: 16.8ms
76: learn: 0.1536761 total: 54.1ms remaining: 16.2ms
77: learn: 0.1524447 total: 54.9ms remaining: 15.5ms
78: learn: 0.1509846 total: 56ms remaining: 14.9ms
79: learn: 0.1498490 total: 56.9ms remaining: 14.2ms
80: learn: 0.1483403 total: 58.2ms remaining: 13.6ms
81: learn: 0.1471602 total: 59.1ms remaining: 13ms
82: learn: 0.1451741 total: 59.7ms remaining: 12.2ms
83: learn: 0.1438251 total: 60.9ms remaining: 11.6ms
84: learn: 0.1426669 total: 61.5ms remaining: 10.9ms
85: learn: 0.1415382 total: 62.3ms remaining: 10.1ms
86: learn: 0.1403451 total: 64ms remaining: 9.56ms
87: learn: 0.1390236 total: 64.7ms remaining: 8.82ms
88: learn: 0.1378576 total: 65.7ms remaining: 8.12ms
89: learn: 0.1366472 total: 66.5ms remaining: 7.39ms
90: learn: 0.1356060 total: 67.4ms remaining: 6.66ms
91: learn: 0.1346185 total: 68ms remaining: 5.91ms
92: learn: 0.1334777 total: 69.1ms remaining: 5.2ms
93: learn: 0.1324954 total: 69.8ms remaining: 4.45ms
94: learn: 0.1314596 total: 70.4ms remaining: 3.71ms
95: learn: 0.1306033 total: 72.6ms remaining: 3.02ms
96: learn: 0.1295111 total: 73.3ms remaining: 2.27ms
97: learn: 0.1285475 total: 74ms remaining: 1.51ms
98: learn: 0.1273036 total: 74.8ms remaining: 755us
99: learn: 0.1260570 total: 75.6ms remaining: 0us
0: learn: 0.6440574 total: 545us remaining: 54ms
1: learn: 0.6071082 total: 1.1ms remaining: 53.9ms
2: learn: 0.5809191 total: 1.89ms remaining: 61ms
3: learn: 0.5513366 total: 2.48ms remaining: 59.5ms
4: learn: 0.5225763 total: 3.04ms remaining: 57.8ms
5: learn: 0.5004805 total: 3.45ms remaining: 54ms
6: learn: 0.4775513 total: 3.88ms remaining: 51.6ms
7: learn: 0.4622070 total: 4.15ms remaining: 47.7ms
8: learn: 0.4410241 total: 4.61ms remaining: 46.6ms
9: learn: 0.4249438 total: 5.11ms remaining: 46ms
10: learn: 0.4078417 total: 5.63ms remaining: 45.6ms
11: learn: 0.3959296 total: 6.02ms remaining: 44.2ms
12: learn: 0.3861366 total: 6.46ms remaining: 43.2ms
13: learn: 0.3747309 total: 7.41ms remaining: 45.5ms
14: learn: 0.3643013 total: 8.28ms remaining: 46.9ms
15: learn: 0.3549483 total: 9.15ms remaining: 48.1ms
16: learn: 0.3464547 total: 10.2ms remaining: 49.6ms
17: learn: 0.3386374 total: 11ms remaining: 50ms
18: learn: 0.3312841 total: 11.7ms remaining: 50.1ms
19: learn: 0.3235724 total: 12.5ms remaining: 50.1ms
20: learn: 0.3169947 total: 13.5ms remaining: 50.9ms
21: learn: 0.3096418 total: 14.5ms remaining: 51.4ms
22: learn: 0.3042768 total: 15.3ms remaining: 51.3ms
23: learn: 0.2978684 total: 15.7ms remaining: 49.7ms
24: learn: 0.2933065 total: 16.5ms remaining: 49.6ms
25: learn: 0.2842634 total: 17.4ms remaining: 49.4ms
26: learn: 0.2772658 total: 17.9ms remaining: 48.3ms
27: learn: 0.2736495 total: 18.4ms remaining: 47.4ms
28: learn: 0.2698878 total: 21.3ms remaining: 52.1ms
29: learn: 0.2629201 total: 22.8ms remaining: 53.3ms
30: learn: 0.2585210 total: 23.6ms remaining: 52.6ms
31: learn: 0.2557255 total: 24.5ms remaining: 52ms
32: learn: 0.2529692 total: 25.4ms remaining: 51.6ms
33: learn: 0.2489882 total: 26.1ms remaining: 50.8ms
34: learn: 0.2452763 total: 26.8ms remaining: 49.7ms
35: learn: 0.2415789 total: 27.2ms remaining: 48.3ms
36: learn: 0.2387921 total: 28.1ms remaining: 47.8ms
37: learn: 0.2351807 total: 28.8ms remaining: 47ms
38: learn: 0.2320092 total: 30.4ms remaining: 47.5ms
39: learn: 0.2298565 total: 31.2ms remaining: 46.7ms
40: learn: 0.2263351 total: 31.8ms remaining: 45.7ms
41: learn: 0.2238749 total: 33ms remaining: 45.5ms
42: learn: 0.2210930 total: 33.7ms remaining: 44.6ms
43: learn: 0.2183586 total: 34.3ms remaining: 43.7ms
44: learn: 0.2160132 total: 35.1ms remaining: 42.9ms
45: learn: 0.2131334 total: 35.7ms remaining: 41.9ms
46: learn: 0.2119702 total: 36.1ms remaining: 40.7ms
47: learn: 0.2102321 total: 36.8ms remaining: 39.8ms
48: learn: 0.2069067 total: 37.4ms remaining: 39ms
49: learn: 0.2032386 total: 38.4ms remaining: 38.4ms
50: learn: 0.1984367 total: 39.2ms remaining: 37.7ms
51: learn: 0.1962224 total: 39.8ms remaining: 36.8ms
52: learn: 0.1944915 total: 41.1ms remaining: 36.5ms
53: learn: 0.1920147 total: 42ms remaining: 35.8ms
54: learn: 0.1877594 total: 42.8ms remaining: 35ms
55: learn: 0.1842736 total: 43.6ms remaining: 34.2ms
56: learn: 0.1808710 total: 44.8ms remaining: 33.8ms
57: learn: 0.1803822 total: 45.3ms remaining: 32.8ms
58: learn: 0.1770663 total: 46.5ms remaining: 32.3ms
59: learn: 0.1739863 total: 47.4ms remaining: 31.6ms
60: learn: 0.1712656 total: 48.4ms remaining: 31ms
61: learn: 0.1696193 total: 49.4ms remaining: 30.3ms
62: learn: 0.1672587 total: 50.4ms remaining: 29.6ms
63: learn: 0.1652099 total: 51.8ms remaining: 29.1ms
64: learn: 0.1630193 total: 52.7ms remaining: 28.4ms
65: learn: 0.1608429 total: 53.3ms remaining: 27.5ms
66: learn: 0.1586667 total: 54.4ms remaining: 26.8ms
67: learn: 0.1572770 total: 55.1ms remaining: 25.9ms
68: learn: 0.1556950 total: 55.8ms remaining: 25.1ms
69: learn: 0.1546852 total: 56.5ms remaining: 24.2ms
70: learn: 0.1521633 total: 57.6ms remaining: 23.5ms
71: learn: 0.1501132 total: 58.4ms remaining: 22.7ms
72: learn: 0.1482866 total: 59.5ms remaining: 22ms
73: learn: 0.1465397 total: 60.1ms remaining: 21.1ms
74: learn: 0.1447200 total: 60.8ms remaining: 20.3ms
75: learn: 0.1429513 total: 61.5ms remaining: 19.4ms
76: learn: 0.1409943 total: 63.4ms remaining: 18.9ms
77: learn: 0.1390590 total: 64.2ms remaining: 18.1ms
78: learn: 0.1372216 total: 64.8ms remaining: 17.2ms
79: learn: 0.1361539 total: 65.8ms remaining: 16.5ms
80: learn: 0.1356446 total: 66.6ms remaining: 15.6ms
81: learn: 0.1339764 total: 67.2ms remaining: 14.8ms
82: learn: 0.1327106 total: 67.9ms remaining: 13.9ms
83: learn: 0.1309381 total: 69ms remaining: 13.1ms
84: learn: 0.1295160 total: 69.7ms remaining: 12.3ms
85: learn: 0.1285081 total: 70.4ms remaining: 11.5ms
86: learn: 0.1277653 total: 71.2ms remaining: 10.6ms
87: learn: 0.1263663 total: 72.1ms remaining: 9.83ms
88: learn: 0.1246948 total: 72.9ms remaining: 9.01ms
89: learn: 0.1235159 total: 73.6ms remaining: 8.18ms
90: learn: 0.1218639 total: 74.4ms remaining: 7.35ms
91: learn: 0.1211715 total: 75.1ms remaining: 6.53ms
92: learn: 0.1202462 total: 76ms remaining: 5.72ms
93: learn: 0.1191105 total: 76.8ms remaining: 4.9ms
94: learn: 0.1179845 total: 77.5ms remaining: 4.08ms
95: learn: 0.1172394 total: 78.2ms remaining: 3.26ms
96: learn: 0.1156975 total: 78.9ms remaining: 2.44ms
97: learn: 0.1149354 total: 79.6ms remaining: 1.62ms
98: learn: 0.1139428 total: 80.2ms remaining: 810us
99: learn: 0.1126096 total: 80.8ms remaining: 0us
0: learn: 0.6461176 total: 611us remaining: 60.5ms
1: learn: 0.6074389 total: 1.08ms remaining: 52.8ms
2: learn: 0.5663744 total: 1.57ms remaining: 51ms
3: learn: 0.5338644 total: 2.12ms remaining: 50.8ms
4: learn: 0.5036071 total: 2.7ms remaining: 51.3ms
5: learn: 0.4810570 total: 3.78ms remaining: 59.2ms
6: learn: 0.4610225 total: 4.45ms remaining: 59.1ms
7: learn: 0.4382829 total: 5ms remaining: 57.5ms
8: learn: 0.4184979 total: 5.7ms remaining: 57.6ms
9: learn: 0.4040368 total: 6.77ms remaining: 60.9ms
10: learn: 0.3879856 total: 7.63ms remaining: 61.7ms
11: learn: 0.3755899 total: 7.97ms remaining: 58.5ms
12: learn: 0.3652345 total: 8.53ms remaining: 57.1ms
13: learn: 0.3522070 total: 9.44ms remaining: 58ms
14: learn: 0.3402084 total: 10.3ms remaining: 58.4ms
15: learn: 0.3334455 total: 11.3ms remaining: 59.1ms
16: learn: 0.3269060 total: 12.2ms remaining: 59.4ms
17: learn: 0.3184363 total: 12.9ms remaining: 58.7ms
18: learn: 0.3122238 total: 13.6ms remaining: 58ms
19: learn: 0.3061585 total: 14.1ms remaining: 56.3ms
20: learn: 0.2989587 total: 15.1ms remaining: 57ms
21: learn: 0.2926460 total: 16.5ms remaining: 58.4ms
22: learn: 0.2872523 total: 17.3ms remaining: 57.9ms
23: learn: 0.2818606 total: 18.2ms remaining: 57.5ms
24: learn: 0.2739065 total: 19ms remaining: 57ms
25: learn: 0.2690171 total: 20ms remaining: 57ms
26: learn: 0.2649473 total: 20.8ms remaining: 56.2ms
27: learn: 0.2615369 total: 21.9ms remaining: 56.3ms
28: learn: 0.2582622 total: 22.8ms remaining: 55.7ms
29: learn: 0.2524443 total: 23.8ms remaining: 55.6ms
30: learn: 0.2490535 total: 25.3ms remaining: 56.2ms
31: learn: 0.2465288 total: 26.5ms remaining: 56.3ms
32: learn: 0.2423547 total: 27ms remaining: 54.8ms
33: learn: 0.2383376 total: 27.7ms remaining: 53.7ms
34: learn: 0.2348481 total: 28.4ms remaining: 52.8ms
35: learn: 0.2314557 total: 30ms remaining: 53.4ms
36: learn: 0.2293154 total: 30.8ms remaining: 52.4ms
37: learn: 0.2269631 total: 31.2ms remaining: 51ms
38: learn: 0.2233239 total: 32ms remaining: 50.1ms
39: learn: 0.2219447 total: 32.9ms remaining: 49.4ms
40: learn: 0.2197608 total: 33.8ms remaining: 48.6ms
41: learn: 0.2179991 total: 34.5ms remaining: 47.7ms
42: learn: 0.2153602 total: 35.8ms remaining: 47.4ms
43: learn: 0.2128584 total: 37ms remaining: 47.1ms
44: learn: 0.2110075 total: 37.8ms remaining: 46.2ms
45: learn: 0.2076444 total: 38.6ms remaining: 45.4ms
46: learn: 0.2059115 total: 39.4ms remaining: 44.4ms
47: learn: 0.2039299 total: 40.3ms remaining: 43.7ms
48: learn: 0.2021505 total: 41.5ms remaining: 43.2ms
49: learn: 0.1995858 total: 42.4ms remaining: 42.4ms
50: learn: 0.1972183 total: 43.1ms remaining: 41.4ms
51: learn: 0.1960866 total: 43.9ms remaining: 40.5ms
52: learn: 0.1931811 total: 44.5ms remaining: 39.5ms
53: learn: 0.1902610 total: 45.3ms remaining: 38.5ms
54: learn: 0.1884566 total: 46.1ms remaining: 37.7ms
55: learn: 0.1871578 total: 47.1ms remaining: 37ms
56: learn: 0.1854270 total: 47.7ms remaining: 36ms
57: learn: 0.1833199 total: 48.6ms remaining: 35.2ms
58: learn: 0.1806623 total: 49.2ms remaining: 34.2ms
59: learn: 0.1789954 total: 50ms remaining: 33.4ms
60: learn: 0.1769297 total: 51ms remaining: 32.6ms
61: learn: 0.1751763 total: 51.8ms remaining: 31.7ms
62: learn: 0.1715643 total: 52.5ms remaining: 30.8ms
63: learn: 0.1690372 total: 53.3ms remaining: 30ms
64: learn: 0.1671563 total: 54ms remaining: 29.1ms
65: learn: 0.1649238 total: 54.7ms remaining: 28.2ms
66: learn: 0.1626491 total: 55.4ms remaining: 27.3ms
67: learn: 0.1605330 total: 56.1ms remaining: 26.4ms
68: learn: 0.1587867 total: 56.8ms remaining: 25.5ms
69: learn: 0.1570920 total: 57.6ms remaining: 24.7ms
70: learn: 0.1558258 total: 58.6ms remaining: 23.9ms
71: learn: 0.1538087 total: 59.3ms remaining: 23.1ms
72: learn: 0.1516208 total: 60.1ms remaining: 22.2ms
73: learn: 0.1499587 total: 60.7ms remaining: 21.3ms
74: learn: 0.1483190 total: 61.6ms remaining: 20.5ms
75: learn: 0.1466576 total: 62.5ms remaining: 19.7ms
76: learn: 0.1451862 total: 63.4ms remaining: 18.9ms
77: learn: 0.1434428 total: 64.3ms remaining: 18.1ms
78: learn: 0.1417734 total: 65.2ms remaining: 17.3ms
79: learn: 0.1403245 total: 66.2ms remaining: 16.5ms
80: learn: 0.1384813 total: 67ms remaining: 15.7ms
81: learn: 0.1365771 total: 68.1ms remaining: 14.9ms
82: learn: 0.1351516 total: 68.9ms remaining: 14.1ms
83: learn: 0.1336962 total: 69.7ms remaining: 13.3ms
84: learn: 0.1328596 total: 70.6ms remaining: 12.5ms
85: learn: 0.1312092 total: 71.3ms remaining: 11.6ms
86: learn: 0.1295276 total: 72.2ms remaining: 10.8ms
87: learn: 0.1286613 total: 73.1ms remaining: 9.96ms
88: learn: 0.1275535 total: 73.9ms remaining: 9.13ms
89: learn: 0.1261839 total: 74.8ms remaining: 8.31ms
90: learn: 0.1249200 total: 75.6ms remaining: 7.47ms
91: learn: 0.1236530 total: 76.6ms remaining: 6.66ms
92: learn: 0.1227653 total: 77.5ms remaining: 5.83ms
93: learn: 0.1218493 total: 78.3ms remaining: 5ms
94: learn: 0.1205549 total: 79.1ms remaining: 4.16ms
95: learn: 0.1197403 total: 79.9ms remaining: 3.33ms
96: learn: 0.1187410 total: 80.7ms remaining: 2.5ms
97: learn: 0.1178329 total: 81.6ms remaining: 1.67ms
98: learn: 0.1166346 total: 82.4ms remaining: 832us
99: learn: 0.1154614 total: 83.1ms remaining: 0us
0: learn: 0.6584029 total: 759us remaining: 75.2ms
1: learn: 0.6158136 total: 1.3ms remaining: 63.7ms
2: learn: 0.5789053 total: 1.88ms remaining: 60.9ms
3: learn: 0.5476814 total: 2.56ms remaining: 61.4ms
4: learn: 0.5177018 total: 3.21ms remaining: 60.9ms
5: learn: 0.4899133 total: 3.95ms remaining: 61.9ms
6: learn: 0.4664993 total: 4.62ms remaining: 61.4ms
7: learn: 0.4507562 total: 5.94ms remaining: 68.4ms
8: learn: 0.4301009 total: 7.69ms remaining: 77.7ms
9: learn: 0.4091667 total: 8.76ms remaining: 78.8ms
10: learn: 0.3923362 total: 9.57ms remaining: 77.4ms
11: learn: 0.3813501 total: 10.7ms remaining: 78.7ms
12: learn: 0.3700872 total: 11.2ms remaining: 75.1ms
13: learn: 0.3595115 total: 11.6ms remaining: 71.5ms
14: learn: 0.3496026 total: 13ms remaining: 73.8ms
15: learn: 0.3430776 total: 13.6ms remaining: 71.4ms
16: learn: 0.3324861 total: 14.4ms remaining: 70.4ms
17: learn: 0.3249253 total: 15.8ms remaining: 72ms
18: learn: 0.3175635 total: 16.8ms remaining: 71.7ms
19: learn: 0.3094936 total: 17.6ms remaining: 70.2ms
20: learn: 0.3045673 total: 18.1ms remaining: 68.1ms
21: learn: 0.2987827 total: 19.3ms remaining: 68.6ms
22: learn: 0.2937094 total: 20.5ms remaining: 68.6ms
23: learn: 0.2869886 total: 21.4ms remaining: 67.8ms
24: learn: 0.2814709 total: 22.9ms remaining: 68.6ms
25: learn: 0.2753705 total: 23.8ms remaining: 67.8ms
26: learn: 0.2707435 total: 24.5ms remaining: 66.3ms
27: learn: 0.2654776 total: 25.7ms remaining: 66.1ms
28: learn: 0.2604934 total: 26.5ms remaining: 65ms
29: learn: 0.2571748 total: 26.8ms remaining: 62.5ms
30: learn: 0.2529820 total: 27.8ms remaining: 61.9ms
31: learn: 0.2471451 total: 28.7ms remaining: 60.9ms
32: learn: 0.2430683 total: 29.6ms remaining: 60.1ms
33: learn: 0.2399410 total: 30.1ms remaining: 58.5ms
34: learn: 0.2359258 total: 30.8ms remaining: 57.3ms
35: learn: 0.2337736 total: 31.8ms remaining: 56.5ms
36: learn: 0.2311579 total: 32.6ms remaining: 55.5ms
37: learn: 0.2289602 total: 33.6ms remaining: 54.8ms
38: learn: 0.2259417 total: 34.5ms remaining: 54ms
39: learn: 0.2229631 total: 35.2ms remaining: 52.8ms
40: learn: 0.2201452 total: 36ms remaining: 51.8ms
41: learn: 0.2183124 total: 37.2ms remaining: 51.4ms
42: learn: 0.2167107 total: 38.1ms remaining: 50.5ms
43: learn: 0.2145715 total: 39.2ms remaining: 49.9ms
44: learn: 0.2138454 total: 40.1ms remaining: 49.1ms
45: learn: 0.2116082 total: 41.7ms remaining: 49ms
46: learn: 0.2103168 total: 42.5ms remaining: 48ms
47: learn: 0.2066677 total: 43.6ms remaining: 47.2ms
48: learn: 0.2035156 total: 45ms remaining: 46.8ms
49: learn: 0.2011849 total: 46.4ms remaining: 46.4ms
50: learn: 0.1984099 total: 47.2ms remaining: 45.4ms
51: learn: 0.1956163 total: 48.3ms remaining: 44.5ms
52: learn: 0.1921853 total: 49.8ms remaining: 44.1ms
53: learn: 0.1902531 total: 51.1ms remaining: 43.5ms
54: learn: 0.1875254 total: 52.3ms remaining: 42.8ms
55: learn: 0.1860239 total: 53.8ms remaining: 42.3ms
56: learn: 0.1837295 total: 54.7ms remaining: 41.2ms
57: learn: 0.1814652 total: 55.8ms remaining: 40.4ms
58: learn: 0.1800774 total: 56.4ms remaining: 39.2ms
59: learn: 0.1786451 total: 57.5ms remaining: 38.3ms
60: learn: 0.1767313 total: 58.6ms remaining: 37.5ms
61: learn: 0.1743092 total: 59.4ms remaining: 36.4ms
62: learn: 0.1714425 total: 60.5ms remaining: 35.5ms
63: learn: 0.1700936 total: 61.6ms remaining: 34.6ms
64: learn: 0.1672716 total: 62.7ms remaining: 33.8ms
65: learn: 0.1661238 total: 63.7ms remaining: 32.8ms
66: learn: 0.1643815 total: 64.7ms remaining: 31.9ms
67: learn: 0.1629536 total: 65.9ms remaining: 31ms
68: learn: 0.1613830 total: 67.4ms remaining: 30.3ms
69: learn: 0.1592482 total: 68.5ms remaining: 29.4ms
70: learn: 0.1580802 total: 69.6ms remaining: 28.4ms
71: learn: 0.1566359 total: 71ms remaining: 27.6ms
72: learn: 0.1547821 total: 71.8ms remaining: 26.6ms
73: learn: 0.1531087 total: 73.1ms remaining: 25.7ms
74: learn: 0.1517356 total: 74.3ms remaining: 24.8ms
75: learn: 0.1501831 total: 75.6ms remaining: 23.9ms
76: learn: 0.1490114 total: 76.7ms remaining: 22.9ms
77: learn: 0.1475740 total: 77.7ms remaining: 21.9ms
78: learn: 0.1463319 total: 78.8ms remaining: 20.9ms
79: learn: 0.1452089 total: 80ms remaining: 20ms
80: learn: 0.1441933 total: 80.9ms remaining: 19ms
81: learn: 0.1430868 total: 82.5ms remaining: 18.1ms
82: learn: 0.1414999 total: 83.6ms remaining: 17.1ms
83: learn: 0.1404724 total: 85ms remaining: 16.2ms
84: learn: 0.1391252 total: 87.4ms remaining: 15.4ms
85: learn: 0.1381679 total: 94.1ms remaining: 15.3ms
86: learn: 0.1376379 total: 96.6ms remaining: 14.4ms
87: learn: 0.1361129 total: 100ms remaining: 13.6ms
88: learn: 0.1355066 total: 106ms remaining: 13.1ms
89: learn: 0.1340172 total: 111ms remaining: 12.4ms
90: learn: 0.1329855 total: 113ms remaining: 11.1ms
91: learn: 0.1321255 total: 114ms remaining: 9.89ms
92: learn: 0.1315768 total: 115ms remaining: 8.69ms
93: learn: 0.1312144 total: 117ms remaining: 7.46ms
94: learn: 0.1299591 total: 120ms remaining: 6.31ms
95: learn: 0.1294979 total: 121ms remaining: 5.04ms
96: learn: 0.1287087 total: 122ms remaining: 3.78ms
97: learn: 0.1276809 total: 123ms remaining: 2.51ms
98: learn: 0.1260381 total: 124ms remaining: 1.25ms
99: learn: 0.1248557 total: 125ms remaining: 0us
0: learn: 0.6619530 total: 765us remaining: 75.8ms
1: learn: 0.6283297 total: 2.03ms remaining: 99.7ms
2: learn: 0.5960954 total: 3.13ms remaining: 101ms
3: learn: 0.5627508 total: 4.66ms remaining: 112ms
4: learn: 0.5359547 total: 5.82ms remaining: 111ms
5: learn: 0.5055974 total: 7.17ms remaining: 112ms
6: learn: 0.4784684 total: 8.65ms remaining: 115ms
7: learn: 0.4624911 total: 9.36ms remaining: 108ms
8: learn: 0.4376804 total: 10.2ms remaining: 103ms
9: learn: 0.4158659 total: 10.8ms remaining: 97.1ms
10: learn: 0.3980852 total: 12.1ms remaining: 98.2ms
11: learn: 0.3829023 total: 12.9ms remaining: 94.2ms
12: learn: 0.3654256 total: 13.7ms remaining: 91.9ms
13: learn: 0.3569122 total: 14.4ms remaining: 88.4ms
14: learn: 0.3479229 total: 15.3ms remaining: 86.9ms
15: learn: 0.3372314 total: 16ms remaining: 84.1ms
16: learn: 0.3236670 total: 16.7ms remaining: 81.4ms
17: learn: 0.3132976 total: 17.4ms remaining: 79.3ms
18: learn: 0.3066007 total: 18.3ms remaining: 78ms
19: learn: 0.2992773 total: 19.2ms remaining: 76.9ms
20: learn: 0.2903921 total: 20ms remaining: 75.3ms
21: learn: 0.2843295 total: 21.1ms remaining: 74.7ms
22: learn: 0.2770463 total: 23.4ms remaining: 78.4ms
23: learn: 0.2721898 total: 24.3ms remaining: 76.9ms
24: learn: 0.2671702 total: 24.9ms remaining: 74.7ms
25: learn: 0.2615728 total: 25.9ms remaining: 73.7ms
26: learn: 0.2559444 total: 26.8ms remaining: 72.3ms
27: learn: 0.2522468 total: 27.7ms remaining: 71.1ms
28: learn: 0.2466611 total: 28.7ms remaining: 70.3ms
29: learn: 0.2428671 total: 29.5ms remaining: 68.9ms
30: learn: 0.2399610 total: 29.9ms remaining: 66.5ms
31: learn: 0.2362310 total: 30.8ms remaining: 65.4ms
32: learn: 0.2325890 total: 31.6ms remaining: 64.3ms
33: learn: 0.2291176 total: 32.6ms remaining: 63.2ms
34: learn: 0.2243576 total: 33.6ms remaining: 62.4ms
35: learn: 0.2212875 total: 34.5ms remaining: 61.4ms
36: learn: 0.2175959 total: 35.4ms remaining: 60.3ms
37: learn: 0.2138491 total: 36.2ms remaining: 59.1ms
38: learn: 0.2101711 total: 37ms remaining: 57.9ms
39: learn: 0.2061384 total: 37.7ms remaining: 56.5ms
40: learn: 0.2041421 total: 38.4ms remaining: 55.3ms
41: learn: 0.2009166 total: 39ms remaining: 53.9ms
42: learn: 0.1967390 total: 39.7ms remaining: 52.6ms
43: learn: 0.1937388 total: 40.4ms remaining: 51.5ms
44: learn: 0.1912718 total: 41.2ms remaining: 50.3ms
45: learn: 0.1891110 total: 41.9ms remaining: 49.2ms
46: learn: 0.1865737 total: 42.5ms remaining: 48ms
47: learn: 0.1845496 total: 43.3ms remaining: 46.9ms
48: learn: 0.1824034 total: 44.1ms remaining: 45.9ms
49: learn: 0.1796541 total: 44.9ms remaining: 44.9ms
50: learn: 0.1768214 total: 45.9ms remaining: 44.1ms
51: learn: 0.1746723 total: 46.7ms remaining: 43.1ms
52: learn: 0.1728966 total: 47.6ms remaining: 42.2ms
53: learn: 0.1711244 total: 48.5ms remaining: 41.3ms
54: learn: 0.1682982 total: 49.3ms remaining: 40.4ms
55: learn: 0.1660184 total: 50ms remaining: 39.3ms
56: learn: 0.1630847 total: 50.9ms remaining: 38.4ms
57: learn: 0.1606168 total: 51.8ms remaining: 37.5ms
58: learn: 0.1578809 total: 53.8ms remaining: 37.4ms
59: learn: 0.1553022 total: 54.8ms remaining: 36.5ms
60: learn: 0.1528066 total: 55.5ms remaining: 35.5ms
61: learn: 0.1500472 total: 56.3ms remaining: 34.5ms
62: learn: 0.1468517 total: 57ms remaining: 33.5ms
63: learn: 0.1449483 total: 57.8ms remaining: 32.5ms
64: learn: 0.1431338 total: 58.5ms remaining: 31.5ms
65: learn: 0.1413144 total: 59.3ms remaining: 30.6ms
66: learn: 0.1392154 total: 60.2ms remaining: 29.7ms
67: learn: 0.1365813 total: 61.2ms remaining: 28.8ms
68: learn: 0.1350982 total: 62ms remaining: 27.8ms
69: learn: 0.1329210 total: 62.8ms remaining: 26.9ms
70: learn: 0.1313183 total: 63.6ms remaining: 26ms
71: learn: 0.1291210 total: 64.4ms remaining: 25ms
72: learn: 0.1267639 total: 65.2ms remaining: 24.1ms
73: learn: 0.1252026 total: 66.1ms remaining: 23.2ms
74: learn: 0.1230517 total: 67ms remaining: 22.3ms
75: learn: 0.1213910 total: 67.9ms remaining: 21.5ms
76: learn: 0.1199263 total: 68.7ms remaining: 20.5ms
77: learn: 0.1181724 total: 69.3ms remaining: 19.6ms
78: learn: 0.1165284 total: 70.2ms remaining: 18.7ms
79: learn: 0.1143646 total: 71ms remaining: 17.8ms
80: learn: 0.1130815 total: 71.7ms remaining: 16.8ms
81: learn: 0.1109337 total: 72.5ms remaining: 15.9ms
82: learn: 0.1099033 total: 73.4ms remaining: 15ms
83: learn: 0.1083300 total: 74.3ms remaining: 14.2ms
84: learn: 0.1066926 total: 75.1ms remaining: 13.3ms
85: learn: 0.1048807 total: 76ms remaining: 12.4ms
86: learn: 0.1029395 total: 76.7ms remaining: 11.5ms
87: learn: 0.1017061 total: 77.6ms remaining: 10.6ms
88: learn: 0.1006564 total: 78.3ms remaining: 9.68ms
89: learn: 0.0991601 total: 79.1ms remaining: 8.79ms
90: learn: 0.0979843 total: 79.8ms remaining: 7.89ms
91: learn: 0.0971031 total: 80.5ms remaining: 7ms
92: learn: 0.0962520 total: 81.3ms remaining: 6.12ms
93: learn: 0.0949233 total: 82.2ms remaining: 5.24ms
94: learn: 0.0935646 total: 83ms remaining: 4.37ms
95: learn: 0.0924970 total: 83.8ms remaining: 3.49ms
96: learn: 0.0913099 total: 84.5ms remaining: 2.61ms
97: learn: 0.0901472 total: 85.5ms remaining: 1.75ms
98: learn: 0.0890628 total: 86.4ms remaining: 873us
99: learn: 0.0883427 total: 87.3ms remaining: 0us
0: learn: 0.6581386 total: 846us remaining: 83.8ms
1: learn: 0.6167948 total: 1.41ms remaining: 69.3ms
2: learn: 0.5790476 total: 2.29ms remaining: 74.2ms
3: learn: 0.5488848 total: 3.03ms remaining: 72.8ms
4: learn: 0.5230009 total: 3.96ms remaining: 75.2ms
5: learn: 0.4964807 total: 4.83ms remaining: 75.6ms
6: learn: 0.4712568 total: 5.57ms remaining: 74ms
7: learn: 0.4526747 total: 6.26ms remaining: 71.9ms
8: learn: 0.4333489 total: 7.08ms remaining: 71.6ms
9: learn: 0.4163775 total: 7.95ms remaining: 71.6ms
10: learn: 0.4031902 total: 8.68ms remaining: 70.2ms
11: learn: 0.3862230 total: 9.55ms remaining: 70ms
12: learn: 0.3749004 total: 9.74ms remaining: 65.2ms
13: learn: 0.3627256 total: 10.6ms remaining: 65ms
14: learn: 0.3513545 total: 11.3ms remaining: 63.9ms
15: learn: 0.3412036 total: 12ms remaining: 63.2ms
16: learn: 0.3338192 total: 12.8ms remaining: 62.7ms
17: learn: 0.3247744 total: 13.7ms remaining: 62.3ms
18: learn: 0.3152335 total: 14.3ms remaining: 60.8ms
19: learn: 0.3101617 total: 14.9ms remaining: 59.6ms
20: learn: 0.3029026 total: 15.6ms remaining: 58.8ms
21: learn: 0.2956389 total: 16.3ms remaining: 57.8ms
22: learn: 0.2879109 total: 17.3ms remaining: 57.8ms
23: learn: 0.2820702 total: 17.8ms remaining: 56.5ms
24: learn: 0.2771588 total: 18.4ms remaining: 55.2ms
25: learn: 0.2726471 total: 19.1ms remaining: 54.3ms
26: learn: 0.2659988 total: 19.9ms remaining: 53.8ms
27: learn: 0.2600717 total: 20.7ms remaining: 53.2ms
28: learn: 0.2585335 total: 21.2ms remaining: 51.8ms
29: learn: 0.2543809 total: 21.8ms remaining: 50.9ms
30: learn: 0.2510589 total: 22.6ms remaining: 50.2ms
31: learn: 0.2476309 total: 23.2ms remaining: 49.2ms
32: learn: 0.2446868 total: 23.9ms remaining: 48.5ms
33: learn: 0.2413541 total: 24.5ms remaining: 47.6ms
34: learn: 0.2375438 total: 25.3ms remaining: 47ms
35: learn: 0.2350143 total: 25.8ms remaining: 45.9ms
36: learn: 0.2316015 total: 26.7ms remaining: 45.4ms
37: learn: 0.2283235 total: 27.5ms remaining: 44.8ms
38: learn: 0.2246981 total: 28.3ms remaining: 44.3ms
39: learn: 0.2227796 total: 29.1ms remaining: 43.6ms
40: learn: 0.2213225 total: 29.6ms remaining: 42.6ms
41: learn: 0.2192394 total: 30.2ms remaining: 41.7ms
42: learn: 0.2178576 total: 30.3ms remaining: 40.2ms
43: learn: 0.2155307 total: 31.2ms remaining: 39.7ms
44: learn: 0.2119431 total: 32.3ms remaining: 39.4ms
45: learn: 0.2100281 total: 33.1ms remaining: 38.9ms
46: learn: 0.2069576 total: 34ms remaining: 38.4ms
47: learn: 0.2036339 total: 35.1ms remaining: 38.1ms
48: learn: 0.2008020 total: 36.1ms remaining: 37.6ms
49: learn: 0.1984541 total: 37.3ms remaining: 37.3ms
50: learn: 0.1966141 total: 38.2ms remaining: 36.7ms
51: learn: 0.1949719 total: 39.1ms remaining: 36.1ms
52: learn: 0.1928572 total: 39.9ms remaining: 35.4ms
53: learn: 0.1901771 total: 40.8ms remaining: 34.7ms
54: learn: 0.1891596 total: 41.4ms remaining: 33.9ms
55: learn: 0.1870296 total: 42.2ms remaining: 33.2ms
56: learn: 0.1859773 total: 43.1ms remaining: 32.5ms
57: learn: 0.1840133 total: 43.9ms remaining: 31.8ms
58: learn: 0.1817740 total: 44.8ms remaining: 31.1ms
59: learn: 0.1794985 total: 45.4ms remaining: 30.3ms
60: learn: 0.1771761 total: 46.3ms remaining: 29.6ms
61: learn: 0.1756084 total: 47ms remaining: 28.8ms
62: learn: 0.1737502 total: 47.6ms remaining: 28ms
63: learn: 0.1726263 total: 48.5ms remaining: 27.3ms
64: learn: 0.1700217 total: 49.3ms remaining: 26.6ms
65: learn: 0.1682406 total: 50.2ms remaining: 25.9ms
66: learn: 0.1659991 total: 50.8ms remaining: 25ms
67: learn: 0.1640786 total: 51.8ms remaining: 24.4ms
68: learn: 0.1633586 total: 52.5ms remaining: 23.6ms
69: learn: 0.1616415 total: 53.4ms remaining: 22.9ms
70: learn: 0.1595020 total: 54ms remaining: 22.1ms
71: learn: 0.1584082 total: 54.7ms remaining: 21.3ms
72: learn: 0.1566504 total: 55.7ms remaining: 20.6ms
73: learn: 0.1552144 total: 56.6ms remaining: 19.9ms
74: learn: 0.1535309 total: 57.4ms remaining: 19.1ms
75: learn: 0.1515302 total: 58.3ms remaining: 18.4ms
76: learn: 0.1507256 total: 59ms remaining: 17.6ms
77: learn: 0.1493253 total: 59.8ms remaining: 16.9ms
78: learn: 0.1475329 total: 60.7ms remaining: 16.1ms
79: learn: 0.1463985 total: 61.5ms remaining: 15.4ms
80: learn: 0.1450349 total: 62.6ms remaining: 14.7ms
81: learn: 0.1434335 total: 63.5ms remaining: 13.9ms
82: learn: 0.1421749 total: 64.2ms remaining: 13.2ms
83: learn: 0.1411376 total: 65.1ms remaining: 12.4ms
84: learn: 0.1399847 total: 65.9ms remaining: 11.6ms
85: learn: 0.1385208 total: 66.7ms remaining: 10.9ms
86: learn: 0.1370404 total: 67.5ms remaining: 10.1ms
87: learn: 0.1355310 total: 68.2ms remaining: 9.3ms
88: learn: 0.1341731 total: 68.9ms remaining: 8.52ms
89: learn: 0.1330891 total: 69.7ms remaining: 7.75ms
90: learn: 0.1323124 total: 70.6ms remaining: 6.98ms
91: learn: 0.1311611 total: 71.5ms remaining: 6.21ms
92: learn: 0.1301685 total: 72.3ms remaining: 5.44ms
93: learn: 0.1288261 total: 73ms remaining: 4.66ms
94: learn: 0.1274803 total: 73.7ms remaining: 3.88ms
95: learn: 0.1264066 total: 74.4ms remaining: 3.1ms
96: learn: 0.1253705 total: 75.4ms remaining: 2.33ms
97: learn: 0.1246544 total: 76.3ms remaining: 1.56ms
98: learn: 0.1238340 total: 77.1ms remaining: 778us
99: learn: 0.1229835 total: 77.9ms remaining: 0us
0: learn: 0.6450008 total: 646us remaining: 64ms
1: learn: 0.5991432 total: 1.45ms remaining: 71.2ms
2: learn: 0.5618368 total: 2.27ms remaining: 73.5ms
3: learn: 0.5318991 total: 3.39ms remaining: 81.3ms
4: learn: 0.5072157 total: 4.36ms remaining: 82.8ms
5: learn: 0.4774205 total: 5.09ms remaining: 79.7ms
6: learn: 0.4542422 total: 5.71ms remaining: 75.8ms
7: learn: 0.4335540 total: 6.47ms remaining: 74.4ms
8: learn: 0.4192534 total: 7.09ms remaining: 71.7ms
9: learn: 0.4003498 total: 8.05ms remaining: 72.5ms
10: learn: 0.3843162 total: 8.76ms remaining: 70.9ms
11: learn: 0.3682167 total: 9.5ms remaining: 69.7ms
12: learn: 0.3542211 total: 9.81ms remaining: 65.7ms
13: learn: 0.3399096 total: 10.4ms remaining: 64ms
14: learn: 0.3288582 total: 11ms remaining: 62.5ms
15: learn: 0.3173883 total: 12.3ms remaining: 64.5ms
16: learn: 0.3096656 total: 13.2ms remaining: 64.4ms
17: learn: 0.3012150 total: 14.1ms remaining: 64ms
18: learn: 0.2922121 total: 14.6ms remaining: 62.2ms
19: learn: 0.2846976 total: 15.4ms remaining: 61.4ms
20: learn: 0.2777799 total: 16.9ms remaining: 63.4ms
21: learn: 0.2716195 total: 17.5ms remaining: 62ms
22: learn: 0.2661617 total: 18.2ms remaining: 60.9ms
23: learn: 0.2633045 total: 19.7ms remaining: 62.5ms
24: learn: 0.2569821 total: 20.3ms remaining: 60.8ms
25: learn: 0.2508427 total: 21.2ms remaining: 60.2ms
26: learn: 0.2446129 total: 21.9ms remaining: 59.2ms
27: learn: 0.2398813 total: 22.8ms remaining: 58.7ms
28: learn: 0.2347845 total: 23.4ms remaining: 57.4ms
29: learn: 0.2305331 total: 24.5ms remaining: 57.2ms
30: learn: 0.2248650 total: 25.1ms remaining: 55.8ms
31: learn: 0.2210912 total: 26.3ms remaining: 55.8ms
32: learn: 0.2184215 total: 27ms remaining: 54.9ms
33: learn: 0.2132052 total: 27.9ms remaining: 54.2ms
34: learn: 0.2106714 total: 28.9ms remaining: 53.7ms
35: learn: 0.2080676 total: 29.6ms remaining: 52.6ms
36: learn: 0.2050681 total: 30.7ms remaining: 52.2ms
37: learn: 0.2022172 total: 31.4ms remaining: 51.2ms
38: learn: 0.1983147 total: 32.2ms remaining: 50.4ms
39: learn: 0.1956122 total: 32.9ms remaining: 49.4ms
40: learn: 0.1929652 total: 33.7ms remaining: 48.4ms
41: learn: 0.1906725 total: 34.3ms remaining: 47.4ms
42: learn: 0.1876575 total: 35.2ms remaining: 46.6ms
43: learn: 0.1857988 total: 35.7ms remaining: 45.5ms
44: learn: 0.1830635 total: 36.4ms remaining: 44.5ms
45: learn: 0.1815324 total: 37.2ms remaining: 43.7ms
46: learn: 0.1788223 total: 37.8ms remaining: 42.7ms
47: learn: 0.1760001 total: 38.6ms remaining: 41.9ms
48: learn: 0.1747437 total: 39.5ms remaining: 41.1ms
49: learn: 0.1737366 total: 39.7ms remaining: 39.7ms
50: learn: 0.1715262 total: 40.4ms remaining: 38.8ms
51: learn: 0.1698449 total: 41.4ms remaining: 38.2ms
52: learn: 0.1674135 total: 42.2ms remaining: 37.4ms
53: learn: 0.1648188 total: 42.8ms remaining: 36.4ms
54: learn: 0.1626409 total: 44.3ms remaining: 36.2ms
55: learn: 0.1603856 total: 45ms remaining: 35.3ms
56: learn: 0.1586729 total: 45.7ms remaining: 34.5ms
57: learn: 0.1565485 total: 47ms remaining: 34ms
58: learn: 0.1546119 total: 47.7ms remaining: 33.1ms
59: learn: 0.1524888 total: 48.4ms remaining: 32.3ms
60: learn: 0.1507917 total: 49.7ms remaining: 31.8ms
61: learn: 0.1488992 total: 51ms remaining: 31.2ms
62: learn: 0.1472201 total: 51.6ms remaining: 30.3ms
63: learn: 0.1455391 total: 53.1ms remaining: 29.9ms
64: learn: 0.1439199 total: 53.9ms remaining: 29ms
65: learn: 0.1418087 total: 54.8ms remaining: 28.2ms
66: learn: 0.1394577 total: 55.5ms remaining: 27.4ms
67: learn: 0.1369945 total: 56.6ms remaining: 26.6ms
68: learn: 0.1355219 total: 57.3ms remaining: 25.8ms
69: learn: 0.1339997 total: 58.1ms remaining: 24.9ms
70: learn: 0.1327667 total: 58.9ms remaining: 24.1ms
71: learn: 0.1314610 total: 60.5ms remaining: 23.5ms
72: learn: 0.1300831 total: 61.2ms remaining: 22.6ms
73: learn: 0.1287133 total: 62.2ms remaining: 21.9ms
74: learn: 0.1273395 total: 63.1ms remaining: 21ms
75: learn: 0.1260626 total: 64.1ms remaining: 20.2ms
76: learn: 0.1248523 total: 65.2ms remaining: 19.5ms
77: learn: 0.1233681 total: 66.3ms remaining: 18.7ms
78: learn: 0.1222555 total: 67.3ms remaining: 17.9ms
79: learn: 0.1205128 total: 68.1ms remaining: 17ms
80: learn: 0.1195078 total: 68.8ms remaining: 16.1ms
81: learn: 0.1182419 total: 69.5ms remaining: 15.3ms
82: learn: 0.1171108 total: 70.4ms remaining: 14.4ms
83: learn: 0.1161304 total: 71.7ms remaining: 13.7ms
84: learn: 0.1148082 total: 72.4ms remaining: 12.8ms
85: learn: 0.1135977 total: 73.3ms remaining: 11.9ms
86: learn: 0.1124611 total: 74.1ms remaining: 11.1ms
87: learn: 0.1113552 total: 75.3ms remaining: 10.3ms
88: learn: 0.1108045 total: 76ms remaining: 9.4ms
89: learn: 0.1099158 total: 77ms remaining: 8.56ms
90: learn: 0.1085172 total: 77.8ms remaining: 7.69ms
91: learn: 0.1072930 total: 78.6ms remaining: 6.84ms
92: learn: 0.1061358 total: 79.5ms remaining: 5.98ms
93: learn: 0.1051470 total: 80.3ms remaining: 5.12ms
94: learn: 0.1041503 total: 80.9ms remaining: 4.26ms
95: learn: 0.1029558 total: 81.8ms remaining: 3.41ms
96: learn: 0.1017439 total: 83.8ms remaining: 2.59ms
97: learn: 0.1008766 total: 84.6ms remaining: 1.73ms
98: learn: 0.1001565 total: 85.3ms remaining: 861us
99: learn: 0.0994854 total: 86.2ms remaining: 0us
0: learn: 0.6424707 total: 755us remaining: 74.8ms
1: learn: 0.6093615 total: 1.91ms remaining: 93.6ms
2: learn: 0.5728187 total: 2.54ms remaining: 82.3ms
3: learn: 0.5402520 total: 3.15ms remaining: 75.7ms
4: learn: 0.5081451 total: 3.83ms remaining: 72.7ms
5: learn: 0.4814130 total: 4.43ms remaining: 69.4ms
6: learn: 0.4585083 total: 5.16ms remaining: 68.6ms
7: learn: 0.4367014 total: 5.69ms remaining: 65.5ms
8: learn: 0.4201252 total: 6.66ms remaining: 67.3ms
9: learn: 0.4062633 total: 7.18ms remaining: 64.6ms
10: learn: 0.3902766 total: 7.54ms remaining: 61ms
11: learn: 0.3733889 total: 8.34ms remaining: 61.2ms
12: learn: 0.3591882 total: 8.94ms remaining: 59.9ms
13: learn: 0.3489420 total: 9.67ms remaining: 59.4ms
14: learn: 0.3384339 total: 10.2ms remaining: 58ms
15: learn: 0.3286939 total: 10.8ms remaining: 56.7ms
16: learn: 0.3207029 total: 11.3ms remaining: 55.1ms
17: learn: 0.3121512 total: 11.8ms remaining: 53.8ms
18: learn: 0.3071762 total: 13.2ms remaining: 56.2ms
19: learn: 0.3015041 total: 13.7ms remaining: 54.7ms
20: learn: 0.2945076 total: 14.6ms remaining: 54.8ms
21: learn: 0.2897907 total: 15.4ms remaining: 54.6ms
22: learn: 0.2811226 total: 16.3ms remaining: 54.7ms
23: learn: 0.2738252 total: 17.1ms remaining: 54.1ms
24: learn: 0.2675024 total: 17.6ms remaining: 53ms
25: learn: 0.2637681 total: 18.4ms remaining: 52.4ms
26: learn: 0.2604760 total: 19.3ms remaining: 52.1ms
27: learn: 0.2545391 total: 19.9ms remaining: 51.3ms
28: learn: 0.2506947 total: 20.4ms remaining: 50ms
29: learn: 0.2453228 total: 20.9ms remaining: 48.9ms
30: learn: 0.2412747 total: 21.6ms remaining: 48.1ms
31: learn: 0.2382940 total: 22.8ms remaining: 48.4ms
32: learn: 0.2333589 total: 23.4ms remaining: 47.6ms
33: learn: 0.2309363 total: 23.8ms remaining: 46.3ms
34: learn: 0.2267844 total: 24.3ms remaining: 45.1ms
35: learn: 0.2240942 total: 24.9ms remaining: 44.3ms
36: learn: 0.2214865 total: 26ms remaining: 44.3ms
37: learn: 0.2187099 total: 26.4ms remaining: 43ms
38: learn: 0.2172117 total: 26.9ms remaining: 42.1ms
39: learn: 0.2149843 total: 27.7ms remaining: 41.5ms
40: learn: 0.2123648 total: 28.5ms remaining: 41ms
41: learn: 0.2094861 total: 29.1ms remaining: 40.1ms
42: learn: 0.2062167 total: 29.6ms remaining: 39.3ms
43: learn: 0.2044708 total: 30.5ms remaining: 38.8ms
44: learn: 0.2019504 total: 31ms remaining: 37.9ms
45: learn: 0.1990791 total: 31.7ms remaining: 37.2ms
46: learn: 0.1979083 total: 32.4ms remaining: 36.5ms
47: learn: 0.1945802 total: 33.2ms remaining: 36ms
48: learn: 0.1920564 total: 33.9ms remaining: 35.3ms
49: learn: 0.1900021 total: 34.7ms remaining: 34.7ms
50: learn: 0.1868044 total: 35.5ms remaining: 34.1ms
51: learn: 0.1845077 total: 36.2ms remaining: 33.4ms
52: learn: 0.1836097 total: 36.7ms remaining: 32.6ms
53: learn: 0.1824432 total: 37.3ms remaining: 31.8ms
54: learn: 0.1811255 total: 38ms remaining: 31.1ms
55: learn: 0.1782458 total: 39ms remaining: 30.7ms
56: learn: 0.1762718 total: 39.7ms remaining: 30ms
57: learn: 0.1740662 total: 40.6ms remaining: 29.4ms
58: learn: 0.1722127 total: 41.3ms remaining: 28.7ms
59: learn: 0.1707521 total: 42.1ms remaining: 28.1ms
60: learn: 0.1693590 total: 43ms remaining: 27.5ms
61: learn: 0.1674723 total: 43.7ms remaining: 26.8ms
62: learn: 0.1664121 total: 44.3ms remaining: 26ms
63: learn: 0.1637733 total: 45.2ms remaining: 25.4ms
64: learn: 0.1620715 total: 46ms remaining: 24.7ms
65: learn: 0.1600542 total: 46.6ms remaining: 24ms
66: learn: 0.1585586 total: 47.3ms remaining: 23.3ms
67: learn: 0.1566883 total: 48.2ms remaining: 22.7ms
68: learn: 0.1547028 total: 49ms remaining: 22ms
69: learn: 0.1530539 total: 50.2ms remaining: 21.5ms
70: learn: 0.1507842 total: 51.3ms remaining: 21ms
71: learn: 0.1488977 total: 52.5ms remaining: 20.4ms
72: learn: 0.1473102 total: 53.7ms remaining: 19.9ms
73: learn: 0.1451056 total: 54.7ms remaining: 19.2ms
74: learn: 0.1436516 total: 55.7ms remaining: 18.6ms
75: learn: 0.1414487 total: 56.5ms remaining: 17.8ms
76: learn: 0.1397309 total: 57.4ms remaining: 17.1ms
77: learn: 0.1384802 total: 58.3ms remaining: 16.5ms
78: learn: 0.1366119 total: 59.3ms remaining: 15.8ms
79: learn: 0.1349292 total: 60.2ms remaining: 15ms
80: learn: 0.1332396 total: 61.3ms remaining: 14.4ms
81: learn: 0.1319711 total: 62ms remaining: 13.6ms
82: learn: 0.1303424 total: 63ms remaining: 12.9ms
83: learn: 0.1293609 total: 63.8ms remaining: 12.2ms
84: learn: 0.1280313 total: 65.3ms remaining: 11.5ms
85: learn: 0.1267555 total: 66.4ms remaining: 10.8ms
86: learn: 0.1253989 total: 67.3ms remaining: 10.1ms
87: learn: 0.1244104 total: 68.2ms remaining: 9.29ms
88: learn: 0.1234635 total: 69.1ms remaining: 8.54ms
89: learn: 0.1225559 total: 70.2ms remaining: 7.79ms
90: learn: 0.1215327 total: 71.3ms remaining: 7.05ms
91: learn: 0.1202572 total: 72.3ms remaining: 6.28ms
92: learn: 0.1189924 total: 73.1ms remaining: 5.5ms
93: learn: 0.1180539 total: 74.1ms remaining: 4.73ms
94: learn: 0.1170686 total: 75ms remaining: 3.95ms
95: learn: 0.1163058 total: 76.3ms remaining: 3.18ms
96: learn: 0.1153647 total: 77.3ms remaining: 2.39ms
97: learn: 0.1145492 total: 78.1ms remaining: 1.59ms
98: learn: 0.1133373 total: 78.8ms remaining: 796us
99: learn: 0.1123185 total: 79.6ms remaining: 0us
0: learn: 0.6442230 total: 542us remaining: 53.7ms
1: learn: 0.6048965 total: 1.23ms remaining: 60ms
2: learn: 0.5655809 total: 1.78ms remaining: 57.6ms
3: learn: 0.5350456 total: 2.25ms remaining: 54.1ms
4: learn: 0.5072746 total: 2.73ms remaining: 51.9ms
5: learn: 0.4781308 total: 3.13ms remaining: 49.1ms
6: learn: 0.4601451 total: 3.52ms remaining: 46.8ms
7: learn: 0.4348007 total: 3.82ms remaining: 44ms
8: learn: 0.4156452 total: 4.34ms remaining: 43.9ms
9: learn: 0.3961522 total: 5.01ms remaining: 45.1ms
10: learn: 0.3820850 total: 5.15ms remaining: 41.6ms
11: learn: 0.3679453 total: 5.7ms remaining: 41.8ms
12: learn: 0.3552289 total: 6.7ms remaining: 44.8ms
13: learn: 0.3430151 total: 7.65ms remaining: 47ms
14: learn: 0.3345714 total: 8.54ms remaining: 48.4ms
15: learn: 0.3238381 total: 9.19ms remaining: 48.3ms
16: learn: 0.3166062 total: 10ms remaining: 48.9ms
17: learn: 0.3115804 total: 10.6ms remaining: 48.5ms
18: learn: 0.3034827 total: 11.4ms remaining: 48.4ms
19: learn: 0.2976907 total: 12ms remaining: 48.1ms
20: learn: 0.2906328 total: 13.1ms remaining: 49.2ms
21: learn: 0.2840429 total: 14.1ms remaining: 49.9ms
22: learn: 0.2768625 total: 14.7ms remaining: 49.2ms
23: learn: 0.2716351 total: 15.3ms remaining: 48.4ms
24: learn: 0.2664229 total: 16ms remaining: 48ms
25: learn: 0.2614586 total: 16.5ms remaining: 47.1ms
26: learn: 0.2561204 total: 17.3ms remaining: 46.8ms
27: learn: 0.2513686 total: 17.9ms remaining: 46.1ms
28: learn: 0.2468270 total: 18.7ms remaining: 45.7ms
29: learn: 0.2422397 total: 19.6ms remaining: 45.8ms
30: learn: 0.2389905 total: 20.5ms remaining: 45.6ms
31: learn: 0.2367266 total: 21.5ms remaining: 45.7ms
32: learn: 0.2344441 total: 22ms remaining: 44.6ms
33: learn: 0.2309710 total: 22.6ms remaining: 43.8ms
34: learn: 0.2294099 total: 22.8ms remaining: 42.3ms
35: learn: 0.2260055 total: 23.3ms remaining: 41.4ms
36: learn: 0.2219051 total: 24.5ms remaining: 41.7ms
37: learn: 0.2194785 total: 25.6ms remaining: 41.8ms
38: learn: 0.2168878 total: 26.6ms remaining: 41.6ms
39: learn: 0.2138163 total: 27.4ms remaining: 41ms
40: learn: 0.2104126 total: 28.1ms remaining: 40.5ms
41: learn: 0.2084811 total: 31.2ms remaining: 43.1ms
42: learn: 0.2062840 total: 32ms remaining: 42.5ms
43: learn: 0.2034978 total: 33.3ms remaining: 42.4ms
44: learn: 0.2009875 total: 34.6ms remaining: 42.3ms
45: learn: 0.1979982 total: 35.9ms remaining: 42.2ms
46: learn: 0.1964768 total: 36.6ms remaining: 41.3ms
47: learn: 0.1949001 total: 37.4ms remaining: 40.5ms
48: learn: 0.1921654 total: 38.6ms remaining: 40.2ms
49: learn: 0.1907001 total: 39.7ms remaining: 39.7ms
50: learn: 0.1896775 total: 40.4ms remaining: 38.9ms
51: learn: 0.1873726 total: 41.4ms remaining: 38.2ms
52: learn: 0.1853846 total: 42.4ms remaining: 37.6ms
53: learn: 0.1833178 total: 43.6ms remaining: 37.1ms
54: learn: 0.1820920 total: 44.3ms remaining: 36.2ms
55: learn: 0.1801462 total: 45.6ms remaining: 35.8ms
56: learn: 0.1782076 total: 46.2ms remaining: 34.8ms
57: learn: 0.1758381 total: 46.8ms remaining: 33.9ms
58: learn: 0.1738434 total: 47.6ms remaining: 33.1ms
59: learn: 0.1725397 total: 48.3ms remaining: 32.2ms
60: learn: 0.1703515 total: 48.9ms remaining: 31.3ms
61: learn: 0.1686117 total: 49.8ms remaining: 30.5ms
62: learn: 0.1673932 total: 50.5ms remaining: 29.7ms
63: learn: 0.1645258 total: 51.2ms remaining: 28.8ms
64: learn: 0.1628199 total: 52ms remaining: 28ms
65: learn: 0.1611516 total: 52.7ms remaining: 27.2ms
66: learn: 0.1595761 total: 53.4ms remaining: 26.3ms
67: learn: 0.1575756 total: 54.1ms remaining: 25.4ms
68: learn: 0.1553832 total: 54.7ms remaining: 24.6ms
69: learn: 0.1535890 total: 55.5ms remaining: 23.8ms
70: learn: 0.1523458 total: 56.1ms remaining: 22.9ms
71: learn: 0.1504340 total: 56.7ms remaining: 22.1ms
72: learn: 0.1490783 total: 57.3ms remaining: 21.2ms
73: learn: 0.1474185 total: 58.1ms remaining: 20.4ms
74: learn: 0.1457551 total: 58.7ms remaining: 19.6ms
75: learn: 0.1440618 total: 59.3ms remaining: 18.7ms
76: learn: 0.1424414 total: 59.9ms remaining: 17.9ms
77: learn: 0.1405599 total: 60.8ms remaining: 17.1ms
78: learn: 0.1388193 total: 61.4ms remaining: 16.3ms
79: learn: 0.1371279 total: 62.1ms remaining: 15.5ms
80: learn: 0.1359502 total: 62.8ms remaining: 14.7ms
81: learn: 0.1348950 total: 63.6ms remaining: 14ms
82: learn: 0.1335539 total: 64.3ms remaining: 13.2ms
83: learn: 0.1323967 total: 65ms remaining: 12.4ms
84: learn: 0.1311411 total: 65.8ms remaining: 11.6ms
85: learn: 0.1302400 total: 66.7ms remaining: 10.9ms
86: learn: 0.1295010 total: 67.6ms remaining: 10.1ms
87: learn: 0.1282550 total: 68.4ms remaining: 9.33ms
88: learn: 0.1268703 total: 69.1ms remaining: 8.54ms
89: learn: 0.1261818 total: 69.7ms remaining: 7.75ms
90: learn: 0.1247559 total: 70.8ms remaining: 7.01ms
91: learn: 0.1237127 total: 71.7ms remaining: 6.23ms
92: learn: 0.1221777 total: 72.4ms remaining: 5.45ms
93: learn: 0.1211794 total: 73.2ms remaining: 4.67ms
94: learn: 0.1201006 total: 74.1ms remaining: 3.9ms
95: learn: 0.1192069 total: 74.7ms remaining: 3.11ms
96: learn: 0.1179311 total: 75.4ms remaining: 2.33ms
97: learn: 0.1169919 total: 76.1ms remaining: 1.55ms
98: learn: 0.1158125 total: 76.8ms remaining: 775us
99: learn: 0.1147522 total: 77.5ms remaining: 0us
0: learn: 0.6447617 total: 732us remaining: 72.5ms
1: learn: 0.6051152 total: 1.42ms remaining: 69.3ms
2: learn: 0.5714000 total: 2.59ms remaining: 83.8ms
3: learn: 0.5414568 total: 3.13ms remaining: 75.2ms
4: learn: 0.5109503 total: 4.02ms remaining: 76.5ms
5: learn: 0.4857484 total: 4.98ms remaining: 78.1ms
6: learn: 0.4643829 total: 5.54ms remaining: 73.7ms
7: learn: 0.4466605 total: 6.17ms remaining: 71ms
8: learn: 0.4343357 total: 6.69ms remaining: 67.6ms
9: learn: 0.4134473 total: 7.97ms remaining: 71.7ms
10: learn: 0.4020706 total: 8.57ms remaining: 69.4ms
11: learn: 0.3879785 total: 9.52ms remaining: 69.8ms
12: learn: 0.3722573 total: 10.3ms remaining: 68.9ms
13: learn: 0.3585459 total: 11.5ms remaining: 70.9ms
14: learn: 0.3485039 total: 12.1ms remaining: 68.8ms
15: learn: 0.3390197 total: 13.2ms remaining: 69ms
16: learn: 0.3285231 total: 13.5ms remaining: 66ms
17: learn: 0.3189429 total: 14.7ms remaining: 66.8ms
18: learn: 0.3085253 total: 15.2ms remaining: 64.6ms
19: learn: 0.3030259 total: 15.7ms remaining: 62.8ms
20: learn: 0.2977019 total: 16.2ms remaining: 61.1ms
21: learn: 0.2911362 total: 18ms remaining: 63.8ms
22: learn: 0.2851606 total: 18.7ms remaining: 62.5ms
23: learn: 0.2788896 total: 19.3ms remaining: 61ms
24: learn: 0.2728411 total: 19.8ms remaining: 59.5ms
25: learn: 0.2678508 total: 20.7ms remaining: 59ms
26: learn: 0.2639602 total: 21.4ms remaining: 57.8ms
27: learn: 0.2581845 total: 22.4ms remaining: 57.5ms
28: learn: 0.2549563 total: 23.6ms remaining: 57.8ms
29: learn: 0.2512371 total: 25.4ms remaining: 59.2ms
30: learn: 0.2482787 total: 25.7ms remaining: 57.2ms
31: learn: 0.2454422 total: 27.5ms remaining: 58.4ms
32: learn: 0.2425382 total: 28.1ms remaining: 57.1ms
33: learn: 0.2382371 total: 28.9ms remaining: 56ms
34: learn: 0.2352638 total: 29.8ms remaining: 55.4ms
35: learn: 0.2323701 total: 30.4ms remaining: 54.1ms
36: learn: 0.2306640 total: 31ms remaining: 52.8ms
37: learn: 0.2270014 total: 31.7ms remaining: 51.7ms
38: learn: 0.2227674 total: 32.2ms remaining: 50.4ms
39: learn: 0.2198379 total: 32.8ms remaining: 49.2ms
40: learn: 0.2175182 total: 33.8ms remaining: 48.6ms
41: learn: 0.2151720 total: 34.6ms remaining: 47.8ms
42: learn: 0.2130668 total: 35.5ms remaining: 47.1ms
43: learn: 0.2090422 total: 36.1ms remaining: 45.9ms
44: learn: 0.2073105 total: 37.2ms remaining: 45.4ms
45: learn: 0.2044258 total: 37.6ms remaining: 44.1ms
46: learn: 0.2008996 total: 38.6ms remaining: 43.5ms
47: learn: 0.1985224 total: 39.3ms remaining: 42.6ms
48: learn: 0.1963995 total: 39.9ms remaining: 41.5ms
49: learn: 0.1938656 total: 40.5ms remaining: 40.5ms
50: learn: 0.1919269 total: 41.2ms remaining: 39.6ms
51: learn: 0.1899985 total: 41.9ms remaining: 38.7ms
52: learn: 0.1885458 total: 42.7ms remaining: 37.9ms
53: learn: 0.1862410 total: 44.1ms remaining: 37.6ms
54: learn: 0.1831951 total: 44.7ms remaining: 36.6ms
55: learn: 0.1807429 total: 45.6ms remaining: 35.8ms
56: learn: 0.1784140 total: 46.4ms remaining: 35ms
57: learn: 0.1771711 total: 47ms remaining: 34ms
58: learn: 0.1746945 total: 47.8ms remaining: 33.2ms
59: learn: 0.1733170 total: 48.9ms remaining: 32.6ms
60: learn: 0.1715161 total: 49.5ms remaining: 31.7ms
61: learn: 0.1700899 total: 50.5ms remaining: 31ms
62: learn: 0.1680760 total: 51.2ms remaining: 30.1ms
63: learn: 0.1656692 total: 52.1ms remaining: 29.3ms
64: learn: 0.1640975 total: 52.9ms remaining: 28.5ms
65: learn: 0.1623665 total: 53.9ms remaining: 27.8ms
66: learn: 0.1605779 total: 54.6ms remaining: 26.9ms
67: learn: 0.1591345 total: 57.5ms remaining: 27ms
68: learn: 0.1573873 total: 58.1ms remaining: 26.1ms
69: learn: 0.1559917 total: 59.2ms remaining: 25.4ms
70: learn: 0.1544555 total: 60ms remaining: 24.5ms
71: learn: 0.1529701 total: 60.6ms remaining: 23.6ms
72: learn: 0.1518744 total: 61.3ms remaining: 22.7ms
73: learn: 0.1501712 total: 62.4ms remaining: 21.9ms
74: learn: 0.1484246 total: 63ms remaining: 21ms
75: learn: 0.1470604 total: 65ms remaining: 20.5ms
76: learn: 0.1455514 total: 65.9ms remaining: 19.7ms
77: learn: 0.1439995 total: 66.8ms remaining: 18.8ms
78: learn: 0.1423412 total: 67.4ms remaining: 17.9ms
79: learn: 0.1412214 total: 68.5ms remaining: 17.1ms
80: learn: 0.1400708 total: 69.3ms remaining: 16.3ms
81: learn: 0.1389393 total: 70.1ms remaining: 15.4ms
82: learn: 0.1368149 total: 70.7ms remaining: 14.5ms
83: learn: 0.1356276 total: 71.9ms remaining: 13.7ms
84: learn: 0.1341433 total: 72.6ms remaining: 12.8ms
85: learn: 0.1328493 total: 73.5ms remaining: 12ms
86: learn: 0.1316228 total: 74.2ms remaining: 11.1ms
87: learn: 0.1304642 total: 74.9ms remaining: 10.2ms
88: learn: 0.1290179 total: 75.7ms remaining: 9.36ms
89: learn: 0.1278740 total: 77ms remaining: 8.55ms
90: learn: 0.1265734 total: 77.7ms remaining: 7.68ms
91: learn: 0.1256661 total: 78.5ms remaining: 6.83ms
92: learn: 0.1242486 total: 79.3ms remaining: 5.97ms
93: learn: 0.1233889 total: 80.3ms remaining: 5.12ms
94: learn: 0.1222500 total: 81ms remaining: 4.26ms
95: learn: 0.1212914 total: 81.7ms remaining: 3.4ms
96: learn: 0.1206818 total: 82.7ms remaining: 2.56ms
97: learn: 0.1198218 total: 83.5ms remaining: 1.7ms
98: learn: 0.1185186 total: 84.6ms remaining: 854us
99: learn: 0.1175654 total: 85.4ms remaining: 0us
##################################
# Defining a function to
# generate a unique hash for the selected optimal model
##################################
final_model_hash = generate_run_hash(best_combo, mccv_seed=987654321)
existing_final_run_id = find_existing_run_by_hash("boosted_cb_experiment", final_model_hash)
if existing_final_run_id:
print(f"Duplicate final model found (hash={final_model_hash}). Removing previous run: {existing_final_run_id}")
delete_run(existing_final_run_id)
else:
print(f"No duplicate final model found. Proceeding to log new final model.")
Duplicate final model found (hash=09ee965f4973379bda050e0af8b07bb7). Removing previous run: 9bf4e5467a3f4ffe8738f70cb38c4770
##################################
# Retraining the most optimal CatBoost model
# on the full training data
##################################
print("Optimal Hyperparameter Combination for CatBoost:", best_combo)
final_model = clone(boosted_cb_pipeline)
final_model.set_params(**best_combo)
final_model.fit(X_preprocessed_train, y_preprocessed_train_encoded)
Optimal Hyperparameter Combination for CatBoost: {'boosted_cb_model__iterations': 50, 'boosted_cb_model__learning_rate': 0.01, 'boosted_cb_model__max_depth': 3, 'boosted_cb_model__num_leaves': 8}
0: learn: 0.6891722 total: 666us remaining: 32.6ms
1: learn: 0.6834783 total: 1.14ms remaining: 27.4ms
2: learn: 0.6782963 total: 1.37ms remaining: 21.5ms
3: learn: 0.6734680 total: 2.53ms remaining: 29.1ms
4: learn: 0.6687357 total: 3.53ms remaining: 31.8ms
5: learn: 0.6634680 total: 3.98ms remaining: 29.2ms
6: learn: 0.6585557 total: 4.63ms remaining: 28.5ms
7: learn: 0.6543455 total: 5.19ms remaining: 27.2ms
8: learn: 0.6494274 total: 5.52ms remaining: 25.2ms
9: learn: 0.6445245 total: 6.07ms remaining: 24.3ms
10: learn: 0.6403235 total: 6.24ms remaining: 22.1ms
11: learn: 0.6356199 total: 6.5ms remaining: 20.6ms
12: learn: 0.6312758 total: 6.99ms remaining: 19.9ms
13: learn: 0.6272985 total: 7.63ms remaining: 19.6ms
14: learn: 0.6234670 total: 8.42ms remaining: 19.6ms
15: learn: 0.6188170 total: 8.8ms remaining: 18.7ms
16: learn: 0.6149020 total: 9.22ms remaining: 17.9ms
17: learn: 0.6107420 total: 9.72ms remaining: 17.3ms
18: learn: 0.6069101 total: 9.91ms remaining: 16.2ms
19: learn: 0.6029967 total: 10.4ms remaining: 15.6ms
20: learn: 0.5990690 total: 10.8ms remaining: 14.9ms
21: learn: 0.5950791 total: 11.4ms remaining: 14.5ms
22: learn: 0.5910606 total: 11.9ms remaining: 13.9ms
23: learn: 0.5872759 total: 12.3ms remaining: 13.3ms
24: learn: 0.5831229 total: 12.8ms remaining: 12.8ms
25: learn: 0.5800303 total: 13.4ms remaining: 12.3ms
26: learn: 0.5767067 total: 14ms remaining: 11.9ms
27: learn: 0.5733769 total: 14.5ms remaining: 11.4ms
28: learn: 0.5702532 total: 14.8ms remaining: 10.7ms
29: learn: 0.5673687 total: 15.1ms remaining: 10.1ms
30: learn: 0.5645879 total: 15.8ms remaining: 9.7ms
31: learn: 0.5613671 total: 16.1ms remaining: 9.03ms
32: learn: 0.5583988 total: 16.7ms remaining: 8.61ms
33: learn: 0.5553886 total: 17.1ms remaining: 8.06ms
34: learn: 0.5518851 total: 17.4ms remaining: 7.46ms
35: learn: 0.5491829 total: 17.9ms remaining: 6.95ms
36: learn: 0.5464052 total: 18.3ms remaining: 6.44ms
37: learn: 0.5437216 total: 18.9ms remaining: 5.96ms
38: learn: 0.5410767 total: 19.3ms remaining: 5.45ms
39: learn: 0.5383734 total: 19.7ms remaining: 4.91ms
40: learn: 0.5354526 total: 20.2ms remaining: 4.43ms
41: learn: 0.5326437 total: 20.6ms remaining: 3.93ms
42: learn: 0.5296890 total: 21.1ms remaining: 3.44ms
43: learn: 0.5267096 total: 21.5ms remaining: 2.93ms
44: learn: 0.5244612 total: 22.1ms remaining: 2.45ms
45: learn: 0.5216877 total: 22.5ms remaining: 1.95ms
46: learn: 0.5186363 total: 23ms remaining: 1.47ms
47: learn: 0.5158441 total: 23.4ms remaining: 976us
48: learn: 0.5132195 total: 23.9ms remaining: 488us
49: learn: 0.5104675 total: 24.2ms remaining: 0us
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_cb_model',
<catboost.core.CatBoostClassifier object at 0x00000203ACB62150>)])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('categorical_preprocessor',
ColumnTransformer(force_int_remainder_cols=False,
remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking',
'Physical_Examination',
'Adenopathy', 'Focality',
'Risk', 'T', 'Stage',
'Response'])])),
('boosted_cb_model',
<catboost.core.CatBoostClassifier object at 0x00000203ACB62150>)])ColumnTransformer(force_int_remainder_cols=False, remainder='passthrough',
transformers=[('cat', OrdinalEncoder(),
['Gender', 'Smoking', 'Physical_Examination',
'Adenopathy', 'Focality', 'Risk', 'T',
'Stage', 'Response'])])['Gender', 'Smoking', 'Physical_Examination', 'Adenopathy', 'Focality', 'Risk', 'T', 'Stage', 'Response']
OrdinalEncoder()
['Age']
passthrough
<catboost.core.CatBoostClassifier object at 0x00000203ACB62150>
##################################
# Evaluating the most optimal CatBoost model
# on the validation data
##################################
y_validation_pred = final_model.predict(X_preprocessed_validation)
validation_metrics = evaluate_binary_metrics(y_preprocessed_validation_encoded, y_validation_pred)
##################################
# Consolidating the optimal hyperparameter combination
# and its corresponding MCCV and validation metrics
##################################
consolidated_table_1 = pd.DataFrame({
'hyperparameter': list(best_combo.keys()),
'mccv.optimal.value': list(best_combo.values())
})
consolidated_table_2 = pd.DataFrame({
'metric': list(best_metrics.keys()),
'mccv': [round(v, 4) for v in best_metrics.values()],
'validation': [round(validation_metrics[k], 4) for k in best_metrics.keys()]
})
print(f"{experiment_name} optimal hyperparameter combination:")
display(consolidated_table_1)
print(f"{experiment_name} optimal hyperparameter combination mccv versus validation performance metrics:")
display(consolidated_table_2)
boosted_cb_experiment optimal hyperparameter combination:
| hyperparameter | mccv.optimal.value | |
|---|---|---|
| 0 | boosted_cb_model__iterations | 50.00 |
| 1 | boosted_cb_model__learning_rate | 0.01 |
| 2 | boosted_cb_model__max_depth | 3.00 |
| 3 | boosted_cb_model__num_leaves | 8.00 |
boosted_cb_experiment optimal hyperparameter combination mccv versus validation performance metrics:
| metric | mccv | validation | |
|---|---|---|---|
| 0 | accuracy | 0.8923 | 0.9130 |
| 1 | sensitivity | 0.8800 | 0.9000 |
| 2 | specificity | 0.8977 | 0.9184 |
| 3 | f1 | 0.8349 | 0.8571 |
##################################
# Decoding the true and predicted labels
# for artifact storage
##################################
y_validation_true_decoded = y_encoder.inverse_transform(y_preprocessed_validation_encoded.reshape(-1, 1)).ravel()
y_validation_pred_decoded = y_encoder.inverse_transform(y_validation_pred.reshape(-1, 1)).ravel()
##################################
# Saving the artifacts from the CatBoost model
# hyperparameter tuning exercise
##################################
mccv_csv_path = os.path.join(artifact_dir, f"{model_name}_MCCV_metrics.csv")
cm_path = os.path.join(artifact_dir, f"{model_name}_validation_confusion_matrix.png")
report_path = os.path.join(artifact_dir, f"{model_name}_validation_classification_report.txt")
df_all_combos = pd.DataFrame(all_combos_metrics)
df_all_combos.to_csv(mccv_csv_path, index=False)
log_confusion_matrix(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=cm_path)
log_classification_report(y_validation_true_decoded, y_validation_pred_decoded, labels=class_labels, output_path=report_path)
pickle_model_path = os.path.join(artifact_dir, f"{model_name}_final_model.pkl")
with open(pickle_model_path, "wb") as f:
pickle.dump(final_model, f)
train_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_train_dataset.csv")
thyroid_cancer_preprocessed_train.to_csv(train_dataset_csv_path, index=False)
validation_dataset_csv_path = os.path.join(artifact_dir, f"{model_name}_validation_dataset.csv")
thyroid_cancer_preprocessed_validation.to_csv(validation_dataset_csv_path, index=False)
##################################
# Logging the most optimal CatBoost model
# developed from the full training data
# and evaluated on the validation data
##################################
# Casting integer columns in preprocessed training data to float64
X_logged_input = X_preprocessed_train.astype({
col: 'float64' for col in X_preprocessed_train.select_dtypes(include='int').columns
})
# Preparing the input signature for model logging
input_example = X_logged_input.sample(n=10, random_state=987654321)
signature = infer_signature(input_example, final_model.predict(input_example))
final_run_name = f"CB_HyperparameterCombo_{best_combo_index + 1}_Optimal"
with mlflow.start_run(run_name=final_run_name, description="Retrained on Full Training Set"):
# Storing the hash in MLflow for lookup
mlflow.set_tag("run_hash", final_model_hash)
mlflow.set_tag("model_type", "final_boosted_cb")
mlflow.set_tag("best_combo_index", best_combo_index)
mlflow.log_params(best_combo)
# Logging the average MCCV metrics computed previously
mlflow.log_metrics({f"avg_mccv_{k}": v for k, v in best_metrics.items()})
# Logging validation set metrics
mlflow.log_metrics({f"validation_{k}": v for k, v in validation_metrics.items()})
# Logging the model with input signature
mlflow.sklearn.log_model(
sk_model=final_model,
name="best_boosted_cb_model",
input_example=input_example,
signature=signature
)
# Logging artifacts
mlflow.log_artifact(mccv_csv_path)
mlflow.log_artifact(cm_path)
mlflow.log_artifact(report_path)
mlflow.log_artifact(pickle_model_path)
mlflow.log_artifact(train_dataset_csv_path)
mlflow.log_artifact(validation_dataset_csv_path)
# Logging datasets with metadata (MLflow dataset tracking)
train_dataset = mlflow.data.from_pandas(
pd.read_csv(train_csv_path),
name="mccv_data"
)
mlflow.log_input(train_dataset, context="training")
print("Final CB Model Trained and Logged with Validation Set Evaluation.")
Final CB Model Trained and Logged with Validation Set Evaluation.
1.7.7 Experiment Organization¶
- The MLflow Tracking User Interface (UI) was launched locally at http://127.0.0.1:5000, enabling systematic comparison of all conducted experiments and identification of the optimal run for each ensemble model family
- The bagged_rf_experiment consisted of 16 total runs with the highest model performance metric achieved for RF_HyperparameterCombo_10_Optimal
- avg_mccv_f1 (MCCV F1 Score) = 0.8310
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8778
- avg_mccv_specificity (MCCV Specificity) = 0.8948
- avg_mccv_accuracy (MCCV Accuracy) = 0.8896
- The boosted_ab_experiment consisted of 8 total runs with the highest model performance metric achieved for AB_HyperparameterCombo_3_Optimal
- avg_mccv_f1 (MCCV F1 Score) = 0.8403
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8757
- avg_mccv_specificity (MCCV Specificity) = 0.9069
- avg_mccv_accuracy (MCCV Accuracy) = 0.8974
- The boosted_gb_experiment consisted of 16 total runs with the highest model performance metric achieved for GB_HyperparameterCombo_11_Optimal
- avg_mccv_f1 (MCCV F1 Score) = 0.7850
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.7705
- avg_mccv_specificity (MCCV Specificity) = 0.9181
- avg_mccv_accuracy (MCCV Accuracy) = 0.8729
- The boosted_xgb_experiment consisted of 16 total runs with the highest model performance metric achieved for XGB_HyperparameterCombo_1_Optimal
- avg_mccv_f1 (MCCV F1 Score) = 0.8264
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8442
- avg_mccv_specificity (MCCV Specificity) = 0.9125
- avg_mccv_accuracy (MCCV Accuracy) = 0.8916
- The boosted_lgbm_experiment consisted of 16 total runs with the highest model performance metric achieved for LGBM_HyperparameterCombo_3_Optimal
- avg_mccv_f1 (MCCV F1 Score) = 0.8025
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8000
- avg_mccv_specificity (MCCV Specificity) = 0.9162
- avg_mccv_accuracy (MCCV Accuracy) = 0.8806
- The boosted_cb_experiment consisted of 16 total runs with the highest model performance metric achieved for CB_HyperparameterCombo_1_Optimal
- avg_mccv_f1 (MCCV F1 Score) = 0.8348
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8799
- avg_mccv_specificity (MCCV Specificity) = 0.8976
- avg_mccv_accuracy (MCCV Accuracy) = 0.8922
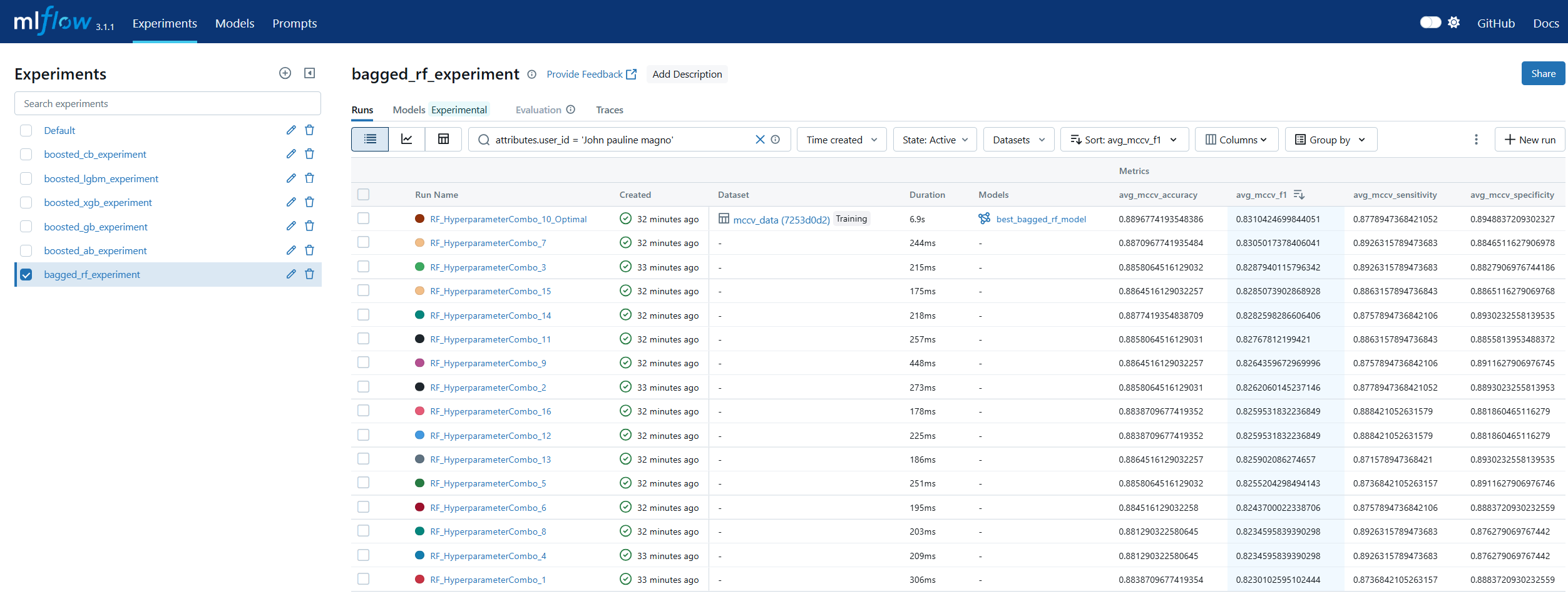
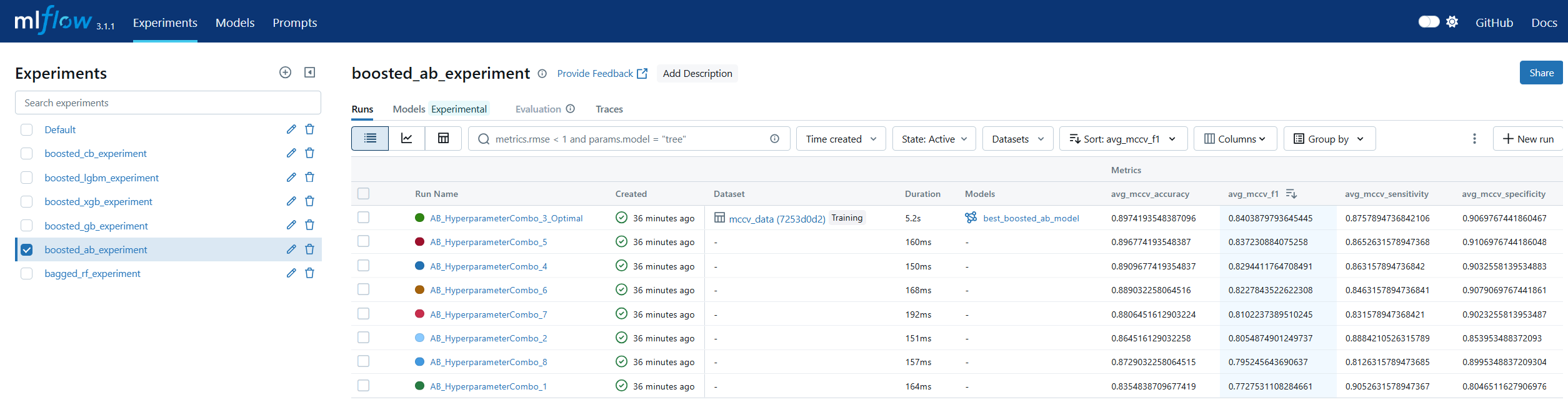
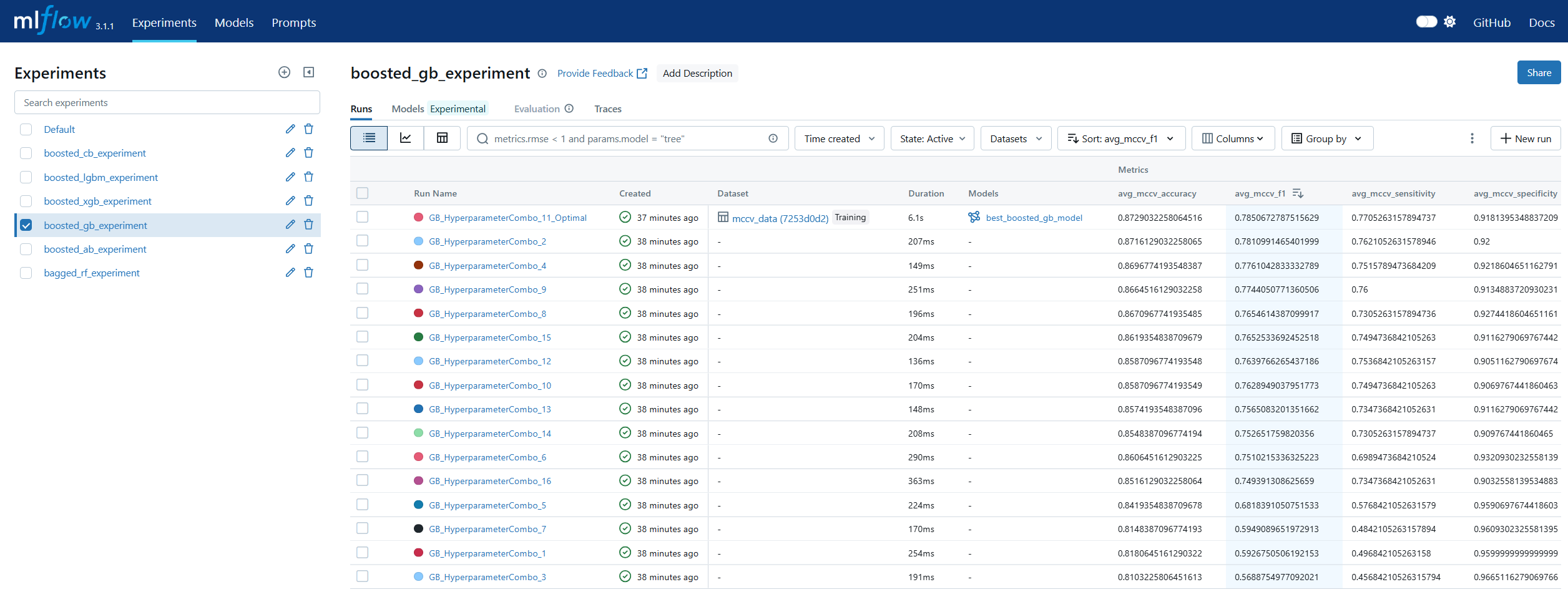
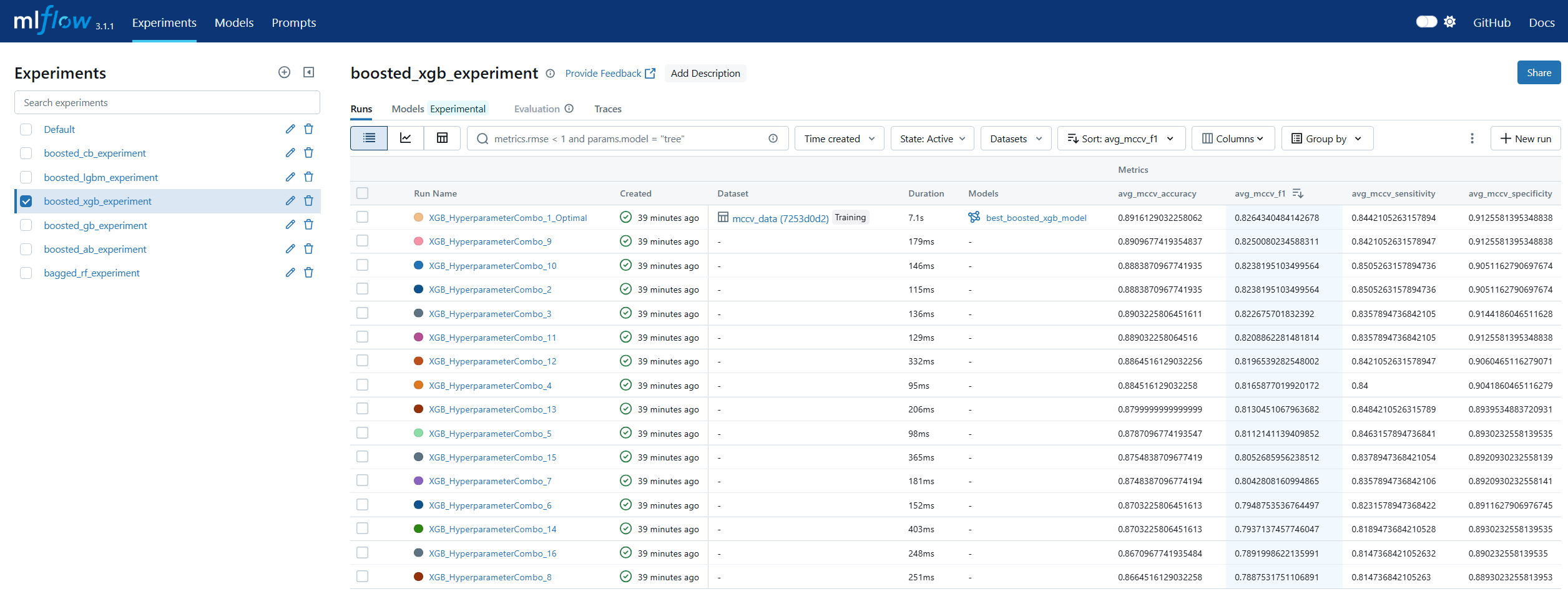
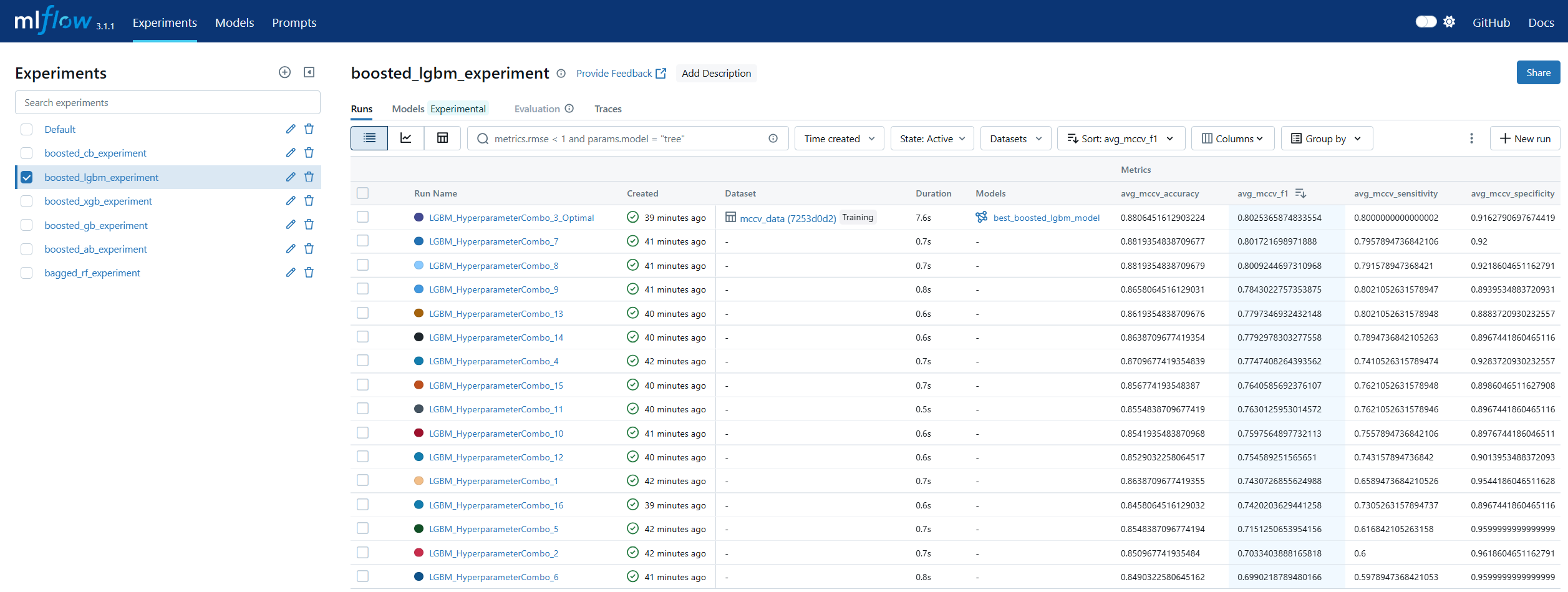
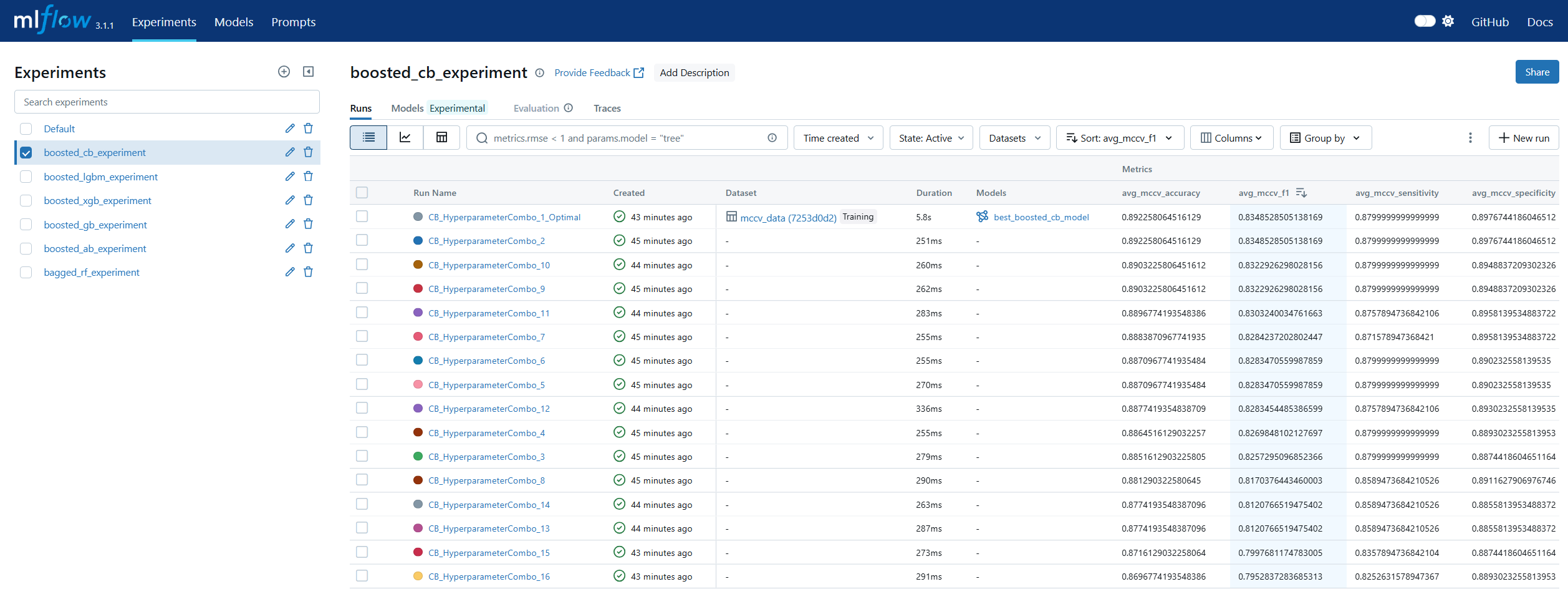

1.7.8 Run Comparison¶
- For the bagged_rf_experiment, the highest avg_mccv_f1 (MCCV F1 Score) was 0.8310 and achieved by the following hyperparameter combination:
- criterion = entropy (against gini)
- max_depth = 3 (against 6)
- min_samples_leaf = 5 (against 10)
- n_estimators = 200 (against 100)
- For the boosted_ab_experiment, the highest avg_mccv_f1 (MCCV F1 Score) was 0.8403 and achieved by the following hyperparameter combination:
- estimator_max_depth = 1 (against 2)
- learning_rate = 0.10 (against 0.01)
- n_estimators = 50 (against 100)
- For the boosted_ab_experiment, the highest avg_mccv_f1 (MCCV F1 Score) was 0.7850 and achieved by the following hyperparameter combination:
- learning_rate = 0.10 (against 0.01)
- max_depth = 3 (against 6)
- min_samples_leaf = 10 (against 5)
- n_estimators = 50 (against 100)
- For the boosted_ab_experiment, the highest avg_mccv_f1 (MCCV F1 Score) was 0.8264 and achieved by the following hyperparameter combination:
- learning_rate = 0.01 (against 0.10)
- max_depth = 3 (against 6)
- gamma 0.10 (against 0.20)
- n_estimators = 50 (against 100)
- For the boosted_ab_experiment, the highest avg_mccv_f1 (MCCV F1 Score) was 0.8025 and achieved by the following hyperparameter combination:
- learning_rate = 0.01 (against 0.10)
- min_child_samples = 3 (against 6)
- num_leaves 8 (against 16)
- n_estimators = 100 (against 200)
- For the boosted_ab_experiment, the highest avg_mccv_f1 (MCCV F1 Score) was 0.8348 and achieved by the following hyperparameter combination:
- learning_rate = 0.01 (against 0.10)
- max_depth = 3 (against 6)
- num_leaves = 8 (against 16)
- iterations = 50 (against 100)
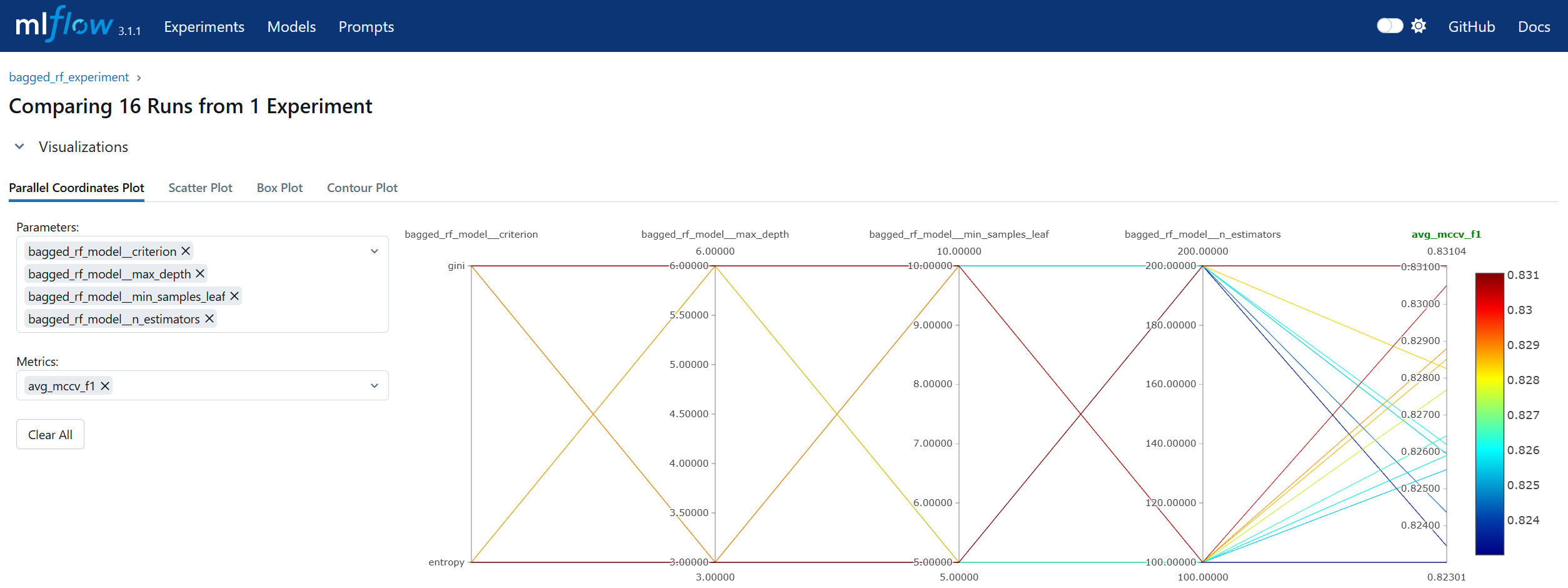
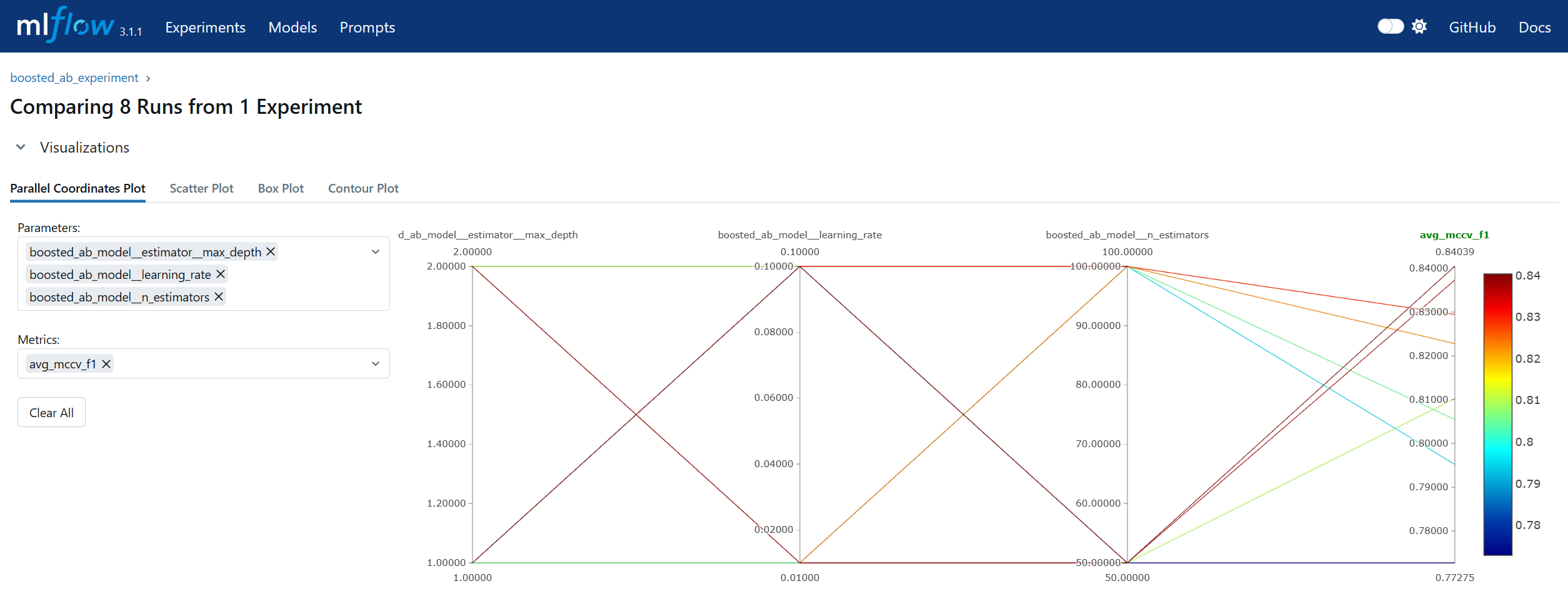
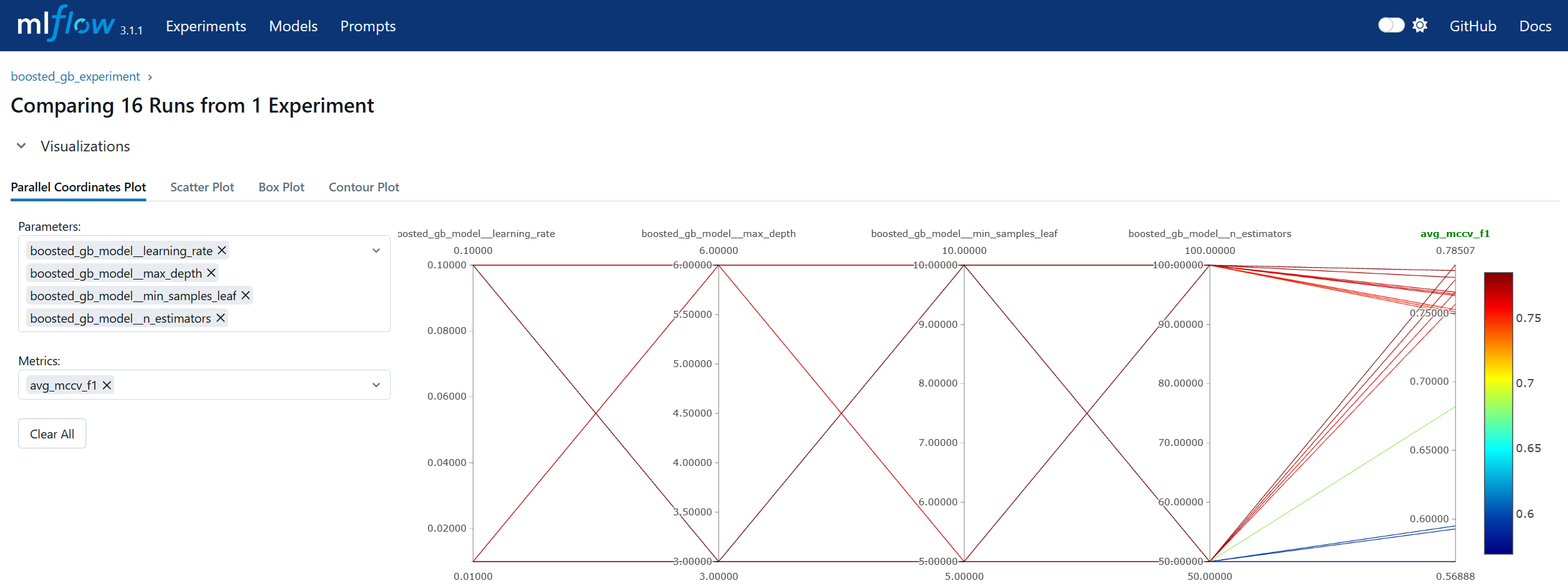
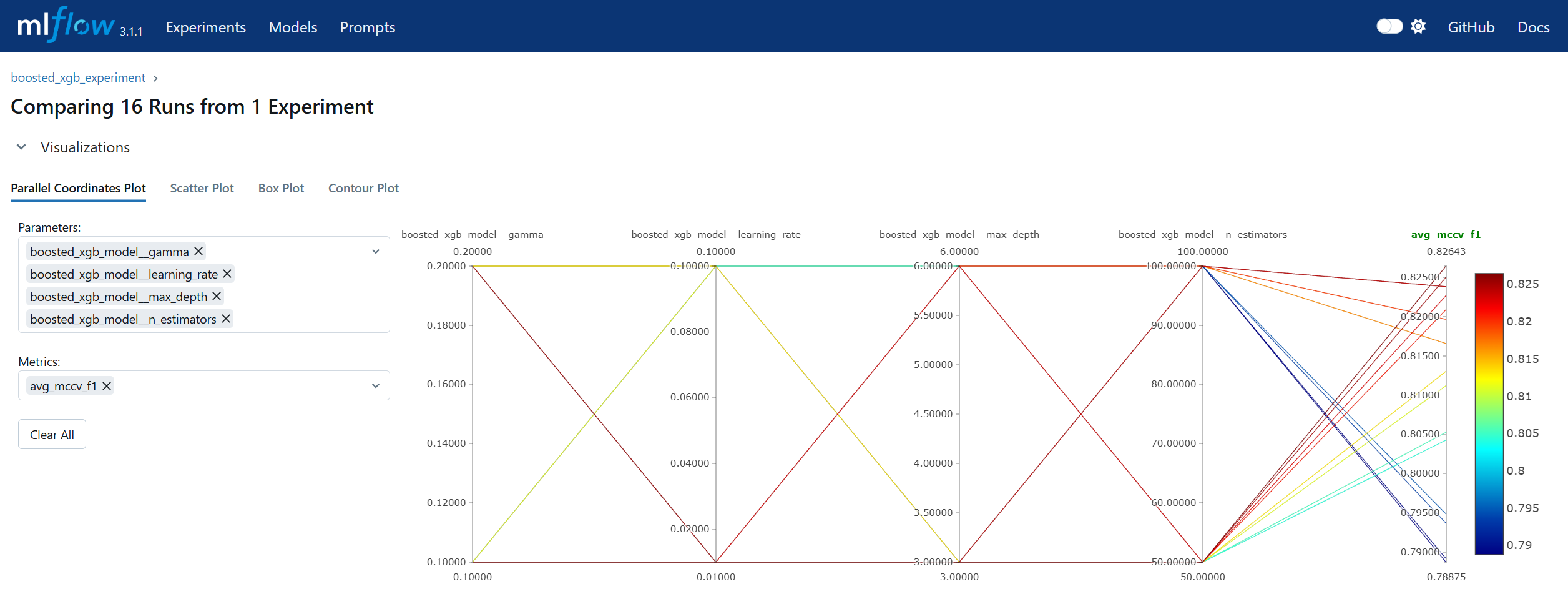
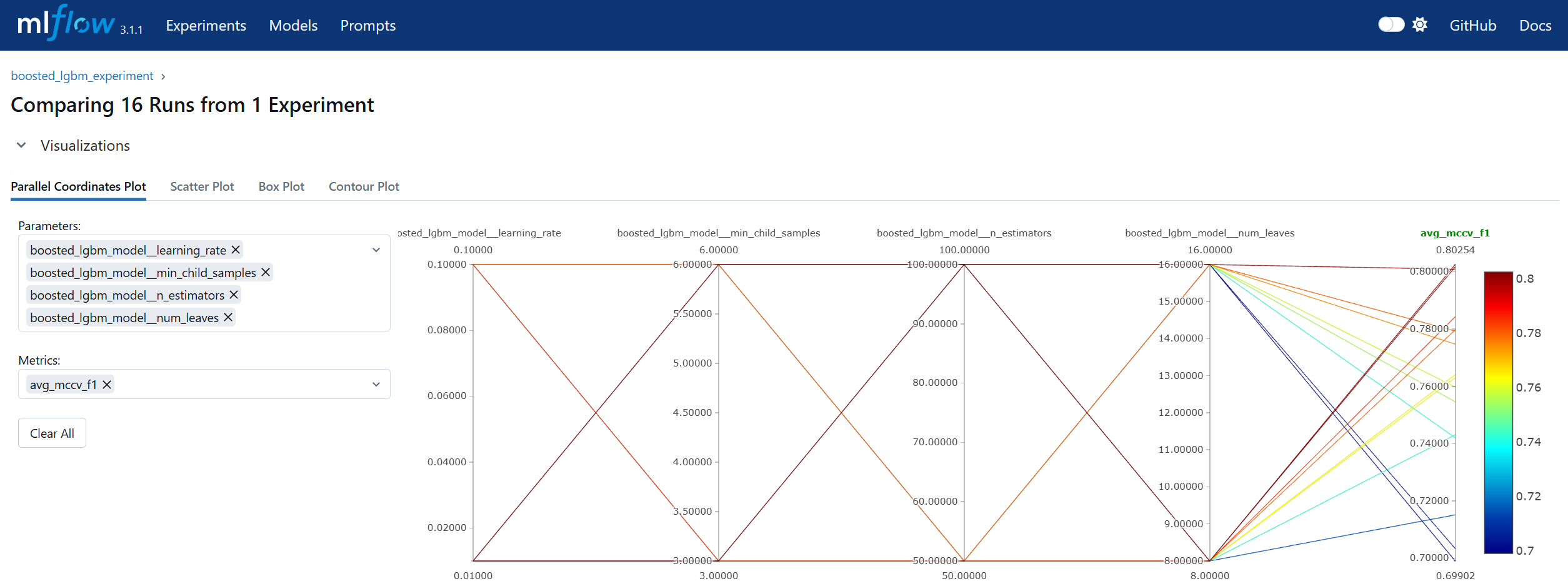
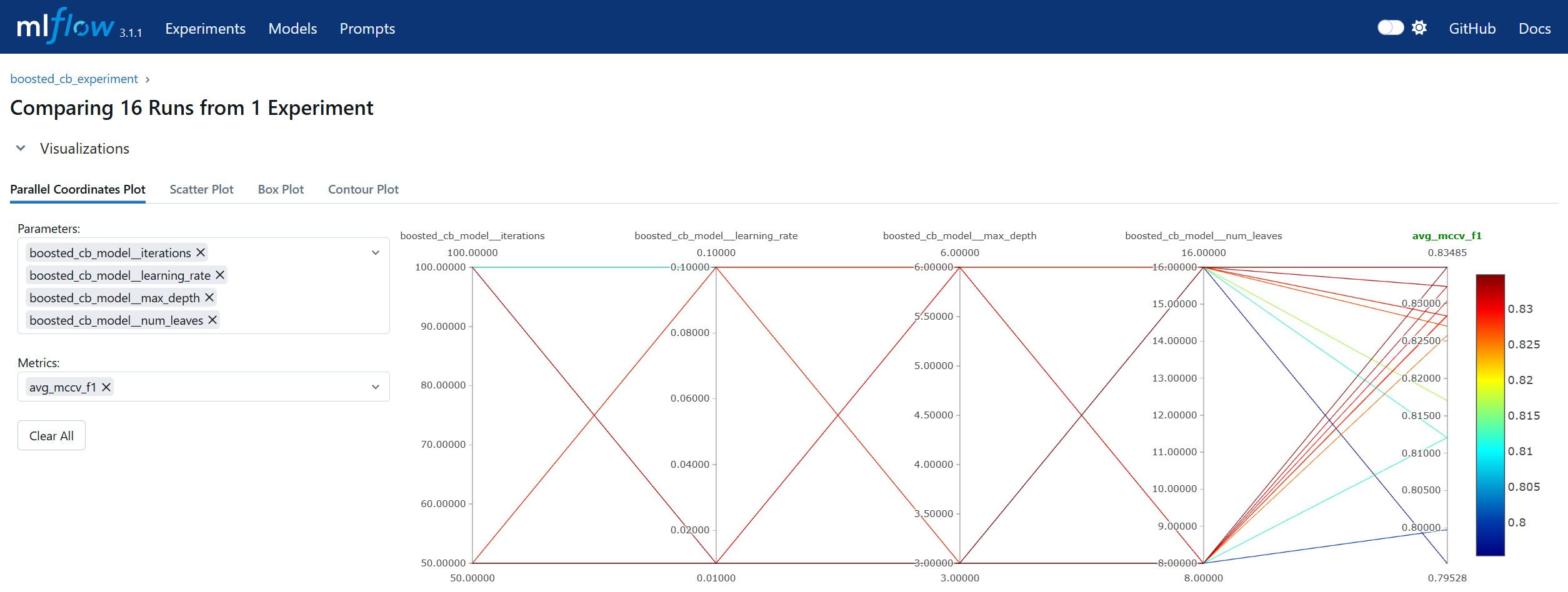
1.7.9 Artifact Storage¶
- For each ensemble algorithm, hyperparameter tuning was performed over a predefined search space. The most performant model from each ensemble experiment was systematically retrained on the full training dataset and logged to MLflow. Model selection was driven by MCCV performance and confirmed via hold-out validation. Each selected model was logged under the convention best_{model_name}_model:
- bagged_rf_experiment: RF_HyperparameterCombo_10_Optimal = best_bagged_rf_model
- criterion = entropy (against gini)
- max_depth = 3 (against 6)
- min_samples_leaf = 5 (against 10)
- n_estimators = 200 (against 100)
- avg_mccv_f1 (MCCV F1 Score) = 0.8310
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8778
- avg_mccv_specificity (MCCV Specificity) = 0.8948
- avg_mccv_accuracy (MCCV Accuracy) = 0.8896
- validation_f1 (Validation F1 Score) = 0.8571
- validation_sensitivity (Validation Sensitivity) = 0.9000
- validation_specificity (Validation Specificity) = 0.9183
- validation_accuracy (Validation Accuracy) = 0.9130
- boosted_ab_experiment: AB_HyperparameterCombo_3_Optimal = best_boosted_ab_model
- estimator_max_depth = 1 (against 2)
- learning_rate = 0.10 (against 0.01)
- n_estimators = 50 (against 100)
- avg_mccv_f1 (MCCV F1 Score) = 0.8403
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8757
- avg_mccv_specificity (MCCV Specificity) = 0.9069
- avg_mccv_accuracy (MCCV Accuracy) = 0.8974
- validation_f1 (Validation F1 Score) = 0.8571
- validation_sensitivity (Validation Sensitivity) = 0.9000
- validation_specificity (Validation Specificity) = 0.9183
- validation_accuracy (Validation Accuracy) = 0.9130
- boosted_gb_experiment: GB_HyperparameterCombo_11_Optimal = best_boosted_gb_model
- learning_rate = 0.10 (against 0.01)
- max_depth = 3 (against 6)
- min_samples_leaf = 10 (against 5)
- n_estimators = 50 (against 100)
- avg_mccv_f1 (MCCV F1 Score) = 0.7850
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.7705
- avg_mccv_specificity (MCCV Specificity) = 0.9181
- avg_mccv_accuracy (MCCV Accuracy) = 0.8729
- validation_f1 (Validation F1 Score) = 0.8292
- validation_sensitivity (Validation Sensitivity) = 0.8500
- validation_specificity (Validation Specificity) = 0.9183
- validation_accuracy (Validation Accuracy) = 0.8985
- boosted_xgb_experiment: XGB_HyperparameterCombo_1_Optimal = best_boosted_xgb_model
- learning_rate = 0.01 (against 0.10)
- max_depth = 3 (against 6)
- gamma 0.10 (against 0.20)
- n_estimators = 50 (against 100)
- avg_mccv_f1 (MCCV F1 Score) = 0.8264
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8442
- avg_mccv_specificity (MCCV Specificity) = 0.9125
- avg_mccv_accuracy (MCCV Accuracy) = 0.8916
- validation_f1 (Validation F1 Score) = 0.8571
- validation_sensitivity (Validation Sensitivity) = 0.9000
- validation_specificity (Validation Specificity) = 0.9183
- validation_accuracy (Validation Accuracy) = 0.9130
- boosted_lgbm_experiment: LGBM_HyperparameterCombo_3_Optimal = best_boosted_lgbm_model
- learning_rate = 0.01 (against 0.10)
- min_child_samples = 3 (against 6)
- num_leaves 8 (against 16)
- n_estimators = 100 (against 200)
- avg_mccv_f1 (MCCV F1 Score) = 0.8025
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8000
- avg_mccv_specificity (MCCV Specificity) = 0.9162
- avg_mccv_accuracy (MCCV Accuracy) = 0.8806
- validation_f1 (Validation F1 Score) = 0.8292
- validation_sensitivity (Validation Sensitivity) = 0.8500
- validation_specificity (Validation Specificity) = 0.9183
- validation_accuracy (Validation Accuracy) = 0.8985
- boosted_cb_experiment: CB_HyperparameterCombo_1_Optimal = best_boosted_cb_model
- learning_rate = 0.01 (against 0.10)
- max_depth = 3 (against 6)
- num_leaves = 8 (against 16)
- iterations = 50 (against 100)
- avg_mccv_f1 (MCCV F1 Score) = 0.8348
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8799
- avg_mccv_specificity (MCCV Specificity) = 0.8976
- avg_mccv_accuracy (MCCV Accuracy) = 0.8922
- validation_f1 (Validation F1 Score) = 0.8571
- validation_sensitivity (Validation Sensitivity) = 0.9000
- validation_specificity (Validation Specificity) = 0.9183
- validation_accuracy (Validation Accuracy) = 0.9130
- bagged_rf_experiment: RF_HyperparameterCombo_10_Optimal = best_bagged_rf_model
- As part of the ensemble model evaluation and selection process, a structured artifact saving pipeline was implemented for each ensemble model for improved reproducibility and traceability. For each model, the following artifacts were generated and saved to both the The MLflow Tracking UI (launched locally at http://127.0.0.1:5000) and the designated directory:
- MCCV_metrics.csv
- Purpose: Stores cross-validation performance metrics for all hyperparameter combinations explored during tuning.
- Contents: Includes classification metrics (Accuracy, Sensitivity, Specificity, F1 Score) for each parameter set.
- File name pattern: {model_name}_MCCV_metrics.csv
- final_model.pkl
- Purpose: Contains the serialized (pickled) optimal model corresponding to the hyperparameter combination that achieved the highest MCCV F1 score.
- Contents: Enables rapid reloading for inference or deployment without retraining.
- File name pattern: {model_name}_final_model.pkl
- validation_classification_report.txt
- Purpose: Captures a full classification report of the optimal model evaluated on the validation set.
- Contents: Precision, Recall, F1-score, Support (per class), Overall Accuracy, Macro Average, and Weighted Average.
- File name pattern: {model_name}_validation_classification_report.txt
- validation_confusion_matrix.png
- Purpose: Visualizes classification results on the validation set via a confusion matrix, providing insight into true positives, true negatives, false positives, and false negatives.
- Contents: PNG image for easy inspection or inclusion in presentations and reports.
- File name pattern: {model_name}_validation_confusion_matrix.png
- MCCV_metrics.csv
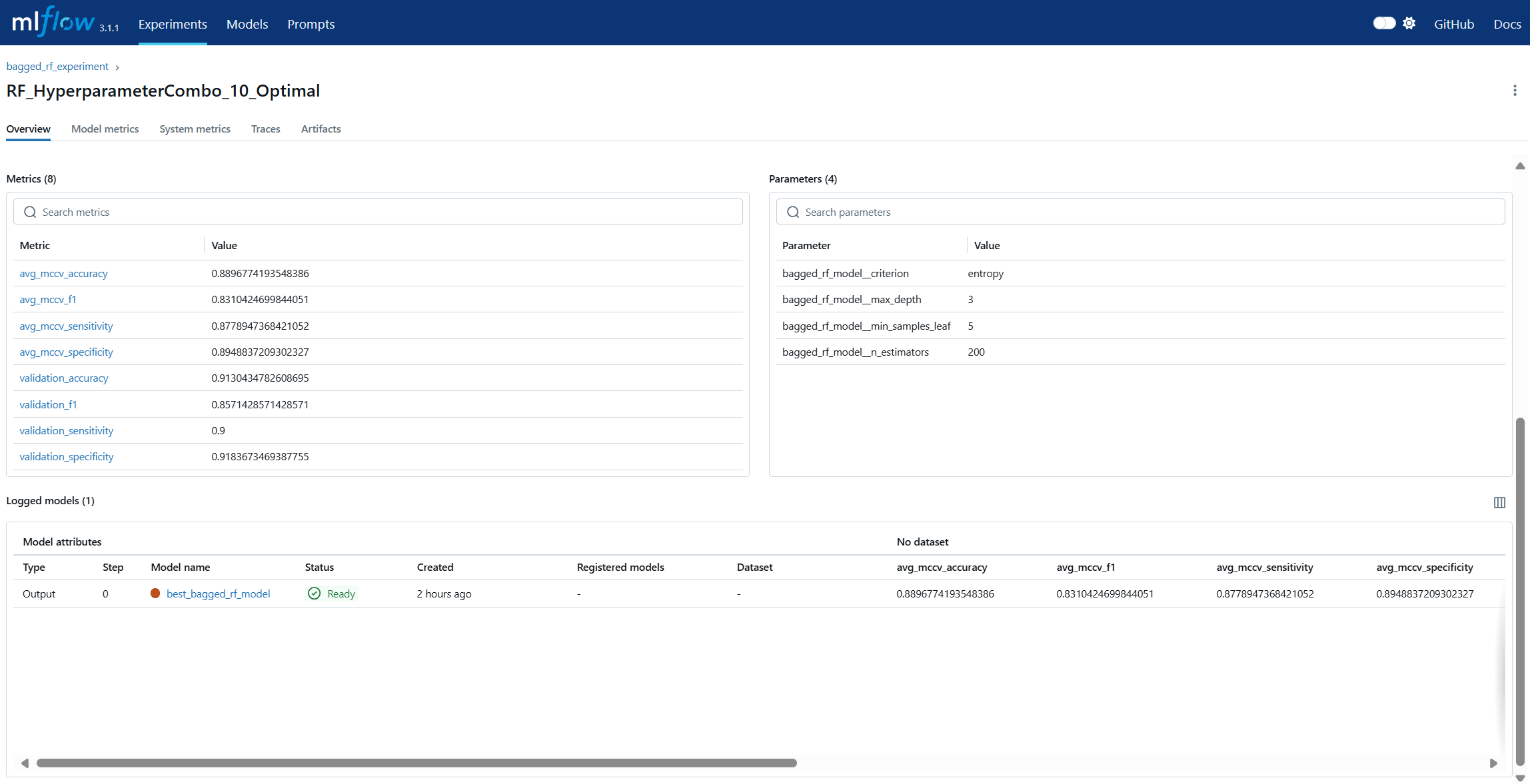
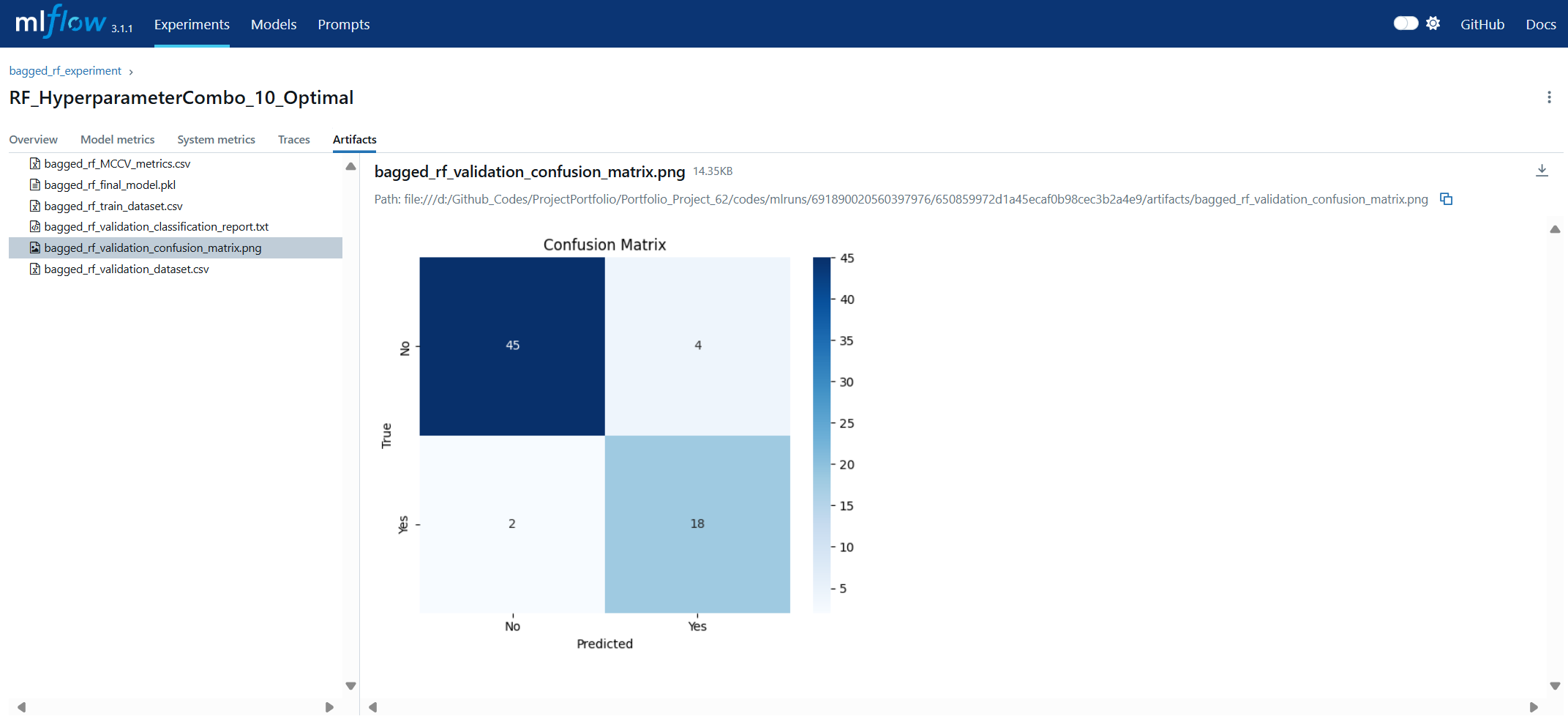
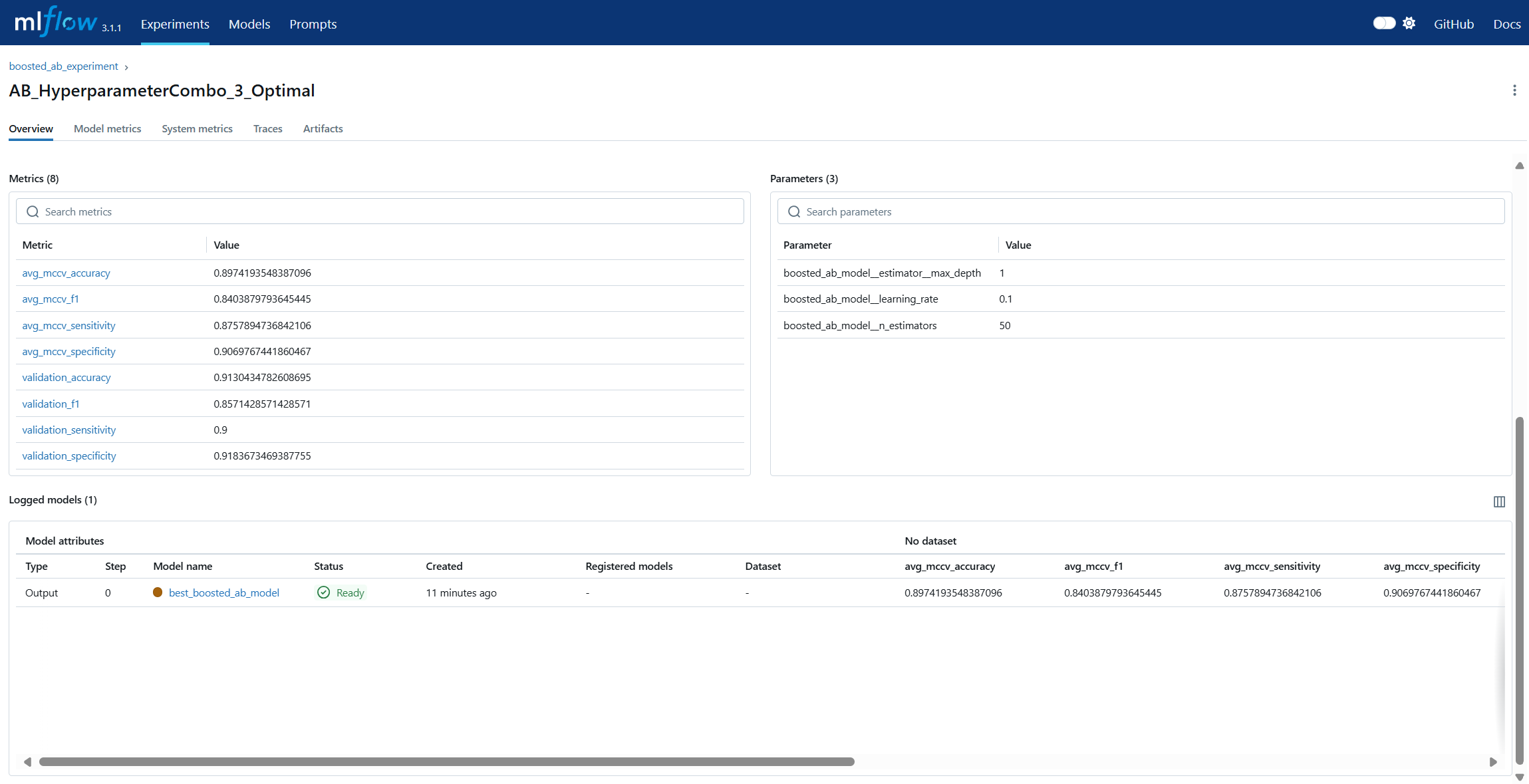
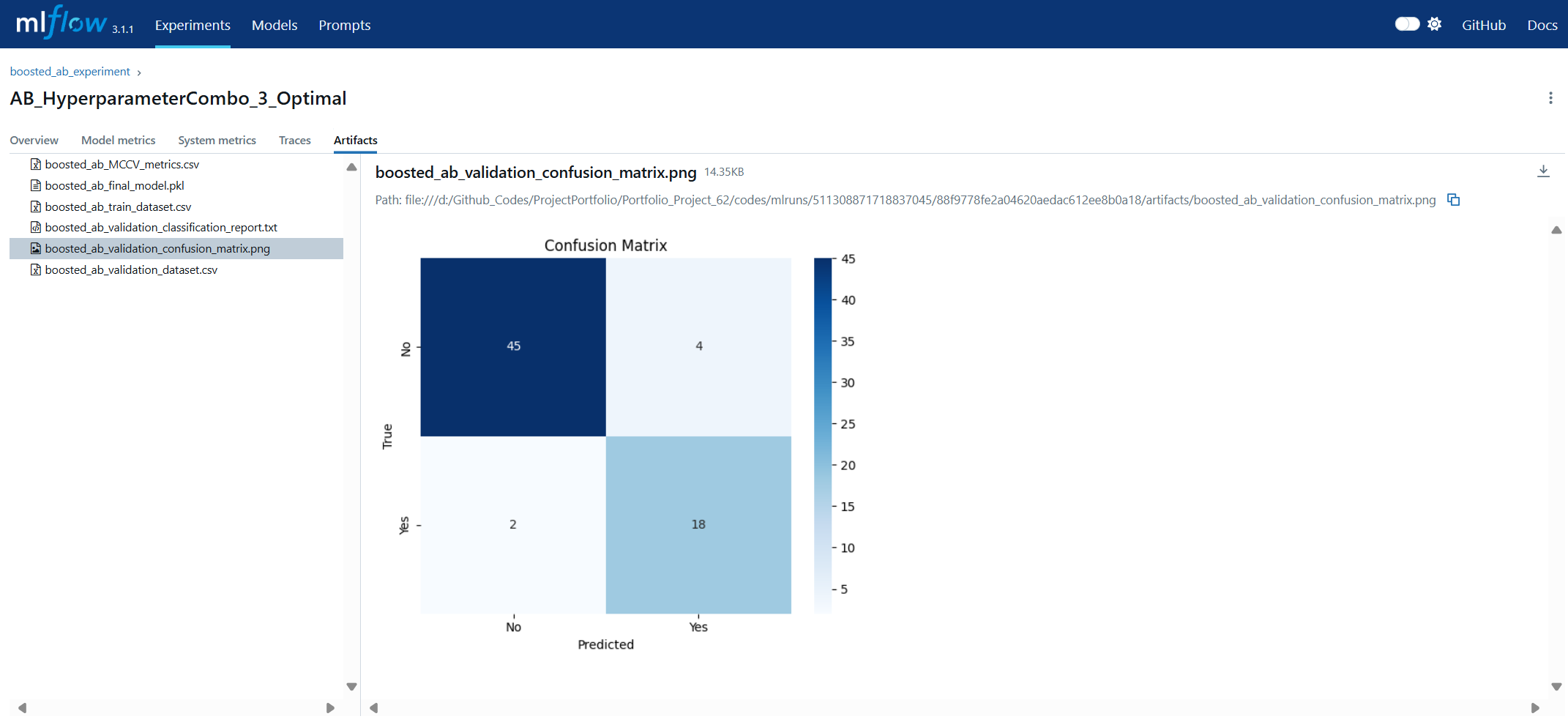
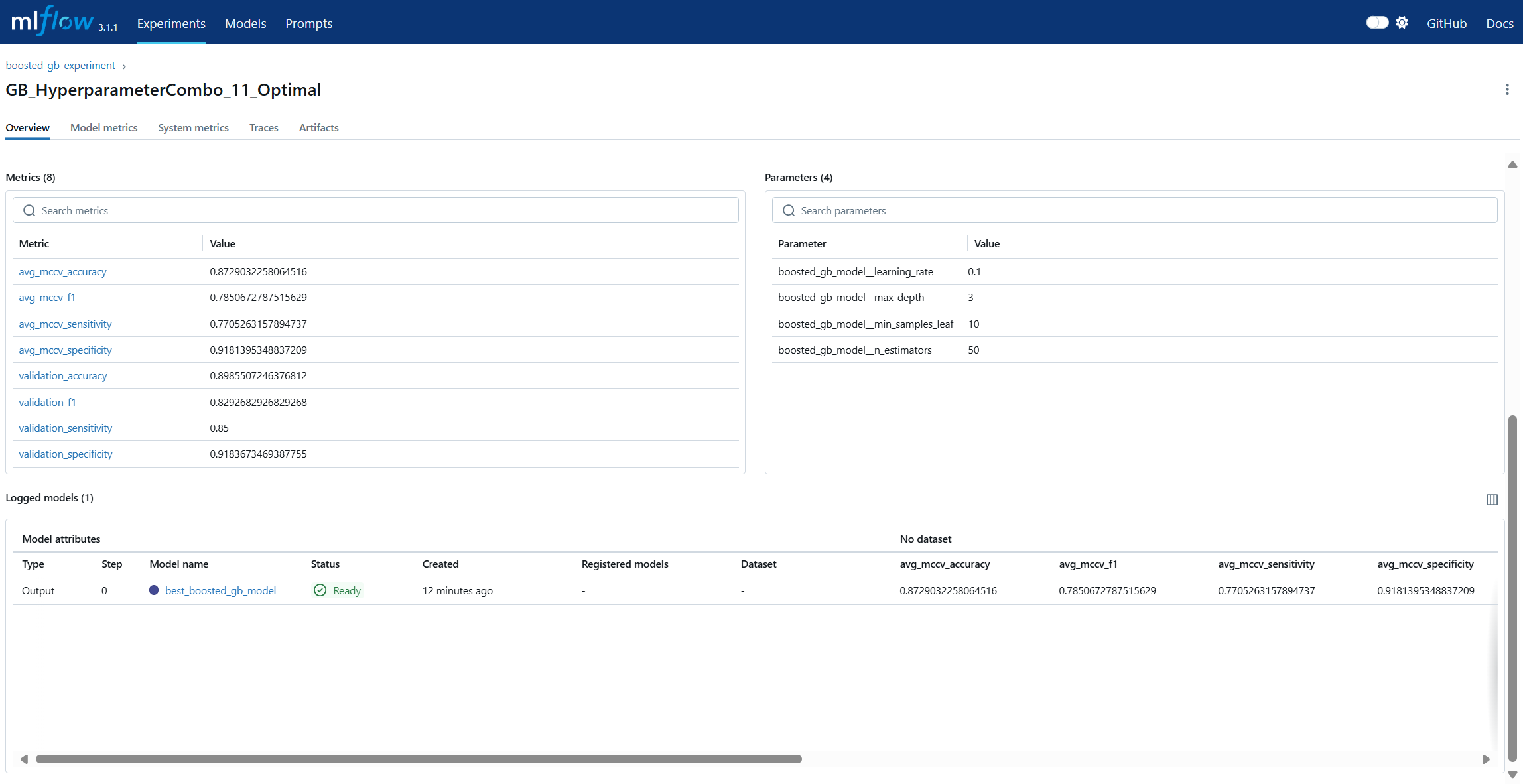
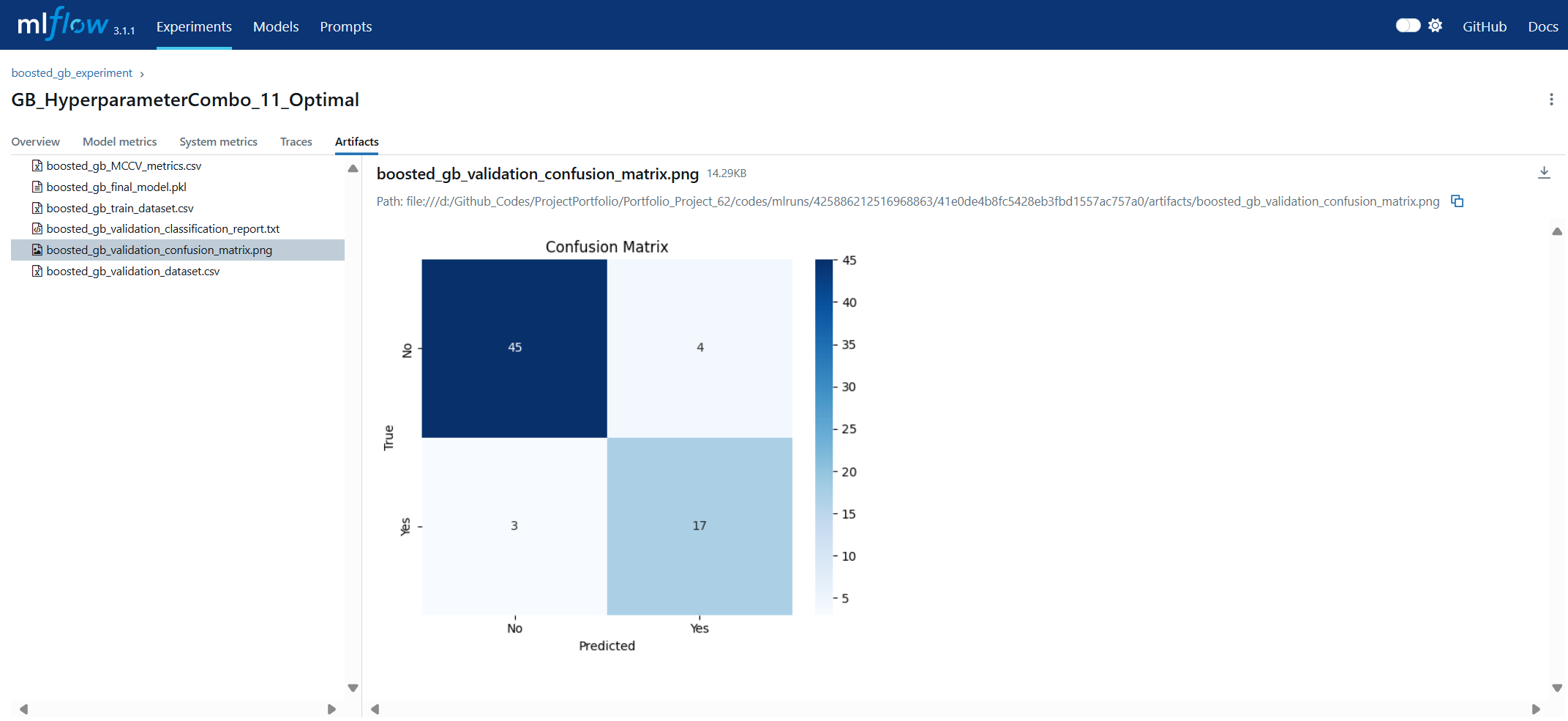
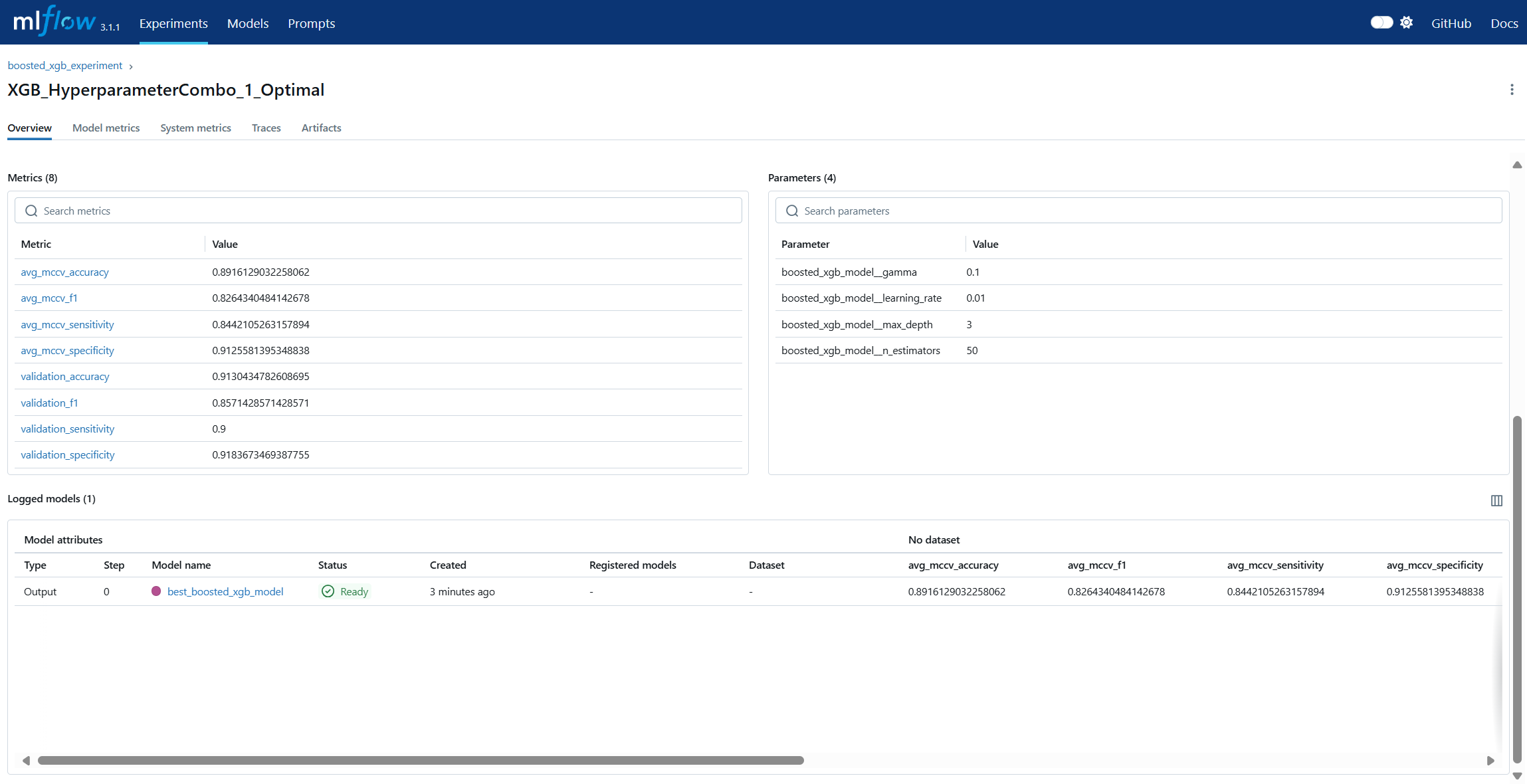
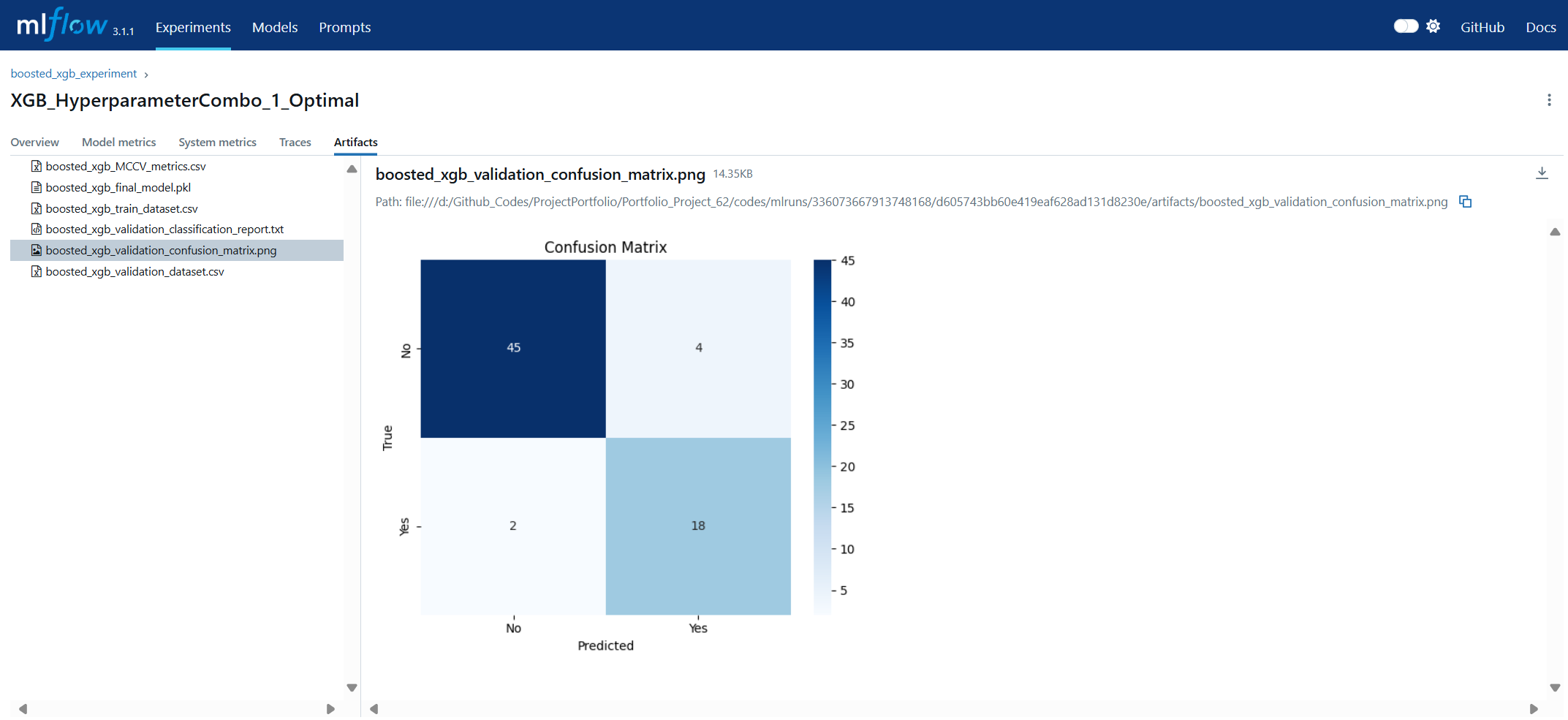
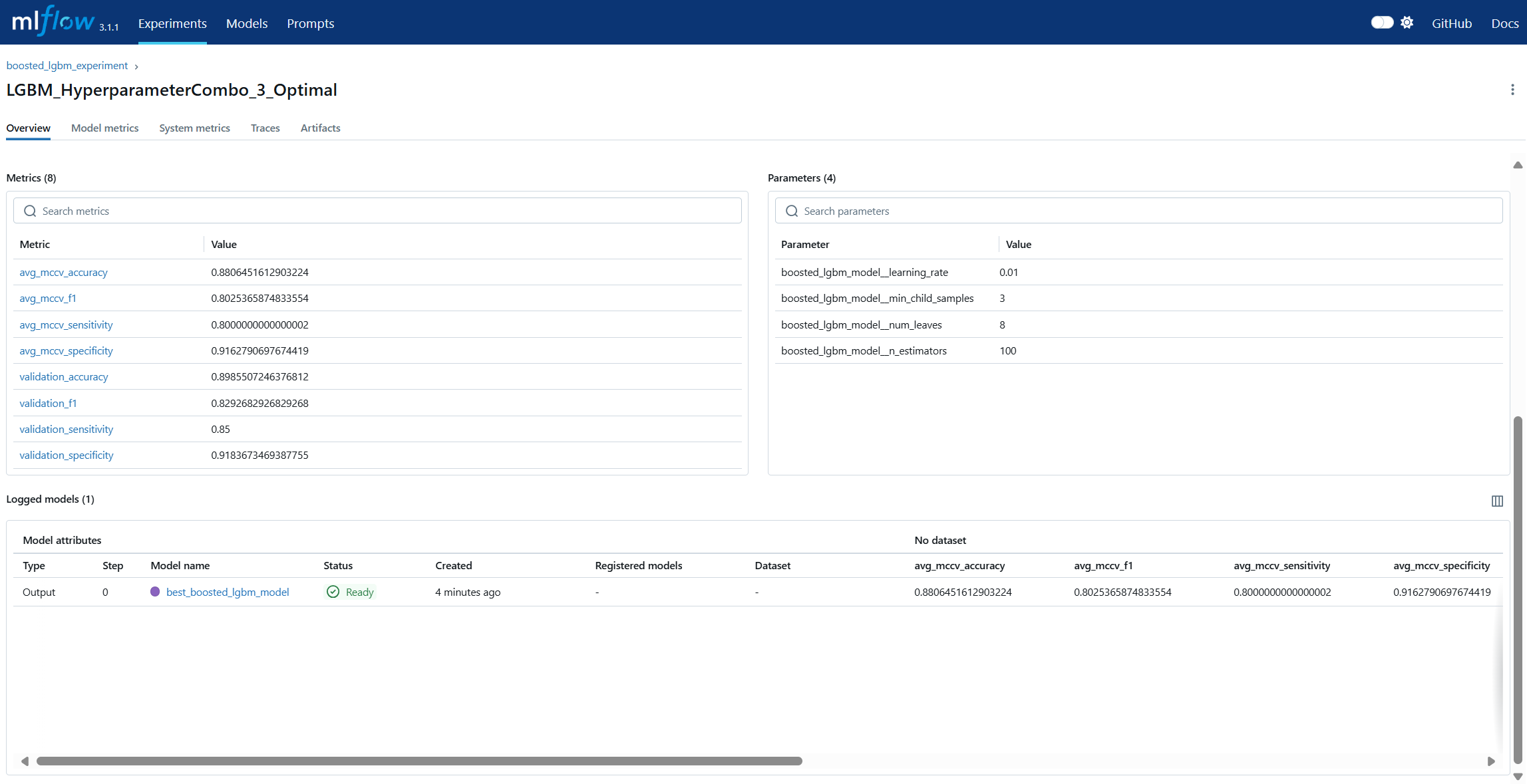

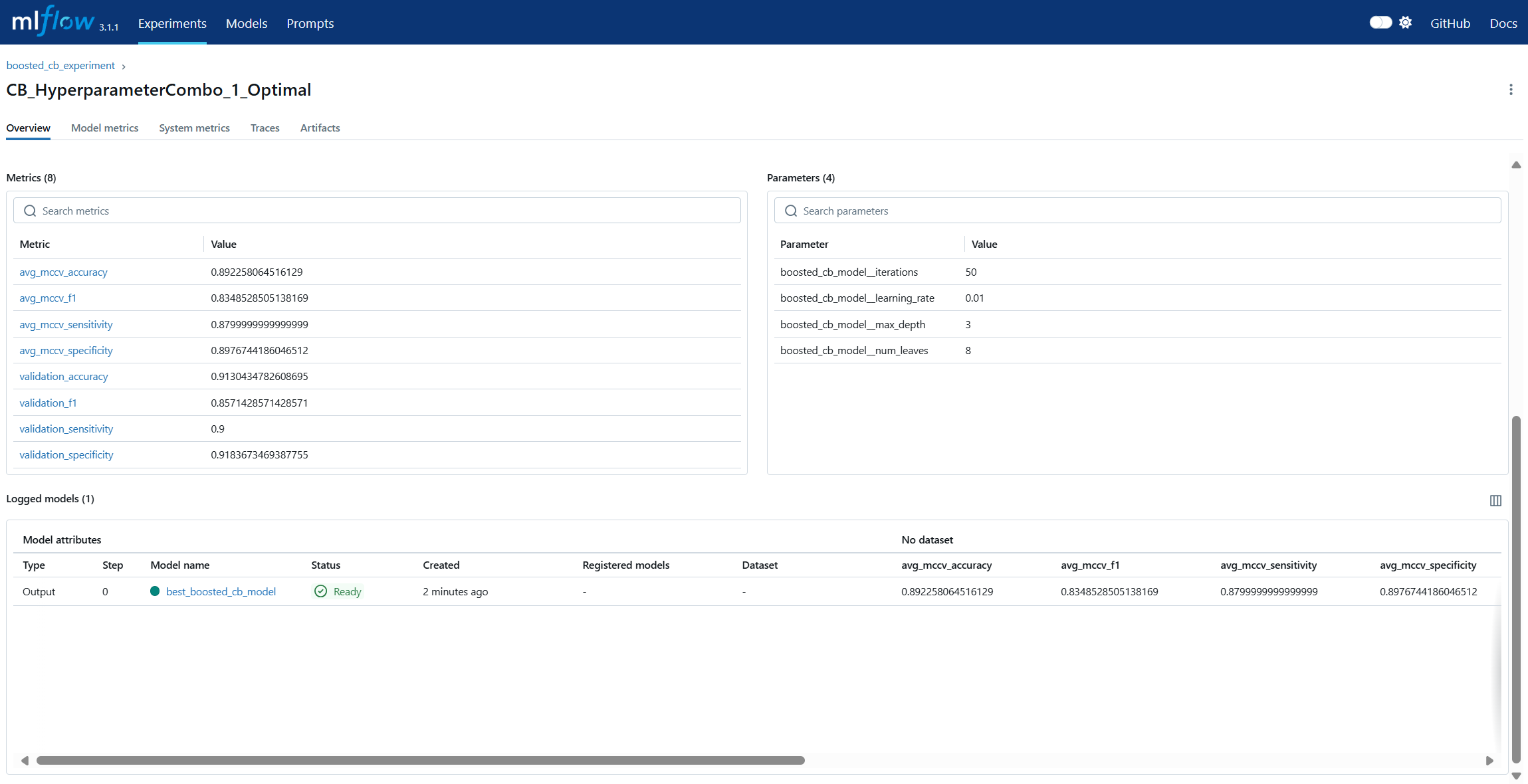
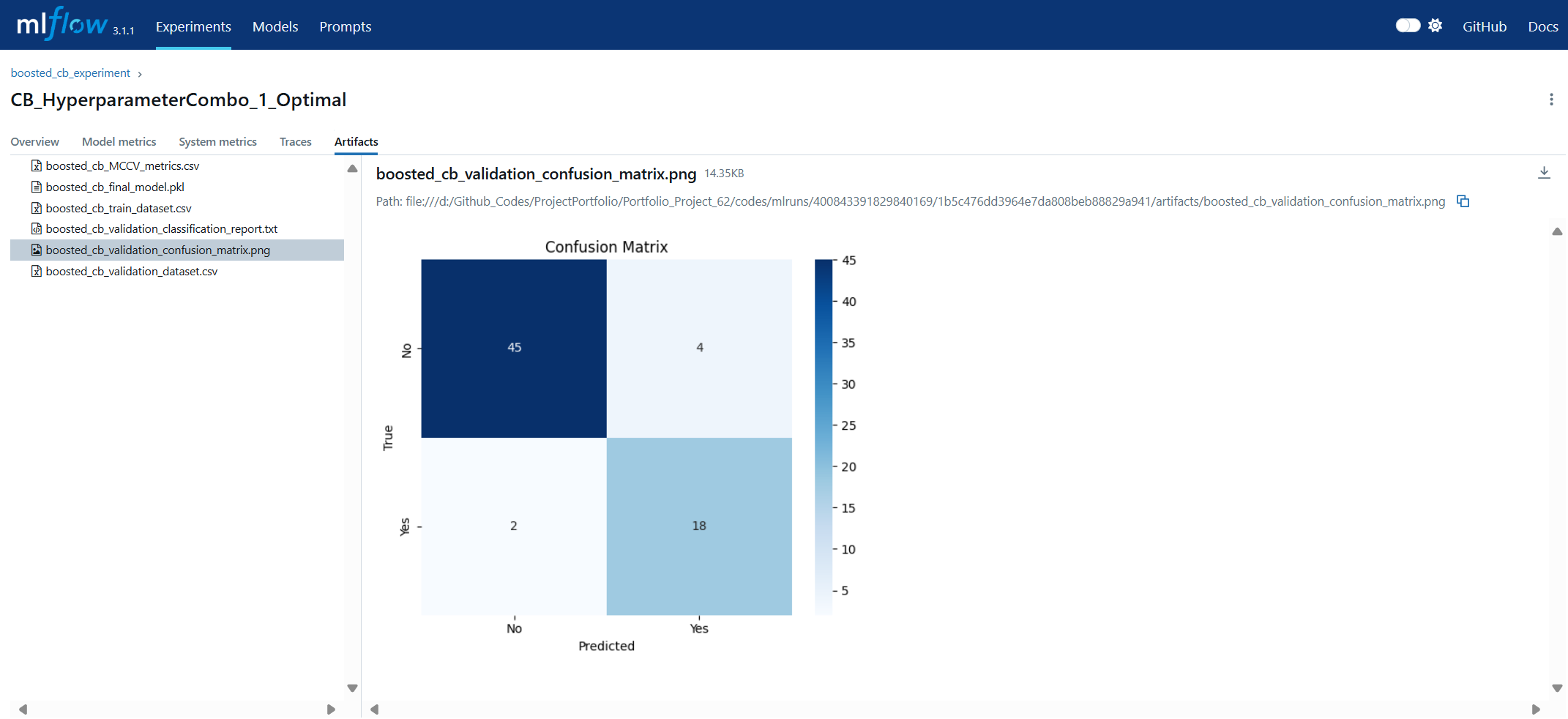
1.7.10 Model Registry¶
- The best model run from each of the six ensemble experiments was identified using the MLflow Tracking API:
- bagged_rf_experiment: RF_HyperparameterCombo_10_Optimal = best_bagged_rf_model
- boosted_ab_experiment: AB_HyperparameterCombo_3_Optimal = best_boosted_ab_model
- boosted_gb_experiment: GB_HyperparameterCombo_11_Optimal = best_boosted_gb_model
- boosted_xgb_experiment: XGB_HyperparameterCombo_1_Optimal = best_boosted_xgb_model
- boosted_lgbm_experiment: LGBM_HyperparameterCombo_3_Optimal = best_boosted_lgbm_model
- boosted_cb_experiment: CB_HyperparameterCombo_1_Optimal = best_boosted_cb_model
- For each candidate, both validation performance and Monte Carlo Cross-Validation (MCCV) metrics were retrieved and compared. The primary metric for selection was validation F1 Score, with secondary sorting on the remaining metrics to resolve ties. This composite scoring mechanism ensures the final model is both accurate and well-balanced in class sensitivity:
- validation_f1 (Validation F1 Score)
- validation_sensitivity (Validation Sensitivity)
- validation_specificity (Validation Specificity)
- validation_accuracy (Validation Accuracy)
- avg_mccv_f1 (MCCV F1 Score)
- avg_mccv_sensitivity (MCCV Sensitivity)
- avg_mccv_specificity (MCCV Specificity)
- avg_mccv_accuracy (MCCV Accuracy)
- The best_boosted_ab_model was determined to have the highest cumulative ranking across the validation and MCCV metrics, and was registered directly from its logged run using mlflow.register_model, with the name selected_final_model. Metadata including the experiment name, experiment ID, run ID, model ID, model path and model URI was captured for downstream use.
- validation_f1 (Validation F1 Score) = 0.8571
- validation_sensitivity (Validation Sensitivity) = 0.9000
- validation_specificity (Validation Specificity) = 0.9183
- validation_accuracy (Validation Accuracy) = 0.9130
- avg_mccv_f1 (MCCV F1 Score) = 0.8403
- avg_mccv_sensitivity (MCCV Sensitivity) = 0.8757
- avg_mccv_specificity (MCCV Specificity) = 0.9069
- avg_mccv_accuracy (MCCV Accuracy) = 0.8974
##################################
# Setting the MLflow Client and selecting the final model
# from the optimal candidate models from the previous experiments
# based on the validation and MCCV performance metrics
##################################
client = MlflowClient()
# Listing all experiments for each model type
experiment_names = [
"bagged_rf_experiment",
"boosted_ab_experiment",
"boosted_gbm_experiment",
"boosted_lgbm_experiment",
"boosted_cb_experiment"
]
best_model_info = None
best_evaluation = {
'validation_f1': -1,
'validation_sensitivity': -1,
'validation_specificity': -1,
'validation_accuracy': -1,
'mccv_f1': -1,
'mccv_sensitivity': -1,
'mccv_specificity': -1,
'mccv_accuracy': -1
}
# Evaluating the optimal candidate models to determine the final model
for exp_name in experiment_names:
exp = client.get_experiment_by_name(exp_name)
if exp is None:
continue
runs = client.search_runs(
experiment_ids=[exp.experiment_id],
filter_string="tags.model_type LIKE 'final_%'",
order_by=[
"metrics.validation_f1 DESC",
"metrics.validation_sensitivity DESC",
"metrics.validation_specificity DESC",
"metrics.validation_accuracy DESC",
"metrics.avg_mccv_f1 DESC",
"metrics.avg_mccv_sensitivity DESC",
"metrics.avg_mccv_specificity DESC",
"metrics.avg_mccv_accuracy DESC"
],
max_results=1
)
if not runs:
continue
run = runs[0]
# Extracting model classification performance metrics
val_f1 = run.data.metrics.get("validation_f1", 0)
val_sens = run.data.metrics.get("validation_sensitivity", 0)
val_spec = run.data.metrics.get("validation_specificity", 0)
val_acc = run.data.metrics.get("validation_accuracy", 0)
mccv_f1 = run.data.metrics.get("avg_mccv_f1", 0)
mccv_sens = run.data.metrics.get("avg_mccv_sensitivity", 0)
mccv_spec = run.data.metrics.get("avg_mccv_specificity", 0)
mccv_acc = run.data.metrics.get("avg_mccv_accuracy", 0)
current_model_score = (val_f1, val_sens, val_spec, val_acc, mccv_f1, mccv_sens, mccv_spec, mccv_acc)
best_model_score = tuple(best_evaluation.values())
if current_model_score > best_model_score:
# Obtaining experiment artifact location and normalizing path
artifact_uri = exp.artifact_location
parsed_uri = urlparse(artifact_uri)
if parsed_uri.scheme != "file":
raise RuntimeError(f"Artifact store for experiment {exp.experiment_id} is not local: {artifact_uri}")
exp_path = parsed_uri.path
if re.match(r"^/[a-zA-Z]:", exp_path): # Windows drive letter fix
exp_path = exp_path[1:]
exp_path = os.path.normpath(exp_path)
# Accessing the run's folder inside the experiment folder
run_folder = os.path.join(exp_path, run.info.run_id)
if not os.path.exists(run_folder):
raise FileNotFoundError(f"Run folder not found: {run_folder}")
# Accessing the 'outputs' folder inside the run folder
outputs_folder = os.path.join(run_folder, "outputs")
if not os.path.exists(outputs_folder):
raise FileNotFoundError(f"No 'outputs' folder found in: {run_folder}")
# Dynamically detecting the model ID (m-<uuid> pattern) inside the outputs folder
entries = os.listdir(outputs_folder)
model_id = None
for entry in entries:
if re.match(r"m-[0-9a-f]{32}", entry):
model_id = entry
break
if not model_id:
raise FileNotFoundError(f"No model ID folder found in outputs: {outputs_folder}")
# Building the directory path to models/<model_id>
models_folder = os.path.join(exp_path, "models", model_id)
if not os.path.exists(models_folder):
raise FileNotFoundError(f"Model folder not found: {models_folder}")
model_path = f"file:{models_folder}"
best_model_info = {
"experiment_name": exp_name,
"experiment_id": exp.experiment_id,
"run_id": run.info.run_id,
"model_id": model_id,
"model_path": model_path,
"model_name": "selected_final_model",
"model_uri": f"runs:/{run.info.run_id}/best_{exp_name.split('_')[0]+"_"+exp_name.split('_')[1]}_model"
}
best_evaluation = {
'validation_f1': val_f1,
'validation_sensitivity': val_sens,
'validation_specificity': val_spec,
'validation_accuracy': val_acc,
'mccv_f1': mccv_f1,
'mccv_sensitivity': mccv_sens,
'mccv_specificity': mccv_spec,
'mccv_accuracy': mccv_acc,
}
print("selected final model information:")
display(best_model_info)
selected final model information:
{'experiment_name': 'boosted_ab_experiment',
'experiment_id': '511308871718837045',
'run_id': 'e949c9a21e5946d4b33b5431efc7c65f',
'model_id': 'm-e4f4e70e2c5542188483c4d1a29872fc',
'model_path': 'file:d:\\Github_Codes\\ProjectPortfolio\\Portfolio_Project_62\\codes\\mlruns\\511308871718837045\\models\\m-e4f4e70e2c5542188483c4d1a29872fc',
'model_name': 'selected_final_model',
'model_uri': 'runs:/e949c9a21e5946d4b33b5431efc7c65f/best_boosted_ab_model'}
##################################
# Registering the final model
##################################
logging.getLogger("mlflow.tracking._model_registry.fluent").setLevel(logging.ERROR)
if best_model_info:
model_name = best_model_info["model_name"]
model_uri = best_model_info["model_uri"]
# Deleting the existing registered model (if exists)
try:
client.delete_registered_model(name=model_name)
print(f"Deleted model '{model_name}' to reset to version 1.")
except RestException as e:
print(f"Model did not exist or couldn't be deleted: {e}")
# Registering the new best model
result = mlflow.register_model(
model_uri=model_uri,
name=model_name
)
print(f"Registered best model from {best_model_info['experiment_name']} as '{result.name}', version {result.version}")
Deleted model 'selected_final_model' to reset to version 1.
Successfully registered model 'selected_final_model'.
Registered best model from boosted_ab_experiment as 'selected_final_model', version 1
Created version '1' of model 'selected_final_model'.
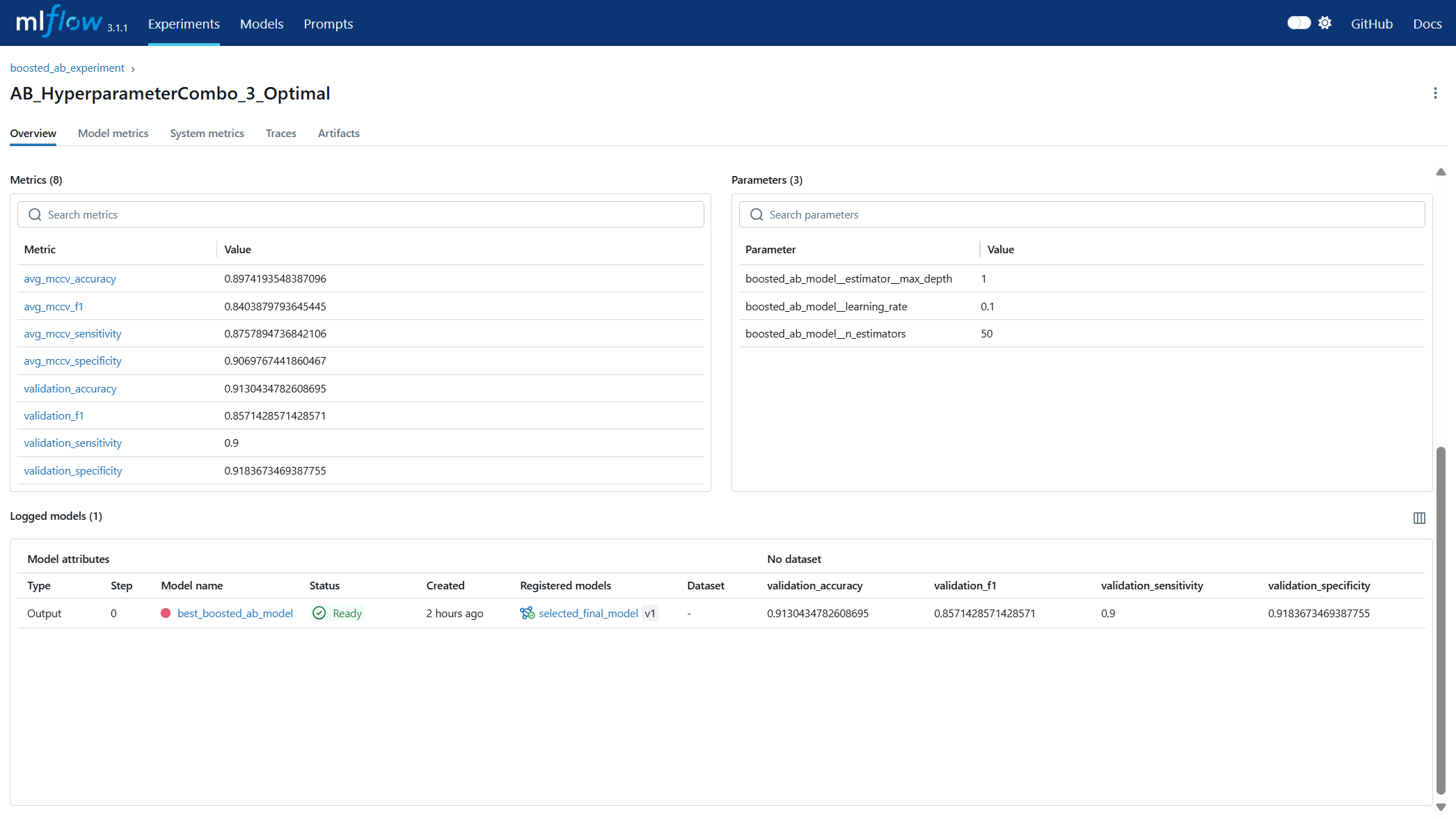
1.7.11 Model Inference¶
- The most performant model, previously registered under the name selected_final_model, was loaded dynamically from the MLflow Model Registry.
- To ensure compatibility with the model input schema:
- The test dataset was cast to the appropriate types (float64 for numeric and object for categorical features).
- Column types were aligned to match those used during training and logged as part of the model's input signature.
- Predictions were generated using the MLflow-wrapped model via .predict(...).
- The ground truth labels were encoded using an OrdinalEncoder trained on the training labels to maintain consistent class encoding.
- Performance metrics were computed using a custom binary evaluation utility, capturing the following metrics:
- test_f1 (Test F1 Score)
- test_sensitivity (Test Sensitivity)
- test_specificity (Test Specificity)
- test_accuracy (Test Accuracy)
- The final evaluation involving batch inference on the test set confirms the selected model’s ability to generalize to unseen data.
- test_f1 (Test F1 Score) = 0.8518
- test_sensitivity (Test Sensitivity) = 0.8518
- test_specificity (Test Specificity) = 0.9375
- test_accuracy (Test Accuracy) = 0.9120
##################################
# Loading the latest version of the registered model
##################################
model_name = "selected_final_model"
final_model = mlflow.pyfunc.load_model(model_uri=f"models:/{model_name}/latest")
# Casting the test data to match the original schema
X_preprocessed_test = X_preprocessed_test.astype({
'Age': 'float64',
'Gender': 'object',
'Smoking': 'object',
'Physical_Examination': 'object',
'Adenopathy': 'object',
'Focality': 'object',
'Risk': 'object',
'T': 'object',
'Stage': 'object',
'Response': 'object'
})
# Evaluating the final model on the test data
y_test_pred = final_model.predict(X_preprocessed_test)
# Encoding the response variables for model evaluation
y_encoder = OrdinalEncoder()
y_encoder.fit(y_preprocessed_train.values.reshape(-1, 1))
y_preprocessed_train_encoded = y_encoder.transform(y_preprocessed_train.values.reshape(-1, 1)).ravel()
class_labels = list(y_encoder.categories_[0])
y_preprocessed_test_encoded = y_encoder.transform(y_preprocessed_test.values.reshape(-1, 1)).ravel()
test_metrics = evaluate_binary_metrics(y_preprocessed_test_encoded, y_test_pred)
# Extracting the test metrics
test_f1 = test_metrics.get("f1", 0)
test_sensitivity = test_metrics.get("sensitivity", 0)
test_specificity = test_metrics.get("specificity", 0)
test_accuracy = test_metrics.get("accuracy", 0)
##################################
# Consolidating the optimal hyperparameter combination
# and its corresponding MCCV and validation metrics
##################################
consolidated_table_final = pd.DataFrame({
'metric': ['accuracy','sensitivity','specificity','f1'],
'test': [test_accuracy, test_sensitivity, test_specificity, test_f1],
})
print("final selected model test performance metrics:")
display(consolidated_table_final)
final selected model test performance metrics:
| metric | test | |
|---|---|---|
| 0 | accuracy | 0.912088 |
| 1 | sensitivity | 0.851852 |
| 2 | specificity | 0.937500 |
| 3 | f1 | 0.851852 |
1.7.12 Model Deployment¶
- The selected_final_model and its latest version (version 1) was loaded from the MLflow Model Registry and was promoted for production by assigning the alias prod to reference it symbolically for future model loads.
- The aliased production model was successfully loaded as final verification.
##################################
# Promoting the selected_final_model (version 1) to production
# by assigning an alias for symbolic referencing
##################################
if best_model_info:
model_name = best_model_info["model_name"]
version = result.version # version from the registration step
try:
# Assigning an alias to the newly registered model version
client.set_registered_model_alias(
name=model_name,
alias="prod", # or use "champion", or any name you prefer
version=str(version)
)
print(f"Alias 'prod' now points to model version {version} of '{model_name}'")
except RestException as e:
print(f"Error setting alias for model version {version}: {e}")
Alias 'prod' now points to model version 1 of 'selected_final_model'
##################################
# Loading the selected_final_model (version 1) to Production stage
# using the model URI and the assigned alias
##################################
mlflow.pyfunc.load_model(model_uri=f"models:/{model_name}@prod")
mlflow.pyfunc.loaded_model: artifact_path: file:///d:/Github_Codes/ProjectPortfolio/Portfolio_Project_62/codes/mlruns/511308871718837045/models/m-e4f4e70e2c5542188483c4d1a29872fc/artifacts flavor: mlflow.sklearn run_id: e949c9a21e5946d4b33b5431efc7c65f

1.8. Consolidated Findings ¶
- This project explored the use of MLflow as an end-to-end MLOps tool for experiment organization, run comparison, artifact storage, model registry management, model inference, and model deployment. The goal was to establish a reproducible workflow for hyperparameter tuning, optimal model selection, and production deployment of an ensemble classification model.
- The project execution process and significant MLflow capabilities leveraged are summarized as follows:
- Experiment Organization (MLflow Tracking UI)
- The MLflow Tracking UI was launched locally to log, organize, and visualize experiments across multiple ensemble algorithms.
- Each experiment captured multiple runs from hyperparameter tuning, enabling systematic review and selection of top-performing candidates per model family.
- Run Comparison (MLflow Search API)
- The MLflow Tracking API was used to query and compare runs based on multiple performance metrics.
- Sorting by primary and secondary metrics allowed precise identification of the optimal run for each ensemble method.
- Artifact Storage (MLflow Logging)
- A structured artifact-saving pipeline was implemented for reproducibility and traceability.
- MLflow stored cross-validation metrics, serialized model objects, validation classification reports, and confusion matrix visualizations alongside each logged run.
- This ensured all necessary model training and evaluation artifacts were versioned and retrievable.
- Model Registry (MLflow Model Registry API)
- The best-performing model from all experiments was programmatically identified and registered in the MLflow Model Registry.
- Metadata such as experiment ID, run ID, model URI, and artifact paths were stored to facilitate future retrieval and reproducibility.
- Model Inference (MLflow PyFunc Models)
- The registered model was dynamically loaded from the registry using its model URI.
- Test datasets were schema-aligned with the training data, and inference was performed directly via the MLflow PyFunc interface.
- Post-inference evaluation confirmed the model’s generalization capability on unseen data.
- Model Deployment (MLflow Model Aliases)
- The final production model version was assigned an alias for symbolic referencing.
- This allowed future production loads via a stable alias URI simplifying CI/CD pipelines and avoiding hardcoded version numbers.
- Experiment Organization (MLflow Tracking UI)
- The end-to-end MLflow pipeline successfully demonstrated how to manage the full lifecycle of machine learning models — from experiment tracking to production deployment — in a reproducible and automated manner:
- MLflow Tracking UI provided intuitive experiment organization and visualization.
- Search API enabled systematic run comparison and metric-based ranking.
- Artifact Logging ensured reproducibility and auditability.
- Model Registry offered centralized version control for candidate and final models.
- PyFunc interfaces simplified inference and integration into downstream applications.
- Model Aliases delivered a modern, stage-free approach to production deployment.
2. Summary ¶
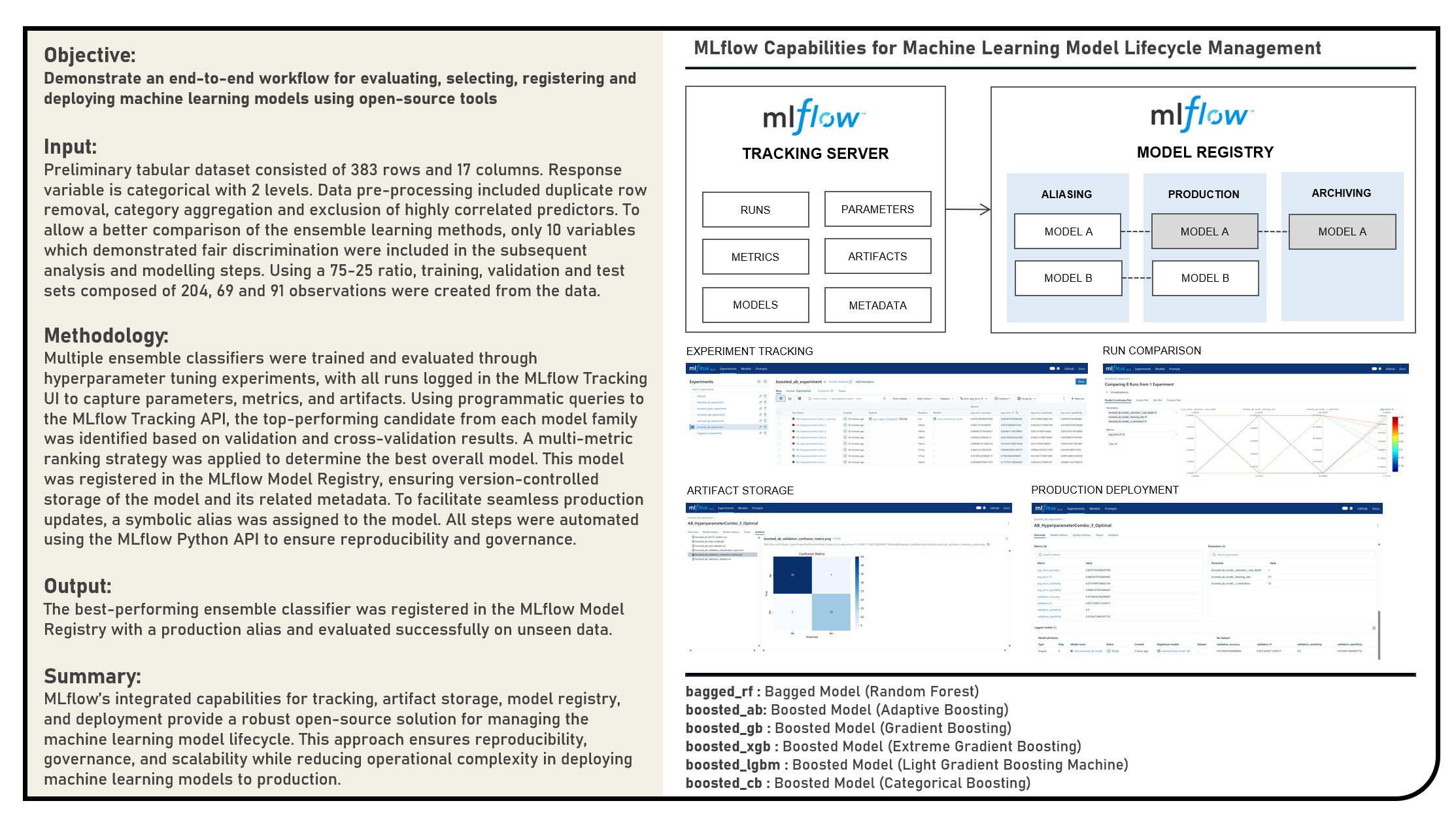
3. References ¶
- [Book] Machine Learning Engineering with MLflow: Manage the End-to-End Machine Learning Life Cycle with MLflow by Natu Lauchande
- [Book] Practical Deep Learning at Scale with MLflow: Bridge the Gap Between Offline Experimentation and Online Production by Yong Liu and Matei Zaharia
- [Book] Implementing MLOps in the Enterprise by Yaron Haviv and Noah Gift
- [Book] Beginning MLOps with MLflow: Deploy Models in AWS SageMaker, Google Cloud, and Microsoft Azure by Sridhar Alla and Suman Kalyan Adari
- [Book] Practical Machine Learning on Databricks: Seamlessly Transition ML Models and MLOps on Databricks by Debu Sinha
- [Book] Ensemble Methods for Machine Learning by Gautam Kunapuli
- [Book] Applied Predictive Modeling by Max Kuhn and Kjell Johnson
- [Book] An Introduction to Statistical Learning by Gareth James, Daniela Witten, Trevor Hastie and Rob Tibshirani
- [Book] Ensemble Methods: Foundations and Algorithms by Zhi-Hua Zhou
- [Book] Effective XGBoost: Optimizing, Tuning, Understanding, and Deploying Classification Models (Treading on Python) by Matt Harrison, Edward Krueger, Alex Rook, Ronald Legere and Bojan Tunguz
- [Python Library API] MLflow by MLflow Team
- [Python Library API] NumPy by NumPy Team
- [Python Library API] pandas by Pandas Team
- [Python Library API] seaborn by Seaborn Team
- [Python Library API] matplotlib.pyplot by MatPlotLib Team
- [Python Library API] itertools by Python Team
- [Python Library API] sklearn.experimental by Scikit-Learn Team
- [Python Library API] sklearn.preprocessing by Scikit-Learn Team
- [Python Library API] scipy by SciPy Team
- [Python Library API] sklearn.tree by Scikit-Learn Team
- [Python Library API] sklearn.ensemble by Scikit-Learn Team
- [Python Library API] sklearn.metrics by Scikit-Learn Team
- [Python Library API] xgboost by XGBoost Team
- [Python Library API] lightgbm by LightGBM Team
- [Python Library API] catboost by CatBoost Team
- [Python Library API] StatsModels by StatsModels Team
- [Python Library API] SciPy by SciPy Team
- [Article] MLflow: A Tool for Managing the Machine Learning Lifecycle by MlFlow Team (MLflow.Org)
- [Article] Experiment Tracking with MLflow in 10 Minutes by Kay Jan Wong (Towards Data Science)
- [Article] Getting Started with MLflow Experiment Tracking by DataMapu (Medium)
- [Article] Experiment Tracking and Model Registry with Mlflow by John Thuo (Medium)
- [Article] Machine Learning Experiment tracking using MLflow by Avikumar Talaviya (Medium)
- [Article] A Comprehensive Guide to MLflow for Data Scientists: Real-World Applications and Examples by Radouane El Mahfoud(Medium)
- [Article] End-to-End ML Pipelines with MLflow: Tracking, Projects & Serving by Antons Tocilins-Ruberts (Towards Data Science)
- [Article] Tracking ML Experiments using MLflow by Hafidz Zulkifli (Medium)
- [Article] A Comprehensive Comparison of ML Experiment Tracking Tools by Eryk Lewinson (Towards Data Science)
- [Article] Introduction to DVC and MLflow for Experiment tracking by Haythem Tellili (Medium)
- [Article] How to Track and Visualize Machine Learning Experiments using MLflow by Gurami Keretchashvili (Medium)
- [Article] Experiment Tracking Template with Keras and Mlflow by Lucas Soares (Towards Data Science)
- [Article] Model and Data Versioning: An Introduction to mlflow and DVC by Anant Kumar (Medium)
- [Article] How to Track and Visualize Machine Learning Experiments using MLflow by Gurami Keretchashvili (Medium)
- [Article] Weights and Biases : Experiment Tracking by Yi Yi Chan Myae Win Shein (Medium)
- [Article] A Guide To ML Experiment Tracking – With Weights & Biases by Yash Prakash (Towards Data Science)
- [Article] Weights & Biases: The ML Experiment Tracking Tool That Won’t Make You Cry by Jose S. Gomez-Olea (Medium)
- [Article] MLOps: Weights & Biases Experiments Tracking by Muhammad Faizan (Medium)
- [Article] Intro to MLOps: Machine Learning Experiment Tracking by Weights and Biases Team (Wandb.AI)
- [Article] Ensemble: Boosting, Bagging, and Stacking Machine Learning by Jason Brownlee (MachineLearningMastery.Com)
- [Article] Stacking Machine Learning: Everything You Need to Know by Ada Parker (MachineLearningPro.Org)
- [Article] Ensemble Learning: Bagging, Boosting and Stacking by Edouard Duchesnay, Tommy Lofstedt and Feki Younes (Duchesnay.GitHub.IO)
- [Article] Stack Machine Learning Models: Get Better Results by Casper Hansen (Developer.IBM.Com)
- [Article] GradientBoosting vs AdaBoost vs XGBoost vs CatBoost vs LightGBM by Geeks for Geeks Team (GeeksForGeeks.Org)
- [Article] A Gentle Introduction to the Gradient Boosting Algorithm for Machine Learning by Jason Brownlee (MachineLearningMastery.Com)
- [Article] The Ultimate Guide to AdaBoost Algorithm | What is AdaBoost Algorithm? by Ashish Kumar (MyGreatLearning.Com)
- [Article] A Gentle Introduction to Ensemble Learning Algorithms by Jason Brownlee (MachineLearningMastery.Com)
- [Article] Ensemble Methods: Elegant Techniques to Produce Improved Machine Learning Results by Necati Demir (Toptal.Com)
- [Article] The Essential Guide to Ensemble Learning by Rohit Kundu (V7Labs.Com)
- [Article] Develop an Intuition for How Ensemble Learning Works by by Jason Brownlee (Machine Learning Mastery)
- [Article] Mastering Ensemble Techniques in Machine Learning: Bagging, Boosting, Bayes Optimal Classifier, and Stacking by Rahul Jain (Medium)
- [Article] Ensemble Learning: Bagging, Boosting, Stacking by Ayşe Kübra Kuyucu (Medium)
- [Article] Ensemble: Boosting, Bagging, and Stacking Machine Learning by Aleyna Şenozan (Medium)
- [Article] Boosting, Stacking, and Bagging for Ensemble Models for Time Series Analysis with Python by Kyle Jones (Medium)
- [Article] Different types of Ensemble Techniques — Bagging, Boosting, Stacking, Voting, Blending by Abhishek Jain (Medium)
- [Article] Mastering Ensemble Techniques in Machine Learning: Bagging, Boosting, Bayes Optimal Classifier, and Stacking by Rahul Jain (Medium)
- [Article] Understanding Ensemble Methods: Bagging, Boosting, and Stacking by Divya bhagat (Medium)
- [Video Tutorial] MLflow Tutorial | ML Ops Tutorial by CodeBasics (YouTube)
- [Video Tutorial] Introduction To MLflow | Track Your Machine Learning Experiments | MLOps by DSWithBappy (YouTube)
- [Video Tutorial] Complete Tutorial For MLflow | Experiment Tracking by Vikash Das (YouTube)
- [Video Tutorial] Weights and Biases End-To-End Demo by Weights & Biases (YouTube)
- [Video Tutorial] BAGGING vs. BOOSTING vs STACKING in Ensemble Learning | Machine Learning by Gate Smashers (YouTube)
- [Video Tutorial] What is Ensemble Method in Machine Learning | Bagging | Boosting | Stacking | Voting by Data_SPILL (YouTube)
- [Video Tutorial] Ensemble Methods | Bagging | Boosting | Stacking by World of Signet (YouTube)
- [Video Tutorial] Ensemble (Boosting, Bagging, and Stacking) in Machine Learning: Easy Explanation for Data Scientists by Emma Ding (YouTube)
- [Video Tutorial] Ensemble Learning - Bagging, Boosting, and Stacking explained in 4 minutes! by Melissa Van Bussel (YouTube)
- [Video Tutorial] Introduction to Ensemble Learning | Bagging , Boosting & Stacking Techniques by UncomplicatingTech (YouTube)
- [Video Tutorial] Machine Learning Basics: Ensemble Learning: Bagging, Boosting, Stacking by ISSAI_NU (YouTube)
- [Course] DataCamp Python Data Analyst Certificate by DataCamp Team (DataCamp)
- [Course] DataCamp Python Associate Data Scientist Certificate by DataCamp Team (DataCamp)
- [Course] DataCamp Python Data Scientist Certificate by DataCamp Team (DataCamp)
- [Course] DataCamp Machine Learning Engineer Certificate by DataCamp Team (DataCamp)
- [Course] DataCamp Machine Learning Scientist Certificate by DataCamp Team (DataCamp)
- [Course] IBM Data Analyst Professional Certificate by IBM Team (Coursera)
- [Course] IBM Data Science Professional Certificate by IBM Team (Coursera)
- [Course] IBM Machine Learning Professional Certificate by IBM Team (Coursera)